Abstract
Investigating DNA methylation (DNAm) in cardiac tissues is vital for epigenetic research in cardiovascular diseases (CVDs). During cardiac surgery, biopsies may not be immediately stored due to a lack of human or technical resources at the collection site. Assessing DNAm stability in cardiac samples left in suboptimal conditions is crucial for applying DNAm analysis. We investigated the stability of DNAm in human cardiac tissues kept at 4 °C and 22 °C for periods of 1, 7, 14, and 28 days (exposed samples) using the Illumina Infinium MethylationEPIC v1.0 BeadChip Array. We observed high correlations between samples analysed immediately after tissue collection and exposed ones (R2 > 0.992). Methylation levels were measured as β-values and median absolute β-value differences (|∆β|) ranged from 0.0093 to 0.0119 in all exposed samples. Pairwise differentially methylated position (DMP) analysis revealed no DMPs under 4 °C (fridge temperature) exposure for up to 28 days and 22 °C (room temperature) exposure for one day, while 3,437, 6,918, and 3,824 DMPs were observed for 22 °C samples at 7, 14, and 28 days, respectively. This study provides insights into the stability of genome-wide DNAm, showing that cardiac tissue can be used for reliable DNAm analysis even when stored suboptimally after surgery.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76027-3.
Keywords: DNA methylation, Stability, Epigenetics, Heart tissue, Illumina MethylationEPIC array, Storage conditions
Subject terms: Epigenomics, Medical research
Introduction
DNA methylation (DNAm) is an epigenetic modification that involves the addition of a methyl group to the fifth carbon of the cytosine nucleobase. Typically, this process occurs at CpG sites, where a cytosine nucleobase is followed by a guanine. This dynamic and reversible process is mediated by two enzyme families: DNA methyltransferases and ten–eleven translocation methylcytosine dioxygenases1. Alterations in DNAm levels have been associated with cardiovascular diseases (CVDs). DNAm regulates various inflammatory genes and pathways correlated with CVD risk factors, such as atherosclerosis, inflammation, hypertension, and diabetes2–4. Blood is commonly used due to its easy accessibility and reflection of multiple metabolic and inflammatory pathways. However, DNAm is tissue-specific5,6, and analysis in cardiac tissue could provide further insight into CVD pathogenesis. Conducting DNAm studies on cardiac tissues from living individuals or postmortem is difficult due to the inaccessibility of such samples. Therefore, cardiac tissues from surgeries and autopsies are often stored for subsequent bulk analysis or retrospective studies. During surgery, the delay between tissue sample collection and the subsequent analysis or long-term storage can affect the stability of molecular markers7,8. Additionally, molecular analysis of postmortem cardiac tissues, which could provide insights into cases of Sudden Cardiac Death (SCD), may be affected by the Postmortem Interval (PMI) and the temperature to which the body was exposed before sample collection. While the stability of DNA9,10, RNA11,12, and proteins13,14 has been extensively studied under different storage conditions and tissues, the DNAm stability is much less explored15–20. Studies have shown that DNAm remains stable in blood samples kept at -4 °C, -20 °C, and − 80 °C for years15,16. However, storing blood samples at room temperature longer than three days could compromise DNAm stability17. Researchers investigated postmortem tissues with PMI for up to 72 h and found stable DNAm levels18,19, but the effect of longer PMI is still unknown.
Given the tissue-specific properties of DNAm and its possible impact on CVDs, it is essential to assess DNAm in cardiac tissue to ensure an unbiased downstream analysis and comparability between studies. However, research on DNAm stability is lacking. Therefore, we aimed to investigate the genome-wide DNAm stability in human cardiac tissue from surgeries exposed to different temperatures and time intervals. We tested the DNAm stability in cardiac tissue samples by keeping them in the fridge (4 °C) and at room temperature (22 °C) for up to 28 days before conducting genome-wide DNAm analysis using the Illumina Infinium MethylationEPIC v1.0 BeadChip Array.
