ABSTRACT
Early stages of Alzheimer's disease (AD) are associated with volume reductions in specific subregions of the medial temporal lobe (MTL). Using a manual segmentation method—the Olsen–Amaral–Palombo (OAP) protocol—previous work in healthy older adults showed that reductions in grey matter volumes in MTL subregions were associated with lower scores on the Montreal Cognitive Assessment (MoCA), suggesting atrophy may occur prior to diagnosis of mild cognitive impairment, a condition that often progresses to AD. However, current “gold standard” manual segmentation methods are labour intensive and time consuming. Here, we examined the utility of Automatic Segmentation of Hippocampal Subfields (ASHS) to detect volumetric differences in MTL subregions of healthy older adults who varied in cognitive status as determined by the MoCA. We trained ASHS on the OAP protocol to create the ASHS‐OAP atlas and then examined how well automated segmentation replicated manual segmentation. Volumetric measures obtained from the ASHS‐OAP atlas were also contrasted against those from the ASHS‐PMC atlas, a widely used atlas provided by the ASHS team. The pattern of volumetric results was similar between the ASHS‐OAP atlas and manual segmentation for anterolateral entorhinal cortex and perirhinal cortex, suggesting that ASHS‐OAP is a viable alternative to current manual segmentation methods for detecting group differences based on cognitive status. Although ASHS‐OAP and ASHS‐PMC produced varying volumes for most regions of interest, they both identified early signs of neurodegeneration in CA2/CA3/DG and identified marginal differences in entorhinal cortex. Our findings highlight the utility of automated segmentation methods but still underscore the need for a unified and harmonized MTL segmentation atlas.
Keywords: Alzheimer's disease, automatic segmentation, medial temporal lobe, mild cognitive impairment, Montreal Cognitive Assessment, neurodegeneration
We examined the utility of automated segmentation (ASHS) to measure medial temporal lobe (MTL) subregions in older adults who varied in cognitive status. We compared a custom atlas (ASHS‐OAP) to an existing atlas. Our findings highlight the utility of ASHS but underscore the need for a harmonized MTL segmentation atlas.
1. Introduction
In Alzheimer's disease (AD), neurodegeneration occurs years before clinical diagnosis. Pathological aggregates of amyloid‐β and hyperphosphorylated forms of tau protein can accumulate in the brains of cognitively healthy older adults decades before symptom onset (Bennett et al. 2005). In particular, neurofibrillary tangles associated with tau pathology have been closely linked to cognition and memory performance in AD and normal aging (Nelson et al. 2012; Marks et al. 2017). Neurofibrillary tangles first appear in the transentorhinal region of the medial temporal lobe (MTL), which likely overlaps with Brodmann area 35 (BA35) of the perirhinal cortex (PRC) and potentially overlaps with the anterolateral entorhinal cortex (alERC) (Braak and Braak 1995). Over time, neurofibrillary tangles affect hippocampal subfields such as the cornu ammonis (CA1) and later spread to the subiculum (SUB) and other brain regions beyond the MTL (Braak and Braak 1995; Braak and Del Tredici 2020).
The pattern of progression of tau pathology in the MTL is similar to the progression of brain atrophy associated with AD and normal aging, which suggests an association between tau pathology and atrophy in the MTL (de Flores et al. 2020; Jack et al. 2002; de Souza et al. 2012). The CA1 subfield of the hippocampus, alERC, and PRC (or more precisely BA35) exhibits the earliest signs of atrophy in individuals with preclinical AD (Yushkevich et al. 2015b; Pini et al. 2016; Xie et al. 2019). The parahippocampal cortex (PHC) and the rest of the hippocampal subfields tend to display greater atrophy as AD progresses (Braak and Braak 1995; Khan et al. 2014). Identifying the earliest signs of neurodegeneration prior to clinical diagnosis of mild cognitive impairment (MCI), a condition that often progresses to AD, is critical to improving early detection of impending AD (Petersen 2004; Sperling et al. 2011).
