Abstract
A fast and direct method for the monitoring of enzymatic DNA hydrolysis was developed using electrospray ionization mass spectrometry. We incorporated the use of a robotic chip-based electrospray ionization source for increased reproducibility and throughput. The mass spectrometry method allows the detection of DNA fragments and intact non-covalent protein–DNA complexes in a single experiment. We used the method to monitor in real-time single-stranded (ss) DNA hydrolysis by colicin E9 DNase and to characterize transient non-covalent E9 DNase–DNA complexes present during the hydrolysis reaction. The mass spectra showed that E9 DNase interacts with ssDNA in the absence of a divalent metal ion, but is strictly dependent on Ni2+ or Co2+ for ssDNA hydrolysis. We demonstrated that the sequence selectivity of E9 DNase is dependent on the ratio protein:ssDNA or the ssDNA concentration and that only 3′-hydroxy and 5′-phosphate termini are produced. It was also shown that the homologous E7 DNase is reactive with Zn2+ as transition metal ion and that this DNase displays a different sequence selectivity. The method described is of general use to analyze the reactivity and specificity of nucleases.
INTRODUCTION
The detection of DNA fragments resulting from enzymatic DNA hydrolysis is often carried out using radiolabeled DNA. In such a sensitive experiment, 32P-end-labeled DNA substrate is incubated with the hydrolytic enzyme (1–3). Alternatively, fluorescence techniques using fluorescent dye–DNA covalent complexes can be used to study DNA hydrolysis (4–6). Mass spectrometry has also been used in some studies to analyze DNA and RNA, and to observe the degradation of antisense oligonucleotides (7–10). The detection of protein–DNA interactions is often carried out using DNA foot-printing assays (11) or gel-shift assays (12). Recently, an electrochemical method was developed to probe protein–DNA interactions (13). In this approach, DNA probes are prepared through the chemical bonding of an oligonucleotide to a polymer film bearing carboxylate groups. Alternatively, structural biology techniques such as NMR, X-ray crystallography and more recently native mass spectrometry are well suited to detect and characterize protein–DNA complexes. In this paper, we introduce a new technique to monitor enzymatic DNA hydrolysis by using electrospray ionization mass spectrometry (ESI-MS). The method allows the characterization of DNA fragments and non-covalent protein–DNA complexes in a single experiment.
Since the introduction of the gentle ionization technique of ESI (14), ESI-MS has been developed into a method that is well suited to investigate non-covalent interactions between proteins, and between proteins and ligands. (15–18). By maintaining non-covalent interactions during the transfer from solution into gas-phase, direct measurement of the mass of a complex reveals the identity and stoichiometry of interacting components. Compared to the total number of publications describing intact protein complexes, relatively few reports have described the interactions between proteins and nucleic acids (19–23). This low number of publications may reflect practical difficulties when measuring these interactions. These difficulties include the need for high salt concentrations, the heterogeneity of oligonucleotides, the binding of cations to nucleic acids and the possibility to observe non-specific interactions.
The protein used in this study is the Escherichia coli colicin E9 DNase. Colicin E9 is a plasmid-encoded protein antibiotic expressed during times of nutrient or environmental stress that kills target cells through degradation of bacterial DNA. In the target cell, the lethal effect is accomplished through the action of a 15 kDa C-terminal DNase domain, termed colicin E9 DNase (24). The enzyme is a member of the HNH family of endonucleases, which are widespread in biology (25), and for which it is still unclear how they hydrolyze DNA substrates. Pommer et al. (3) have studied the mechanism and specificity by which E9 DNase hydrolyzes double-stranded (ds) DNA using radiolabeled DNA substrates. In their report, it was shown that, in the presence of the divalent metal ion Mg2+, E9 DNase preferentially hydrolyzes dsDNA at thymine bases, produces 3′-hydroxy and 5′-phosphate DNA termini and does not cleave smaller substrates, such as dinucleotides. In addition, they have reported the hydrolysis of a 23mer single-stranded (ss) DNA by E9 DNase; however, the sequence selectivity of this reaction was not reported. Ni2+ appeared to be the most efficient cofactor for ssDNA hydrolysis, whereas Mg2+ was only a poor cofactor. E9 DNase is thus a metallo-dependent enzyme, which binds a single divalent metal ion. Dissociation constants have been determined, showing a decrease in affinity in the order Zn2+ > Ni2+, Co2+ ≫ Mg2+ (26). The issue of the metal ion as a catalytic cofactor has proven controversial in the field of colicin DNase mechanism, with Chak and co-workers (27) reporting that the homologous E7 DNase (which has an identical HNH motif to the E9 DNase) is a Zn2+-dependent enzyme.
In recent studies, we have demonstrated by ESI-MS and other biophysical methods that upon divalent metal ion binding, the conformation of E9 DNase dramatically shifts from a rather loosely packed structure into a very compact structure (28). In addition, we showed that specific phosphate ion binding to E9 DNase only occurs in the presence of Zn2+ and not in the presence of Ni2+ or Mg2+ (29). We anticipated that this finding might also be relevant for the binding of DNA to E9 DNase.
