Abstract
The Southern Ocean microbial ecosystem, with its pronounced seasonal shifts, is vulnerable to the impacts of climate change. Since viruses are key modulators of microbial abundance, diversity, and evolution, we need a better understanding of the effects of seasonality on the viruses in this region. Our comprehensive exploration of DNA viral diversity in the Southern Ocean reveals a unique and largely uncharted viral landscape, of which 75% was previously unidentified in other oceanic areas. We uncover novel viral taxa at high taxonomic ranks, expanding our understanding of crassphage, polinton-like virus, and virophage diversity. Nucleocytoviricota viruses represent an abundant and diverse group of Antarctic viruses, highlighting their potential as important regulators of phytoplankton population dynamics. Our temporal analysis reveals complex seasonal patterns in marine viral communities (bacteriophages, eukaryotic viruses) which underscores the apparent interactions with their microbial hosts, whilst deepening our understanding of their roles in the world’s most sensitive and rapidly changing ecosystem.
Subject terms: Metagenomics, Microbial biooceanography, Bacteriophages, Metagenomics, Cryospheric science
The study reveals that the Southern Ocean hosts a unique and largely underexplored DNA viral community. It explores the diversity of viral communities infecting all domains of life and their seasonality, emphasizing their role in ecosystem dynamics.
Introduction
Viruses are prevalent and numerically dominant agents of microbial mortality, as well as drivers of biodiversity1–3. They play important roles in shaping microbial communities and biogeochemical cycles in the oceans, where two-thirds of living biomass is microbial4. Understanding the ecological significance of viruses requires a comprehensive insight into their diversity. Viral metagenomics has revolutionized marine viral ecology5–7, revealing viral community structure, biogeochemical roles, evolutionary trajectories, and taxonomic diversity8–13. Still, key oceanic regions, such as the Southern Ocean, remain understudied14.
The Southern Ocean, pivotal in global ocean circulation15, absorbs over 40% of anthropogenic carbon dioxide emissions and hence, plays a crucial role in moderating climate change16. This process is significantly driven by microbial activity17, emphasizing the need for studying the impact of viruses on these microbial hosts. The short polar productive season results in pronounced temporal dynamics in (micro)biological activity and community composition18–21. Substantial seasonal variation was also reported for viral lysis of Antarctic marine microorganisms22–26. Yet, our understanding of the diversity and dynamics of the virus communities remains limited. The few studies that addressed their metagenomic characterization provide a restricted view due to a limited sample number and narrow sampling period23,27–30.
The present study delves into the intricate seasonal variations in Antarctic marine dsDNA virus diversity. It has high temporal resolution (24 sampling days) over the full productive season in Marguerite Bay, located in the Western Antarctic Peninsula. We advance and expand the catalog of marine viral diversity by combining multiple virus detection and classification approaches tailored to specific viral taxa. Moreover, an improved understanding of seasonal variations in virus-host dynamics highlights the ecological importance of viruses in the Southern Ocean as drivers of diversity and succession. Therefore, this study establishes an essential foundation for understanding the dynamics of this region, which is particularly sensitive to global climate change.
Results
Antarctic viral communities harbor a diverse range of taxa
To explore the dynamics of Antarctic marine viruses and interactions with their hosts, we generated paired metagenomes from viral (< 0.22 µm and free-DNA digestion) and cellular (> 0.22 µm) size fractions. Viral metagenomes, totaling 887 million reads, were co-assembled into 161,651 viral scaffolds, of which 8045 were mid- to high-quality (>10 kb or > 70% CheckV31 completeness, Supplementary data 1). The viral reads represented 59% of the viral fraction metagenomes, and on average 4% of the reads did not map to the co-assembly (Supplementary Fig. 1a). The sample rarefaction and the species accumulation curves show that we have sequenced deeply enough and with enough sample coverage of the site (Supplementary Fig. 2a, b). The viral fraction metagenomes displayed clear enrichment in viral sequences and depletion in cellular markers (Supplementary Fig. 1a, b). The mid- to high-quality viral sequences clustered into 7957 vOTUs. When comparing the mid- to high-quality viral scaffolds to the Global and Southern Ocean Viromes8,29 (GOV and SOV), we found 75% of viral sequences to be novel at the species rank.
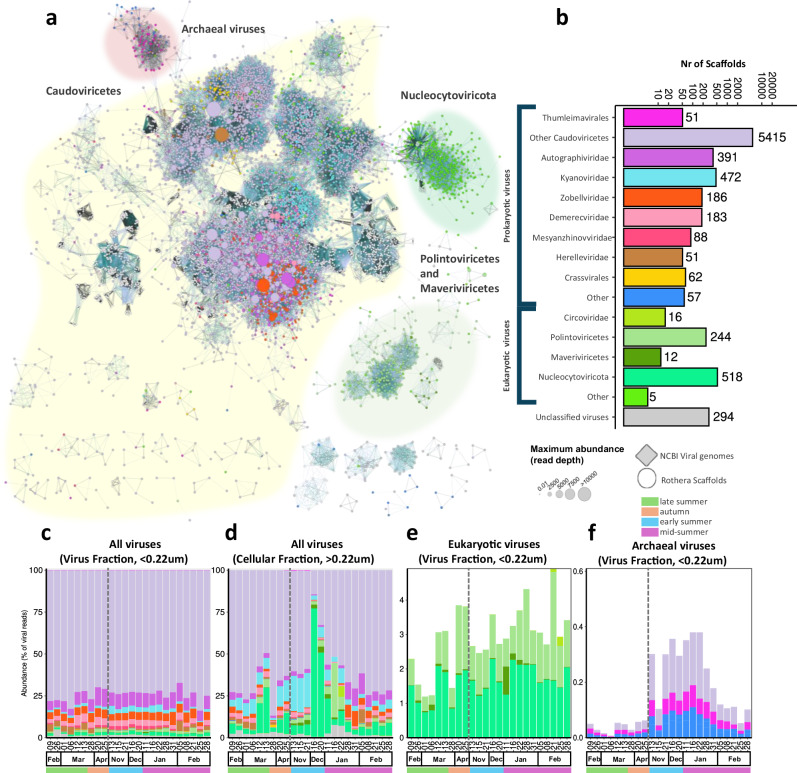
The most frequent and abundant viruses were the tailed dsDNA bacteriophages of the class Caudoviricetes (Fig. 1a–c), outnumbering other viruses by 53-fold and making up 75% of all scaffolds (Fig. 1b, c). We further categorized Caudoviricetes sequences to family rank using PhaGCN232, except the order Crassvirales which was characterized using a targeted approach. Seasonal patterns in compositional viral abundance could be discerned for various Caudoviricetes taxa, even more pronounced in the cellular fraction (Fig. 1c, d). Differing abundance patterns in the viral and cellular fraction can be the result of seasonal dynamics in lytic and lysogenic viral infection, as the viral reads from the cellular fraction can reflect both actively replicating and lysogenic viral infection while the virus fraction is composed of free viral particles. For example, kyanoviruses displayed different occurrence patterns in the viral and cellular metagenomes, with less reads in the virus fraction than in the cellular metagenome. Similarly, the Autographiviridae family sequences were the most abundant (on average 11% viral fraction and 3.5% cellular fraction reads), and displayed increased virus productivity in summer, reflected by the higher abundance in the cellular fraction from mid-January to February (Fig. 1c, d). Few viruses (0.36% reads or 279 scaffolds) were identified as archaeal viruses, which showed higher abundance from November to January (Fig. 1e) and were many classified as Thumleimavirales (51 scaffolds).
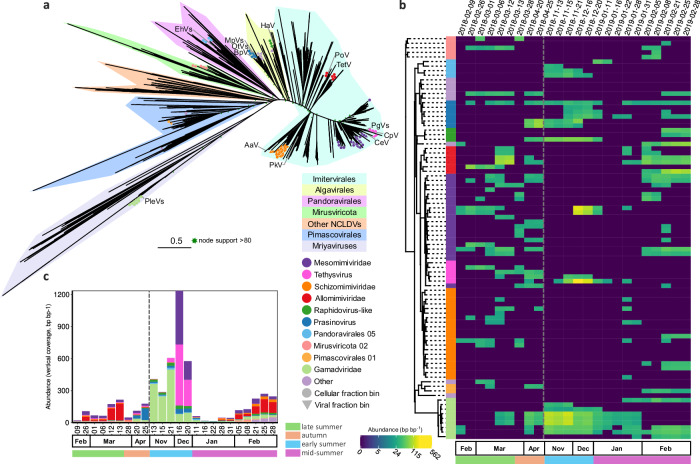
Fig. 1. Taxonomic diversity of Antarctic dsDNA viruses over the productive season.
a Gene sharing network of mid to high-quality viral contigs (>10 kb or 70% complete) coloured by taxonomic classification (see panel b for legend). The node size represents the maximum read abundance over the whole virome dataset. The shaded cluster was determined manually by overlaying the cenote-taker2 taxonomy. Family level taxonomy for Caudoviricetes predicted using PhaGCN2. Nucleocytoviricota, Maveriviricetes, Polintonviricetes, and Crassvirales were classified with a costume approach. b Number of scaffolds classified to each taxonomic rank (other represents ranks with less than 50 scaffolds). c–f Read abundance for viral taxa, reads were mapped to all scaffolds and abundance normalized to the total viral reads for all viruses in the viral fraction(<0.22 µm) (c) and cellular fraction (>0.22 µm) (d) and for archaeal viruses (f) and eukaryotic viruses (e) in the viral fraction (< 0.22 µm). Note that the are compositional and the total virus particle abundances (Fig. 3c). The dashed vertical line indicates the winter sampling gap between April and November 2018.
Abundant eukaryotic viruses belonged to the phylum Nucleocytoviricota (originally nucleocytoplasmic large DNA viruses), the classes Polintoviricetes (polinton-like viruses or PLVs) and Maveriviricetes (virophages), and the family Circoviridae (ssDNA; Fig. 1b, c, f). The Nucleocytoviricota viruses (NCVs) are the most abundant eukaryotic viruses, accounting for 12.4% and 1.6% of the cellular and viral fraction reads, respectively. NCV abundance in the cellular fraction spiked in March and December 2018, accounting for up to 27% and 77% of viral reads (respectively), implying heightened NCV production.
PLVs form an important fraction of the viral diversity, accounting for 1678 scaffolds. Of these, 53 had a CheckV completeness higher than 50%, and six contained terminal repeats. In total 243 exceeded 10 kb, which likely represent near-complete genomes, as their length is typically around 20 kb33,34. PLVs also constitute a significant proportion of reads, averaging 3.5% and 1.2% of cellular and viral fractions respectively (Fig. 1d, e). A maximum of 27% of the viral reads in the cellular fraction during January 2019 probably reflects an increase in the cellular host abundance rather than lytic viral infection, as these are prevalent endogenous viral elements of eukaryotes33. Meanwhile, their abundance and detection in the viral fraction likely indicates a shift to an active infectious state.
Though the used standard Illumina library preparation method is biased against ssDNA viruses35, we identified five scaffolds assigned to the Microviridae family (non-enveloped bacteriophages), two belonging to the filamentous Inoviridae family (both infecting bacteria), and 16 to the eukaryotic specific Circoviridae family36. We do not expect ssDNA viruses to be a significant portion of the DNA viral communities, as previous surveys using methods biased towards ssDNA viruses still found dsDNA viruses to dominate28.