Manual segmentation has been considered the standard technique for measuring structural volume changes in brain regions associated with AD. Using one such method, the Olsen–Amaral–Palombo (OAP) manual segmentation protocol, Olsen et al. (2017) found that certain regions of the MTL exhibited grey matter volume declines in healthy community‐dwelling older adults who did not have subjective memory complaints but who nonetheless exhibited signs of cognitive decline associated with impending MCI, as measured by the Montreal Cognitive Assessment (MoCA; Nasreddine et al. 2005). The MoCA is a brief neuropsychological assessment tool that exhibits excellent specificity and sensitivity in detecting MCI (Markwick, Zamboni, and de Jager 2012; Nasreddine et al. 2005) and predicting future conversion to AD (Julayanont et al. 2014). Specifically, in their study, the participants who failed the MoCA (half of the study sample) showed reduced grey matter volumes in the alERC, PRC, and CA1 subfield of the hippocampus—the very same MTL structures where tau pathology consistently accumulates in early AD. Other work has found that structural changes in these aforementioned regions are related to cognitive decline as assessed via other behavioral measures beyond the MoCA (Yeung et al. 2017, 2019). Collectively, these studies have shown that lower MoCA scores in healthy older adults were related to smaller volumes in specific MTL subregions that are affected in AD, highlighting the potential sensitivity of both the MoCA and manual segmentation methods in detecting subtle AD‐related changes in grey matter.
Although manual segmentation of hippocampal subfields and extrahippocampal regions remain the standard in the field of neuroimaging, this method is labor intensive and time consuming, especially when large amounts of complex imaging datasets must be analyzed. Establishing inter‐rater reliability can also often pose a significant challenge. In addition, manual segmentation requires specialized expertise, which can result in equity issues due to differences in funding and personnel across research labs. By contrast, automated segmentation is time efficient, requires limited operator training, and provides consistent results with repeated iterations on any given dataset (Bender et al. 2018).
There are several automated segmentation software tools currently available for use by the scientific community, including FreeSurfer (Iglesias et al. 2015), MAGeT‐Brain (Pipitone et al. 2014), HippUnfold (DeKraker et al. 2022), and Automatic Segmentation of Hippocampal Subfields (ASHS; Yushkevich et al. 2015b). These tools are different in terms of the MR image they use for segmentation: FreeSurfer is deployed on T1‐weighted images or T1‐weighted images with an optional additional T2‐weighted image for segmentation of hippocampal subfields, MAGeT‐Brain is run on either T1‐weighted or T2‐weighted images, HippUnfold uses either T1‐weighted or T2‐weighted images, and ASHS uses T1‐weighted images or T1‐weighted images with an additional T2‐weighted image (depending on the atlas) for subfield delineation. Although T1‐weighted scans with ~1 millimeter (mm) isotropic resolution may suffice for certain segmentation goals, previous research has suggested that images with submillimetric (e.g., 0.4 × 0.4) in‐plane resolution are the most appropriate image type for segmenting hippocampal subfields (Wisse et al. 2021). Of the four tools mentioned above, FreeSurfer relies on volumetric registration using a single atlas (Iglesias et al. 2015; Wisse et al. 2021); ASHS, and MAGeT‐Brain make use of multi‐atlas registration (Yushkevich et al. 2015b; Pipitone et al. 2014); HippUnfold uses topological registration and U‐Net tissue classification based on a large training set for hippocampal subfield segmentation (DeKraker et al. 2022).
In the current study, we were particularly interested in using ASHS as our preferred automated segmentation tool for two main reasons. First, ASHS can be applied using a segmentation atlas provided by the ASHS team (e.g., ASHS‐PMC: Penn Memory Center T2‐weighted 3T MRI Atlas; ASHS‐PMC‐T1: Penn Memory Center T1‐Only Atlas for T1‐weighted 3T MRI; ASHS‐Utrecht: Utrecht Medical Center 7T MRI Atlas) or other existing atlases (e.g., Neuroimaging Tools & Resources Collaboratory: https://www.nitrc.org/frs/?group_id=370). Alternatively, ASHS can be “trained” based on a segmentation protocol of a user's own choice (i.e., the user can design their own custom atlas package to label MRI scans). Second, ASHS provides atlases that are trained on MR images that are similar to MR images available for use in the current work in terms of their resolution (i.e., ASHS‐PMC: Penn Memory Center T2‐weighted 3T MRI Atlas). Given these factors, ASHS emerged as the preferred choice for our current study.
We trained ASHS on the manual segmentation protocol (OAP protocol) developed by our lab and used in our previous work (Olsen et al. 2017, 2013) to determine how well the automated segmentation (ASHS‐OAP) approximated the MTL volumes that were generated by the “gold standard” manual segmentations in Olsen et al. (2017). By using the same MRI dataset of 40 healthy older adult brains collected by Olsen et al. (2017), we were able to determine whether the ASHS‐OAP atlas could be used to detect the earliest signs of neurodegeneration in hippocampal subfields and extrahippocampal regions of an older adult sample. We examined whether volumetric measures obtained from ASHS‐OAP would relate to MoCA performance in the same way as they did when manual segmentation was used (Olsen et al. 2017).