Here, we describe an ESI-MS method to monitor ssDNA fragments and non-covalent protein–ssDNA complexes in a single experiment. For these studies, we used a robotic chip-based ESI source for increased reproducibility and throughput. We applied this method to study the ssDNA hydrolysis by E.coli E9 DNase and E7 DNase. We established that E9 DNase is strictly Ni2+ or Co2+ dependent, with no reactivity in the apo-form or in the presence of Zn2+ or Mg2+, has preferred sites for ssDNA hydrolysis and forms transient protein–ssDNA complexes during catalysis. In addition, we demonstrated that E7 DNase has a different cofactor specificity.
MATERIALS AND METHODS
DNA hydrolysis
17mer ssDNA (Biolegio, The Netherlands) was mixed with E7 DNase or E9 DNase, and a transition metal ion (Ni2+, Co2+, Mg2+ or Zn2+) in aqueous 50 mM ammonium acetate (pH 6.7) to final concentrations of 50–800, 10 and 20 μM for ssDNA, DNase and metal ion, respectively. The reaction mixtures were incubated at 20°C from 0 up to 120 min. For low concentrations of ssDNA (50–100 μM), the reaction was measured in real-time by ESI-MS. For high concentrations of ssDNA (>100 μM), the reaction was stopped at time intervals by diluting part of the reaction mixture 20- to 40-fold using aqueous 50 mM ammonium acetate, pH 6.7 and rapid freezing. The reaction mixture was then analyzed by ESI-MS. The sequence of the ssDNA used was 5′-GTAAAACGACGGCCAGT-3′ and did not contain phosphate moieties at the 5′- and 3′-termini. The average molecular masses of the ssDNA, E9 DNase and E7 DNase as measured by ESI-MS was 5228.3, 15 087.1 and 15 375.5 Da, which is in close agreement with the calculated average masses of 5228.5, 15 088.0 and 15 374.4 Da. E7 DNase and E9 DNase were purified according to a previously established method (30). Metal-free apo-DNase was produced by EDTA treatment and extensive dialysis as described before (26). Protein concentrations were determined by absorbance at 280 nm (31).
Mass spectrometry
A Micromass ESI-TOF mass spectrometer (LCT, Waters) equipped with a robotic chip-based ESI source (Nanomate, Advion Biosciences) was used. The ESI source was programmed to aspirate 2 μl of sample into the conductive pipette tip and then to deliver the sample to the inlet side of the ESI chip. This chip consists of 10 × 10 nozzles in a silicon wafer and a channel extends from the nozzle to an inlet side at the opposite face of the chip. Electrospray is initiated by applying an electric voltage of 1850 V and a head pressure of 0.2 psi to the sample in the pipette tip and a cone voltage of 30 V. All samples were measured in positive ion mode. The real-time samples were measured continuously for 45 min, whereas the other samples were measured for 2 min with a 2 s scan time. Repeating the experiments with standard borosilicate capillaries instead of the automated ESI source did not alter the outcome of the experiments.
Data analysis
We semi-quantified our data to determine the initial reaction velocity and to determine the concentrations of ssDNA fragments formed. We determined that the total ion intensity of a ssDNA fragment is linearly dependent on the concentration of ssDNA up to 75 μM. To correct for the discrimination factor of large ssDNA fragments, we focused on the conditions in which only one hydrolysis reaction occurred (Equation 1). The mass spectra showed that up to 240 min only two ssDNA fragments were formed (F1 and F2), which consequently must have identical concentrations (Equation 2). The total concentration of DNA during the reaction was constant (Equation 3). Mass spectra were averaged over 2 min for better signal-to-noise ratio, smoothed and centered, thereby using the areas option in MassLynx 4.0. The total areas were divided by the molecular masses of the ssDNA fragments (964.6 and 4281.9 Da, respectively) after which the intensity of the overestimated small ssDNA fragment was corrected with a normalizing factor R (R = 2.5) (Equation 4). This, together with the calibration curve of 17mer ssDNA, allowed us to calculate the concentrations of the ssDNA fragments formed (Equation 3). The molecular mass and normalizing factor were used to calculate the concentrations after 240 min of reaction. This procedure was also used to calculate ssDNA concentrations for reactions in which multiple hydrolysis reactions occurred. For this, we divided the ssDNA fragments formed into two classes, namely 3–7 nucleic acids (F1) and 10–14 nucleic acids (F2). Only F1 was corrected with the factor R. According to the calculated total intensities, our error using this procedure is ∼20%. We validated our data with synthetic oligonucleotides having 4, 11 and 17 nucleic acids.
| 1 |
| 2 |
| 3 |
| 4 |
where, Mw is the molecular mass; I, the ion intensity; R, the normalization factor; F1, the fragment of 3–7 nucleic acids and F2, the fragment of 10–14 nucleic acids.