Antarctic virus diversity responds to seasonal changes
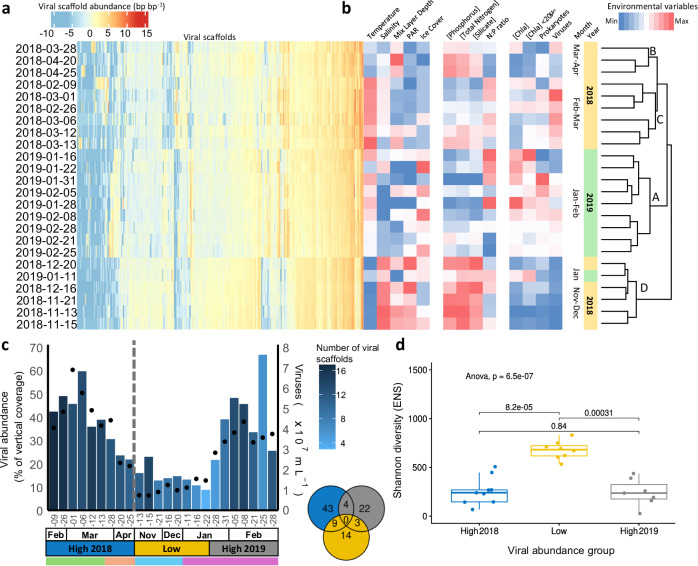
Viral communities changed over the sampling period and could be delineated into four distinct clusters (Fig. 2a, b), each representing specific periods in the productive season (November to March) with distinct environmental conditions. The early summer (cluster D) is characterized by low water temperature, high salinity, high dissolved inorganic nutrient concentrations, low chlorophyll a (Chl-a) concentration, and low bacterial and viral abundances. During mid-summer (cluster A), increased temperature led to sea-ice melting and subsequently lower salinity and stronger stratification (shallower mixed layer depth). Accordingly, higher photosynthetic active radiation (PAR) coincided with peak Chl-a concentrations and nutrient depletion, while bacterial and viral abundances increased. Late-summer (cluster C) showed reducing Chl-a concentrations and prokaryote abundance and the peak in viral abundance. This cluster also coincided with increased temperature, diminished sea-ice cover, and relatively low nutrients. By the onset of autumn (cluster B), higher wind activity and lowered temperature led to increased mixing and sea-ice formation (with replenished nutrient concentrations and increased salinity) and declining microbial productivity (low Chl-a and microbial abundances)37. February 2018 and 2019 came in two separate clusters (C and A respectively), indicating some interannual variability.
Fig. 2. Seasonal dynamics of viral diversity.
a Heatmap per sampling day of the clr-transformed scaffold vertical coverage (read abundance normalised by scaffold length) for mid to high quality scaffolds (> 10 kb or 70% complete). b changes of relevant environmental variables, including temperature, salinity, mix layer depth, photosynthetic active radiation (PAR), ice cover for physical variables, phosphorus, total nitrogen, silicate, and nitrogen to phosphorus ratio (N:P ratio) for chemical variables, and total Chlorophyll a (Chl-a), < 20 µm Chlorophyll a, prokaryotic cell counts, and viral particle counts for biological variables. The heatmap (a, b) are sorted vertically according to the hierarchical clustering of the clr-transformed viral abundances (dendrogram on the right). The letters on the dendrogram highlight the separate clusters referred to in the main text. c Compositional cumulative viral vertical coverage of dominant viral scaffolds (with abundance > 0.1%) for mid to high quality scaffolds. Bar colours represent the number of dominant viral scaffolds per day and black dots represent the flow cytometry viral particle counts for the same day. The Venn diagram shows the number of shared and unique dominant viral scaffolds between different periods. The grey line indicates the winter sampling gap between April and November 2018. d Shannon diversity expressed in effective number of species (ENS) for the samples (n = 24) in the high and low viral-scaffold abundance periods (box plot represent median ± quartiles and the whiskers the point range), these periods are denoted on (c), p-values for an ANOVA and pair-wise two-sided Wilcoxon tests are shown.
We evaluated environmental determinants of virus community composition using a multivariate regression model. Temperature and salinity explained 31% of the community variance (p-value = 0.001 and 0.004, respectively), while another 23% was explained by inorganic phosphate (p-value = 0.001), nitrogen-to-phosphorus ratio (p-value = 0.016), and PAR (p-value = 0.077). The correlation between viral and microbial community compositions (Supplementary Fig. 3) was examined by comparing the pairwise-Aitchison distances. The overall Microbial community composition explained 68% of the variation in the viral community when controlling for the environmental variables (partial Mantel r-statistic=0.6842, p-value = 0.01). Meanwhile, for the prokaryotic community composition and phage community alone the partial Mantel r-statistic was 0.3411 with a p-value of 0.002.
The abundances and temporal dynamics of the dominant viruses mirrored the total virus count by flow cytometry (Fig. 2c; R2 = 0.82 and p-value = 9.5 × 10−7), and followed the bacterial (bloom) dynamics (Fig. 3a). Viral communities in late summer were directed by a higher contribution (and number) of the dominant viruses (96% Caudoviricetes; Supplementary data 1) than in November to mid-January (Fig. 2c). Consequently, during periods of high viral abundance, the viral communities were heavily skewed towards this small subset of viruses, leading to a decrease in alpha diversity (Fig. 2d and Supplementary Fig. 2c–e). Overall, these results highlight distinct viral communities during different periods in the season (Fig. 2c).
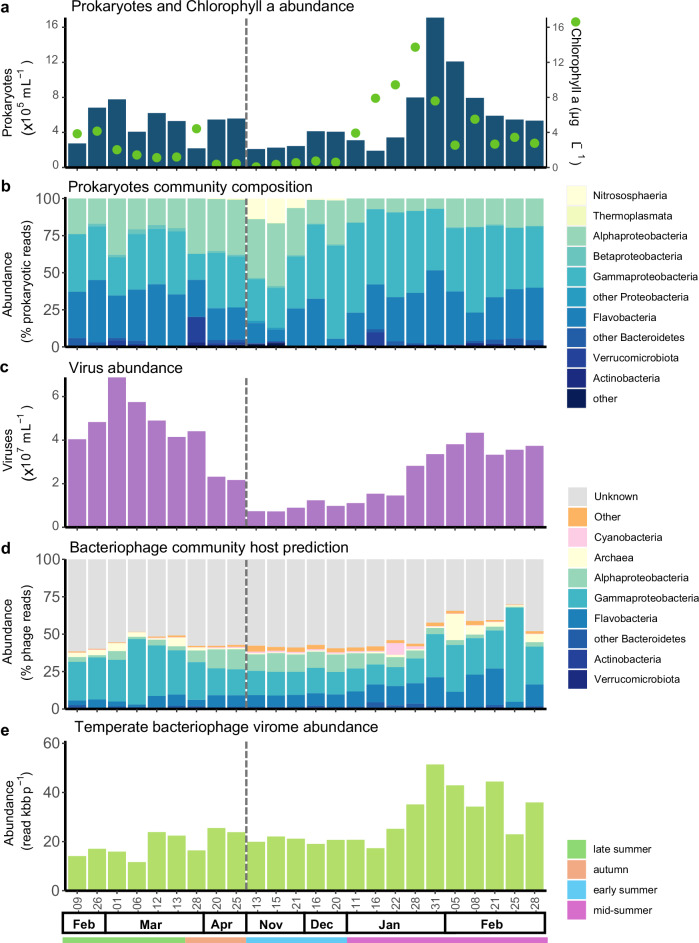
Fig. 3. Bacteriophage and prokaryotic host dynamics.
a Absolute prokaryotic abundances determined by flow cytometry, and chlorophyll a concentration. b Prokaryotic relative abundance (% of prokaryotic reads, > 0.2 µm) determined by PhyloFlash assembled 16S rRNA gene assembly at the class rank. c Absolute viral abundance determined by flow cytometry. d bacteriophage relative abundance (% of bacteriophage reads, < 0.2 µm) of mid to high-quality prokaryotic virus (longer than 10 kb or 70% complete) coloured according to the predicted host class. e abundance of viral scaffolds containing integrase expressed in vertical coverage (read bases per scaffold bases) for the viral fraction (< 0.22 µm). The grey dashed line indicates the winter sampling gap.
Abundant Antarctic bacteriophages infect co-abundant bacterial taxa
We predicted the hosts of 2412 viral scaffolds using the integrated bacteriophage host prediction tool iPHoP38. These scaffolds covered 48% of the reads mapping to mid to high-quality bacteriophage genomes (Fig. 3d) and were consistent with the vContact2 gene-sharing network analysis (Supplementary Fig. 4). Most bacteriophages targeted one of the three predominant bacterial classes (Fig. 3): Gammaproteobacteria (n = 769), Flavobacteria (n = 545), and Alphaproteobacteria (n = 348), which are important components of coastal Antarctic bacterial communities39–41. iPHoP predicted 59 viruses to infect Archaea which peaked in February (Fig. 3c), contrasting with the seasonal abundance of Archaea (Fig. 3a) and archaeal cluster viruses (Fig. 1e).
Gammaproteobacteria-infecting bacteriophages represented 25% of the dominant phages (Supplementary data 1), suggesting a significant role in seasonal virus production. In December, there was a noticeable rise in Gammaproteobacteria, predominantly Oceanospirillales, ahead of their bacteriophages (Fig. 3b–d). In late-summer (March), Gammaproteobacteria bacteriophage abundances were relatively high and increasing, while host abundances were not particularly high (Fig. 3d). In contrast, Flavobacteria-infecting bacteriophages increased steadily with their host during December-January (mostly Polaribacter). Alphaproteobacteria-infecting bacteriophages were most abundant at the end of the season (April) and were still relatively high at the start of the next season (November), coinciding with the highest abundances of their predicted host (mostly SAR11). Thereafter, from December to January, the host population declined more rapidly than that of the bacteriophages.
Temperate bacteriophage reactivation with bacterial bloom onset
High prevalence of lysogenic infection has been reported throughout the Southern Ocean22,23,25, and is potentially involved in the overwintering survival of Antarctic bacteriophages23. To elucidate the seasonal dynamics of temperate bacteriophages, we monitored the abundance of viral scaffolds containing genes involved in lysogenization and prophage reactivation (Fig. 3e and Supplementary Fig. 5), specifically integrase (n = 2502), excisionase (n = 40), and bacteriophage repressor proteins (n = 134). Of the mid to high-quality phage sequences, 386 had at least one of the lysogeny genes. All genes displayed similar seasonal patterns in the cellular and viral fractions (Supplementary Fig. 5). With an average vertical coverage (read abundance normalized by scaffold length) of 25,157 bp bp−1, the scaffolds containing integrases were most abundant (2.4% of total viral vertical coverage). These viruses were most abundant during mid and late-summer (Supplementary Fig. 5), coinciding with elevated bacterial counts (Fig. 3a).
