Abstract
Background
Multiple Sulfatase Deficiency (MSD) is an ultra-rare autosomal recessive disorder characterized by deficient enzymatic activity of all known sulfatases. MSD patients frequently carry two loss of function mutations in the SUMF1 gene, encoding a formylglycine-generating enzyme (FGE) that activates 17 different sulfatases. MSD patients show common features of other lysosomal diseases like mucopolysaccharidosis and metachromatic leukodystrophy, including neurologic impairments, developmental delay, and visceromegaly. There are currently no approved therapies for MSD patients. Hematopoietic stem cell transplant (HSCT) has been applied with success in the treatment of certain lysosomal diseases. In HSCT, donor-derived myeloid cells are a continuous source of active sulfatase enzymes that can be taken up by sulfatase-deficient host cells. Thus, HSCT could be a potential approach for the treatment of MSD.
Methods
To test this hypothesis, we used a clinically relevant mouse model for MSD, B6-Sumf1(S153P/S153P) mice, engrafted with bone marrow cells, Sumf1+/+, from B6-PtprcK302E mice (CD45.1 immunoreactive).
Results
After 10 months post-transplant, flow cytometric analysis shows an average of 90% of circulating leukocytes of donor origin (Sumf1(+/+)). Enzymatic activity for ARSA, ARSB, and SGSH is significantly increased in spleen of B6-Sumf1(S153P/S153P) recipient mice. In non-lymphoid organs, only liver and heart show a significant correction of sulfatase activity and GAG accumulation. Frequency of inflammatory cells and lysosomal pathology is significantly reduced in liver and heart, while no significant improvement is detected in brain.
Conclusions
Our results indicate that HSCT could be a suitable approach to treat MSD-pathology affecting peripheral organs, however that benefit to CNS pathology might be limited.
Subject terms: Haematopoietic stem cells, Neonatal brain damage
Plain Language Summary
Multiple Sulfatase Deficiency (MSD) is a rare genetic disorder caused by loss-of-function variations in the SUMF1 gene. This deficiency results in the accumulation of toxic compounds, leading to developmental delays and neurological impairments. In a bone marrow transplant (BMT), donor cells are infused into the patient and secrete active proteins that can help remove those toxic compounds. We carried out BMT in a mouse model for MSD and saw beneficial effects on peripheral organs, such as the liver and heart, but less change in neurological symptoms. Our results will be useful for the design of potential cell therapy approaches that could be used clinically to treat MSD.
Presa et al. use a mouse model of multiple sulfatase deficiency (MSD) to assess efficacy of bone marrow transplantation to increase sulfatase activity. Sulfatase activity is partially restored in some peripheral tissues such as spleen, liver and heart, but no significant benefit is observed in the brain.
Introduction
Multiple sulfatase deficiency is an ultra-rare, autosomal recessive lysosomal storage disorder (LSD). MSD is characterized by a severe reduction of all sulfatase activities1,2. Sulfatases catalyze the hydrolysis of sulfate esters in macromolecules including glycosaminoglycans (GAGs) and sulfolipids. Formylglycine-generating enzyme (FGE) converts an active-site cysteine residue to C-α-formylglycine, a post-translational modification required for sulfatase activity. MSD patients have mutations in the sulfatase modifying factor 1 (SUMF1) gene, which encodes FGE, leading to deficiencies in sulfatase activation and function3,4. There are 17 identified cellular sulfatases in the human genome, most of which are located in the lysosomes5,6. The lack of sulfatase activities results in the accumulation of sulfatase substrates in the lysosomes7, ultimately leading to impaired lysosomal function and a widespread inflammatory response that was identified in a Sumf1 knock-out mouse model for MSD by the presence of highly vacuolated macrophages, which are the primary location of lysosomal storage8.
MSD is estimated to occur in one in every 500,000 individuals2,9. Most patients carry hypomorphic SUMF1 variants, allowing a variable degree of residual sulfatase activities10. The clinical manifestations—including orthopedic disease, vision and hearing loss, cardiac involvement, and progressive, lethal neurologic deterioration—result from the additive effects of each individual sulfatase deficiency1,2,11. Currently, there are no approved disease-modifying therapies.
Many treatments under development for LSDs are based on the biological phenomenon of cross-correction, whereby functional enzymes are secreted by cells, taken up, and sorted to the lysosomes of neighboring enzyme-deficient cells12–14. Based on this phenomenon, therapies aim to provide a source of functional enzymes. One such approach is allogeneic hematopoietic stem cell transplant (HSCT), in which metabolically-competent cells from healthy donors are transplanted into patients. These transplanted cells and their progeny can ameliorate disease by providing a permanent source of functional enzymes that cross-correct patients’ cells15. Importantly, donor-derived monocytes have been shown to cross the blood-brain barrier, engraft into the brain, and differentiate into resident microglia16. Therefore, HSCT has the potential to treat patients with severe CNS involvement as functional enzymes can be secreted from microglia-like cells after engraftment.
Additionally, healthy macrophages and microglia derived from donor hematopoietic stem cells could directly modulate local inflammation that occurs in the diseased state. Neuroinflammation, associated with microglial and astrocyte activation, is a hallmark of LSDs with CNS involvement and is thought to be a key factor leading to neurodegeneration17. The restoration of normal microglial activity after HSCT may act on neuroinflammation and augment outcomes18.
HSCT has shown a marked benefit in some LSDs, most notably mucopolysaccharidosis type I19–21. Because of this success, we sought to determine if this treatment would improve disease phenotypes in MSD. Although MSD is caused by FGE dysfunction, the rationale to apply HSCT to this disease is that healthy donor cells will serve as sources of activated sulfatases in patients8. In fact, HSCT has recently been trialed in two human patients with MSD. Full reports of outcomes are pending22. To thoroughly investigate the utility of this approach for MSD in a formalized preclinical setting, we performed HSCT on a clinically relevant mouse model of MSD, Sumf1(S153P)23. We used CD45.1+ bone marrow cells derived from JaxBoys mice as source for Sumf1(+/+) cells, and Sumf1(S153P) mice as recipients. Our results indicate that sulfatase activity is partially restored in some peripheral tissues such as spleen, liver and heart, while not significant benefit is observed in brain. Overall, our data support the use of HSCT for the treatment of attenuated MSD cases with mild CNS pathology.
Methods
Mice
All animal experiments and care procedures adhere to OLAW guidelines and were approved by the Institutional Animal Care and Use Committee at The Jackson Laboratory. The Sumf1(S153P)23 mice (stock number 31558, C57BL/6J-Sumf1em8Lutzy/Mmjax) were maintained as homozygous colony at The Jackson Laboratory, and used as MSD model. We used as Sumf1 wild type recipients, C57BL/6J mice (stock number 664). As Sumf1(+/+) donor strain, we used JAXBoy mice (stock number 33076, C57BL/6J-Ptprcem6Lutzy/J), a strain carrying the variant p.Lys302Glu in the Ptprc gene, responsible for the Ptprca allele (CD45.1)24. The Sumf1(A277V)23 mice (stock number 31423) were maintained as homozygous colony at The Jackson Laboratory, and utilized for sulfatide analysis. All animals were maintained in an SPF facility, housed in individually and positively ventilated polysulfonate cages with HEPA filtered air at a density of 3–4 mice per cage. Filtered tap water, acidified to a pH of 2.5 to 3.0, and normal rodent chow will be provided ad libitum. Animals were weekly checked for welfare and individuals presenting more than 15% of maximum body weight loss were considered at humane endpoint and euthanized.
Bone marrow transplants
Each cohort of recipient mice (CD45.2+) consisted of 10 mice by sex. For preconditioning, 4 week-old recipient mice received 750 cGy in a Cs137 gamma irradiator25. Donor cells (CD45.1+) were collected from long bones, and iliac crest from 8 to 10 week-old and sex matched JaxBoys. After purification by ficoll gradient, the cell concentration was adjusted to inject 10 million live bone marrow cells by mouse, using i.v. delivery, via retro-orbital injection at 24 h after irradiation. Donor and recipient mice were sex matched. The experimental groups were Sumf1(S153P) (CD45.2) receiving Sumf1(+/+) (CD45.1) bone marrow (BM), Sumf1(S153P) (CD45.2) receiving Sumf1(S153P) (CD45.2) BM, Sumf1(+/+) (CD45.2) receiving Sumf1(+/+) (CD45.1) BM, and no BMT controls Sumf1(+/+) and Sumf1(S153P).