Materials and methods
Study design and population
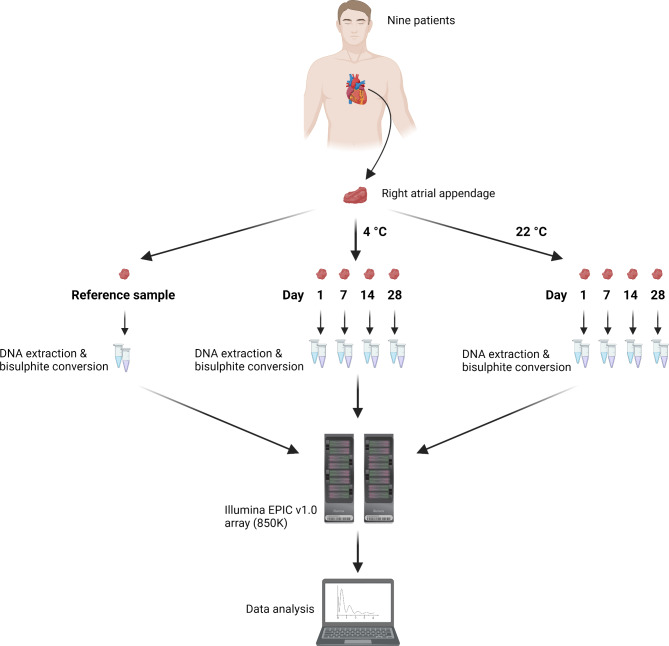
Figure 1 illustrates the experimental design of this study. Right Atrial Appendage (RAA) tissues were collected from nine male patients undergoing cardiac surgery at the Department of Cardiothoracic Surgery, Rigshospitalet, Copenhagen, Denmark. Each RAA sample was sectioned into nine pieces (~ 3 × 3 × 3 mm). One sample piece was analysed immediately after tissue collection (reference sample), and the remaining eight pieces were stored in 2 ml Eppendorf tubes with 10 µl isotonic Phosphate-Buffered Saline (PBS) to maintain the osmotic and ion concentrations similar to those in the human body. The samples were placed in cardboard boxes and kept at 4–22 °C for 1, 7, 14, and 28 days. The clinical characteristics of the nine patients who donated the cardiac samples are presented in Supplementary Table 1.
Fig. 1.
Study design (created with Biorender.com).
DNA preparation
DNA extraction from cardiac tissues was conducted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), following the manufacturer’s guidelines. Subsequently, the DNA quality of each sample was assessed with the Quantifiler® Trio DNA Quantification kit (Thermo Fisher Scientific, Waltham, MA, USA) utilising the ABI 7900 instrument (Thermo Fisher Scientific, Waltham, MA, USA) and following the manufacturer’s protocol. The Degradation Index (DI), calculated as the ratio between the smaller and larger autosomal amplicons, was used to measure the DNA quality. The DNA (250 ng) was bisulfite-converted using the EZ DNA Methylation™ Kit (Zymo Research, Irvine, CA, USA) and eluted in 10 µl, following the supplier’s instructions.
DNA methylation profiling
Genome-wide DNAm was assessed using the Infinium MethylationEPIC kit v1.0 (Illumina, San Diego, CA, USA), which covers around 850 K CpG sites, following the manufacturer’s protocol. Samples were randomly distributed on the EPIC chips and chip positions (rows 1–8). The arrays were scanned using the iScan System (Illumina, San Diego, CA, USA) to obtain raw Intensity Data files (.idat). Due to the capacity limitations of the EPIC array kit, which accommodates a maximum of eight samples per array, three of our collected samples (Sample 3 exposed at 4 °C for 28 days, Sample 5 exposed at 4 °C for 28 days, and Sample 7 exposed at 22 °C for 28 days) were not analysed. This decision was based on the consideration that acquiring an additional kit solely for these three samples was not cost-effective and would not significantly impact the overall conclusions of the study.
Data quality control
Data analysis was conducted in the R statistical environment (version 4.3.1)21, employing packages from the Bioconductor project (version 3.18). Raw idat files were imported into R and processed using the Bioconductor package SeSAMe (version 1.20.0)22. β-values and quality metrics were obtained using the SeSAMe function openSesame() with its default parameters (details in the package’s manual23). In brief, the selected pre-processing functions qualityMask(), inferInfiniumIChannel(), dyeBiasNL(), pOOBAH(), and noob() masked the probes of poor-quality design (105,454 probes) previously identified by Zhou et al.24 , inferred the channel of Infinium-I probes, corrected for dye bias, removed probes with low-quality signal (detection p-value ≥ 0.05), and implemented a background subtraction. Quality control (QC) was performed using the standard SeSAMe QC quality metrics. Based on these results, we decided to include the data from all the samples. The 59 SNP probes included in the EPIC array were used to verify the identity of the samples.
Correction of methylation microarray batch effects
To identify potential batch effects in the DNAm data, principal component analysis (PCA) was performed using the prcomp() function. The associations of the first 10 principal components (PCs) with technical and non-technical variables were evaluated using the plomix R-package25. For PCA and downstream analysis, only data from probes with no missing values in all investigated samples (657,859 probes) were used. Due to the observed correlations between PCs and technical variables, the dataset was normalised regarding chip ID and chip position. M-values were calculated from β-values using the formula M = log2((β)/(1-β))26 and normalised using the Combat() function from the sva package (version 3.50.0)27. Following normalisation, the M-values were converted back to β-values using the formula β = 2M/(2M + 1). PCA and association analysis were then rerun after batch correction. PCA plots were generated using the ggplot2 R-library (version 3.4.4).