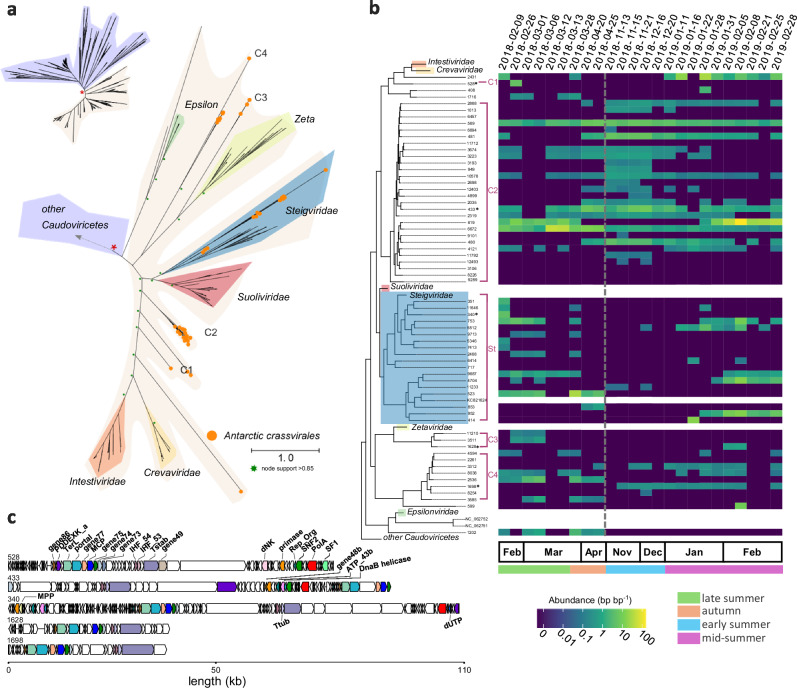
Antarctic waters harbor novel and diverse Crassvirales bacteriophages
Our target analysis identified 62 novel crassphage sequences (order Crassvirales). Of these, 17 were longer than 50 kb, 18 were at least 50% complete according to CheckV and seven had a completeness higher than 95%, two with direct terminal repeats. The terminase large subunit (TerL) phylogeny (Fig. 4a) validates the phylogenetic placement of these sequences within the Crassvirales order and their further classification at the family rank. Most scaffolds clustered into novel clades within the known Crassvirales diversity, labeled as C1 through C4 (Fig. 4a, b) and prospective new crassphage families. Additionally, 20 scaffolds belong to the Steigviridae family (Clade St, Fig. 4), which encompasses the Cellulophaga phage phi14:2 that infects marine free-living Flavobacteria42,43. We further confirmed the family placement with the phylogeny of the portal protein (Portal, n = 51) and major capsid protein (MCP, n = 37, Supplementary Fig. 6a, b). These crassphages presented a typical gene content and organization for members of the Crassvirales (Fig. 4c), with all the 30 core genes44 identified in at least one of the Antarctic Crassvirales scaffolds (Supplementary Fig. 6c). Clade C2 displayed the highest diversity and prevalence, with several viruses persisting throughout the season and others only detectable during autumn (April) and early summer (November to January).
Fig. 4. Diversity and dynamics of Antarctic Crassvirales.
a Unrooted maximum likelihood phylogenetic tree of the terminase large subunit (TerL) of Antarctic Crassvirales scaffolds and reference crassviruses. Crassvirales found in this study are highlighted with the orange circles (n = 62) and shaded area corresponds to the Crassvirales order. b The Crassvirales section of the phylogenetic tree is shown on the left. Reference sequences from the five existing families have been collapsed, except for the Steigviridae that is interspersed with sequences from the Antarctic samples. The vertical coverage (read abundance normalised by scaffold length) for the viral fraction ( < 0.22 µm) are shown as a heatmap.The grey dashed line indicates the winter sampling gap. c Genome map with functional gene annotations for the longest crassviruses of each of the 5 clades, noted in (b) with *. The annotated genes are coloured consistently across the genome maps. Protein abbreviations as follows, major capsid protein (MCP), portal protein, putative structural proteins (genes 73–75, 77 and 86), integration host factor subunits (IHF 53 and 54), tail stabilization protein (Tstab), PD-(D/E)XK family nuclease (PDDEXK), crassvirus uncharacterized gene 49, DnaG family primase, replisome organizer protein (Rep_Org), SNF2 helicase, DNA polymerase family A (PolA), SF1 helicase, AAA domain ATPase (ATP_43b), phage replicative helicase (DnaB), metallophosphatase (MPP), tail tubular protein (Ttub), dUTPase (dUTP).
Nucleocytoviricota viruses are a diverse and abundant group of Antarctic eukaryotic viruses
Considering the large genomes and particle sizes of many NCVs45, we characterized their diversity using metagenomic binning in both the viral and cellular fractions. This resulted in 88 bins (Fig. 5a, Supplementary data 2), that were phylogenetically placed in the orders Imitervirales (n = 51), Algavirales (n = 15), candidate Pandoravirales46 (n = 4) and Pimascovirales (n = 4), and candidate class Mriyaviricetes47 (n = 9). Additionally, our analysis recovered five bins belonging to the candidate phylum Mirusviricota, which shares some phylogenetic marker genes with NCVs48.
Fig. 5. Diversity and seasonal dynamics of Antarctic NCVs.
a Unrooted maximum likelihood phylogenetic tree of Antarctic NCVs, Mirusviricota and reference genomes from isolates and marine metagenomic surveys based on 7 concatenated marker genes. b Temporal dynamics of sequencing vertical abundance (read abundance normalised by bin length, log-scaled) for the cellular fraction metagenomes (> 0.22 um). The heatmap is organized phylogenetically (rooted between mirusviruses and NCVs) and the first column represents the taxonomy (see panel a). c temporal dynamics at the family level for the cellular fraction metagenomes in bin vertical coverage (read abundance normalised by bin length). The grey dash line indicates the winter sampling gap between April and November 2018. Virus abbreviations as follows, Phaeocystis globosa viruses (PgVs), Chrysochromulina ericina and C. parva viruses (CeV and CpV), Prymnesium kappa virus (PkV), Pyramimonas orientalis virus (PoV) Tetraselmis sp. Virus (TetV), Heterosigma akashiwo virus (HaV), Bathycoccus prasinos (BpV), Ostreococcus tauri (OtV), Micromonas pusilla virus (MpV), Aureococcus anophagefferens virus (AaV) and Emiliania huxleyi viruses (EhVs).
Most Imitervirales bins belonged to the Mesomimiviridae family (n = 25), of which five belonged to the genus Tethysvirus which includes viruses infecting the prymnesiophytes Phaeocystis globosa and Chrysochromulina ericina49,50. The December peak of one prevalent Tethysvirus (Fig. 5b, c), coincided with a decline in P. antarctica (Supplementary Fig. 3c), the most abundant prymnesiophyte (> 99% of Prymnesiophyceae rRNA gene reads, except on 9 February and 6 March 2018). Of these, bin 2-643264.cc.b128 was the most abundant of all NCVs and closely related to P. globosa viruses. Twenty of the remaining Imitervirales-related bins belonged to the Schizomimiviridae family, with their peak abundance during late summer (Fig. 5b), particularly in the viral fraction (Supplementary Fig. 7). The remaining six Imitervirales viruses belonged to the Allomimiviridae family and occurred in late-summer (Feb-March; Fig. 5b, c).
Most Algavirales viruses belonged to the candidate family Prasinoviridae AG_01 (n = 10), of which six belonged to genus Prasinovirus. Additionally, one belonged to family AG_3 and three to AG_04 (Raphidovirus-like), which were predominantly abundant in the cellular fraction during the autumn and early and mid-summer (Fig. 5b, c). Prasinovirus bin vRhyme.bin.4783 and AG_01 bin vRhyme.bin.4827 were persistent throughout the productive season in the viral fraction (Supplementary Fig. 7).
The nine Mriyaviruses all belong to the candidate family Gamadviridae and were found inserted into larger bins recovered from the cellular fraction. Eight of these bins had high Eukaryotic content, representing two unclassified Eukaryotes and six classified as Phaeocystis antarctica (Supplementary data 2). We hypothesize that these viruses were binned together with their host, representing temperate or persistent infection (Supplementary data 2), consistent with their presence inside the genome assemblies of Phaeocystis51 and other eukaryotes47. Additionally, five bins in the candidate phylum Mirusviricota48 were identified in the cellular fraction (Fig. 5b, c). Lastly, we detected eight NCVs of which four belonged to the order of the Pimascovirales (which includes the genera Pithovirus, and Marseillevirus) and four to the proposed Pandoravirales46. The cellular fraction NCV composition co-varied with the Eukaryotic composition (partial Mantel r-statistic 0.2888, p-value 0.001), while controlling for environmental variables.
Antarctic virophages and polinton-like viruses expand the known diversity of these groups
By targeting their distinct yet diverse52 MCP phylogeny, we identified 1678 PLV and 57 virophage scaffolds, and delineated them at the family rank. Most PLVs and virophages possessed virus hallmarks other than MCP, e.g., packaging ATPase or one replication related gene such as polymerase B, primase-helicase, or helicase (Fig. 6c, d). Their gene-sharing network clustering agrees with the MCP clades (Supplementary Fig. 8), while viruses of the Omnilimnoviroviridae seem to share more genes with TsV and PgVV group PLVs than with other virophages. Similarly, Chi group PLVs seem to share more genes with virophages than with other PLVs, reflecting their complex evolutionary relationships53.
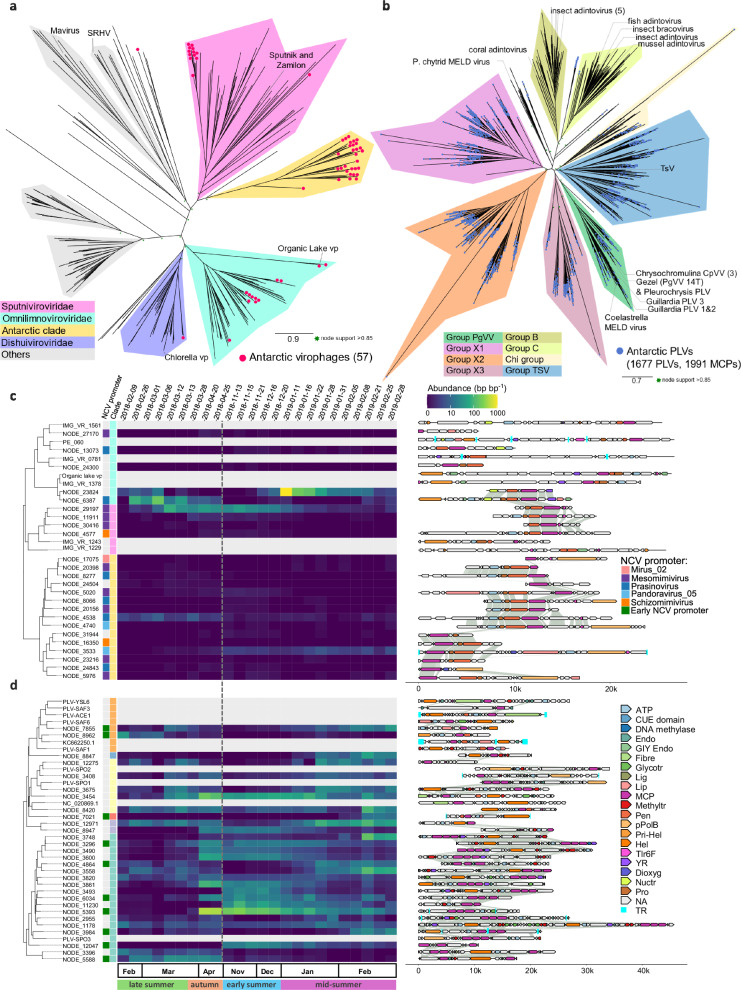
Fig. 6. Antarctic virophages and polinton-like viruses.