Flow cytometry
For in vivo PBL analysis at 4 and 8 weeks post-transplant, 100–200 µl of blood was collected and mixed with 10 mM EDTA. RBC were lysed using Gey’s Buffer. PBL’s were washed using PBS with 2% FBS (FACS Buffer), and finally resuspended in 50 µl of FACS buffer for further antibody staining. For bone marrow analysis at end point (10 months post-transplant), long bones were crushed in a mortar and flushed with FACS Buffer, then strained through a 35 µm filter, in 1 ml of FACS Buffer. For spleen cell staining, spleens were mashed with forceps and filtered through nylon mesh in 2 ml of FACS Buffer. RBC were lysed using Gey’s buffer, and splenocytes washed using FACS Buffer. Aliquots of respective cells were stained with each panel respectively (Fig. S1B). After 30 min of incubation at 4 °C in the dark, the cells were washed with 2 ml FACS Buffer. All cells were resuspended in 200 µl of FACS buffer and stained with DAPI. All samples were run on a BD FACSymphony A5 SE (BD Biosciences). All data was analyzed using FlowJo (version 10.8.0). Gating strategy is shown in the supplemental figures (S.1-A for Spleen and S.1.B for bone marrow).
Electroretinography
Electroretinography (ERG) assesses the electrical responsiveness of the rods and cones of the retina in reaction to flashes of light as a measure of visual function. Mice are dark-adapted for a minimum of 120 min within a light-tight and ventilated adaptation chamber. The eyes are dilated with an ophthalmic solution and mice are anesthetized by an intraperitoneal injection of a ketamine/xylazine mixture.
When fully anesthetized, the mouse is placed on the heated platform of the ERG system.
Two electrodes (i.e., sub-dermal needles) are placed: The ground electrode is placed under the skin at the base of the tail. The positive reference electrode is placed under the skin on the bridge of the nose between the eyes. The recording electrode, (a loop of fine gold wire) is placed in contact with the corneal surface through the thin layer of methylcellulose or hypromellose.
The rod test is performed first. The eyes are subjected to light flashes of varying intensity. Following a10-min light adaptation interval, the cone test is performed with the system’s dome light on. Cones are tested with brighter light flashes than are the rods.
The test duration ranges from 30 to 60 min.
Sulfatase analysis
Arylsulfatase A (ARSA) was assessed in tissue homogenates using the Sulfatase assay kit from Sigma Aldrich (MAK276) following vendor instructions. Briefly, frozen tissue was homogenized at 50 mg/ml in PBS supplemented with 1x HALT protease inhibitor (Cat# 78425, ThermoFisher Scientific), incubated in ice 15 min, and centrifuged 5 min at 4 °C and 16800 × g. Total protein concentration was assessed in tissue lysates by DC assay (BioRad). For ARSA assay, 10 µl of tissue lysate were mixed with 90 µl of assay buffer, followed by 2 h incubation at 37 °C. The hydrolysis, at pH 5, of a sulfate ester from 4-nitrocatechol sulfate, produces 4-nitrocatechol, which is detected measuring absorbance at 515 nm in a microplate reader (SpectraMaxi3x, Molecular Devices). One unit of ARSA activity is defined as the amount of enzyme that generates 1.0 mole of 4-nitrocatechol per minute at pH 5 at 37 °C. ARSA activity in each assay was normalized to the average Sumf1(+/+) activity and represented as percentage of Sumf1(+/+). For Arylsulfatase B (ARSB) analysis, 2 µg of protein were adjusted to 10 µl with water and mixed with 10 µl of 6.25 mM 4-MU sulfate (4-methylumbelliferly, Sigma M7133) in substrate buffer (0.375 mM AgNO3, 0.1 M NaOAc, pH 5). After 3 h of incubation at 37 °C, the reaction was stopped by adding glycine-carbonate buffer pH 10.7. The 4-MU fluorescence was measured at 365 nm excitation and 559 nm emission in a fluorometer (SpectraMax i3). All samples were subtracted the background of non-sample control and ARSB activity calculated as the amount of 4-MU nmol produced in 1 h by 1 mg of protein. For measurement of SGSH enzymatic activity, 2 µg of protein were adjusted to 10 µl with water and mixed with 20 µl of 10 mM substrate 4MU alpha -N-sulpho-D-Glucosaminide (Carbosynth, EM06602) and incubated at 37 °C for 17 h. The reaction was stopped by adding 6 of Pi/Ci buffer (0.2 M Na2HPO4/0.1 M citric acid, pH 6.7). The reaction followed with addition of 10 µl of α-Glucosidase 10 U/ml (G5003, Sigma) and incubate at 37 °C for 24 h. The final reaction was stopped by addition of 200 µl stop buffer (0.025% Triton X-100, in 0.5 M NaHCO3/0.5 M Na2CO3 pH 10.7). Fluorescence was read on a SpectraMax i3 microplate reader, using excitation 365 nm and emission of 559 nm. SGSH activity was defined as the amount of 4-MU nmol produced in 1 h by 1 mg of protein. All samples were analyzed in duplicates. enzymatic activity data was represented as percentage of average Sumf1(+/+) activity.
GAG analysis
For GAG analysis, we used the Glycosaminoglycans Assay Kit (Cat#6022 Chondrex) following vendor protocols. This kit is optimized for the extraction and quantification of sulfated Glycosaminoglycans (sGAGs), using the cationic dye 1,9-dimethylmethylene blue (DMB) which binds to sulfated GAGs. A standard curve with chondroitin sulfate was used for quantification. DNA was extracted from tissue lysates by ice cold 100 % ethanol precipitation and concentration determined in a spectrophotometer measuring absorbance to 260 nm. The GAG content in tissues was normalized to DNA amount and represented as µg sGAG/µDNA.
Sulfatides analysis
Sulfatides were extracted based on protocol from Mirzaian et al.26 with spike-in N-octadecanoyl-D3-sulfatide at 10 pmol/mL for sulfatides and N-acetylsulfatide at 100 pmol/mL for lysosulfatides. Extracted samples were analyzed in triplicate for 14 endogenous sulfatides. For each sample replicate, the sum of sulfatide peak intensities was determined. Finally, strain means were calculated among sample groups after calculating a sample mean across replicates.
The ratio of the sulfatide peak intensity to the spike-in intensity and the spike-in concentration was used to compute the concentrations of sulfatide and lysosulfatide.
Histology
Tissues were fixed in 10% NBF during 24–48 h, then processed for paraffin embedding. Brains were cut in 6 µm thickness sagittal sections, and slides were processed in a Leica Bond auto-staining system (Leica Biosystems Deer Park, IL). The following primary antibodies were utilized: anti-CD68 (Abcam, ab125212), anti-GFAP (Abcam, ab16997), anti-Iba1 (Abcam, ab178846), anti-Neun (Abcam, ab104225).
The slides were digitalized using a Nanozoomer S210 (Hamamatsu) at 40x magnification. For the brain image analysis, two regions of interest (ROI) were selected at 40x magnification for the CA1 region of hippocampus and at 20x magnification for the motor cortex and visual cortex. Three ROI were taken at 10x magnification for the heart and liver. CellProfiler (version 4.2.4), a free open-source software, was used to import the ROI and analyze them through a pipeline to evaluate the number of positive cells body or total stained area. The percentage of Iba-1+ was reported on the total of nuclei stained in hematoxylin, and total stained area. The percentage of surface occupied by CD68, GFAP and LAMP1 staining was measured using a threshold parameter reported on total surface of the ROI. To prevent analysis bias, the analysis was performed in a double-blinded manner.