Correlations analysis
The correlations between the reference samples and samples subjected to different times and temperatures (hereafter termed exposed samples) were calculated using squared Pearson correlation coefficients (R2) with the cor() function. To investigate the impact of time and temperature on different DNAm levels, we calculated R2 using subsets of the data: CpG sites with β ≤ 0.333, 0.333 < β < 0.666, and β ≤ 0.666. A paired t-test was applied to compare R2-values between temperatures at each time point. P-values were adjusted for multiple testing using the R function p.adjust() and applying the Benjamini-Hochberg method28 to control the False Discovery Rate (FDR) at a threshold of p = 0.05.
The absolute differences in beta values (|∆β|) between the reference and exposed samples were calculated for all CpG sites. The median, interquartile range (IQR), and percentage of CpG sites with |∆β| ≥ 0.05 and 0.1 were calculated from the distribution of |∆β| per sample. Cumulative frequency plots were generated using the ggplot2 function stat_ecdf().
Differentially methylated positions
Differentially methylated positions (DMPs), also known as differentially methylated loci (DML), were detected using the SeSAMe DML() function. Two different approaches were used in the DMP analyses: (1) pairwise comparisons between the reference and exposed samples, and (2) two linear regression models for 4 and 22 °C, respectively, with time as a continuous variable to identify systematically deviating CpG positions indicating hypo- and hyper-methylation with increased storage time (Supplementary Fig. 1). Multiple testing correction of p-values was carried out using the R function p.adjust(), applying the Benjamini-Hochberg method28.
The criteria for defining DMPs was an adjusted p-value < 0.05 and a |∆β| > 0.1. Specifically, in the pairwise comparison (approach 1), DMPs with changes in β-values were identified using |∆β| thresholds of 0.1 (10%), 0.2 (20%), and 0.3 (30%). In approach 2, the detection of DMPs was based on the model slope coefficient. The threshold to identify DMPs was calculated by multiplying the slope coefficient with the maximum number of days (28 days). A slope coefficient of 0.0035, corresponding to a |∆β| of ~ 0.1 between day 0 (reference sample) and day 28 (|slope| * days = 0.0035 * 28 = 0.098), was used as threshold.
We additionally performed a DMP analysis using non-ComBat corrected data and M-values, with chip ID and chip position as covariates, to validate the robustness of our initial results using ComBat corrected data.
Differentially methylated positions annotated to cardiac genes, CpG island, and gene context
DMPs were annotated to cardiac genes using the annotation file developed by Zhou et al. (GENCODEv36)29. Subsequently, genes were filtered based on a curated list of cardiac genes linked to (likely) pathogenic genetic variants (Supplementary Table 2). This list contains genes associated with “Cardiovascular” and “Cardiovascular metabolic” phenotypes from The American College of Medical Genetics and Genomics (ACMG) SF v3.2 list30. Relationship to CpG island was assigned to each DMPs using the Illumina EPIC annotation file with the R/Bioconductor annotation package IlluminaHumanMethylationEPICanno.ilm10b4.hg19 (version 0.6.0). Gene context annotation was performed using the R/Bioconductor packages TxDb.Hsapiens.UCSC.hg19.knownGene (version 3.2.2) and ChIPseeker (version 1.38.0)31.
Results
To study the stability of DNAm levels in cardiac tissue samples, we investigated genome-wide methylation levels in RAA samples kept at 4 °C and 22 °C and various time intervals (1, 7, 14, and 28 days) and compared the results to those obtained from RAA samples analysed immediately after tissue collection (reference RAA samples).
QC and correction of methylation microarray batch effects
Supplementary Figs. 2–5 show QC metrics. The number of detected CpG sites ranged from 773,844 to 856,313 (median: 848,287), and the bisulfite conversion efficiency ranged from 0.992 to 1 (median: 0.998). DI was calculated for all samples to assess DNA quality; however, no statistically significant difference was found between the reference and exposed samples (data not shown). We employed PCA in combination with linear models to evaluate the association between variation in the data with two technical variables (chip ID and chip position) and four non-technical variables (time, temperature, age, and individual). Supplementary Fig. 6A-C shows PCA and correlation analyses of PC1-10 with time, temperature, individual ID, age, chip ID, and chip position before batch correction. Chip ID and chip position were correlated with the PCs of the dataset (R2 of 0.301 and 0.584 for chip ID and chip position, respectively). Therefore, a methylation data correction for chip ID and chip position was conducted (batch correction). After methylation data correction, we eliminated the effct of chip ID and chip position (Supplementary Fig. 6D-F). We found that time, temperature, and individual ID had the highest correlation with the genome-wide DNAm variation (R22 of 0.321 and 0.166 for temperature and time, respectively).