Shown are major capsid protein (MCP) phylogenies for (a) virophages (vp, Maverivicetes) and (b) polinton-like viruses (PLV, Polintoviricetes). Virophage family and PLV clade names according to refs. 29,37,38. Phylogeny for (c) virophages longer than 5 kb and (d) top 30 most abundant PLVs longer than 10 kb with the corresponding heatmap of metagenome abundance in the viral fraction (< 0.22 µm) and annotated genome map. Protein abbreviations as follows, FtsK-HerA family DNA-packaging ATPase (ATP), dioxygenase (Dioxyg), endonuclease (Endo), GIY-YIG nuclease domains (GIY), fibre-like protein (Fibre), glycosyltransferase (Glycotr), ligase (Lig), lipase (Lip), methyltransferase (Mthyltr), nucleotidyltransferase (Nuctr), penton (Pen), protein-primed polymerase B (pPolB), primase-helicase (Prim-Hel), helicase (Hel), maturation Cysterine Protease (Pro), uncharacterized protein (Tlr6F), putative tyrosine recombinase (YR). The dash grey line indicates the winter sampling gap.
For the virophages, we applied the recently updated taxonomic framework52. Most sequences belonged to the families Sputniviroviridae (n = 13), and Omnilimnoviroviridae (n = 12). The most abundant virophage (NODE_23824) belonged to the latter family and is closely related to the Antarctic organic lake virophage54. Notably, a distinct MCP clade of 30 virophages was identified, likely forming a novel virophage family (Fig. 6a). Within this clade, the virophage sequence NODE_3533 has a presumably complete 23.9 kb genome with core genes and direct terminal repeats (Fig. 6c). This sequence occurred mostly in January, together with NODE_23824, whereas NODE_6367 and _4538 were most prevalent in March. Only NODE_29197 dominated at the start and end of the season.
We identified NCVs putatively associated with these virophages by matching the promoter motif of the virophages with the late transcription promoter motifs of the co-occurring NCVs (Supplementary data 3 and Fig. 6c), as these are often shared55,56. Antarctic virophages from the Sputniviroviridae matched with two mesomimiviruses, and a schizomimivirus. In the Omnilimnoviroviridae, one matched with mesomimivirus, and two with prasinoviruses. The novel clade matched mostly with prasinoviruses, and mesomimiviruses with one match to a schizomimivirus, a mirusvirus, and a pandoravirus.
We classified PLVs based on previous surveys on this group34,57, as most clades have no formal taxonomic classification. Remarkably, Antarctic PLVs populated most MCP phylogenetic groups (Fig. 6b), except for clades A and B which mostly contain MCPs endogenous to Opisthokonta genomes, and animal adintoviruses (Fig. 6b). Another three separate groups (X1, X2, and X3) could be defined, with reference viruses from the previous clade X40 and cluster TriMCP57. These groups seem to arise from single PLVs exhibiting a highly divergent MCP gene triplication (Fig. 6b and Supplementary Fig. 9a). MCPs from the same PLV are generally more closely related in TSV than in group X, with their phylogenetic distance being on average 2.4 times greater than in group X (p-value < 2.2E-16, t-test). Additionally, this distance is on average 2.26 times smaller than between MCPs from different PLVs in the TsV group (p-value < 2.2E-16, t-test). In group X, shorter sequences with less than three MCPs are likely incomplete (Supplementary Fig. 9). Many of these PLVs MCP (n = 1387) make-up novel clades within these three groups. Most of the 291 remaining Antarctic PLVs belong to the Chi, TsV, and PgVV groups, with several forming distinct Antarctic clades (Fig. 6b). Of the 30 most abundant PLVs (>10 kb), about half were prevalent during February, and the others during the autumn and early summer (Fig. 6d). We investigated the possibility of these viruses parasitizing on NCVs by matching their promoter motif with the NCV early transcription promoter motif ‘WWWWWTGWWWWW’, as was found for Gezel-14T (Supplementary data 3 and Fig. 6d). This resulted in matches for 19% of the clade PgVV, 11% of the clades X1-3 and Chi and 6% of the clade TsV longer than 5 kb.
Discussion
This study provides an in-depth characterization of highly unique marine DNA viral diversity in Antarctic surface waters, where 75% of viral species were not present in the GOV8 and SOV29 datasets. The limited overlap with SOV indicates a high degree of variability within the Antarctic virome. Overall, these findings underscore the Southern Ocean as an underexplored region regarding viral diversity, emphasizing the need for further comprehensive studies in this region. Beyond identifying a high proportion of new viral species, our comprehensive approach uncovered novel diversity at higher taxonomic ranks. The discovery of four novel Crassvirales families, C2 and C4 with many representatives greatly expands our understanding of crassphage diversity, and their environmental range, beyond the primate gut microbiome43,58. The new-found diversity of crassphages in the Antarctic calls for further investigation of this class regarding diversity and activity in both the marine and other non-host-associated environments. Mining public data repositories may reveal additional members of these newly discovered lineages. These viruses might similarly59 infect Antarctic Bacteroidetes, a significant component of Marguerite Bay bacterial communities (classes Flavobacteria, Sphingobacteriia, Saprospiria and Cytophagia). Similarly, we discovered a novel virophage family and successfully recovered a complete genome of this family. Virophages replicate with co-infecting NCVs and may, in some cases such as mavirus, provide their hosts’ population-level protection against NCVs60,61. Since virophages parasitize the transcription machinery of their associated NCVs, the gene promoter motifs between giant viruses and virophages often share detectable similarity. In our analysis, we attempted to match virophages to their co-occurring NCVs by their putative promoters. Surprisingly, we found promoter signals that were shared between virophage and prasinovirus sequences, suggesting that virophages can depend on NCVs outside the Imitervirales order52. Our findings reveal a remarkably high diversity of PLVs, a highly understudied group34,57 that was recently found prevalent in Eukaryotic genomes33. The notable prominence of PLVs in our dataset (1,678 sequences) raises the question of whether this is Antarctic-specific or more widespread in the ocean, as has been found for freshwater environments57. We found 154 PgVV-clade PLVs34, with the one known isolate displaying a virophage-like lifestyle (Gezel-14T51). These PLVs had the highest proportion of matches to the NCV early transcription promoter motif, suggesting that, similarly to Gezel-14T, they may also depend on NCVs. While five sequences related to Gezel-14T have been previously reported in Antarctic virome samples27, our study substantially broadens the known diversity of Antarctic PLVs beyond the PgVV group. Additionally, 360 PLVs belonged to the TsV group that includes PLV Tetraselmis viridis virus S1 that does not require an NCV co-infection62. Notably, most PLVs (1387) were related to those in group X34 which our analysis shows to form three MCP clades, many representing one virus with duplicate and triplicate MCPs. The high divergence between the group X MCPs suggests this to be an early evolutionary event, contrasting with the triplication in group TsV which tends to have more closely related triplicates. In summary, these findings paint a picture of PLVs as an active and diverse group of viruses infecting Antarctic eukaryotes.
Another highlight of this study is the improved insight into the intricate relationship between host and virus. The NCVs were the dominant eukaryotic viruses and most belonged to the families Allo-, Meso-, and Schizomimiviridae, as well as the order Algavirales, of which all isolated members infect photosynthetic protists. The prevalence and temporal dynamics of these NCVs reinforces their active role in affecting Antarctic phytoplankton population dynamics24,26, ranging from preventive to reductive viral control63. Indeed, some of the prasinoviruses, for which all isolates are known to infect picophytoplankton in the order Mamiellales64, and capable of maintaining stable coexistence with their hosts65, were remarkably persistent suggesting a more continuous lysis pressure. PgV-like viruses from the genus Tethysvirus have previously been identified as an important member of Antarctic viral communities27,66. Our study’s temporal resolution sheds light on their role in impacting bloom decline of the co-occurring P. antarctica host (reductive control), aligning with recent findings of viral lysis being a major mortality factor of this Antarctic phytoplankter24. The steep increase in PgV-like viruses at the end of a P. antarctica bloom in December corresponded with the increase of Gammaproteobacteria. This bacterial group has been reported to quickly respond to viral infection of Phaeocystis67, reinforcing the importance of viral lysis in fueling the microbial loop. Overall, this study provides a comprehensive characterization of the Bamfordvirae kingdom diversity at the study site. This characterization expands the known PLV and virophage diversity, includes the recovery of novel NCVs together with those belonging to the candidate class Mriyaviricetes47, and also identifies members of the recently described candidate phylum Mirusviricota48.
The comprehensive seasonal coverage and enhanced sampling resolution of this study reveals that timing of sampling affects the likelihood of virus detection and discovery. Temporal sampling improves the understanding of the dynamics of the different viruses, i.e., NCVs, virophages, PLVs and phages in our study. It offered a deeper insight into prokaryotic bacteriophage-host interactions, with group-specific responses. For instance, Gammaproteobacteria displayed a typical predator-prey relationship illustrated by the decoupling between the host and bacteriophages abundance during the low sea-ice period. This pattern, characterized by high viral activity and low host abundance, was also found by viral activity assays in the ice-free areas of the Weddell Sea22. Such observations gain significance in the context of global warming, which is leading to the expansion of ice-free zones and longer ice-free periods around the Antarctic Peninsula68. Moreover, we found closely coupled dynamics of Flavobacteria with their bacteriophages, particularly during the phytoplankton bloom in January (cluster A). The increasing abundance of Flavobacteria bacteriophages coincided with the bacterial bloom, whereas Gammaproteobacteria bacteriophages were most dominant after the bloom (cluster C). Such successional pattern can be explained by the association of Antarctic Flavobacteria with Chl-a fluorescence, and their degradation of diverse complex organic compounds69,70. They make labile compounds available to other bacteria such as the Gammaproteobacteria and Alphaproteobacteria69. In our study, Alphaproteobacteria and their bacteriophages became more prominent during the colder periods, at the end of the season (cluster B) and pre-bloom phase (cluster D), consistent with this group's affinity with low temperature71.
The early summer marks an important transition for viral communities, where the onset of microbial production leads to increased bacteriophage production and abundance25. Our analysis shows this period is characterized by the emergence of a few dominant bacteriophages that account for a substantial portion of viral activity. A pronounced increase in bacteriophage activity during these months in the same study area led to a shift in carbon flow towards the microbial loop rather than higher trophic levels25. Concurrently, there is an apparent activation of temperate bacteriophages during the bacterial bloom, which is indicative of a refugium-to-activation dynamics72, and aligns with earlier research focusing on Antarctic prophage induction22,23,25.
This study reports for the first time the strong seasonal variations in diversity and dynamics for the Antarctic viral community. Our phylogeny-informed characterization of viral diversity underscores the distinctness of these communities. Finally, the time-resolved and co-sampling of host communities offers a unique perspective, shedding light on the seasonality and abundance of Antarctic marine viruses and providing a more complete understanding of their diversity in relation to microbial hosts.