Statistics and reproducibility
The sample size (n) is indicated in each figure, as well as in supplemental material. Each mouse is considered a biological replicate. Two to three technical replicates were used in biochemical assays. Comparisons between multiple groups were performed by Two-way ANOVA followed by correction for multiple comparisons test, using GraphPad Prism version 9.2.0 for Windows (GraphPad Software, San Diego, CA). A p < 0.05 was considered statistically significant. ROUT test was applied to analyze outliers.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Efficient engraftment rate and long-term stability in Sumf1(S153P) mice
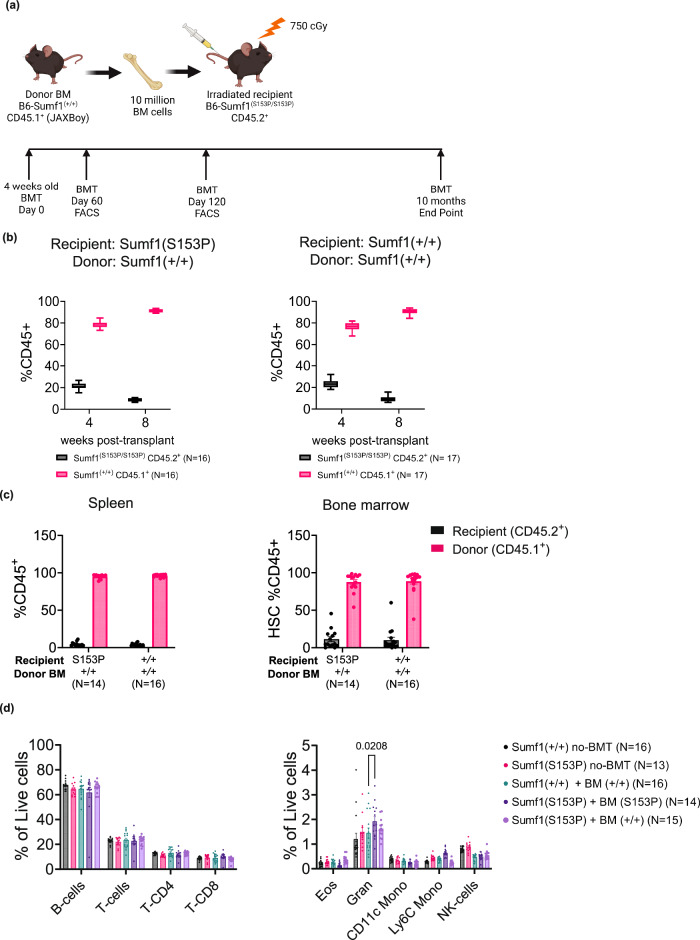
The Sumf1 deficient mice present a severe neonatal onset and very aggressive disease development, characteristics that make them a very difficult model to test HSCT as potential therapy for MSD. Thus, recently developed mice models carrying the clinical mutations p.Ser155Pro (equivalent to mouse p.Ser153Pro) and p.Ala279Val (equivalent to mouse p.Ala277Val), do not show the severe neonatal lethality observed in a complete Sumf1 null mouse. However, they still present a severe reduction in sulfatase activity, and cellular histopathology associated to lysosomal disfunction, macrophage, microglia and astrocytes hyperactivation23. These characteristics make them very useful to assess efficacy of HSCT therapy. To this end, we selected as HSCT recipient, homozygous Sumf1(S153P) mice and C57BL/6J mice (Sumf1(+/+)). We selected as donor Sumf1(+/+) HSC, a recently stablished Ptprc KI mouse strain carrying the CD45.1 allele (Ptprc(K305E/K305E), also known as JAxBoy)24. Since both recipient strains are homozygous for the CD45.2 allele (Ptprc(K305K/K305K)), common to all C57BL/6 \J derived stocks, this system allows us to track in vivo by flow cytometry the engraftment efficiency (Fig. 1a). Ten recipient mice by sex at 4 weeks of age were sublethal irradiated with 750 cGy in a cesium irradiator. The following day, the recipients were engrafted via intravenous (i.v.) retro-orbital injection with 10 million bone marrow cells, derived from sex matched Sumf1(+/+) JaxBoy donors (CD45.1+) or Sumf1(S153P) mice for syngeneic control (Fig. 1a). Engraftment efficiency was tested by flow cytometry at 2- and 4-months post-transplant in blood of Sumf1(S153P) and Sumf1(+/+) mice engrafted with CD45.1+ Sumf1(+/+) bone marrow cells (Supplementary Fig. 1a, b). The frequency of circulating donor cells increased from 80 % at 4 weeks to 90% after 8 weeks post-transplant in both Sumf1(S153P) and Sumf1(+/+) recipients (Fig. 1b). Engraftment levels remained elevated after 10 months post bone marrow transplant (BMT). Donor cells were detected in bone marrow and spleen at 90% and 95% of all resident leukocytes respectively (Fig. 1c). Donor hematopoietic stem cells not only engrafted efficiently in recipient mice but also produced normal frequencies of the most common leukocyte populations (Fig. 1d).
Fig. 1. High engraftment efficiency and stability after HSCT.
a Experimental design for HSCT strategy, Created with BioRender.com. Recipient mice at 4 weeks of age, N = 10 by sex and group were irradiated in a Cs137 irradiator at 750 cGy. Mice received 10 million bone marrow cells isolated from Sumf1(+/+) CD45.1+ (JaxBoy) or Sumf1(S153P) for syngeneic control. b At one and 2 months post-transplant, the engraftment efficiency was tested by FACS on PBL’s from Sumf1(S153P) and Sumf1(+/+) recipients. The frequency of circulating leukocytes CD45.1+ and CD45.2+ is reported as percentage of total CD45+ cells. c After 10 months post-transplant, engraftment efficiency was analyzed by FACS in spleen and bone marrow. d Major leukocyte populations were analyzed in spleen. Data points represent individual mouse, and bar graphs display mean ± SEM. Multiple comparisons test using Kruskal-Wallis method. Exact P values are in Supplementary Data 1.
Partial restoration of sulfatase activity and reduction of GAG accumulation after HSCT
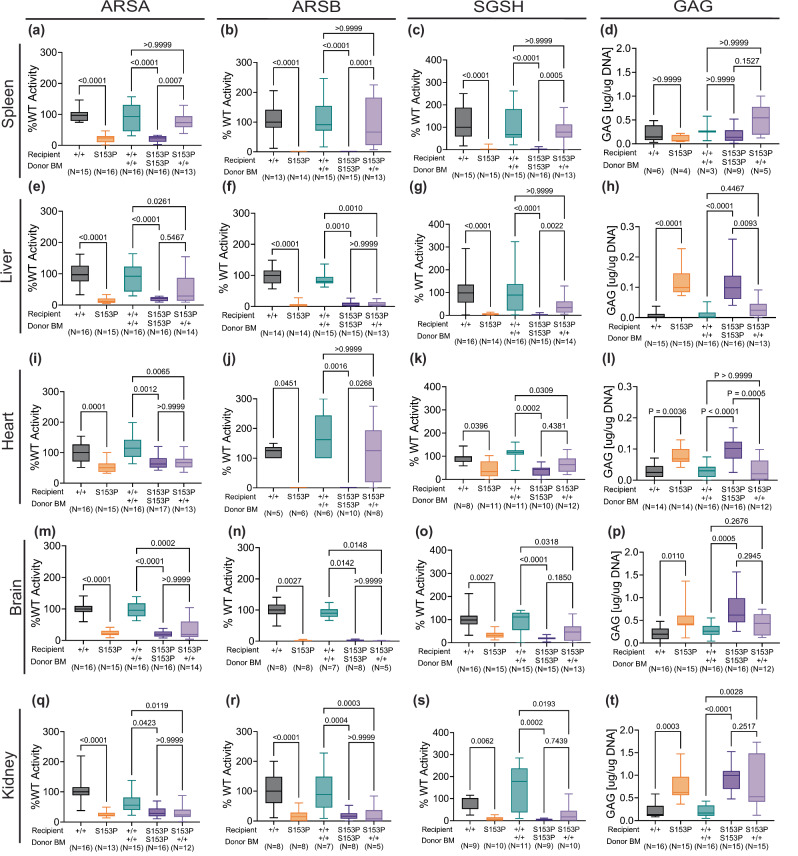
Sumf1(S153P) mice have a severe reduction in enzymatic activity of several sulfatases23. To assess efficacy of HSCT and the level of cross-correction, we tested at 10 months post-transplant the enzymatic activity levels for three common sulfatases, arylsulfatase A (ARSA), arylsulfatase B (ARSB) and sulfamidase (SGSH). ARSA levels were significantly restored to wild type levels in spleen of Sumf1(S153P) mice receiving Sumf1(+/+) bone marrow (Fig. 2a). In contrast, ARSA activity levels were only slightly increased in liver and brain of Sumf1(S153P) mice receiving Sumf1(+/+) bone marrow (Fig. 2e, m), while no significant changes were detected in heart and kidney (Fig. 2l, q). ARSB activity was barely detected in Sumf1(S153P) mice without HSCT or in the syngeneic control group engrafted with Sum1(S153P) bone marrow, while Sumf1(S153P) mice receiving Sumf1(+/+) bone marrow, showed normal ARSB activity levels in spleen, and heart (Fig. 2b, j), but unfortunately not in liver, brain, and kidney (Fig. 2f, n, r). Sulfamidase activity levels were also extremely low at base line in Sumf1(S153P) mice without HSCT and in the syngeneic control group. Sumf1(S153P) mice engrafted with Sumf1(+/+) bone marrow showed a significant increase in SGSH levels in spleen and liver (Fig. 2c, g), while heart, brain, and kidney only showed a mild increase (Fig. 2k, o, s).