Correlation of methylation levels between reference and exposed samples
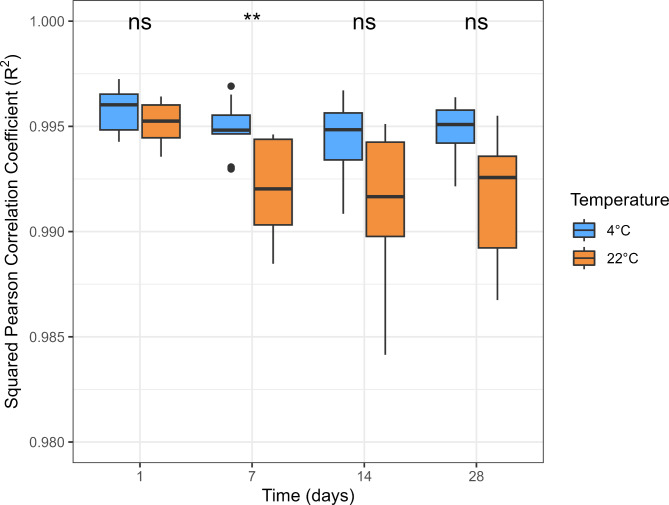
We calculated paired R2 of β-values between the reference and exposed samples (Fig. 2 and Supplementary Table 3). All exposed samples exhibited high correlations (R2 > 0.984) with the reference samples. The median R2 values at 4 °C were consistently high over time, with R2 = 0.996 on day 1 and 0.995 on days 7, 14, and 28. At 22 °C, median R2 values were slightly lower, starting with R2 = 0.995 on day 1, followed by 0.992 on days 7 and 14, and 0.993 on day 28. We further calculated R2 in different β-value intervals (β ≤ 0.333, 0.333 < β < 0.666, and β ≤ 0.666). The R2 values were lower in the intermediate β-value interval (0.333 < β < 0.666) than in the extreme intervals (β ≤ 0.333 and β ≥ 0.666) (Supplementary Fig. 7). All correlation tests had an adjusted p-value below 0.05 (data not shown).
Fig. 2.
Paired squared Pearson correlation coefficients (R2) between the reference and exposed sample. *: p-value < 0.05; **: p-value < 0.01; ***: p-value < 0.001; ns: not significant.
Pairwise absolute β-value differences (|∆β|) between reference and exposed samples were calculated for all CpG sites. Table 1 provides quantile metrics of the |∆β| and the percentage of CpG sites with |∆β| higher than 0.05 and 0.1 for all exposed samples. The lowest median and IQR values were found for samples kept at 4 °C for one day (Median = 0.0093, IQR = 0.0175). Samples kept at 22 °C for 7, 14, and 28 days showed higher median, IQR, and percentage of CpG sites with a |∆β| greater than 0.05 and 0.1 compared to those stored at 4 °C for the same duration. Supplementary Fig. 8 shows cumulative plots for |∆β| for the different conditions. The cumulative frequency curve of samples kept at 4 °C for one day increased more rapidly than the other exposed samples, indicating an overall lower |∆β| for 4 °C on day one compared with the other exposed samples. Samples kept at 22 °C for 7, 14, and 28 days increased less rapidly than all other conditions. Supplementary File 1 contains the |∆β| for each CpG site detected with the EPIC v1.0 array for all time and temperature conditions.
Table 1.
Absolute β-value differences (|∆β|) between reference and exposed samples.
| Temperature | Time (days) | Q2 (Median) | IQR | |Δβ| ≥ 0.05 (%) | |Δβ| ≥ 0.10 (%) | |
|---|---|---|---|---|---|---|
| 4 °C | 1 | 0.0093 | 0.0175 | 4.92 | 0.30 | |
| 4 °C | 7 | 0.0103 | 0.0197 | 6.68 | 0.46 | |
| 4 °C | 14 | 0.0104 | 0.0200 | 7.24 | 0.63 | |
| 4 °C | 28 | 0.0101 | 0.0193 | 6.60 | 0.50 | |
| 22 °C | 1 | 0.0099 | 0.0189 | 6.03 | 0.38 | |
| 22 °C | 7 | 0.0121 | 0.0252 | 11.61 | 1.43 | |
| 22 °C | 14 | 0.0115 | 0.0246 | 11.80 | 1.95 | |
| 22 °C | 28 | 0.0119 | 0.0250 | 11.65 | 1.68 | |
IQR = Interquartile range.