RESULTS AND DISCUSSION
E9 DNase–ssDNA complexes
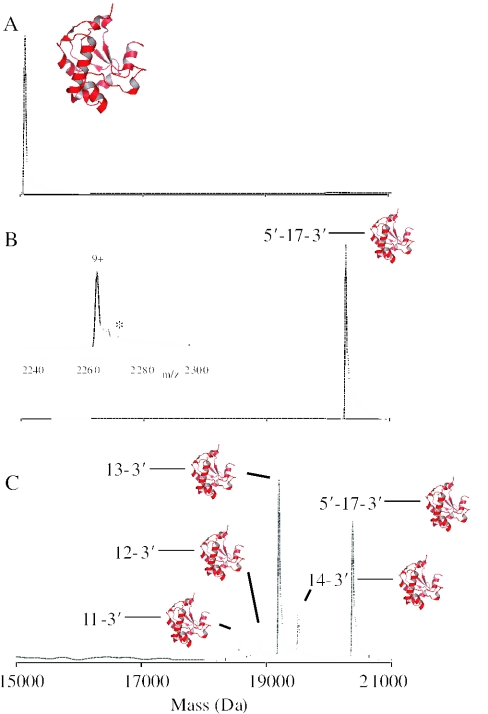
We first examined the ability of E9 DNase to interact with a 17mer ssDNA (5′-GTAAAACGACGGCCAGT-3′). When we incubated E9 DNase (10 μM) with a 10-fold excess of ssDNA, we only observed ion series of free ssDNA and a stoichiometric non-covalent E9 DNase–DNA complex having molecular masses of 5228.3 Da and 20 313.6 Da, respectively. Similarly, when we added a 10-fold excess of Mg2+ to this mixture, we only observed a binary E9 DNase–DNA complex. On the other hand, when we added a 2-fold excess of Zn2+, Ni2+ or Co2+ we primarily observed the ternary complex between E9 DNase, 17mer ssDNA and the transition metal ion having a measured molecular mass of 20 379.6, 20 375.6 and 20 375.1 Da, respectively (Figure 1). The ESI-MS method thus allows the analysis and characterization of non-covalent protein–DNA interactions and can easily discriminate between apo-protein and metal-bound protein having average mass differences of 65, 59 and 59 Da for Zn2+, Ni2+ and Co2+, respectively. These results also demonstrate that a divalent metal ion is not required for ssDNA binding to E9 DNase. This is in line with data of the homologous E7 DNase for which it has been demonstrated that it can interact with 27mer dsDNA in the absence of any transition metal ion (27).
Figure 1.
Deconvoluted ESI-MS spectra of E9 DNase:ssDNA complexes. E9 DNase (10 μM) was mixed with a transition metal ion (20 μM) and 17mer ssDNA (100 μM) and incubated for 10 min at 20°C. (A) E9 DNase, (B) E9 DNase in the presence of Zn2+ and 17mer ssDNA revealing the ternary complex between protein, ssDNA and Zn2+ and (C) E9 DNase in the presence of Ni2+ and 17mer ssDNA revealing the ternary complex between protein, ssDNA and Ni2+, and ternary complexes comprising hydrolyzed ssDNA products. All complexes with hydrolyzed ssDNA contained a phosphate moiety at the 5′ terminus of the ssDNA. The inset in (B) shows the raw data of the 9+ ion of E9 DNase in complex with Zn2+ and 17mer ssDNA. The low amount of the complex between E9 DNase and the two molecules of Zn2+ is indicated by the peak labeled with the asterisk. These data show that the E9 DNase–Zn2+ interaction is specific.
The spectra of the binary E9 DNase–ssDNA and ternary E9 DNase–Zn2+–ssDNA complexes did not change in time. Even at long incubation times (120 min), no other protein–DNA complexes were detected in the mass spectra. The ternary complex comprising the metal ion Ni2+ or Co2+, however, did degrade in time. After 10 min of incubation, the intensity of the original ternary complex was decreased and other complexes with lower molecular masses appeared (Figure 1), indicating the enzymatic hydrolysis of ssDNA.
ssDNA hydrolysis by E9 DNase
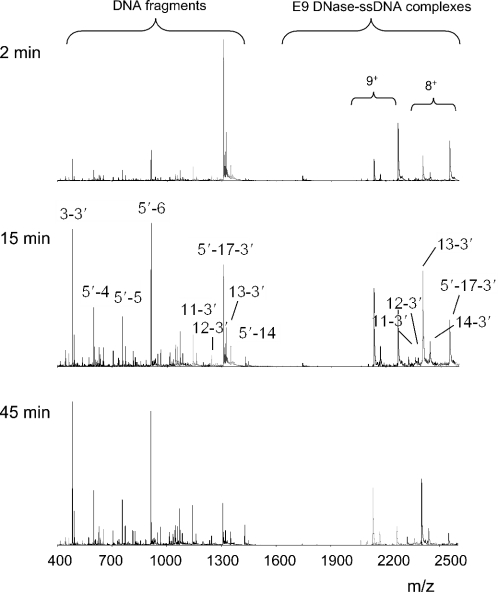
The results described in the previous section encouraged us to monitor in real-time the Ni2+-induced hydrolysis of the 17mer ssDNA substrate. At first, the concentrations of E9 DNase (10 μM) and ssDNA (50 μM) were chosen such that we could monitor online both the ssDNA fragments produced and the protein–ssDNA complexes present during the hydrolysis reaction. Within 45 min, the full-length 17mer ssDNA was nearly completely hydrolyzed as can be seen from the low intensities of the ions of the free 17mer ssDNA (m/z 1308.2, 4+; m/z 1 743.9, 3+) and the E9 DNase: Ni2+–17mer ssDNA complex (m/z 2 265.1, 9+; m/z 2548.1, 8+) in Figure 2. A listing of all detected ion signals of ssDNA fragments and non-covalent E9 DNase–ssDNA complexes, their molecular masses and assignments is given in Table 1. We could assign peaks corresponding to 14 different DNA fragmentation products. The determined molecular masses of the DNA fragments allowed us to conclude that all hydrolysis products contain exclusively 5′-phosphate and 3′-hydroxy termini, which is in line with the data of Pommer et al. (3), albeit for dsDNA. We found fragments with both intact 3′ and 5′ ends and we found exclusively pairs of DNA fragments, i.e. when we detected ions of 5′-AGT-3′ (abbreviated here as 3-3′; three nucleic acids long and intact 3′-terminus present) we also detected ions of 5′-GTAAAACGACGGCC-3′ (5′-14; 14 nucleic acids long and intact 5′-terminus present), together forming the 17mer ssDNA substrate (Table 1).