Fig. 2. HSCT results in partial restoration of sulfatase activity.
Enzymatic activity for Arylsulfatase-A (ARSA), Arylsulfatase-B (ARSB), N-Sulfoglucosamine Sulfohydrolase (SGSH) and Glycosaminoglycans accumulation (GAG) was assessed in spleen (a–d) liver (e–h), heart (i–l), brain (m–p) and kidney (q–t) tissues, 10 months post-BMT. Activity is represented as percentage of average WT activity. GAG amounts in µg is normalized to µg of DNA content. Recipient and donor Sumf1 genotype is indicated as WT (+/+), or Sumf1(S153P) homozygous (S153P). Data is represented as box plots with median, min and max. Kruskal-Wallis tests and Dunn’s multiple comparisons test were used. Exact P values are in Supplementary Data 1.
Glycosaminoglycans (GAG) accumulation is a known consequence of the sulfatase deficiency caused by Sumf1 loss of function mutations. Sumf1(S153P) mice showed a significant increase in total GAG accumulation in tissues like liver, heart, brain, and kidney (Fig. 2h, l, p, t). Sumf1(S153P) mice engrafted with Sumf1(+/+) bone marrow showed a significant reduction in liver GAG content in liver and heart (Fig. 2h, l), while a small reduction was detected in brain and kidney (Fig. 2p, t).
ARSA deficiency also results in the accumulation of sulfatides and lysosulfatides which can be detected in plasma of MLD patients and mouse models26 and MSD patients1,2. To assess the potential use of sulfatides as an additional biomarker, we quantified total sulfatides and lysosulfatides in plasma from 6 month old MSD-mice, Sumf1(S153P) and Sumf1(A277V). Both strains showed similar levels to age matched Sumf1(+/+) mice (Fig. S2A, C, D). However, the sulfatides profile was altered in both strains, showing a small reduction in C16:0-OH, and C24:1, while presenting a significant increase in C22:0 species (Supplementary Fig. 2b). This findings indicate that the residual ARSA activity in the two Sumf1 mutant mice is sufficient to avoid accumulation of sulfatides in contrast to ARSA(-/-) models26,27. Thus, sulfatides quantification is not a good biomarker in this MSD mouse model, and was omitted from the analysis of MSD-HSCT samples.
Lysosomal and inflammatory pathology is reduced in peripheral organs after HSCT
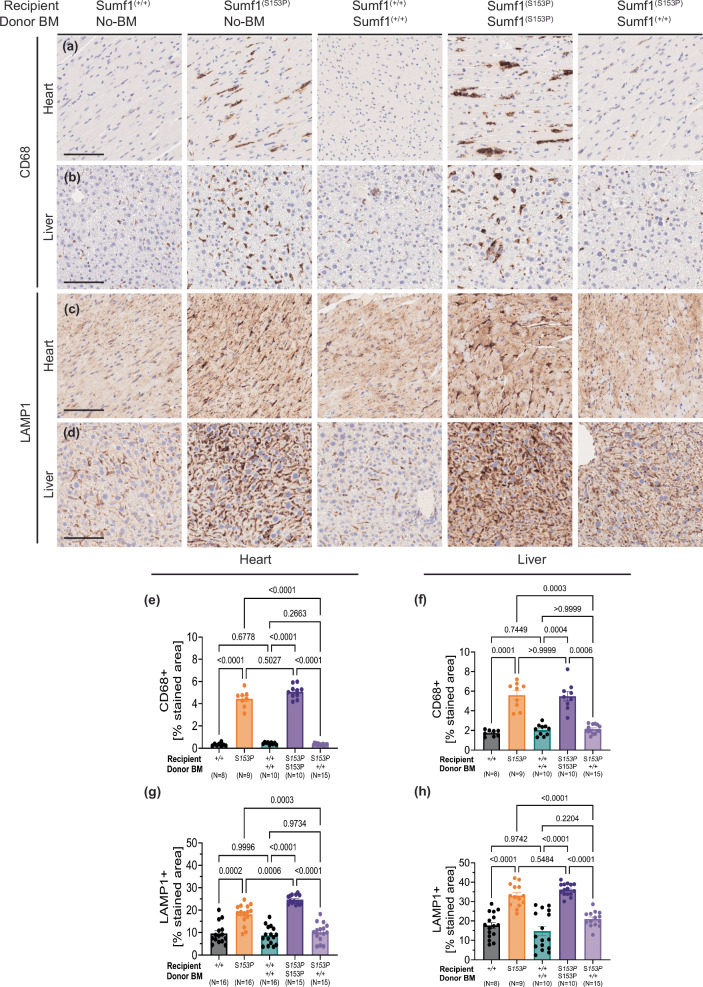
Generalized pro-inflammatory response and lysosomal pathology has been reported in Sumf1 deficient mice8 and recently documented in two MSD-mouse models, Sumf1(S153P) and Sumf1(A277V) mice23. Compared to Sumf1(+/+) mice, Sumf1(S153P) untreated and syngeneic controls showed in heart and liver, a remarkable increase of CD68+ infiltrating cells (macrophages and monocytes) (Fig. 3a, b), accompanied by an increased lysosomal accumulation (Fig. 3c, d). In contrast, Sumf1(S153P) engrafted with Sumf1(+/+) bone marrow, showed a reduction of both CD68+ cells (Fig. 3a, b) as well as lysosome accumulation (Fig. 3c, d). Both the frequencies of inflammatory cells (Fig. 3e, f), as well as the lysosome-stained area (Fig. 3e, f), in Sumf1(S153P) engrafted with Sumf1(+/+) bone marrow, were similar to Sumf1(+/+) mice. Thus, in organs like liver and heart, the reduction in GAG accumulation (Fig. 2h, l) and increased ARSB and SGSH (Fig. 2j, g), correlates with the efficacy detected by histopathology of inflammation and lysosome markers.
Fig. 3. Reduced systemic inflammation and lysosome pathology in Sumf1(S153P) mice after HSCT.
Representative CD68 and LAMP1 immunohistochemistry on Heart (a, c) and Liver (b, d) at 10 months post-BMT. Genotype for recipients and donors is indicated. Image magnification: x10 scale bar, 100 μm. Quantification represents percentage of stained area in Heart (e, g) and Liver (f, h) over total area shown as bar graphs with mean ± SEM, dots represent individual mouse. Multiple comparisons were performed using Brown-Forsythe and Welch ANOVA tests. Exact P values are in Supplementary Data 1.