Methods
Study site and sample collection
Samples were collected from 15 m depth in Marguerite Bay at the site of the Rothera Time-Series73 (RaTS, latitude 67.572°S; longitude 68.231°W). Sampling conditions in the Antarctic are highly variable and not always favorable for sampling, still we were able to collect samples on 24 days over two consecutive productive seasons (seven months period, from February until April 2018 and from November 2018 until February 2019). The water column properties were characterized using a SeaBird SBE 19 + CTD that measured conductivity, temperature, depth, salinity, chlorophyll autofluorescence, and photosynthetically available radiation. Mix layer depth (MLD) was defined as the depth with a water density difference of 0.05 kg m−3 relative to the surface19. Water samples were collected by performing sequential casts with Niskin bottles. These were emptied by gentle siphoning (using clean PC tube) into 20 L pre-rinsed polycarbonate carboys. All carboys and tubing used for sampling were decontaminated by rinsing with Contrad 70™ 2% v/v, followed by MilliQ ultrapure water (UP). Prior to sample collection, carboys were rinsed thrice with sample water. The carboys were protected from direct sunlight using opaque bags. Sampling was performed in the morning between 9:30 and 11:00 AM. Samples were further processed within 4 h from the start of sampling. The lab was kept at in situ temperature (with a minimum of 0.1 °C), and if needed, samples were kept on ice.
For dissolved inorganic nutrient subsampling, seawater was filtered through a 0.2 μm pore-size Supor membrane Acrodisc® filter (Pall, Port Washington, NY 11050, USA) and collected in 3 times sample-rinsed high-density polyethylene pony vials (PerkinElmer, USA). The subsamples for dissolved inorganic nitrogen and phosphorus were stored at −20 °C until analysis, and samples for dissolved inorganic silicate were stored in the dark at 4 °C. Samples for chlorophyll-a (Chl-a) were collected by filtering 1 to 8.5 L of unfiltered (total Chl-a) and 20 µm reverse-sieved (<20 µm fraction) onto GF/F filters (Whatman, UK), after which the filters were each wrapped in aluminum-foil, placed in a plastic bag, flash-frozen in liquid nitrogen, and stored at −80 °C until further analysis. For microbial abundances, 1 mL of seawater was fixed with glutaraldehyde to 0.5% v/v final concentration (25%, EM-grade, Sigma Aldrich, USA) for 30 min at 1 °C (in the dark), after which it was flash-frozen in liquid nitrogen and stored at −80 °C until analysis in the NIOZ laboratory (The Netherlands). Phytoplankton cells were counted on fresh samples (kept on ice) directly upon arrival in the Rothera lab.
Prior to handling the genomic samples, all bench surfaces were cleaned with Contrad 70™ 2% v/v (Decon Labs, East Sussex, UK) and rinsed with UP water. The cellular fraction metagenomic samples were collected by filtering 3 to 15 L (1 h filtering time at in situ temperature) of whole water through a 0.22 µm Sterivex™ (MilliporeSigma, Massachusetts, US). The filters were sealed with parafilm and stored at −80 °C until extraction. The viral fraction metagenomic samples were collected from 20 L seawater using the iron flocculation method according to Hurwitz et al. 2013 (ref. 74). Briefly, particles bigger than 0.22 µm were removed by two subsequential filtrations using a A/E glass fiber filter (142 mm, Whatman, GE Healthcare, UK) followed by a 0.22 µm PES filter (142 mm, MilliporeSigma, Massachusetts, US). The viruses were flocculated with 1 mg L−1 of FeCl3 (1 h incubation) and collected onto a 1 µm PC filter (142 mm, Whatman, GE Healthcare, UK). Filters were stored at 4 °C in the dark until further analysis. The full sampling metadata is available in Supplementary data 4.
Microbial abundances, chlorophyll-a, and nutrient concentrations
Viruses and bacteria were enumerated according to Brussaard et al. (2010, ref. 75) with modification to the TE-buffer (pH 8.2, according to ref. 76). Upon dilution in TE-buffer and staining with SYBRGreen-I (Sigma Aldrich, USA), the samples were enumerated using a FACSCalibur flow cytometer (BD Biosciences, USA) equipped with an air-cooled Argon laser with an excitation wavelength of 488 nm with the trigger on green fluorescence (Supplementary Fig. 10). Total phytoplankton abundances were obtained using a benchtop FACS Celesta flow cytometer (BD Biosciences, USA) with the trigger on red chlorophyll autofluorescence77.
Chlorophyll-a concentration (Chl-a) was determined using HPLC (according to ref. 18). Briefly, filters were freeze dried (48 h) and pigments were extracted using 90% acetone (48 h at 4 °C in the dark)78. Pigments were separated by high performance liquid chromatography (HPLC; Waters model 2690), equipped with a 3.5 μm particle size Zorbax Eclipse XDB-C8 column with a cooled autosampler (4 °C) according to Van Heukelem and Thomas (2001). Pigment detection and quantification was based on comparison with DHI LAB standards at 346 nm (Waters 996 HPLC Photodiode Array Detector)79.
The concentrations of dissolved inorganic macro-nutrients were obtained using a TRAACS 800 autoanalyzer (Bran+Luebbe, Germany), following Grasshoff et al. (1983) for nitrate and nitrite, of Murphy and Riley (1962) for phosphate, and of Strickland and Parsons (1968) for silicate (refs. 80–82). The calibration standards were diluted in low nutrient seawater (LNSW) with salinity levels similar to the samples (35‰). The detection limit was 0.007 µM for nitrate + nitrite, 0.001 µM for nitrite, 0.007 µM for phosphate, and 0.03 µM for silicate.
Viral and cellular fraction metagenomes
The DNA of the cellular fraction (Sterivex filters) was extracted using the Qiagen DNeasy Powersoil kit (Qiagen, Carlsbad, USA), according to the manufacturer’s instructions. The filters were opened and cut into approximately 3 × 3 mm pieces prior to 2 × 10 s bead beating at 3.55 m s−1 with a 30 sec dwell on a Bead Ruptor Elite (Omni Internationals, USA). Viruses from the viral fraction (iron flocculation method) were resuspended at 4 °C overnight in a EDTA (0.1 M) -MgCl2 (0. 2 M) - ascorbate (0.2 M) buffer with pH of 6.0 (1 mL per liter of sampled seawater). The resuspended sample was gently filtered through a 0.45 µm SFCA syringe filter (Sartorius, Göttingen, Germany) and treated with 1 U µL−1 of DNase I (Roche, Basel, Switzerland) for 18 h at 4 °C. DNase I was deactivated by adding EDTA and EGTA (0.1 M final concentration). The viruses were concentrated to a volume of 0.8 to 1 mL using a 100 kDa Amicon (MilliporeSigma, Massachusetts, US). DNA was extracted using Wizard® PCR Preps DNA Purification kit (Promega, Wisconsin, United States) according to manufacturer’s instructions. The DNA was eluted twice in 50 µL of 80 °C molecular grade low TE buffer (pH 8.0, 10 mM Tris and 0.1 mM EDTA).
The libraries were prepared using the TruSeq Nano DNA Kit (Illumina, San Diego, CA) and sequenced with the Illumina platform NovaSeq6000 S2 (2 × 150 bp), according to the manufacturer’s instructions, on average 80 M reads per sample (58–123 M). The read quality was assessed before and after quality trimming using MultiQC v1.883. PhiX reads were removed using bbduk v38.07 (https://sourceforge.net/projects/bbmap/), low complexity (dust filter threshold of 20) and PCR duplicates were removed with prinseq-lite v0.20.484. Illumina adaptors were clipped with trimmomatic v0.3685. To assess the degree of enrichment of our viromes, the abundance of microbial genetic markers was assessed using ViromeQC v1.0.1 in all metagenomes86.
PhyloFlash v3.487 was used to characterize microbial communities by target assembly and classification of the 16/18S rRNA gene. Reads that mapped to plastid 16S rRNA genes were removed.
Virus genome assembly, taxonomy, and host prediction
All reads from the viral fraction metagenomes were co-assembled into scaffolds using metaSpades v3.14.188. Scaffolds of at least 1500 bp long were checked for viral signals using PPR-Meta v1.189, which was found to be the most sensitive virus prediction tool with relatively few false positives in a benchmark work that included some of the samples from this study90. Sequences identified as viral but with no CheckV v0.7.0 viral hallmark genes and cellular hallmark genes31 and classified as non-viral by Cenote-Taker291 were removed. The remaining co-assembled scaffolds are equivalent to viral species-level operational taxonomic units (vOTUS) as clustering at 95% identity over 85% of the sequences8 resulted in the clustering of only 0.039% of the sequences.
We quantified the overlap between Rothera viral sequences and the Global Ocean Virome8 and Southern Ocean Virome29 datasets by performing an all versus all blastn which were then clustered at 95% average nucleotide identity over 80% of the sequence using the CheckV virus clustering.
For viral sequences longer than 5000 bp or with >= 70% completeness (CheckV completeness), coding regions were predicted using prodigal92 (v2.6.3 -p meta). The viral sequences were clustered based on their coding regions using vContact293 using the RefSeq prokaryotic viruses (--db ProkaryoticViralRefSeq94-Merged) together with a custom collection of protist DNA viruses (Supplementary data 5). The clusters were visualized using Cytoscape v3.8.2 with a perfuse force-directed layout94, and scaffold viral taxonomy was overlayed. In the sections below we detail the target identification of Crassvirales, Nucleocytoviricota and Preplasmiviricota (PLVs and virophages), while for other viral sequences we used the Cenote-taker2 taxonomy. The Caudoviricetes (the single class in the Uroviricota Phylum), Nucleocytoviricota, Preplasmiviricota, and archaeal virus groups were manually defined (Supplementary Fig. 11) which always corresponded with the taxonomy of reference genomes within those groups and largely with the Cenote-taker2 taxonomy for the metagenomic viral sequences. Unknown sequences within the above defined groups were attributed the group phylum rank taxonomy. Scaffolds annotated as Caudoviricetes by Cenote-taker2 and all sequences within the Caudoviricetes mega cluster (excluding the archaeal group) were further classified into family rank taxonomy using PhaGCN232. Archaeal group sequences classified as halovirus by cenote-taker were classified as Thumleimavirales. For Crassvirales, Nucleocytoviricota, Maveriviricetes (virophages) and Polintoviricetes (polinton-like viruses) sequences, we performed a targeted identification and taxonomic classification (see sections below).
The host genus was predicted for high and mid-quality bacteriophage sequences (longer than 10 kb or at least 70% complete) using iPHoP v0.938 with the standard cut-off for genus rank prediction and a 0.2 false discovery rate cut-off for class and phylum rank prediction (using All_combined_scores.csv). Conflicting host predictions were removed. The standard genus level output resulted in 744 predictions while the lower prediction threshold resulted in 1668 predictions at the class level.