Figure 2.
ESI-MS spectra of the enzymatic hydrolysis of ssDNA substrate by E9 DNase. E9 DNase (10 μM) was incubated with Ni2+ (20 μM) and 17mer ssDNA (100 μM) at 20°C and monitored in real-time. Selected spectra are shown after incubation times of 2, 15 and 45 min. The spectra show on the left-hand side the formation of several DNA products and on the right-hand side the formation and degradation of E9 DNase–ssDNA complexes. The ions of 17mer ssDNA are 1308.2 (4+) and 1743.9 (3+). The most abundant ions of the E9 DNase–ssDNA complexes are the 8+ and 9+ ions. A listing of all detected ion signals of ssDNA fragments and E9 DNase–ssDNA complexes, their molecular masses and assignments is given in Table 1.
Table 1.
A listing of all detected ion signals of DNA fragments and E9 DNase–DNA complexes, their molecular masses and assignments
| Abbreviation | Ions (m/z) | Mw (Da) | |
|---|---|---|---|
| ssDNA fragments | |||
| GTAAAACGACGGCCAGT | 5′-17-3′ | 1308.2 (4+), 1743.9 (3+) | 5228.5 |
| GTAAAACGACGGCC | 5′-14 | 1071.5 (4+), 1428.3 (3+) | 4281.9 |
| GTAAAACGACGG | 5′-12 | 1235.7 (3+) | 3703.5 |
| GTAAAACGAC | 5′-10 | 1016.1 (3+) | 3045.1 |
| GTAAAAC | 5′-7 | 1057.8 (2+) | 2113.6 |
| GTAAAA | 5′-6 | 913.1 (2+) | 1824.3 |
| GTAAA | 5′-5 | 756.5 (2+) | 1511.0 |
| GTAA | 5′-4 | 599.8 (2+) | 1197.6 |
| AACGACGGCCAGT | 13-3′ | 1013.3 (4+), 1350.5 (3+) | 4048.6 |
| ACGACGGCCAGT | 12-3′ | 1246.2 (3+) | 3735.6 |
| CGACGGCCAGT | 11-3′ | 1141.7 (3+) | 3422.1 |
| GACGGCCAGT | 10-3′ | 1045.4 (3+) | 3133.2 |
| GGCCAGT | 7-3′ | 734.7 (2+) | 2201.4 |
| CCAGT | 5-3′ | 772.5 (2+) | 1543.0 |
| AGT | 3-3′ | 483.2 (2+), 965.3 (1+) | 964.6 |
| E9:DNase–Ni2+–ssDNA complexes | |||
| E9:DNase-Ni2+-5′-17-3′ | 2548.1 (8+), 2265.1 (9+) | 20 375.6 | |
| E9:DNase-Ni2+-14-3′ | 2440.0 (8+), 2169.0 (9+) | 19 512.6 | |
| E9:DNase-Ni2+-13-3′ | 2400.9 (8+), 2134.3 (9+) | 19 199.8 | |
| E9:DNase-Ni2+-12-3′ | 2361.5 (8+), 2099.2 (9+) | 18 886.1 | |
| E9:DNase-Ni2+-11-3′ | 2322.6 (8+), 2064.6 (9+) | 18 572.8 | |
DNA fragments and non-covalent E9 DNase–Ni2+–ssDNA complexes resulting from the enzymatic reaction. E9 DNase (10 μM) was incubated with Ni2+ (20 μM) and 17mer ssDNA (100 μM) at 20°C for 45 min.
The same incubation in the presence of Zn2+ or Mg2+ instead of Ni2+ or in the absence of any transition metal ion did not yield any hydrolyzed DNA fragments of other protein–ssDNA complexes. This clearly shows that the reaction observed in the presence of Ni2+ is an enzymatic reaction catalyzed by the non-covalent E9 DNase–Ni2+ complex. These results are in line with previous data, indicating that hydrolysis of ssDNA is not supported by Zn2+, and that only a very weak activity is associated with Mg2+ (3).