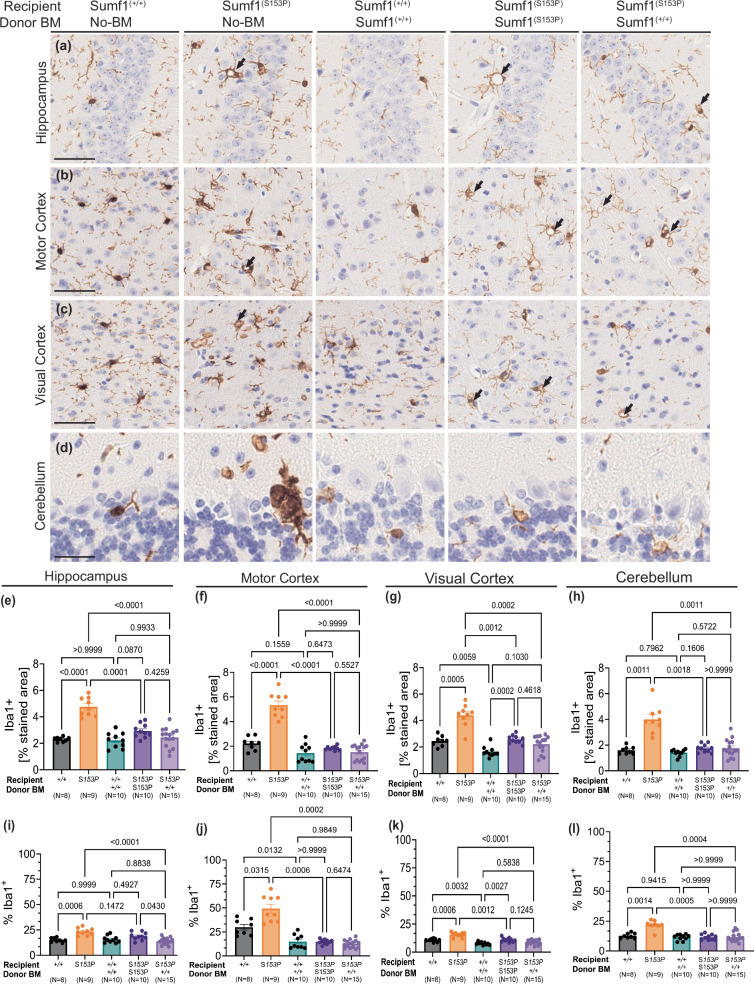
The Sumf1(S153P) mice exhibit a distinctive brain phenotype marked by microgliosis and astrocytosis. Given that hematopoietic stem cells (HSCs) possess the capacity to differentiate into myeloid cells, particularly monocytes capable of traversing the blood-brain barrier and reaching the brain, they serve as a potential source of actively secreted sulfatases for cross-correction. To assess the efficacy of HSCT into the CNS pathology, we performed immunohistochemistry analysis of Iba1 (activated microglia), GFAP (astrocytes), Lamp1 (lysosomes), and Neun (neurons). As expected, severe microgliosis was present in hippocampus, visual and motor cortex, and cerebellum regions of untreated-Sum1(S153P) mice (Fig. 4a–d). Both total Iba1-stained area (Fig. 4e–h) and frequency of Iba1+ cells (Fig. 4i–l) are significantly increased in untreated-Sumf1(S153P) mice compared to Sumf1(+/+) controls. Notably, microgliosis (measured as stained area and frequency Iba1+) was significantly reduced in Sumf1(S153P) mice engrafted with Sum1(S153P) bone marrow (syngeneic control) compared to untreated-Sumf1(S153P) mice (Fig. 4e–l). This is a consequence of the preconditioning regimen of gamma radiation, since even Sumf1(+/+) mice engrafted with Sumf1(+/+) bone marrow showed significant microglia reduction in cortical brain regions (Fig. 4j–k). Similarly, Sum1(S153P) mice engrafted with Sumf1(+/+) bone marrow, showed a significant reduction of microgliosis compared to untreated-Sum1(S153P) mice, but almost identical levels to Sum1(S153P) bone marrow (syngeneic control). Although, microgliosis seems affected by the preconditioning treatment, it is very clear in Sum1(S153P) recipients of either Sumf1(+/+) BM or syngeneic control, the presence of typical cytoplasmic morphology with vacuolization (Fig. 4a–d, black arrows). Since the microglia in both groups shares similar morphology, we might hypothesize that there is a low frequency of donor-derived myeloid cells into the brain, or perhaps insufficient cross-correction.
Fig. 4. Persistent microgliosis after HSCT.
Microglia assessed by Iba-1 immunohistochemistry on brain of Sumf1(+/+) no-BMT, Sumf1(S153P) no-BMT, recipient Sumf1(+/+) + BM-Sumf1(+/+), recipient Sumf1(S153P) + BM-Sumf1(S153P), and recipient Sumf1(S153P) + BM-Sumf1(+/+) at 10 months post-transplant. Representative images from hippocampus (a), motor cortex (b), visual cortex (c), and cerebellum (d). Microgliosis is represented as the percentage of stained area for Iba-1, and as percentage of Iba-1+ cells over total nucleated cells, in hippocampus (CA1) (e, i), motor cortex (f, j), visual cortex (g, k) and cerebellum (h, l) shown as bar graphs with mean ± SEM, dots represent individual mouse. Images (a–c) at x20 magnification, 50 μm scale bar. Images for cerebellum at x40, 25 μm scale bar. The black arrows point to vacuolated Iba-1+ cells. Multiple comparisons using Brown-Forsythe and Welch ANOVA tests. Exact P values are in Supplementary Data 1.
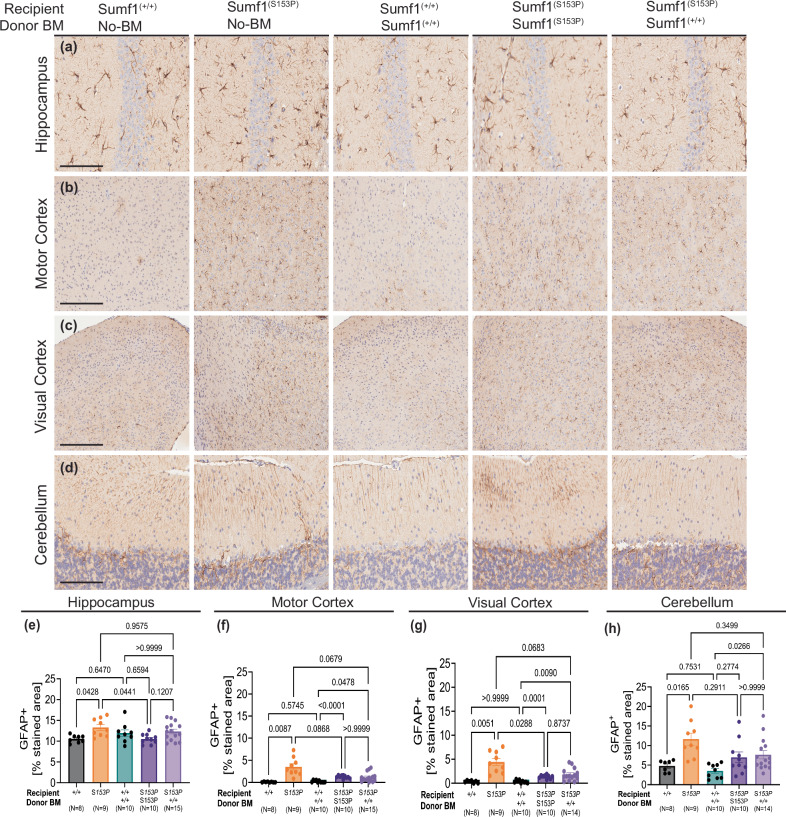
The astrocyte marker GFAP, was significantly increased in hippocampus, cortex (visual and motor), and cerebellum areas of untreated-Sumf1(S153P) mice (Fig. 5a–d). Similarly to microgliosis, astrocytes staining was reduced in Sumf1(S153P) syngeneic control, also consequence of the use of gamma radiation for preconditioning (Fig. 5e–h). The GFAP level in Sumf1(S153P) mice engrafted with Sumf1(+/+) bone marrow was also similar to Sumf1(S153P) syngeneic control but slightly higher than Sumf1(+/+) control group receiving Sumf1(+/+) bone marrow (Fig. 5e–h). This indicates that there is not significant correction of astrocytosis in the brain regions analyzed.