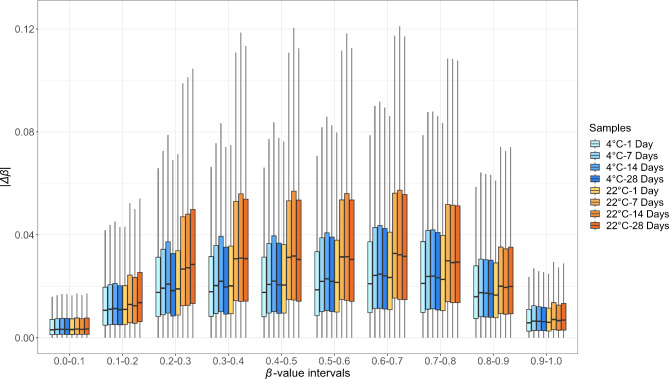
We further investigated |∆β| in CpGs using β-value intervals of 0.1 (Fig. 3 and Supplementary Table 4). We observed higher median |∆β| values and IQR for β-values between 0.1 and 0.9 than for the extreme β-values intervals (0.0-0.1 and 0.9-1.0). Samples kept at 22 °C for 7, 14, and 28 days exhibited higher |∆β| median and IQR values than those kept at 4 °C for all time intervals and 22 °C for one day. This difference was particularly evident for intermediate β-values.
Fig. 3.
Absolute β-value differences (|∆β|) between reference and exposed samples, subdivided into β-value intervals of 0.1.
Differentially methylated positions
We conducted a DMP analysis to estimate the number of DMPs and detect systematic hypo- and hypermethylation DNAm changes with increasing time. DMPs were defined by a threshold of |∆β| > 0.1 and an adjusted p-value < 0.05. However, we also report the number of probes with |∆β| < 0.1, as some studies may apply lower thresholds to detect DNAm changes. Two different approaches were used for the DMP analysis. In the first approach, pairwise comparisons were made between the reference and exposed samples. The number of DMPs identified from this analysis is presented in Table 2. No DMPs were found between the reference samples and samples kept at 4 °C. Only three probe in samples kept at 4 °C for 14 days was statically different from the reference samples, however, the |Δβ| was lower than 0.1. Samples kept at 22 °C for 7, 14, and 28 days had total numbers of probes with |Δβ| lower than 0.1 of 46,694, 68,206, and 42,252, respectively. However, when using a |∆β| threshold of 0.1, the number of DMPs at 22 °C decreased drastically to 3,432 DMPs at day 7, 6,889 DMPs at day 14, and 3,782 DMPs at day 28. When increasing the |Δβ| threshold to 0.2, we found 3, 28, and 40 DMPs on days 7, 14, and 28, respectively. Finally, with a |Δβ| threshold of 0.3, only 2, 1, and 2 DMPs were found at 22 °C for days 7, 14, and 28. Samples kept at 22 °C for one day did not result in any DMP. We also performed DMP analysis using raw data and M-values, with chip ID and chip position included as covariates. No statistically significant probes were identified for samples kept at 4 °C across the different time points, nor for samples kept at 22 °C for 1 day. However, we identified 24,299, 83,181, and 12,764 statistically significant probes for samples stored at 22 °C for 7, 14, and 28 days, respectively. Venn diagrams showing the overlapping probes between the two methods used are shown in Supplementary Fig. 9.
Table 2.
Numbers of differentially methylated CpG positions in reference and exposed samples
| |Δβ| | 4°C | 22°C | ||||||
|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 7 | Day 14 | Day 28 | Day 1 | Day 7 | Day 14 | Day 28 | |
| |Δβ| > 0 | 0 | 0 | 1 (1+) | 0 | 0 | 46,694 (12,360+; 34,334−) | 68,206 (19,480+; 48,726−) | 42,252 (11,388+; 30,864−) |
| |Δβ| > 0.1 | 0 | 0 | 0 | 0 | 0 | 508 (272+; 236−) | 1588 (534+; 1054−) | 488 (196+; 292−) |
| |Δβ| > 0.2 | 0 | 0 | 0 | 0 | 0 | 1 (1+) | 3 (2+; 1−) | 1 (1+) |
| |Δβ| > 0.3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (1+) | 1 (1+) |
+: Hypermethylation. −: Hypomethylation.
The second approach aimed to find CpG sites showing a decreasing or increasing trend in methylation levels over time. We employed a linear regression model for each temperature with time as a continuous variable. An absolute slope coefficient (|β|) threshold of 0.0035 was used to measure a minimum |Δβ| of 0.1 between the reference and day 28 samples. Table 3 displays the number of DMPs with statistically significant p-values (< 0.05). For the 4 °C samples, only six DMPs had a minimum |Δβ| of 0.1. Half of them were hypermethylated, and the other half were hypomethylated. For the 22 °C samples, 165 DMPs had at least a |Δβ| of 0.1, 69 showed hypermethylation, while 96 showed hypomethylation.