There is increased interest in the field for automated methods that delineate hippocampal subfields (Hickling et al. 2024). If ASHS, when trained on the OAP protocol, can produce volumetric results that are similar to the volumetric results produced by manual segmentation on the same sample of healthy older adult MRI images, then the ASHS‐OAP atlas may be a more efficient alternative to manual tracing of the MTL. Moreover, in the current study, we provide the first demonstration of automatic volumetric estimations of entorhinal cortex (ERC) subregions (i.e., alERC and posteromedial entorhinal cortex, pmERC) in any sample. Therefore, this work may be of particular interest for researchers who study brain regions associated with early signs of AD‐related neurodegeneration.
We also compared the volumetric results from the ASHS‐OAP atlas to an atlas provided by the ASHS team (ASHS‐PMC) and commonly used by researchers to identify hippocampal and extrahippocampal atrophy among older adults and those with MCI and/or dementia (Yushkevich et al. 2015b; Xie et al. 2017). Furthermore, we examined whether volumetric measures obtained from ASHS‐PMC would relate to MoCA performance. By comparing the volumetric results from the ASHS‐OAP atlas to the volumetric results from the ASHS‐PMC atlas, we highlight the need for a unified and harmonized MTL segmentation protocol to improve reproducibility. The ASHS‐OAP atlas, along with a README file containing information about the data/file structures and software requirements for using the atlas, are available on Dryad, an open data publishing platform (Mazloum‐Farzaghi et al. 2024; https://doi.org/10.5061/dryad.6djh9w17h).
2. Methods
2.1. Participants
Forty community‐dwelling healthy older adults (as described in Olsen et al. 2017; age range = 59–81 years; mean age = 71.4; mean education = 16.3 years; range = 12–23; 30 female) were recruited from participant databases at the Rotman Research Institute and the University of Toronto. All participants received the MoCA (M score = 26.63, SD = 2.86) (Nasreddine et al. 2005) and were divided to two age‐matched groups that differed solely in their MoCA score. Although a score of 26 is not a definitive threshold for indicating risk of MCI, it is the threshold score that warrants further dementia screening (sensitivity = 98%; Damian et al. 2011). Likewise, in our prior work (D'Angelo et al. 2016; Yeung et al. 2013; Olsen et al. 2017), we found significant cognitive and neural differences between those who scored above versus below the 26 threshold score. Therefore, the two groups in the current study were defined as an “at‐risk” group (N = 20; 17 females; M age = 72.5 years, range = 59–81, M education = 16.2 years, range = 12–22) who scored below 26 (indicating potentially pathologic cognitive impairment and risk for MCI, M score = 23.4, range = 17–25), and a “healthy” group (N = 20; 13 females; M age = 70.3 years, range = 63–77, M education = 16.6 years, range = 12–23) who scored 26 and above (M score = 27.9, range = 26–30). t‐tests showed no difference in age, t(38) = 1.29, p = 0.20, or years of education, t(38) = 0.51, p = 0.61, between the two groups, but there was a significant difference in MoCA score, t(38) = 7.87, p < 0.001 between them.
None of the older adults had any significant subjective memory complaints as measured by the Memory Functioning Questionnaire (MFQ; Gilewski, Zelinski, and Schaie 1990). Moreover, neuropsychological performance in these subjects was in the nonimpaired range (see Olsen et al. (2017) for more details). All participants were fluent English speakers with normal or corrected‐to‐normal vision and were screened for non‐MRI compatible metal implants, color blindness, diabetes, neurologic disorders, stroke, or brain trauma. All participants gave written informed consent and were compensated for their participation. The Research Ethics Board at Baycrest Hospital approved this research.