Fig. 5. HSCT was not able to ameliorate Astrocytosis.
GFAP immunohistochemistry on brain of Sumf1(+/+) no-BMT, Sumf1(S153P) no-BMT, recipient Sumf1(+/+) + BM-Sumf1(+/+), recipient Sumf1(S153P) + BM-Sumf1(S153P), and recipient Sumf1(S153P) + BM-Sumf1(+/+) at 10 months. Representative images from hippocampus (a), motor cortex (b), visual cortex (c), and cerebellum (d), Quantification of astrocytosis as %Stained Area of GFAP+ in Hippocampus (CA1) (e), Motor Cortex (f), Visual Cortex (g) and Cerebellum (h) shown as bar graphs with mean ± SEM, dots represent individual mouse. Representative images (a–c) at x20 magnification, 50 μm scale bar. Cerebellum images at x40, 25 μm scale bar. Multiple comparisons using Brown-Forsythe and Welch ANOVA tests. Exact P values are in Supplementary Data 1.
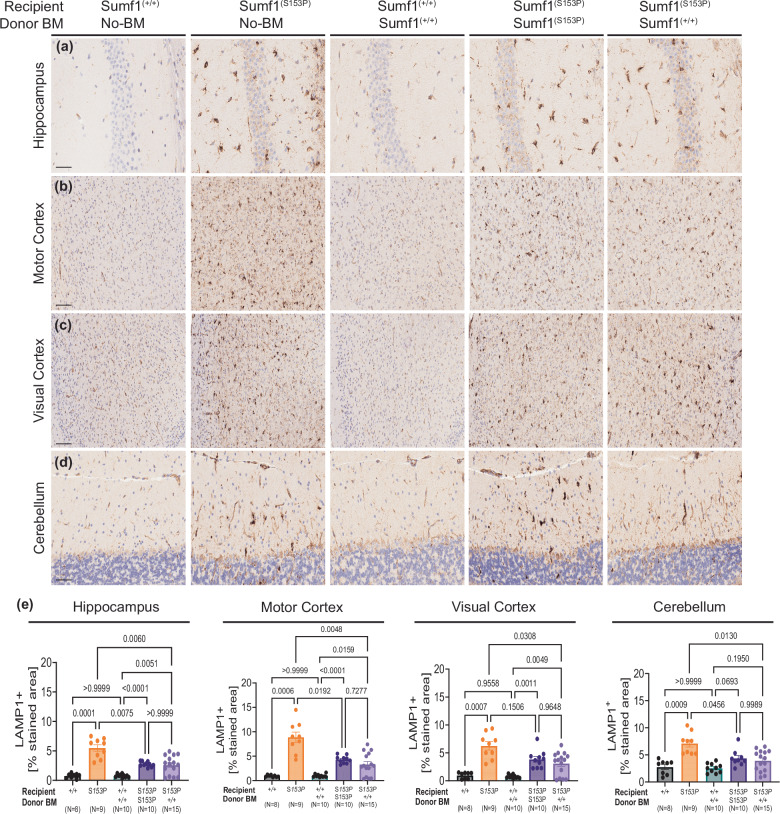
The lysosomal pathology observed in the brains of Sumf1(S153P) mice is characterized by and increased number and size of lysosomal organelles23 Immunohistochemical analysis revealed a significant elevation in the lysosomal marker Lamp1 in brain regions of untreated-Sumf1(S153P) mice compared to Sumf1(+/+) control mice (Fig. 6a–d). However, no significant differences in Lamp1 staining were observed between Sumf1(S153P) mice engrafted with Sumf1(+/+) bone marrow and the Sumf1(S153P) syngeneic control mice (Fig. 6e). Although both transplanted groups exhibited reduced Lamp1 staining compared to untreated-Sumf1(S153P) mice, this reduction appears to be primarily attributed to the decrease in microglia and astrocyte populations resulting from irradiation preconditioning. These findings suggest that despite the decrease in inflammatory myeloid cells following transplantation, the lysosomal pathology remains active and is not significantly influenced by the introduction of Sumf1(+/+) donor-derived myeloid cells. Furthermore, in addition to the ongoing neuroinflammatory phenotype, we did not observe any indications of neuronal loss based on Neun immunohistochemistry analysis (Supplementary Fig. 3a–h).
Fig. 6. Brain lysosomal pathology is not corrected by HSCT.
Lysosomal pathology is assessed by LAMP-1 immunohistochemistry on brain regions of Sumf1+/+, and Sumf1(S153P), receiving bone marrow from Sumf1+/+ (CD45.1+, JaxBoy) or Sumf1(S153P), at 10 months post-transplant (N = 8–15, mixed sex). Representative images of (a) hippocampus (CA1), (b) motor cortex, (c) visual cortex, (d) cerebellum. e The percentage of stained area for LAMP-1 over total area is shown as mean ± SEM, for each brain region. Image magnification: Cerebellum and Hippocampus at x40 magnification, scale bar 50 μm. Motor and visual Cortex at x20 magnification, scale bar 100 μm. Data points represent individual mouse, and bar graphs indicate the mean ± SEM. Statistical significance assessed using correction for multiple comparisons by Brown-Forsythe and Welch ANOVA tests. Exact P values are in Supplementary Data 1.
We previously reported a vision phenotype in Sumf1(S153P) mice, characterized by degeneration of rods and cones23. The electro-retinography analysis at 6 months post-BMT showed a reduction in the A-wave and B-wave response in rods, affecting all the groups subjected to BMT, either Sumf1(+/+) or Sumf1(S153P) recipients (Supplementary Fig. 4a, b). Unfortunately, the preconditioning method affected the base line response of our syngeneic controls, introducing a confounding component in the vision phenotype of Sumf1(S153P) mice.
Discussion
In this study, we evaluated the therapeutic potential of allogeneic bone marrow transplant to ameliorate disease phenotypes in a clinically relevant mouse model of MSD. While recipient mice showed very high, long-term engraftment as well as normal differentiation of CD45+ donor cells, biochemical and pathological markers of MSD only partially improved after transplant. Specifically, transplant with Sumf1(+/+) bone marrow was effective in improving enzyme activity, GAG accumulation, and inflammation in some peripheral organs, but this rescue was not seen in the brain.
Tissue analyses of ARSA, ARSB, and SGSH activities demonstrate partial restoration in an organ-specific manner after bone marrow transplant in MSD mice. Transplant with Sumf1(+/+) bone marrow significantly increased the activity of all three sulfatases in the spleen. However, in non-lymphoid organs, only SGSH activity was increased in liver, and ARSB activity in heart, with no significant changes in other sulfatases tested. Finally, Sumf1(+/+) bone marrow transplant only minimally increased sulfatase activities in the brain. A possible explanation for the organ-specific improvement of sulfatase activities may be the density of donor cells per organ after transplant. Because the spleen is a major homing site of hematopoietic stem cells after bone marrow transplant28, the increase in sulfatase activities in the spleen is a direct result of a high number of functional donor cells there. Furthermore, the biochemical effects of bone marrow transplant on MSD mice might depend on the specific sulfatase analyzed. For example, only SGSH activity significantly increased in the liver after transplant, while ARSA and ARSB activities were unaffected. One caveat of these preclinical studies of MSD is that endogenous SGSH protein levels are significantly greater in mice as compared to the levels of other sulfatases29, a trend not seen in humans. Therefore, Sumf1(+/+) bone marrow may secrete greater amounts of functional SGSH than ARSA or ARSB for cross-correction, a process that may not be recapitulated in other species.