E9 DNase–ssDNA complexes during hydrolysis
The mass spectra in Figure 2 also show ions around m/z 2000–2600. Mass determination revealed that these ions represent the 8+ and 9+ ions of complexes between E9 DNase, Ni2+ and hydrolyzed ssDNA fragments. These DNA fragments ranged in size in between 11 and 14 nucleic acids (Table 1), with the E9 DNase–Ni2+-13-3′ complex being the most abundant. Intriguingly, we only observed non-covalent complexes between E9 DNase and 5′ end hydrolyzed DNA fragments, e.g. 13-3′ (Figure 2). Thus, in spite of the fact that E9-catalyzed hydrolysis yielded fragments with both intact 3′ and 5′ ends (Table 1), only non-covalent complexes with 5′ end hydrolyzed DNA fragments were stable enough to be observed. In addition, short DNA fragments (≤10 nucleic acids) did not form stable complexes with the enzyme. This agrees with previous data that have demonstrated that ssDNA length was the predominant factor for binding to E9 DNase, with >10 nucleic acids being optimal (3). The observation that all 3′ and short 5′ end hydrolyzed DNA fragments did not form stable complexes with E9 DNase may also suggest that they were not further hydrolyzed. Indeed, the mass spectra of the online kinetic measurements of 17mer ssDNA hydrolysis showed that the concentration of the 3′ and short 5′ end hydrolyzed DNA fragments remained constant or increased during the course of the E9 DNase-catalyzed hydrolysis reactions, whereas the concentration of long 5′ end hydrolyzed fragments decreased (Table 2).
Table 2.
Concentration of ssDNA fragments resulting from the E9 DNase-catalyzed reaction in the presence of ssDNA and Ni2+
| ssDNA fragments | Abbreviation | Time (min) | ||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 5 | 10 | 15 | 30 | 45 | ||
| Concentration (μM) | ||||||||
| GTAAAACGACGGCCAGT | 5′-17-3′ | 100 | 38.9 | 23.4 | 12.4 | 9.4 | 4.3 | 1.8 |
| GTAAAACGACGGCC | 5′-14 | 0 | 5.1 | 6.6 | 6.2 | 6.2 | 7.3 | 6.3 |
| GTAAAACGACGG | 5′-12 | 0 | 0.8 | 0.9 | 0.7 | 0.7 | 0.8 | 0.8 |
| GTAAAACGAC | 5′-10 | 0 | 1.7 | 2.3 | 2.3 | 2.4 | 2.2 | 2.6 |
| GTAAAAC | 5′-7 | 0 | 2.5 | 3.0 | 3.2 | 3.5 | 3.7 | 3.7 |
| GTAAAA | 5′-6 | 0 | 9.5 | 12.5 | 14.4 | 15.8 | 17.0 | 16.5 |
| GTAAA | 5′-5 | 0 | 4.0 | 5.0 | 6.1 | 6.6 | 6.9 | 7.1 |
| GTAA | 5′-4 | 0 | 4.8 | 6.6 | 8.9 | 9.9 | 10.7 | 11.3 |
| AACGACGGCCAGT | 13-3′ | 0 | 7.6 | 6.0 | 4.5 | 4.1 | 3.3 | 2.5 |
| ACGACGGCCAGT | 12-3′ | 0 | 3.2 | 2.2 | 1.6 | 1.6 | 1.6 | 1.3 |
| CGACGGCCAGT | 11-3′ | 0 | 5.8 | 6.5 | 6.3 | 6.4 | 6.3 | 5.0 |
| GACGGCCAGT | 10-3′ | 0 | 2.9 | 2.8 | 2.6 | 2.6 | 2.5 | 2.1 |
| GGCCAGT | 7-3′ | 0 | 1.0 | 0.9 | 0.9 | 0.9 | 0.5 | 0.5 |
| CCAGT | 5-3′ | 0 | 2.1 | 2.4 | 2.5 | 2.9 | 3.1 | 3.5 |
| AGT | 3-3′ | 0 | 11.5 | 18.6 | 26.9 | 27.3 | 29.5 | 35.0 |
E9 DNase (10 μM) was incubated with 17mer ssDNA substrate (100 μM) and Ni2+ (20 μM) at 20°C for 45 min.
Sequence selectivity of E9 DNase
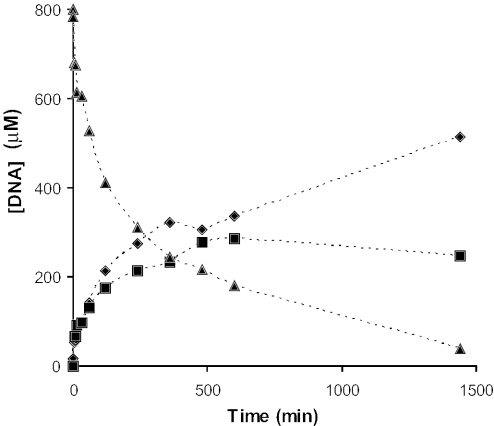
The previous data may suggest that ssDNA hydrolysis by E9 DNase is either somewhat specific for length or specific for hydrolysis in between cytosine and adenine bases, however, some other cleavages occur as well, but at a lower frequency (Table 2). We extended our kinetic experiments with a molar ratio protein:DNA of 1:80. Intriguingly, the resulting mass spectra of these incubations revealed a single cleavage site forming two fragmentation products, namely 3-3′ (m/z 965.4, 1+; m/z 483.2, 2+) and 5′-14 (m/z 1428.3, 3+; m/z 1072.0, 4+). Thus, under these conditions, E9 DNase is much more (length or base) specific for the hydrolysis in between cytosine14 and adenine15. In Figure 3, the time-dependent concentration of the 17mer ssDNA and the two fragmentation products is plotted. From this kinetic plot, we could estimate an apparent turnover rate of 0.71 ± 0.10 min−1. In the initial phase of the reaction, the 14mer degradation product was not further hydrolyzed by E9 DNase as was indicated by equal concentrations of 3-3′ and 5′-14. Only when the concentration of the initial 17mer ssDNA was lower than that of the 5′-14 fragment (after ∼240 min), E9 DNase hydrolysis of the latter fragment became apparent. This hydrolysis was indicated by the relatively lower concentration of the 5′-14 fragment in comparison with the 3-3′ fragment and the occurrence of additional low abundant fragmentation products.