Table 3.
Number of differentially methylated CpG positions showing hypo- or hypermethylation over time for tissues kept at 4 °C and 22 °C.
| |β| | 4 °C | 22 °C |
|---|---|---|
| | β | > 0 | 1,719 (566+; 1,153-) | 14,324 (4325+; 9,999-) |
| | β | > 0.0035 | 6 (3+; 3-) | 165 (69+; 96-) |
| | β | > 0.007 | 0 | 0 |
+: Hypermethylation. -: Hypomethylation.
Applying a higher slope coefficient threshold of 0.007, corresponding to a 0.2 DNAm change between the reference and day 28 samples, resulted in no DMP for both 4 °C and 22 °C. Supplementary Fig. 10 shows scatterplots of the four DMPs (cg06088745, cg06878649, cg07658449, and cg13044675) showing the largest change in DNAm levels between the reference and day 28 samples at 22 °C.
We investigated the genomic distribution of DMPs in Table 2 and observed an enrichment of CpGs in open sea regions (Supplementary Fig. 11A), while the proportion of DMPs within gene contexts remained consistent with the proportion observed for all CpG analysed (Supplementary Fig. 11B).
Differentially methylated positions annotated to cardiac genes
A total of 2,257 CpG sites detected with the EPIC v1.0 array were annotated to CVD genes necessitating the reporting of secondary findings in genetic analyses (see Materials and Methods for more details). For approach 1, we found no DMP in CVD genes for samples kept at 22 °C for one day. For days 7, 14, and 28 at 22 °C, we observed 226, 278 and 140 DMPs linked to CVD genes, respectively. When we applied a |Δβ| threshold of 0.1, we observed only one DMP in CVD genes for day 7 and four DMPs for day 14. These four DMPs were associated with PRKAG2, MYL2, FBN, and LDLR (Supplementary Table 5).
For approach 2, none of the DMPs were associated with CVD genes (Supplementary Table 6).
Discussion
DNAm is an epigenetic mechanism that regulates gene expression and influences various biological processes and disease pathogenesis32. Given the tissue-specific nature of DNAm5,6, analysis of DNAm in cardiac tissue may offer unique insight into cardiac biology and diseases compared to the commonly used analyses of DNA obtained from blood. However, cardiac samples are difficult to obtain. Biopsies from cardiac surgery are not always analysed or stored appropriately immediately after collection due to a lack of human or technical resources at the collection site. We showed that DNAm is stable in improperly stored cardiac tissues and that it can be reliably analysed. Temperatures affected DNAm analysis more than time. We found the lowest β-value correlations between reference samples (samples analysed immediately after tissue collection) and those kept at 22 °C for 7, 14, and 28 days (median R2-values: 0.992, 0.992, and 0.993, respectively). However, all the correlations were still high, and the R2-values were comparable to those obtained by Christiansen et al.33 , who reported high within-laboratory reproducibility of DNAm examinations (median R2: 0.993). We employed the absolute β-value differences (|∆β|) between the reference and exposed samples to assess how much β-values deviated with time and temperature. We found median |∆β| between 0.0093 and 0.0119, indicating a low deviation from the original β-values. We observed that |∆β| were higher within intermediate β-value ranges (0.1–0.9) than in the extreme ranges (0-0.1 and 0.9-1.0) in all exposed samples. These observations suggest a reduced consistency of DNAm measurements in CpG sites with an intermediate methylation state (not fully methylated nor unmethylated). A similar increased variability between replicate analyses of the intermediate methylation states with the EPIC v1.0 array was previously described33. This indicates that the increased variation in DNAm in the intermediate β-value range is an in vitro phenomenon most likely caused by the EPIC v1.0 array. Our results also showed that the intermediate β-values are more sensitive to environmental conditions, including increasing temperature, as the samples exposed to 22 °C exhibited higher |∆β| for the intermediate β-values than those exposed to 4 °C. A relatively high impact of room temperature (22 °C) after one day of exposure was also demonstrated in the DMP analysis. Our findings showed that samples kept at 4 °C for up to 28 days did not show differential methylation, while the effect on the DMP analysis varied with the duration of exposure at room temperature. Single-day storage generated no DMP, while extended storage periods (7, 14, and 28 days) led to the observation of DMPs. We also found systematic hypo- and hypermethylation in some DMPs in the exposed samples. However, these DMPs were not associated with any CVD-associated genes.