The effects of allogeneic bone marrow transplant on GAG accumulation in MSD also differed in an organ-specific manner. Tissue analysis of GAG levels after Sumf1(+/+) bone marrow transplant revealed a significant reduction of GAGs in the liver and heart. However, this rescue was not seen in the brain or kidney. To better understand how the decrease in GAG levels relates to the organ-specific improvement in sulfatase activities, future studies could employ mass spectrometry-based measurement of specific GAG subspecies.
Histological analyses of inflammatory and lysosomal pathology markers further demonstrate the restoration of disease phenotypes in peripheral organs after Sumf1(+/+) bone marrow transplant. Our data show a significant reduction in CD68+ cells in the heart and liver of MSD mice after transplant of Sumf1(+/+) but not Sumf1(S153P) syngeneic control bone marrow, suggesting that Sumf1(+/+) bone marrow is able to reduce the presence of activated macrophages and monocytes in these tissues. Surprisingly, the reduction of inflammatory cells also resulted in an impressive reduction of lysosomal pathology in liver and heart, providing evidence of effective cross-correction. Our findings support previous studies indicating that replenishing the hematopoietic system with healthy donor-derived immune cells after bone marrow transplant may be sufficient to reduce inflammation in the periphery19–21.
On the other hand, improvements in brain neuroinflammation may not be a result of Sumf1(+/+) bone marrow, but rather a result of the preconditioning regimen in the transplant process. Despite the decrease in inflammatory CD68+ cells in the heart and liver of treated mice, the direct effect of Sumf1(+/+) bone marrow transplant on microglia in the brain is less clear. Interestingly, both MSD mice that received Sumf1(+/+) and Sumf1(S153P) bone marrow show a reduction in the number of Iba1+ stained cells in the brain. The similar results between these groups suggest that microglia may be sensitive to the gamma radiation used for preconditioning before transplant. Although ultimately beneficial to the disease state of MSD mice, this preconditioning may add a confounding factor when trying to assess the direct effects of Sumf1(+/+) bone marrow transplant on inflammation in the brain. This confounding effect of irradiation is also seen in our GFAP analysis of astrogliosis in the brain, as both Sumf1(+/+) and Sumf1(S153P) bone marrow reduce levels of GFAP staining. Finally, while there is a reduction in the total amount of Iba1 staining in the brain, microglia of transplanted mice still exhibit an amoeboid morphology characteristic of a neuroinflammatory state after transplant30. The vacuolized morphology of the remaining microglia after transplant may suggest that there is a low engraftment of healthy donor-derived microglia to the brain. As a consequence, the amount of secreted enzymes from microglia may have been insufficient to produce efficient cross-correction.
Another hallmark pathological marker in LSDs, and in MSD in particular, is the accumulation of enlarged lysosomes in diseased tissues23. Analysis of Lamp1 staining in various brain regions of transplanted mice showed a partial reduction of Lamp1 in the hippocampus, motor cortex, visual cortex, and cerebellum of mice that received either Sumf1(+/+) or Sumf1(S153P) bone marrow. As with the neuroinflammation, we observed no differences in Lamp1 levels between mice that received either bone marrow suggesting that the reduction in Lamp1 staining is again due to preconditioning irradiation rather than to the bone marrow transplant itself.
A limitation to our study is the lack of a severe neurobehavioral phenotype exhibited by this mouse model of MSD. We previously generated and characterized a mouse model of MSD that harbors a common human pathogenic variant (p.S155P in patients, p.S153P in mice)23. MSD mice have a vision phenotype of retinopathy as measured by electroretinography. This phenotype matches the retinal abnormalities experienced by patients with MSD and related single-sulfatase disorders1,9. In the current study, both Sumf1(S153P) and Sumf1(+/+) control mice that received bone marrow transplant unexpectedly showed a poor rod response and a decrease in vision performance (Fig. S4A, B). Again, this suggests a secondary, confounding effect of the irradiation received by the mice for transplant preconditioning. Furthermore, in our previous characterization we found that untreated Sumf1(S153P) mice did not show significant differences as compared to Sumf1(+/+) mice in a battery of neurobehavioral tests. While the mild phenotype of this mouse enables testing of novel therapeutics before the early death seen in the Sumf1 knock-out model8, it limits our ability to provide functional endpoints to examine the rescue of the neurological manifestations seen in MSD after therapeutic interventions such as allogeneic bone marrow transplant in the current study.
Overall, allogeneic Sumf1(+/+) bone marrow transplant can provide some benefit to the peripheral organs of MSD mice but provides minimal improvement to the brain. Neurological manifestations of MSD are the most detrimental to patients as CNS complications predominantly contribute to the clinical presentation of patients31. Therefore, it is imperative that treatments target the CNS to improve overall disease outcomes. Our findings that Sumf1(+/+) bone marrow transplant is unable to fully rescue the brain in MSD mice is consistent with previous studies that demonstrate the difficulty of this approach to treat related LSDs with severe CNS involvement and rapid disease progression at the time of transplant32–37. For example, allogeneic bone marrow transplant is not considered a standard treatment for a related single sulfatase disorder, MPS II, due to the inability to effectively treat CNS manifestations38.
The mechanism of action of allogeneic bone marrow transplant to correct neurological manifestations of MSD is the ability of donor-derived monocytes to cross the blood-brain barrier, differentiate into resident microglia, and engraft in the brain. Here, these microglia can serve as sources of functional enzymes that are secreted to cross-correct neighboring neurons16. Reasons that bone marrow transplant is unable to correct brain pathology in our study might be the slow pace of microglia replacement in the brain or an insufficient number of engrafted cells. The development of methods to accurately measure the number of donor-derived cells in the mouse brain in a translational context would better elucidate these mechanisms. In addition, strategies to accelerate microglial turnover and increase the engraftment of functional donor cells into the brain are needed to improve CNS manifestations after transplant.
The poor outcomes in the brain may also be due to insufficient amounts of functional enzymes being secreted from wild-type Sumf1(+/+) donor cells for cross-correction. Our tissue-specific analyses of sulfatase activities revealed that activities were only rescued in tissues with high hematopoietic stem cell homing after transplant such as the spleen. Tissues such as the liver and brain did not exhibit significant improvement. These findings suggest that while cells from healthy donors contain functional enzymes, these cells may not secrete enough enzymes to be taken up into neighboring cells for cross-correction. Expressing supraphysiological levels of FGE through approaches like AAV or ex vivo lentiviral gene therapy may benefit MSD patients.
In this study, we report that sulfatase activities, GAG accumulation, and inflammation are partially restored in some peripheral tissues of MSD mice that receive wild-type bone marrow. However, the functional, wild-type bone marrow itself was not able to improve outcomes in the brain of MSD mice. Instead, the gamma radiation preconditioning regimen may be sufficient to modulate the inflammatory environment and rescue neuroinflammation. Overall, this study supports the potential therapeutic efficacy of allogeneic bone marrow transplant for MSD patients with attenuated forms of the disease and for those who do not have severe CNS involvement.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors are grateful for funding and support from the United MSD Foundation (to C.L. and M.P.). This work was supported by The Center for Precision Genetics at The Jackson Laboratory (NIH grant U54 OD020351 and U54 OD030187) (to C.L.) and the Mouse Mutant Resource and Research Center (NIH grant U42 OD010921) (to C.L.). The authors wish to acknowledge the Scientific Services at The Jackson Laboratory for assistance in flow cytometry, histology and proteomics analysis. These services are supported by NIH grant CA034196. The authors are also grateful to Amber Olsen and Alan Finglas for their endless dedication and discussions in providing the patient perspectives to this work.
Author contributions
Conceptualization: M.P., L.S., T.L., R.C.A., and C.L. Experiments: M.P., S.R., P-A.P., J.R., T.B., and H.C. Analysis: M.P., P-A.P., J. R., and T.B. Writing: M.P. V.P., P-A.P., L.S., T.L., R.C.A., and C.L.