Figure 3.
Apparent turnover rate of E9 DNase with 17mer ssDNA substrate. Progress curve of reaction catalyzed by E9 DNase (10 μM) in the presence of Ni2+ (20 μM) and 17mer ssDNA (800 μM) at 20°C. ssDNA 17mer, 5′-14 fragment and 3-3′ fragment are indicated by triangles, squares and diamonds, respectively. The calculated turnover rate is 0.71 ± 0.10 s−1.
The results show that the sequence selectivity of the enzyme is dependent on the protein:DNA ratio or DNA concentration. Very few examples are described in literature describing such a phenomenon. Zambianchi et al. (32) reported the effect of substrate concentration on the stereospecificity of cyclohexanone monooxygenase. They have proposed a model in which the increased stereospecificity is due to the binding of a second substrate molecule to the protein. Our mass spectrometry experiments, performed in both positive and negative ion mode, did not show the interaction with a second molecule of DNA. However, we cannot completely exclude that a second low affinity site for DNA is not observed by ESI-MS, i.e. the affinity might be too low to be observed by ESI-MS.
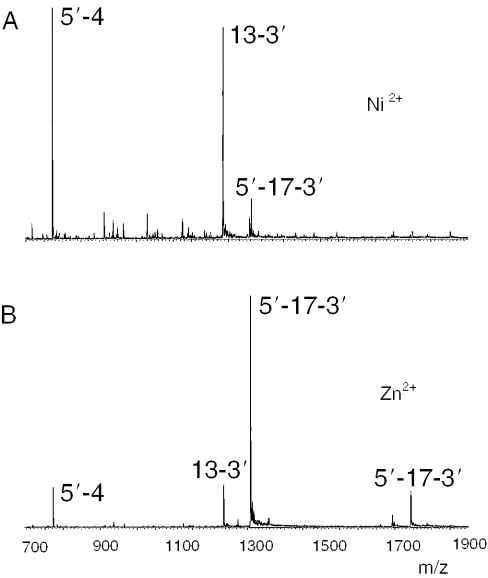
ssDNA hydrolysis by E7 DNase
We also used our mass spectrometry method to investigate the metal-dependency and the sequence selectivity of colicin E7 DNase. Upon mixing E7 DNase (10 μM) with a 50-fold excess of 17mer ssDNA (5′-GTAAAACGACGGCCAGT-3′) and a stoichiometric amount of Ni2+ or Zn2+, we only observed binary complexes between E7 DNase and ssDNA. Both in the presence of Ni2+ and Zn 2+, the binary complexes were degraded in time, indicating the hydrolysis of ssDNA. The mass spectra of the incubation in the presence of Ni2+ or Zn2+ clearly showed a single cleavage site with the formation of two fragmentation products, namely 4-3′ (m/z 772.1, 2+) and 5′-13 (m/z 1235.2, 3+) (Figure 4). However, the turnover rate with Zn2+ was ∼20-fold lower as compared to the rate with Ni2+. As discussed before in this paper, the issue of the transition metal ion as a catalytic cofactor is still under debate in the field of colicin DNases. The present results clearly demonstrate that E7 DNase and E9 DNase show a different reactivity toward transition metal ions. Moreover, E7 DNase and E9 DNase have different cleavage specificities toward ssDNA. These differences are remarkable as E7 DNase and E9 DNase have very similar primary sequences (70% sequence identity) and very homologous X-ray structures (root mean square deviation of 0.65 Å for 131 Cα atoms). In addition, the active site cavities of E7 DNase and E9 DNase are highly conserved and have >80% primary sequence identity. In previous work, Kleanthous and co-workers did not find any Zn2+-dependent activity for E7 DNase. The Kunitz assays used in these studies may not have sufficient sensitivity to detect such activity (33,34) or the high Zn2+ concentrations used may have inhibited the enzyme as has been reported previously (27).
Figure 4.
ESI-MS spectra of the enzymatic hydrolysis of ssDNA substrate by E7 DNase. E7 DNase (10 μM) was incubated with a transition metal ion (10 μM) and 17mer ssDNA (500 μM) at 20°C. (A) ssDNA fragments formed after 15 min of incubation in the presence of Ni2+, (B) ssDNA fragments formed after 45 min of incubation in the presence of Zn2+. The ions of 17mer ssDNA are 1308.2 (4+) and 1743.9 (3+). The most abundant ssDNA fragment ions formed were 4-3′ (m/z 772.1, 2+) and 5′-13 (m/z 1235.2, 3+).
CONCLUSIONS
The results reported here clearly show that ESI-MS is a very powerful technique to study kinetics and sequence selectivity of enzymatic DNA hydrolysis, and to analyze transiently present protein–DNA complexes. In comparison with the methods using 32P-end-labeled or fluorescent dye-labeled DNA to study DNA hydrolysis, the described ESI-MS method is faster and does not require any DNA manipulation or modification. In comparison with the gel-shift assays, DNA foot-printing and electrochemical methods to study protein–DNA complexes, the current method allows full characterization of the complexes and is faster. Moreover, the ESI-MS method allows the simultaneous identification of DNA fragments and non-covalent protein–DNA complexes. The detection limit for ssDNA and protein–ssDNA complexes in our current set-up is ∼200 fmol and 1 pmol, respectively. From the results in this paper, we can conclude that the described method is a general tool to study the reactivity and specificity of nucleases. Detailed kinetic and mechanistic studies of the DNA hydrolysis by different DNases and varying nucleic acid substrates, including ssDNA, dsDNA and RNA are currently ongoing in our laboratory.