All viral sequences, excluding those classified as eukaryotic viruses, were inspected for the presence of the lysogeny genes integrase, excisionase, and bacteriophage repressor protein. The coding regions were predicted using prodigal92 (v2.6.3 -p meta) which were matched by hmmsearch (e-value 1e-5, HMMER v3.1b2) to HMM profiles from the PFAM (34.0), KO (97.0), ncbi COGs (COGS2020) and by diamond search (e-value 1e-10, v0.9.22) to the nrNCBI (downloaded in March 2022). For integrase were considered matches to K03733, K14059, K04763, K21039, COG0582, COG1518, PF00589, PF13495, PF00665, PF13683, PF13102, PF18697, PF09003, PF02899, PF12835, PF02022, PF14659, PF18644, PF13976, PF13333, PF14882, PF12834, PF17921 and those containing the string “integrase” in the diamond top hit excluding those containing the string “CRISPR” and “Transposase”. For excisionase were considered matches to PF06806 and PF0782, and those containing the string “excisionase” in the diamond top hit. For the phage repressor protein were considered matched to PF00717, K22300, K07727, K18830 and COG2932.
Reads from the viral and cellular metagenomes were mapped to all scaffolds using bwa v0.7.17 mem95. Reads mapped with less than 95% identity over 80% of the read length were excluded with CoverM v0.6.196, and the number of reads mapped to each scaffold was calculated with samtools coverage v1.1197. When scaffolds had less than 70% horizontal coverage, their abundance was set to 0. To remove the effect of non-viral contamination, the read relative abundances were calculated by dividing the read counts by the total number of viral reads in each sample. The depth corrected scaffold vertical coverage was calculated by dividing the relative read abundance by the total scaffold length multiplied by the average number of viral read bases. Read abundance was used where assembly genome fragmentation could impact abundance such as for higher taxonomy level compositional abundance. Vertical coverage was used where genome fragmentation was not expected to be an issue, such as for scaffold abundance, cumulative abundance of scaffolds containing a single gene, or the binned NCV genomes.
Antarctic Crassvirales bacteriophages
To identify Crassvirales scaffolds, we used a targeted approach that does not depend on protein predictions as Crassvirales are known to include alternative genetic codes44. First, we used transeq98 to obtain the six-frames translations of the DNA scaffolds (options: -frame 6 -table 11 -clean). HMM profiles of the Crassvirales phylogenetic marker genes (see ref. 44), including the major capsid protein (MCP), the terminase large subunit (TerL), and the portal protein (portal), were used to search for homologs via hmmscan (HMMER v3.1b2). Hmmscan results were then filtered (e-value < 0.05, coverage > 0.3). Scaffolds over 10 kb in length and containing matches to any of these three proteins were put forward for further inspection as candidate Crassvirales (n = 466).
To place the candidate Crassvirales scaffolds within the larger context of Caudoviricetes families, we used the Virus Metadata Resource from the International Committee on Taxonomy of Viruses (VMR_21-221122_MSL37). We selected at random ten isolates (or all if < 10 available) for each of the 43 Caudoviricetes families excluding: i) members of the Crassvirales order (Feb 2023), and ii) isolates from the Helgolandviridae (n = 1), Graaviviridae (n = 2), and Duneviridae (n = 6) from which no sequences could be extracted using our approach. This resulted in 245 species/isolates for which we obtained genomes, encoded proteins, and gene annotations from the NCBI Nucleotide database.
To reconstruct the phylogeny, we retrieved the TerL, MCP, and portal proteins encoded in the reference genomes by extracting the proteins containing in their headers “terminase”, excluding “terminase small”, “major head”, “major capsid”, and “portal”, using grep and seqtk subseq (https://github.com/lh3/seqtk). We were able to extract the phylogenetic markers from most of the sequences (243 TerL (99.2%), 234 MCP (95.5%) and 185 portal (75.5%) proteins from the 245 genomes), no gene duplicates were detected. In addition, we collected 673 Crassvirales genomes and TerL, MCP and portal protein sequences (from refs. 43,44). These sequences were used with those from our candidate Crassvirales to build separate phylogenetic trees of each protein. For the multiple sequence alignment, we used MAFFT99 (v7.475, option: –auto), modeltest-ng100 to determine the amino acids substitution model (option: -d aa), and IQ-TREE2 v2.1.1101 to compute the phylogeny (option: -m LG + G4 + F -B 1000). Candidate Crassvirales scaffolds for which all three proteins (TerL, MCP, and Portal) that fell within the clade formed by the ICTV-approved Crassvirales families were deemed Crassvirales sequences.
The ORFs of candidate scaffolds were detected with Prodigal v2.6.392 using three modes to consider stop codon reassignments (options: standard, -TGA W, and -TAG Q). These were functionally annotated using the marker protein HMMs from Yutin et al. 2021 (ref. 44). The trees and their annotation were created using the software suite ggtree v3.6.2102 and iTOL v5103. The genomic maps were built via gggenomes v0.9.5.9104.
Nucleocytoviricota and Mirusviricota
To identify NCVs, we first binned all assembled scaffolds using vRhyme v1.1.0 for the viral fraction co-assembly and using CONCOCT v1.1.0 for the corresponding cellular fraction assemblies. Scaffolds were examined for Nucleocytoviricota markers by performing a hmmsearch on prodigal predicted proteins92 (v2.6.3 -p meta) against the nine giant virus orthologous groups HMM profiles using ncldv_markersearch46. All bins containing hits to phylogenetic marker genes were further investigated. Bins were decontaminated from likely contaminating cellular scaffolds using viralrecall105 (removing those with score < 0 as per ref. 106). Separate phylogenetic trees for the markers were built by combining the bins with a comprehensive set of NCVs, Mirusviricota, Herpesvirales, Eukaryotic and Archaeal genomes collected by Karki et al. 2024 (ref. 107), including the recently described NCV class Mriyaviricetes47. We further removed scaffolds containing markers falling within the Eukaryotic clades for RNA polymerase, DNA topoisomerase I, and DNA polymerase B.
The phylogenic trees were built for the bins and a reference set containing NCV isolates and metagenomic bins collected from previous metagenomic surveys46–48,106,108 and additional RefSeq NCV isolates as of September 2022 (Supplementary data 2). First, by detecting and aligning the seven genetic markers46 using ncldv_markersearch (VLTF3, A32, TopoII, TFIIB, PolB, RNAPL and SFII). The alignments were trimmed of regions where > 20% of the sequences have a gap and with an entropy score bellow 0.55 using BMGE v1.12111. The trimmed alignments were used to build a maximum likelihood tree using IQ-tree2101 the best model finder option (-m MFP) with ultrafast bootstrap of 1000 replicates. Bins were manually assigned a taxonomy based on the reference set taxonomy.
For the final dataset, only bins containing at least four out of seven markers were kept, except for those classified as Mirusviricota48 and Mriyaviricetes47 where all bins with at least two markers were kept, since these viral groups not possess all NCV marker genes (see Supplementary data 2 for gene presence/non-detection). The Mriyaviruses sequences belonged to large genomic bins which also contained Eukaryotic-like RNA polymerase, DNA topoisomerase II and DNA polymerase B genes. We removed this Eukaryotic host contamination by retaining only the contigs containing the marker genes VLTF3, A32 and MCP47. Bins with more than one duplication of the genes SFII, VLTF3, A32, and PolB were removed to avoid bins with high strain heterogeneity (as per ref. 106). We removed redundant bins by clustering bins at 95% average nucleotide identity or 100% marker gene amino acid identity and selected the best representative (most marker genes or highest N50). Eukaryotic contamination was identified on the NCV bins and the unfiltered Mriyaviruses by using MetaEuk v6.a5d39d9109 to predict (using Pfam reference) and classify (using MMETSP_zenodo_3247846_uniclust90_2018) ORFs.
We used ncldv_markersearch to screen the final set of bins for the seven genetic markers46,and to perform the alignments and concatenation. The alignment was trimmed of regions where >20% of the sequences had a gap and with an entropy score bellow 0.55 using BMGE v1.12110. The trimmed alignment was used to build a maximum likelihood tree using iqtree2101 the best model finder option (-m MFP) with ultrafast bootstrap of 1000 replicates. The best fit model was LG + F + R10. The tree was visualized using FigTree v1.4.4111.
Maveriviricetes and Polintonviricetes viruses
All scaffolds were inspected for virophages (class Maveriviricetes) and polinton-like viruses (PLVs, class Polintonviricetes) major capsid protein (MCP) using a collection HMM built using global, and clade restricted protein alignments. Rothera scaffolds ORFs were predicted and translated to amino acid using prodigal92 (v2.6.3 -p meta).
For the Maveriviricetes, we used a collection of non-redundant isolate and metagenomic MCP sequences52. The global and family rank MCP alignments were built with MAFFT99 (v7.407, -auto), and the HMM profile built with hmmbuild112 2 (HMMER v3.1b2). We used these profiles to identify putative virophage MCPs in the Rothera scaffolds’ amino acid sequences (> 200 aa) using hmmsearch (HMMER v3.1b2). The new putative virophage MCP were joined with the reference sequences and the Maveriviricetes phylogeny was built, and the sequences classified according to the newly proposed taxonomy (ref. 52).
For PLVs, HMM profiles were constructed using the MCP genes from a collection of curated PLV sequences34,57. PLV MCP identified in Yutin et al., 2015 (ref. 34) were used as queries to search the non-redundant NCBI protein database. The hits were collected, clustered with UCLUST113 with a similarity threshold of 0.5 and were iteratively aligned and clustered using the approach described by Wolf et al., 2018 (ref. 114). From the alignment of the cluster containing the query proteins, a tree was built using FastTree115. From the tree topology eight clades were manually inferred for which separate HMM profiles were built. Translated Rothera scaffolds were searched with these HMM profiles and the profiles provided by Bellas and Sommaruga, 2021 (ref. 57). The hits were mixed with reference PLV MCPs (proteins of the HMM profiles). The resulting protein set was iteratively aligned and clustered using the abovementioned approach, and the alignment of the protein cluster containing reference PLV MCPs was used to build a tree using FastTree. Tree clades were named according to the clades as defined in Yutin et al., 2015 and Bellas and Sommaruga, 2021 (refs. 34,57).
For the functional annotation of the other virophage genes, all predicted proteins from putative virophage scaffolds were pooled with proteins predicted from virophage genomes available from Roux et al. 2023 (ref. 52). Proteins were clustered using mmseqs easy-cluster116, and for each cluster, alignments and profiles were generated by MAFFT99 (v7.505, - einsi), hhconsensus117 (v3.3.0) and finally hhmake117 (v3.3.0). All profiles were then searched against the Pfam (http://pfam.xfam.org/) and PDB (https://www.rcsb.org/) databases using hhsearch117 (v3.3.0, -E 1e-4). Matches with at least 90% probability, e-values below 1e-4, clear functional annotation as well as a score that was at least 90% of the best score per cluster were used for annotation. Annotation of PLV scaffolds was performed similarly to the virophages, with all predicted proteins from putative PLV scaffolds being pooled with proteins predicted from available PLV genomes34,57. We additionally searched the profiles against a database consisting of the profiles of clusters from Bellas and Sommaruga 2021 (ref. 57) and transferred annotations. The annotated genomes were visualized using gggenomes104 in combination with ggtree102.