Peer review
Peer review information
Communications Medicine thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
The source data and statistical analysis for main and Supplementary Figs. in this article is organized for each figure and accessible from the Supplementary data file 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s43856-024-00648-y.
References
- 1.Adang, L. A. et al. Natural history of multiple sulfatase deficiency: retrospective phenotyping and functional variant analysis to characterize an ultra-rare disease. J. Inherit. Metab. Dis.43, 1298–1309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlotawa, L., Adang, L. A., Radhakrishnan, K. & Ahrens-Nicklas, R. C. Multiple sulfatase deficiency: a disease comprising mucopolysaccharidosis, sphingolipidosis, and more caused by a defect in posttranslational modification. Int. J. Mol, Sci.21, 3448 (2020). [DOI] [PMC free article] [PubMed]
- 3.Cosma, M. P. et al. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell113, 445–456 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Dierks, T. et al. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human Cα-formylglycine generating enzyme. Cell113, 435–444 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Diez-Roux, G. & Ballabio, A. Sulfatases and human disease. Annu. Rev. Genom. Hum. Genet.6, 355–379 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Parenti, G., Meroni, G. & Ballabio, A. The sulfatase gene family. Curr. Opin. Genet. Dev.7, 386–391 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Dierks, T. et al. Molecular basis of multiple sulfatase deficiency, mucolipidosis II/III and Niemann–Pick C1 disease — Lysosomal storage disorders caused by defects of non-lysosomal proteins. Biochim. et. Biophys. Acta (BBA) Mol. Cell Res.1793, 710–725 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Settembre, C. et al. Systemic inflammation and neurodegeneration in a mouse model of multiple sulfatase deficiency. Proc. Natl Acad. Sci. USA104, 4506–4511 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cappuccio, G., Alagia, M. & Brunetti-Pierri, N. A systematic cross-sectional survey of multiple sulfatase deficiency. Mol. Genet Metab.130, 283–288 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Annunziata, I., Bouchè, V., Lombardi, A., Settembre, C. & Ballabio, A. Multiple sulfatase deficiency is due to hypomorphic mutations of the SUMF1 gene. Hum. Mutat.28, 928 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Hopwood, J. J. & Ballabio, A. Multiple sulfatase deficiency and the nature of the sulfatase family. in The Online Metabolic and Molecular Bases of Inherited Disease (eds. Valle, D. L., Antonarakis, S., Ballabio, A., Beaudet, A.L. & Mitchell, G. A.) (McGraw-Hill Education, 2019).
- 12.Fratantoni, J. C., Hall, C. W. & Neufeld, E. F. Hurler and Hunter syndromes: mutual correction of the defect in cultured fibroblasts. Science162, 570–572 (1968). [DOI] [PubMed] [Google Scholar]
- 13.Hasilik, A., Klein, U., Waheed, A., Strecker, G. & von Figura, K. Phosphorylated oligosaccharides in lysosomal enzymes: identification of alpha-N-acetylglucosamine(1)phospho(6)mannose diester groups. Proc. Natl Acad. Sci. USA77, 7074–7078 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orchard, P. J. et al. Hematopoietic cell therapy for metabolic disease. J. Pediatr.151, 340–346 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Biffi, A. Hematopoietic stem cell gene therapy for storage disease: current and new indications. Mol. Ther.25, 1155–1162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capotondo, A. et al. Brain conditioning is instrumental for successful microglia reconstitution following hematopoietic stem cell transplantation. Proc. Natl Acad. Sci. USA109, 15018–15023 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigante, D., Cipolla, C., Basile, U., Gulli, F. & Savastano, M. C. Overview of immune abnormalities in lysosomal storage disorders. Immunol. Lett.188, 79–85 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Wada, R., Tifft, C. J. & Proia, R. L. Microglial activation precedes acute neurodegeneration in Sandhoff disease and is suppressed by bone marrow transplantation. Proc. Natl Acad. Sci. USA97, 10954–10959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Ru, M. H. et al. Enzyme replacement therapy and/or hematopoietic stem cell transplantation at diagnosis in patients with mucopolysaccharidosis type I: results of a European consensus procedure. Orphanet J. Rare Dis.6, 55 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hobbs, J. R. et al. Reversal of clinical features of Hurler’s disease and biochemical improvement after treatment by bone-marrow transplantation. Lancet2, 709–712 (1981). [DOI] [PubMed] [Google Scholar]
- 21.Kunin-Batson, A. S. et al. Long-term cognitive and functional outcomes in children with Mucopolysaccharidosis (MPS)-IH (Hurler Syndrome) treated with hematopoietic cell transplantation. JIMD Rep.29, 95–102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillai, N. R. et al. Evaluation of the effectiveness of hematopoietic stem cell transplantation in multiple sulfatase deficiency. Mol. Genet. Metab.132, S87 (2021). [Google Scholar]
- 23.Sorrentino, N. C. et al. New mouse models with hypomorphic SUMF1 variants mimic attenuated forms of multiple sulfatase deficiency. J. Inherit. Metabol. Dis.46, 335–347 (2022). [DOI] [PMC free article] [PubMed]
- 24.Mercier, F. E., Sykes, D. B. & Scadden, D. T. Single targeted exon mutation creates a true congenic mouse for competitive hematopoietic stem cell transplantation: the C57BL/6-CD45.1STEM Mouse. Stem Cell Rep.6, 985–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf, D. A. et al. Increased longevity and metabolic correction following syngeneic BMT in a murine model of mucopolysaccharidosis type I. Bone Marrow Transplant.47, 1235–1240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirzaian, M., Kramer, G. & Poorthuis, B. J. H. M. Quantification of sulfatides and lysosulfatides in tissues and body fluids by liquid chromatography-tandem mass spectrometry [S]. J. Lipid Res.56, 936–943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blomqvist, M., Gieselmann, V. & Månsson, J.-E. Accumulation of lysosulfatide in the brain of arylsulfatase A-deficient mice. Lipids Health Dis.10, 28 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapidot, T., Dar, A. & Kollet, O. How do stem cells find their way home? Blood106, 1901–1910 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Xingxuan, He, H. Schuchman, E. & Simonaro, C. A new fluorescent method to detect sulfamidase activity in blood, tissue extracts and dried blood spots. J. Inborn Errors Metab. Screening9, e20200021 (2021).
- 30.Lier, J., Streit, W. J. & Bechmann, I. Beyond activation: characterizing microglial functional phenotypes. Cells10, 2236 (2021). [DOI] [PMC free article] [PubMed]
- 31.Ahrens-Nicklas, R. et al. Complex care of individuals with multiple sulfatase deficiency: Clinical cases and consensus statement. Mol. Genet Metab.123, 337–346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aldenhoven, M. et al. Long-term outcome of Hurler syndrome patients after hematopoietic cell transplantation: an international multicenter study. Blood125, 2164–2172 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Mynarek, M. et al. Allogeneic hematopoietic SCT for alpha-mannosidosis: an analysis of 17 patients. Bone Marrow Transpl.47, 352–359 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Turbeville, S. et al. Clinical outcomes following hematopoietic stem cell transplantation for the treatment of mucopolysaccharidosis VI. Mol. Genet Metab.102, 111–115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welling, L. et al. Early umbilical cord blood-derived stem cell transplantation does not prevent neurological deterioration in mucopolysaccharidosis type III. JIMD Rep.18, 63–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lund, T. C. et al. Outcomes after hematopoietic stem cell transplantation for children with I-cell disease. Biol. Blood Marrow Transpl.20, 1847–1851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solders, M. et al. Hematopoietic SCT: a useful treatment for late metachromatic leukodystrophy. Bone Marrow Transpl.49, 1046–1051 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Selvanathan, A. et al. Effectiveness of early hematopoietic stem cell transplantation in preventing neurocognitive decline in mucopolysaccharidosis Type II: a case series. JIMD Rep.41, 81–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The source data and statistical analysis for main and Supplementary Figs. in this article is organized for each figure and accessible from the Supplementary data file 1.