Acknowledgments
We thank Nadine Kirkpatrick (University of York) for providing purified E7 DNase and E9 DNase and Bruno Godinho (Utrecht University) for excellent mass spectrometry assistance. R.H.H.vdH. was supported by a VENI grant (700.54.402) from the Netherlands Organization for Scientific Research (NWO). P.G. thanks the ‘Fonds National de la Recherche Scientifique’ for financial support. Funding to pay the Open Access publication charges for this article was provided by Utrecht University and the Netherlands Proteomics Centre.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lima W.F., Crooke S.T. Highly efficient endonucleolytic cleavage of RNA by a Cys(2)His(2) zinc-finger peptide. Proc. Natl Acad. Sci. USA. 1999;96:10010–10015. doi: 10.1073/pnas.96.18.10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nomura A., Sugiura Y. Sequence-selective and hydrolytic cleavage of DNA by zinc finger mutants. J. Am. Chem. Soc. 2004;126:15374–15375. doi: 10.1021/ja045663l. [DOI] [PubMed] [Google Scholar]
- 3.Pommer A.J., Cal S., Keeble A.H., Walker D., Evans S.J., Kuhlmann U.C., Cooper A., Connolly B.A., Hemmings A.M., Moore G.R., et al. Mechanism and cleavage specificity of the H–N–H endonuclease colicin E9. J. Mol. Biol. 2001;314:735–749. doi: 10.1006/jmbi.2001.5189. [DOI] [PubMed] [Google Scholar]
- 4.Uhler S.A., Cai D., Man Y., Figge C., Walter N.G. RNA degradation in cell extracts: real-time monitoring by fluorescence resonance energy transfer. J. Am. Chem. Soc. 2003;125:14230–14231. doi: 10.1021/ja036854b. [DOI] [PubMed] [Google Scholar]
- 5.Trubetskoy V.S., Hagstrom J.E., Budker V.G. Self-quenched covalent fluorescent dye-nucleic acid conjugates as polymeric substrates for enzymatic nuclease assays. Anal. Biochem. 2002;300:22–26. doi: 10.1006/abio.2001.5442. [DOI] [PubMed] [Google Scholar]
- 6.Kelemen B.R., Klink T.A., Behlke M.A., Eubanks S.R., Leland P.A., Raines R.T. Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 1999;27:3696–3701. doi: 10.1093/nar/27.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuette J.M., Pieles U., Maleknia S.D., Srivatsa G.S., Cole D.L., Moser H.E., Afeyan N.B. Sequence analysis of phosphorothioate oligonucleotides via matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 1995;13:1195–1203. doi: 10.1016/0731-7085(95)01534-r. [DOI] [PubMed] [Google Scholar]
- 8.Ni J., Pomerantz C., Rozenski J., Zhang Y., McCloskey J.A. Interpretation of oligonucleotide mass spectra for determination of sequence using electrospray ionization and tandem mass spectrometry. Anal. Chem. 1996;68:1989–1999. doi: 10.1021/ac960270t. [DOI] [PubMed] [Google Scholar]
- 9.McCloskey J.A., Whitehill A.B., Rozenski J., Qiu F., Crain P.F. New techniques for the rapid characterization of oligonucleotides by mass spectrometry. Nucleosides Nucleotides. 1999;18:1549–1553. doi: 10.1080/07328319908044782. [DOI] [PubMed] [Google Scholar]
- 10.Griffey R.H., Greig M.J., Gaus H.J., Liu K., Monteith D., Winniman M., Cummins L.L. Characterization of oligonucleotide metabolism in vivo via liquid chromatography/electrospray tandem mass spectrometry with a quadrupole ion trap mass spectrometer. J. Mass Spectrom. 1997;32:305–313. doi: 10.1002/(SICI)1096-9888(199703)32:3<305::AID-JMS482>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Galas D.J., Schmitz A. DNAse footprinting: a simple method for the detection of protein–DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fried M., Crothers D.M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ban C., Chung S., Park D.S., Shim Y.B. Detection of protein–DNA interaction with a DNA probe: distinction between single-strand and double-strand DNA–protein interaction. Nucleic Acids Res. 2004;32:e110. doi: 10.1093/nar/gnh109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenn J.B., Mann M., Meng C.K., Wong S.F., Whitehouse C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 15.Przybylski M., Glocker M.O. Electrospray mass spectrometry of biomacromolecular complexes with noncovalent interactions. New analytical perspectives for supramolecular chemistry and molecular recognition processes. Angew. Chem. Int. Ed. Engl. 1996;35:806–826. [Google Scholar]
- 16.van den Heuvel R.H., Heck A.J. Native protein mass spectrometry: from intact oligomers to functional machineries. Curr. Opin. Chem. Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Wiseman J.M., Takáts Z., Gologan B., Davisson V.J., Cooks R.G. Direct characterization of enzyme–substrate complexes by using electrosonic spray ionization mass spectrometry. Angew. Chem. Int. Ed. Engl. 2005;44:913–916. doi: 10.1002/anie.200461672. [DOI] [PubMed] [Google Scholar]