For all NCV bins, predicted proteins were searched against the GVDB database using HMMER v3.3.2. Upstream regions (50 bp) of genes homologous to Cafeteria roenbergensis virus ‘late’ genes were extracted from the genomes and searched for enriched motifs using the streme tool from the meme-suite v5.5.0 and the protein-coding regions as negative controls. Candidate motifs were filtered for non-AT consensus sites and scores below 1e−3. Similarly, upstream regions of all virophage genes were extracted and searched for enriched occurrences of these candidate motifs using the sea tool (--thresh 1) from the meme-suite, again using the protein-coding regions as negative controls. PLV scaffolds above 5,000 bps were analyzed similarly for occurrences of a consensus ‘early’ NCV promoter motif (WWWWWTGWWWWW).
Viral diversity and correlation with environmental parameters
We performed a multivariate analysis on viral sequences of high and mid quality (longer than 10 kb or at least 70% complete) to avoid highly fragmented genomes. Given the compositional nature of the abundance date, the analysis was performed using compositional data analysis methods118. Viral read abundance was normalized by sequence length and centered-log ratio transformation using microbiome R package v1.20.0 (https://github.com/microbiome/microbiome). We explored the seasonal dynamics of alpha diversity by calculating the species richness and the Shannon and inverse Simpson diversity indexes for each day using hillR v0.5.1119, which are expressed in effective number of species. Simpson evenness was calculated by dividing the inverse Simpson index by the species richness. Additionally, we tracked the number and abundance of dominant viruses, defined as viral scaffolds representing over 0.5% of the viral reads at any given time (n = 203).
We performed a hierarchical cluster analysis based of the Aitchison distance using hclust120 (method centroid), and the normalized abundances visualized in heatmap using the stats package in R.
Microbial abundances and Chl-a concentrations were log-transformed and other environmental metadata was visually checked for normality and z-scored or log transformed to achieve a distribution closest to normal. Pairwise Pearson correlation was computed and from strongly covarying environmental metadata pairs, one was removed (r > 0.8). To find which combination of variables better correlated with changes in the viral community, we fitted a general multivariate regression model to our data using Adonis2 (vegan v2.6-4)121. Variables were sequentially permuted so that the variables were input to the model by order of highest sum of squares, non-significant variables were removed from the model. The association between the phage and bacterial communities were tested by using a partial Mantel test on the Aitchison distance matrixes, while controlling for environmental conditions using the mantel.partial function (Pearson correlation and 999 permutations, Vegan v2.6-4). The control environmental matrix was computed as the Euclidean distances of the non-covarying variables which were significant according to the general multivariate regression model.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was funded by the POMVIDDY project (grant ALWPP.2016.019 awarded to C.P.D.B.) which was supported by the Dutch Research Council (NWO), within the Netherlands Polar Programme. We are grateful to the British Antarctic Survey, Cambridge, UK, and the MMB-NIOZ technical staff for the logistical and technical support, as well as Nisma Abdelmalik and Monika Krolikowski for field assistance and Gosse Metz for the Sterivex DNA extractions. We thank the NIOZ Nutrient Lab on Texel for performing the inorganic nutrient analysis. We are grateful to Alejandro Abdala, Julia Engelmann and Nina Dombrowsky for providing the functional annotation pipeline. This work was supported by the European Research Council (ERC) Consolidator grant 865694: DiversiPHI (B.E.D), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC 2051 – Project-ID 39071386 (B.E.D), the European Union’s Horizon 2020 research and innovation program, under the Marie Skłodowska-Curie Actions Innovative Training Networks grant agreement no. 955974 (VIROINF, B.E.D), the Alexander von Humboldt Foundation in the context of an Alexander von Humboldt-Professorship founded by the German Federal Ministry of Education and Research (B.E.D), and a junior researcher fellowship from the Fonds voor Wetenschappelijk Onderzoek (FWO) [12D8623N, C.L.].
Author contributions
G.J.P., C.P.D.B., and B.E.D. designed the study. E.M.W. and G.J.P. collected all samples. G.J.P. processed and analyzed the data. T.E.G.B. analyzed the environmental metadata. G.J.P., M.E.S, N.Y., and M.G.F. designed and performed the virophage and polinton-like virus analysis. C.L. and M.V.F. designed and performed the crassphage analysis. G.J.P. and L.W. designed and performed the phage host prediction analysis. G.J.P. took the lead in writing. C.P.D.B. conceived the study and helped shape the research, overall direction and writing of the manuscript. B.E.D. helped shape the analyses and provide critical feedback. H.B. and L.W. helped revise the manuscript. All authors provided feedback on the results and manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Metagenomic raw reads have been deposited in the European Nucleotide Archive under the Bioproject PRJEB71789. Figure source data are provided with this paper. The sampling metadata, used HHM profiles, viral scaffold nucleotide sequences, NCV bins, taxonomic and host annotations, viral marker gene protein alignments and phylogenies are available through the NIOZ dataverse 10.25850/nioz/7b.b.wg. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gonçalo J. Piedade, Email: goncalo.piedade@nioz.nl
Corina P. D. Brussaard, Email: corina.brussaard@nioz.nl
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53317-y.
References
- 1.Suttle, C. A. Marine viruses - Major players in the global ecosystem. Nat. Rev. Microbiol.5, 801–812 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Fuhrman, J. A. Marine viruses and their biogeochemical and ecological effects. Nature399, 541–548 (1999). [DOI] [PubMed] [Google Scholar]
- 3.López-García, P. et al. Metagenome-derived virus-microbe ratios across ecosystems. ISME J.17, 1552–1563 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-On, Y. M. & Milo, R. The Biomass Composition of the Oceans: A Blueprint of Our Blue Planet. Cell179, 1451–1454 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Brum, J. R. & Sullivan, M. B. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol.13, 147–159 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Sullivan, M. B., Weitz, J. S. & Wilhelm, S. Viral ecology comes of age. Environ. Microbiol. Rep.9, 33–35 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Breitbart, M. et al. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci.99, 14250–14255 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory, A. C. et al. Marine DNA viral macro- and microdiversity from Pole to Pole. Cell177, 1109–1123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roux, S. et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature537, 689–693 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Luo, E., Leu, A. O., Eppley, J. M., Karl, D. M. & DeLong, E. F. Diversity and origins of bacterial and archaeal viruses on sinking particles reaching the abyssal ocean. ISME J.16, 1627–1635 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ignacio-Espinoza, J. C., Ahlgren, N. A. & Fuhrman, J. A. Long-term stability and Red Queen-like strain dynamics in marine viruses. Nat. Microbiol.5, 265–271 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Coutinho, F. H. et al. Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nat. Commun.8, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassman, N. et al. Oxygen minimum zones harbour novel viral communities with low diversity. Environ. Microbiol.14, 3043–3065 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Heinrichs, M. E. et al. Breaking the Ice: A Review of Phages in Polar Ecosystems. in 31–71. 10.1007/978-1-0716-3549-0_3 (Humana, New York, NY, 2024). [DOI] [PubMed]
- 15.Marshall, J. & Speer, K. Closure of the meridional overturning circulation through Southern Ocean upwelling. Nat. Geosci.5, 171–180 (2012). [Google Scholar]
- 16.Frölicher, T. L. et al. Dominance of the Southern Ocean in Anthropogenic Carbon and Heat Uptake in CMIP5 Models. J. Clim.28, 862–886 (2015). [Google Scholar]
- 17.Huang, Y., Fassbender, A. J. & Bushinsky, S. M. Biogenic carbon pool production maintains the Southern Ocean carbon sink. Proc. Natl Acad. Sci. USA.120, e2217909120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biggs, T. E. G. et al. Antarctic phytoplankton community composition and size structure: importance of ice type and temperature as regulatory factors. Polar Biol.42, 1997–2015 (2019). [Google Scholar]
- 19.Venables, H. J., Clarke, A. & Meredith, M. P. Wintertime controls on summer stratification and productivity at the western Antarctic Peninsula. Limnol. Oceanogr.58, 1035–1047 (2013). [Google Scholar]
- 20.Luria, C. M. et al. Seasonal shifts in bacterial community responses to phytoplankton-derived dissolved organic matter in the Western Antarctic Peninsula. Front. Microbiol.8, 2117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho, F., Kohut, J., Oliver, M. J., Sherrell, R. M. & Schofield, O. Mixing and phytoplankton dynamics in a submarine canyon in the West Antarctic Peninsula. J. Geophys. Res. Ocean.121, 5069–5083 (2016). [Google Scholar]
- 22.Evans, C. & Brussaard, C. P. D. Regional variation in lytic and lysogenic viral infection in the southern ocean and its contribution to biogeochemical cycling. Appl. Environ. Microbiol.78, 6741–6748 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brum, J. R., Hurwitz, B. L., Schofield, O., Ducklow, H. W. & Sullivan, M. B. Seasonal time bombs: Dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J.10, 437–449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biggs, T. E. G., Huisman, J. & Brussaard, C. P. D. Viral lysis modifies seasonal phytoplankton dynamics and carbon flow in the Southern Ocean. ISME J.15, 3615–3622 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans, C. et al. Shift from Carbon Flow through the Microbial Loop to the Viral Shunt in Coastal Antarctic Waters during Austral Summer. Microorganisms9, 460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eich, C. et al. Ecological Importance of Viral Lysis as a Loss Factor of Phytoplankton in the Amundsen Sea. Microorganisms10, 1967 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, Q. et al. Metagenomic Characterization of the Viral Community of the South Scotia Ridge. Viruses11, 95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong, Z. et al. Viral Diversity and Its Relationship With Environmental Factors at the Surface and Deep Sea of Prydz Bay, Antarctica. Front. Microbiol.9, 2981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alarcón-Schumacher, T., Guajardo-Leiva, S., Martinez-Garcia, M. & Díez, B. Ecogenomics and Adaptation Strategies of Southern Ocean Viral Communities. mSystems6, e0039621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Simon, J. et al. Viruses under the Antarctic Ice Shelf are active and potentially involved in global nutrient cycles. Nat. Commun.14, 1–10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nayfach, S. et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol.39, 578–585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang, J.-Z. et al. Virus classification for viral genomic fragments using PhaGCN2. Brief. Bioinform.24, bbac505 (2023). [DOI] [PubMed] [Google Scholar]
- 33.Bellas, C. et al. Large-scale invasion of unicellular eukaryotic genomes by integrating DNA viruses. Proc. Natl Acad. Sci. USA.120, e2300465120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yutin, N., Shevchenko, S., Kapitonov, V., Krupovic, M. & Koonin, E. V. A novel group of diverse Polinton-like viruses discovered by metagenome analysis. BMC Biol.13, 1–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roux, S. et al. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ2016, e2777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breitbart, M., Delwart, E., Rosario, K., Segalés, J. & Varsani, A. ICTV virus taxonomy profile: Circoviridae. J. Gen. Virol.98, 1997–1998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venables, H. et al. Sustained year-round oceanographic measurements from Rothera Research Station, Antarctica, 1997–2017. Sci. Data10, 1–13 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roux, S. et al. iPHoP: An integrated machine learning framework to maximize host prediction for metagenome-derived viruses of archaea and bacteria. PLOS Biol.21, e3002083 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Straza, T. R. A., Ducklow, H. W., Murray, A. E. & Kirchmana, D. L. Abundance and single-cell activity of bacterial groups in Antarctic coastal waters. Limnol. Oceanogr.55, 2526–2536 (2010). [Google Scholar]