Our data suggest that cardiac samples collected from cardiac surgery and not immediately stored at -20 °C and − 80 °C could still be used for DNAm analysis. When the samples were stored in the fridge (4 °C) for up to 28 days, the DNAm analysis was still reliable, and significant changes in DNAm were not observed. Cardiac samples kept at room temperature showed increasingly different β-values with increasing time elapsed from sample collection to DNAm analysis. After one day at room temperature, cardiac samples showed results comparable to those of samples analysed immediately after tissue collection. Short-term DNAm stability at room temperature was also found by Staunstrup et al.34 using DNA immunoprecipitation coupled with deep sequencing in blood samples. However, we found differentially methylated CpG positions in cardiac tissue stored from day seven to 28 at 22 °C. These results are similar to those obtained by Huang et al.17, who observed alteration in DNAm in blood samples kept at room temperature for more than three days. Nevertheless, storing biopsies at room temperature for extended periods is uncommon in cardiac surgery settings.
Further efforts should focus on determining the precise cause of β-value deviations in cardiac samples kept at room temperature for more days, i.e. if it is due to cleavage of DNA chains, chemical changes in cytosine nucleotides, etc.
An alternative method for long-term tissue preservation is formalin-fixation followed by paraffin-embedding (FFPE), primarily used for histological examination. FFPE decreases the DNA quality and probe success rate, and genome-wide DNAm levels are overestimated in cardiac tissue compared to fresh or frozen cardiac tissue20. Furthermore, FFPE samples need additional laboratory steps to recover DNA. Therefore, it is advisable to avoid the FFPE method, and cardiac samples should be kept under suboptimal conditions (fridge and room temperature) as short as possible to obtain DNAm values comparable to those of fresh and frozen samples.
Underlying cardiac diseases cause SCD, and molecular investigations of postmortem cardiac tissue may be one of the keys to revealing the cause of death35–37. Previous studies showed that DNAm is stable in postmortem human brain tissue for 72 h18,19. Our study indicates that DNAm stability is time- and temperature-dependent. This information is relevant to DNAm analysis of postmortem cardiac tissue from deceased individuals who may have been exposed to prolonged PMIs at room temperature before being brought to the morgue that usually have a storage temperature of approximately 4 °C. The biological events caused by the death of an individual may alter the DNAm landscape in a specific manner. Therefore, future studies should focus on studying DNAm stability in postmortem samples subjected to different PMI.
In conclusion, the data showed that the results of DNAm examination of cardiac tissue stored for one day at 22 °C and 28 days at 4 °C were similar to those obtained from fresh cardiac tissue. Minor changes in DNAm were found in cardiac tissue stored at 22 °C for seven days or more.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Anja Ladegaard Jørgensen for assistance with this work.
Author contributions
J.D.A., N.M., S.N.C., J.T.H. and A.V. conceived and designed the experiments; M.H.S. and S.B.J performed the acquisition of the samples; B.P, S.B.J. and M.E.D. carried out the experiments and data analysis; B.P. and J.D.A. wrote the paper. All authors revised critically the manuscript. All authors approved the final version of the manuscript.
Data availability
The datasets are available from the corresponding authors upon reasonable request.
Declarations
Ethics approval and consent to participate
The study conformed to the Declaration of Helsinki and was approved by the Committees of Health Research Ethics in the Capital Region of Denmark (H-20039524). The biobank, where the samples are held, is registered at the University of Copenhagen’s joint records for processing of personal data in research projects and biobanks (514-0528/20-3000). The register complies with the rules of the General Data Protection Regulation (Regulation (EU) 2016/679). Informed written consent was collected from all individuals. Patient material and data were pseudonymised.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Brando Poggiali, Email: brando.poggiali@sund.ku.dk.
Jeppe Dyrberg Andersen, Email: jeppe.dyrberg.andersen@sund.ku.dk.