- 18.van Duijn E., Bakkes P.J., Heeren R.M., van den Heuvel R.H., van Heerikhuizen H., van der Vies S.M., Heck A.J. Monitoring macromolecular complexes involved in the chaperonin-assisted protein folding cycle by mass spectrometry. Nature Methods. 2005;2:371–376. doi: 10.1038/nmeth753. [DOI] [PubMed] [Google Scholar]
- 19.Potier N., Donald L.J., Chernushevich I., Ayed A., Ens W., Arrowsmith C.H., Standing K.G., Duckworth H.W. Study of a noncovalent trp repressor: DNA operator complex by electrospray ionization time-of-flight mass spectrometry. Protein Sci. 1998;7:1388–1395. doi: 10.1002/pro.5560070615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofstadler S.A., Griffey R.H. Analysis of noncovalent complexes of DNA and RNA by mass spectrometry. Chem. Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 21.Dickman M.J., Sedelnikova S.E., Rafferty J.B., Hornby D.P. Rapid analysis of protein–nucleic acid complexes using MALDI TOF mass spectrometry and ion pair reverse phase liquid chromatography. J. Biochem. Biophys. Methods. 2004;58:39–48. doi: 10.1016/s0165-022x(03)00146-5. [DOI] [PubMed] [Google Scholar]
- 22.Hanson C.L., Robinson C.V. Protein–nucleic acid interactions and the expanding role of mass spectrometry. J. Biol. Chem. 2004;279:24907–24910. doi: 10.1074/jbc.R300037200. [DOI] [PubMed] [Google Scholar]
- 23.Akashi S., Osawa R., Nishimura Y. Evaluation of protein–DNA binding affinity by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2005;16:116–125. doi: 10.1016/j.jasms.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 24.James R., Penfold C.N., Moore G.R., Kleanthous C. Killing of E.coli cells by E group nuclease colicins. Biochimie. 2002;84:381–389. doi: 10.1016/s0300-9084(02)01450-5. [DOI] [PubMed] [Google Scholar]
- 25.Chevalier B.S., Stoddard B.L. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pommer A.J., Kuhlmann U.C., Cooper A., Hemmings A.M., Moore G.R., James R., Kleanthous C. Homing in on the role of transition metals in the HNH motif of colicin endonucleases. J. Biol. Chem. 1999;274:27153–27160. doi: 10.1074/jbc.274.38.27153. [DOI] [PubMed] [Google Scholar]
- 27.Ku W.Y., Liu Y.W., Hsu Y.C., Liao C.C., Liang P.H., Yuan H.S., Chak K.F. The zinc ion in the HNH motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis. Nucleic Acids Res. 2002;30:1670–1678. doi: 10.1093/nar/30.7.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Bremer E.T., Jiskoot W., James R., Moore G.R., Kleanthous C., Heck A.J., Maier C.S. Probing metal ion binding and conformational properties of the colicin E9 endonuclease by electrospray ionization time-of-flight mass spectrometry. Protein Sci. 2002;11:1738–1752. doi: 10.1110/ps.0200502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Bremer E.T., Keeble A.H., Kleanthous C., Heck A.J. Metal induced selectivity in phosphate ion binding in E9 DNase. Chem. Commun. 2005;9:1137–1139. doi: 10.1039/b415709e. [DOI] [PubMed] [Google Scholar]
- 30.Garinot-Schneider C., Pommer A.J., Moore G.R., Kleanthous C., James R. Identification of putative active-site residues in the DNase domain of colicin E9 by random mutagenesis. J. Mol. Biol. 1996;260:731–742. doi: 10.1006/jmbi.1996.0433. [DOI] [PubMed] [Google Scholar]
- 31.Wallis R., Reilly A., Barnes K., Abell C., Campbell D.G., Moore G.R., James R., Kleanthous C. Tandem overproduction and characterisation of the nuclease domain of colicin E9 and its cognate inhibitor protein Im9 Eur. J. Biochem. 1994;220:447–454. doi: 10.1111/j.1432-1033.1994.tb18642.x. [DOI] [PubMed] [Google Scholar]
- 32.Zambianchi F., Pasta P., Ottolina G., Carrea G., Colonna S., Gaggero N., Ward J.M. Effect of substrate concentration on the enantioselectivity of cyclohexanone monooxygenase from Acinetobacter calcoaceticus and its rationalization. Tetrahedron. 2000;11:3653–3657. [Google Scholar]
- 33.Keeble A.H., Hemmings A.M., James R., Moore G.R., Kleanthous C. Multistep binding of transition metals to the H–N–H endonuclease toxin colicin E9. Biochemistry. 2002;41:10234–10244. doi: 10.1021/bi020174o. [DOI] [PubMed] [Google Scholar]
- 34.Pommer A.J., Wallis R., Moore G.R., James R., Kleanthous C. Enzymological characterization of the nuclease domain from the bacterial toxin colicin E9 from Escherichia coli. Biochem. J. 1998;334:387–392. doi: 10.1042/bj3340387. [DOI] [PMC free article] [PubMed] [Google Scholar]