- 40.Sow, S. L. S. et al. Biogeography of Southern Ocean prokaryotes: a comparison of the Indian and Pacific sectors. Environ. Microbiol.24, 2449–2466 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luria, C. M., Amaral-Zettler, L. A., Ducklow, H. W. & Rich, J. J. Seasonal succession of free-living bacterial communities in coastal waters of the western antarctic peninsula. Front. Microbiol.7, 1731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner, D. et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol.168, 74 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yutin, N. et al. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol.3, 38–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yutin, N. et al. Analysis of metagenome-assembled viral genomes from the human gut reveals diverse putative CrAss-like phages with unique genomic features. Nat. Commun.12, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abergel, C., Legendre, M. & Claverie, J. M. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol. Rev.39, 779–796 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Aylward, F. O., Moniruzzaman, M., Ha, A. D. & Koonin, E. V. A phylogenomic framework for charting the diversity and evolution of giant viruses. PLOS Biol.19, e3001430 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yutin, N., Mutz, P., Krupovic, M. & Koonin, E. V. Mriyaviruses: small relatives of giant viruses. MBio10.1128/mbio.01035-24 (2024). [DOI] [PMC free article] [PubMed]
- 48.Gaïa, M. et al. Mirusviruses link herpesviruses to giant viruses. Nature616, 783–789 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallot-Lavallée, L. et al. The 474-Kilobase-Pair Complete Genome Sequence of CeV-01B, a Virus Infecting Haptolina (Chrysochromulina) ericina (Prymnesiophyceae). Genome Announc3, e01413–e01415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santini, S. et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl Acad. Sci. USA.110, 10800–10805 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roitman, S. et al. Isolation and infection cycle of a polinton-like virus virophage in an abundant marine alga. Nat. Microbiol.8, 332–346 (2023). [DOI] [PubMed] [Google Scholar]
- 52.Roux, S. et al. Updated Virophage Taxonomy and Distinction from Polinton-like Viruses. Biomolecules13, 204 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yutin, N., Raoult, D. & Koonin, E. V. Virophages, polintons, and transpovirons: A complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol. J.10, 1–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yau, S. et al. Virophage control of antarctic algal host-virus dynamics. Proc. Natl Acad. Sci. USA.108, 6163–6168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischer, M. G. & Suttle, C. A. A virophage at the origin of large DNA transposons. Science.332, 231–234 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Roux, S. et al. Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics. Nat. Commun.8, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bellas, C. M. & Sommaruga, R. Polinton-like viruses are abundant in aquatic ecosystems. Microbiome9, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dutilh, B. E. et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun.5, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edwards, R. A. et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol.4, 1727–1736 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischer, M. G. & Hackl, T. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature540, 288–291 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Koonin, E. V. & Krupovic, M. Polintons, virophages and transpovirons: a tangled web linking viruses, transposons and immunity. Curr. Opin. Virol.25, 7–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pagarete, A., Grébert, T., Stepanova, O., Sandaa, R. A. & Bratbak, G. Tsv-N1: A Novel DNA Algal Virus that Infects Tetraselmis striata. Viruses7, 3937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brussaard, C. P. D. Viral Control of Phytoplankton Populations—a Review1. J. Eukaryot. Microbiol.51, 125–138 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Moreau, H. et al. Marine Prasinovirus Genomes Show Low Evolutionary Divergence and Acquisition of Protein Metabolism Genes by Horizontal Gene Transfer. J. Virol.84, 12555–12563 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yau, S. et al. Virus-host coexistence in phytoplankton through the genomic lens. Sci. Adv.6, eaay2587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alarcón-Schumacher, T., Guajardo-Leiva, S., Antón, J. & Díez, B. Elucidating viral communities during a phytoplankton bloom on the west Antarctic Peninsula. Front. Microbiol.10, 1014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheik, A. R. et al. Responses of the coastal bacterial community to viral infection of the algae Phaeocystis globosa. ISME J.8, 212–225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Massom, R. A. et al. Antarctic ice shelf disintegration triggered by sea ice loss and ocean swell. Nature558, 383–389 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Williams, T. J. et al. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol.15, 1302–1317 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Piontek, J., Meeske, C., Hassenrück, C., Engel, A. & Jürgens, K. Organic matter availability drives the spatial variation in the community composition and activity of Antarctic marine bacterioplankton. Environ. Microbiol.24, 4030–4048 (2022). [DOI] [PubMed] [Google Scholar]
- 71.Tada, Y., Makabe, R., Kasamatsu-Takazawa, N., Taniguchi, A. & Hamasaki, K. Growth and distribution patterns of Roseobacter/Rhodobacter, SAR11, and Bacteroidetes lineages in the Southern Ocean. Polar Biol.36, 691–704 (2013). [Google Scholar]
- 72.Silveira, C. B., Luque, A. & Rohwer, F. The landscape of lysogeny across microbial community density, diversity and energetics. Environ. Microbiol.23, 4098–4111 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Clarke, A., Meredith, M. P., Wallace, M. I., Brandon, M. A. & Thomas, D. N. Seasonal and interannual variability in temperature, chlorophyll and macronutrients in northern Marguerite Bay, Antarctica. Deep Sea Res. Part II Top. Stud. Oceanogr.55, 1988–2006 (2008). [Google Scholar]
- 74.Hurwitz, B. L., Deng, L., Poulos, B. T. & Sullivan, M. B. Evaluation of methods to concentrate and purify ocean virus communities through comparative, replicated metagenomics. Environ. Microbiol.15, 1428–1440 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brussaard, C. P. D., Payet, J. P., Winter, C. & Weinbauer, M. G. Quantification of aquatic viruses by flow cytometry. in Manual of Aquatic Viral Ecology 102–109 10.4319/mave.2010.978-0-9845591-0-7.102 (American Society of Limnology and Oceanography, 2010).
- 76.Mojica, K. D. A., Evans, C. & Brussaard, C. P. D. Flow cytometric enumeration of marine viral populations at low abundances. Aquat. Microb. Ecol.71, 203–209 (2014). [Google Scholar]
- 77.Marie, D., Partensky, F., Vaulot, D. & Brussaard, C. Enumeration of Phytoplankton, Bacteria, and Viruses in Marine Samples. Curr. Protoc. Cytom.10, 11.11.1–11.11.15 (1999). [DOI] [PubMed] [Google Scholar]
- 78.van Leeuwe, M. A., Villerius, L. A., Roggeveld, J., Visser, R. J. W. & Stefels, J. An optimized method for automated analysis of algal pigments by HPLC. Mar. Chem.102, 267–275 (2006). [Google Scholar]
- 79.Van Heukelem, L. & Thomas, C. S. Computer-assisted high-performance liquid chromatography method development with applications to the isolation and analysis of phytoplankton pigments. J. Chromatogr. A910, 31–49 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Strickland, J. D. H. & Parsons, T. R. A Practical Handbook of Seawater Analysis. Ottawa: Fisheries Research Board of Canada, Bulletin 167. Internationale Revue der gesamten Hydrobiologie und Hydrographievol. 55 (1972).
- 81.Murphy, J. & Riley, J. P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta27, 31–36 (1962). [Google Scholar]
- 82.Grasshoff, K., Kremling, K. & Ehrhardt, M. Methods of seawater analysis. (1983).
- 83.Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics32, 3047–3048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmieder, R. & Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zolfo, M. et al. Detecting contamination in viromes using ViromeQC. Nat. Biotechnol. 2019 371237, 1408–1412 (2019). [DOI] [PubMed] [Google Scholar]
- 87.Gruber-Vodicka, H. R., Seah, B. K. B. & Pruesse, E. phyloFlash: Rapid Small-Subunit rRNA Profiling and Targeted Assembly from Metagenomes. mSystems5, e00920 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fang, Z. et al. PPR-Meta: a tool for identifying phages and plasmids from metagenomic fragments using deep learning. Gigascience8, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu, L.-Y. et al. Benchmarking bioinformatic virus identification tools using real-world metagenomic data across biomes.Genome Biol 25, 97. 10.1186/s13059-024-03236-4 (2024). [DOI] [PMC free article] [PubMed]
- 91.Tisza, M. J., Belford, A. K., Dominguez-Huerta, G., Bolduc, B. & Buck, C. B. Cenote-Taker 2 democratizes virus discovery and sequence annotation. Virus Evol. 7, veaa100 (2021). [DOI] [PMC free article] [PubMed]
- 92.Hyatt, D. et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma.11, 1–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bin Jang, H. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol.37, 632–639 (2019). [DOI] [PubMed] [Google Scholar]
- 94.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 10.48550/arxiv.1303.3997 (2013).
- 96.Robbins, S. J. et al. A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat. Microbiol.4, 2090–2100 (2019). [DOI] [PubMed] [Google Scholar]
- 97.Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rice, P., Longden, L. & Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 99.Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol.30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Darriba, D. et al. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol.37, 291–294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Minh, B. Q. et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol.37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu, G., Smith, D. K., Zhu, H., Guan, Y. & Lam, T. T. Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol.8, 28–36 (2017). [Google Scholar]
- 103.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hackl, T., Duponchel, S., Barenhoff, K., Weinmann, A. & Fischer, M. G. Virophages and retrotransposons colonize the genomes of a heterotrophic flagellate. Elife10, e72674 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aylward, F. O. & Moniruzzaman, M. ViralRecall—A Flexible Command-Line Tool for the Detection of Giant Virus Signatures in ‘Omic Data. Viruses13, 150 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moniruzzaman, M., Martinez-Gutierrez, C. A., Weinheimer, A. R. & Aylward, F. O. Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nat. Commun.11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karki, S., Barth, Z. K., Aylward, F. O. Chimeric origin of eukaryotes from Asgard archaea and ancestral giant viruses. bioRxiv 4–6 10.1101/2024.04.22.590592 (2024).
- 108.Schulz, F. et al. Giant virus diversity and host interactions through global metagenomics. Nature578, 432–436 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Levy Karin, E., Mirdita, M. & Söding, J. MetaEuk-sensitive, high-throughput gene discovery, and annotation for large-scale eukaryotic metagenomics. Microbiome8, 1–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Criscuolo, A. & Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol.10, 1–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rambaut, A. FigTree v1.4.4 (2018).
- 112.Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol.7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 114.Wolf, Y. I. et al. Origins and Evolution of the Global RNA Virome. MBio9, e02329–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol.35, 1026–1028 (2017). [DOI] [PubMed] [Google Scholar]
- 117.Steinegger, M. et al. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinforma.20, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V. & Egozcue, J. J. Microbiome datasets are compositional: And this is not optional. Front. Microbiol.8, 294209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li, D. hillR: taxonomic, functional, and phylogenetic diversity and similarity through Hill Numbers. J. Open Source Softw.3, 1041 (2018). [Google Scholar]
- 120.Murtagh, F. & Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif.31, 274–295 (2014). [Google Scholar]
- 121.McArdle, B. H. & Anderson, M. J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology82, 290–297 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Metagenomic raw reads have been deposited in the European Nucleotide Archive under the Bioproject PRJEB71789. Figure source data are provided with this paper. The sampling metadata, used HHM profiles, viral scaffold nucleotide sequences, NCV bins, taxonomic and host annotations, viral marker gene protein alignments and phylogenies are available through the NIOZ dataverse 10.25850/nioz/7b.b.wg. Source data are provided with this paper.