References
- 1.Moore, L. D., Le, T. & Fan, G. D. N. A. Methylation and its basic function. Neuropsychopharmacology. 38, 23–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muka, T. et al. The role of epigenetic modifications in cardiovascular disease: a systematic review. Int. J. Cardiol. 212, 174–183 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Ivanova, A. A., Maksimova, S. V. & Gurazheva, A. A. Role of DNA methylation in Development of Cardiovascular diseases, resulting in a Sudden Cardiac Death (Review). Sovremennye Tehnologii v Med. 14, 83 (2022). [Google Scholar]
- 4.Duan, L., Hu, J., Xiong, X., Liu, Y. & Wang, J. The role of DNA methylation in coronary artery disease. Gene. 646, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Lokk, K. et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 15, 3248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz, M. D. et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 523, 212–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adibi, J. J. et al. Placental biomarkers of phthalate effects on mRNA transcription: application in epidemiologic research. Environ. Health. 8, 20 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong, S. H. et al. Effects of delay in the snap freezing of colorectal cancer tissues on the quality of DNA and RNA. J. Korean Soc. Coloproctol. 26, 316–323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bär, W., Kratzer, A., Mächler, M. & Schmid, W. Postmortem stability of DNA. Forensic Sci. Int. 39, 59–70 (1988). [DOI] [PubMed] [Google Scholar]
- 10.Matange, K., Tuck, J. M. & Keung, A. J. DNA stability: a central design consideration for DNA data storage systems. Nat. Commun. 12, 1358 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira, P. G. et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat. Commun. 9, 490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chheda, U. et al. Factors affecting Stability of RNA—temperature, length, concentration, pH, and buffering species. J. Pharm. Sci. 113, 377–385 (2024). [DOI] [PubMed] [Google Scholar]
- 13.Kocsmár, É. et al. Proteome alterations in human autopsy tissues in relation to time after death. Cell. Mol. Life Sci. 80, 117 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpson, R. J. Stabilization of Proteins for Storage. Cold Spring Harb Protoc pdb.top79 (2010). (2010). [DOI] [PubMed]
- 15.Gosselt, H. R. et al. Global DNA (hydroxy)methylation is stable over time under several storage conditions and temperatures. Epigenetics. 16, 45–53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y. et al. Stability of global methylation profiles of whole blood and extracted DNA under different storage durations and conditions. Epigenomics. 10, 797–811 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Huang, L. H. et al. The effects of storage temperature and duration of blood samples on DNA and RNA qualities. PLoS One. 12, e0184692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrachina, M. & Ferrer, I. D. N. A. Methylation of Alzheimer disease and tauopathy-related genes in Postmortem Brain. J. Neuropathol. Exp. Neurol. 68, 880–891 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Rhein, M. et al. DNA methylation results depend on DNA integrityâ€role of post mortem interval. Front. Genet. 6, (2015). [DOI] [PMC free article] [PubMed]
- 20.Dupont, M. E. et al. Fresh and frozen cardiac tissue are comparable in DNA methylation array β-values, but formalin-fixed, paraffin-embedded tissue may overestimate DNA methylation levels. Sci. Rep. 13, 16381 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Core Team. R: A Language and Environment for Statistical Computing. Preprint at (2021). https://www.R-project.org/
- 22.Zhou, W., Triche, T. J., Laird, P. W. & Shen, H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 10.1093/nar/gky691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.http://bioconductor.statistik.tu-dortmund.de/packages/3.18/data/experiment/manuals/sesameData/man/sesameData.pdf
- 24.Zhou, W., Laird, P. W. & Shen, H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res gkw967 doi: (2016). 10.1093/nar/gkw967 [DOI] [PMC free article] [PubMed]
- 25.Yuan, V. plomics. GitHub repository Preprint at (2019). https://github.com/wvictor14/plomics
- 26.Du, P. et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 11, 587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leek, J. T. et al. sva: Surrogate Variable Analysis. Preprint at (2023). 10.18129/B9.bioc.sva
- 28.Benjamini, Y. & Hochberg, Y. Controlling the false Discovery rate: a practical and powerful Approach to multiple testing. J. R Stat. Soc. Ser. B Stat. Methodol. 57, 289–300 (1995). [Google Scholar]
- 29.InfiniumMethylation BeadChips Annotation. https://zwdzwd.github.io/InfiniumAnnotation
- 30.Miller, D. T. et al. ACMG SF v3.2 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Sci. 25, 100866 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Schübeler, D. Function and information content of DNA methylation. Nature. 517, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Christiansen, S. N. et al. Reproducibility of the Infinium methylationEPIC BeadChip assay using low DNA amounts. Epigenetics. 17, 1636–1645 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staunstrup, N. H. et al. The genome-wide DNA methylation Profile of Peripheral blood is not systematically changed by short-time storage at Room temperature. Epigenomes. 1, 23 (2017). [Google Scholar]
- 35.Christiansen, S. N. et al. Differential methylation in the GSTT1 Regulatory Region in Sudden unexplained death and sudden unexpected death in Epilepsy. Int. J. Mol. Sci. 22, 2790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanova, A. A. et al. ABCA1 gene promoter methylation and sudden cardiac death. Bull. Siberian Med. 19, 80–85 (2021). [Google Scholar]
- 37.Zeppenfeld, K. et al. 2022 ESC guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death. Eur. Heart J. 43, 3997–4126 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets are available from the corresponding authors upon reasonable request.