2.2. Structural Image Acquisition
Neuroimaging was completed on a 3‐Tesla Siemens Trio scanner using a 12‐channel head coil. All participants received a T1‐weighted, magnetization‐prepared, rapid acquisition with gradient echo image (MP‐RAGE) whole‐brain anatomical scan (TE/TR = 2.63 ms/2000 ms, 160 axial slices perpendicular to the AC‐PC line, voxel size = 1 × 1 × 1mm, FOV = 256 mm, TI = 1100 ms). The T1‐weighted MP‐RAGE was used for slice placement during the acquisition of a subsequent high In‐plane resolution T2‐weighted scan in an oblique‐coronal plane, perpendicular to the hippocampal long axis (TE/TR = 68/3000 ms, 20–28 slices depending on head size, 512 × 512 acquisition matrix, voxel size = 0.43 × 0.43 × 3mm, no skip, FOV = 220 mm). For the T2‐weighted scan, the first slice was placed anterior to the collateral sulcus (CS) (including the temporal pole, when possible) and the last slice was placed posterior to the hippocampal tail to ensure full coverage of the entire hippocampus and MTL cortices for all participants.
2.3. Manual Segmentation
Manual segmentation of grey matter was previously performed on the 40 older adult participants' T2‐weighted images using the OAP protocol, which had been employed for previous volumetric investigations of MTL subregions (Olsen et al. 2009, 2013; Palombo et al. 2013; Yushkevich et al. 2015a). The manual segmentations (termed Manual‐OAP) were conducted on participants' native space, on the oblique‐coronal plane perpendicular to the long axis of the hippocampus. A single rater who was blind to the MoCA scores manually delineated regions of interest (ROI), which included three hippocampal subfields (CA1, a region combining dentate gyrus (DG), CA2 and CA3 (CA2/CA3/DG), and SUB) and four MTL cortex subregions (alERC, pmERC, PRC, and PHC) in FSLView (Figure 1A). Hippocampal subfields (CA1, CA2/CA3/DG, SUB) were defined in the body and posterior head of the hippocampus. Note that ROI were also delineated for the tail and anterior head of the hippocampus, but these regions were not further divided into their individual subfields. Currently, there is little consensus as to how to subdivide these regions into subfields using in vivo 3‐Tesla MRI, which is why it has been our practice to combine them into a single ROI, as do other high‐resolution protocols (e.g., see Schlichting and Preston protocol in Yushkevich et al. 2015a). Because the hippocampal subfields within these regions are not segmented into subregions, these regions were excluded from further analysis. A second rater who was also blind to the MoCA scores segmented the same regions to establish inter‐rater reliability. Inter‐rater reliability and intra‐rater reliability results were similar to reliability values reported in previous studies on the manual segmentation of the hippocampus and MTL cortices (Wisse et al. 2012; Yushkevich et al. 2015b). For a detailed description of the segmentation protocol and reliability results see Olsen et al. (2017).
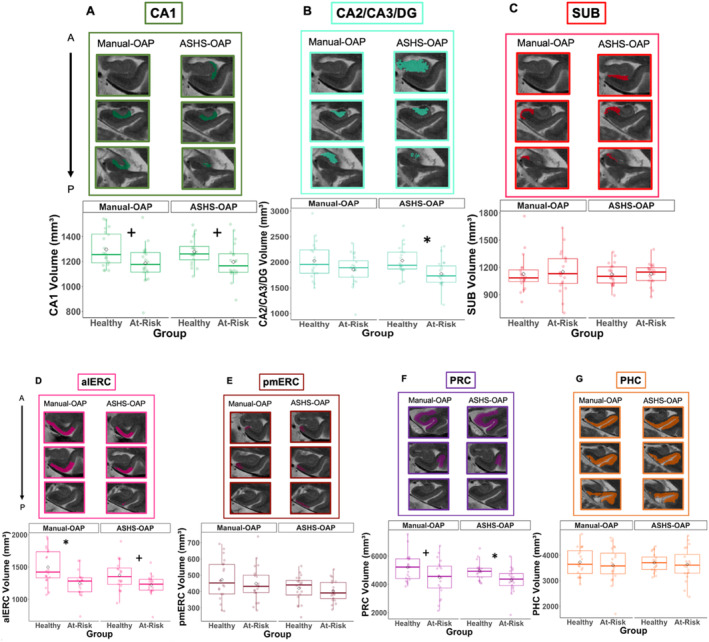
FIGURE 1.

The extent along the anterior–posterior axis (A–P in the figure) of the different anatomical labels included in the Manual‐OAP (A), ASHS‐OAP (C) and ASHS‐PMC (B & D) atlases. Dashed black vertical lines outline MRI slices (the number of slices is variable from subject to subject). Dashed black lines surrounding the ROI (C and D) indicate the combining of regions into a single ROI for comparison across protocols (Modified ASHS‐OAP to Modified ASHS‐PMC) in our analyses. Note the tail and anterior head were not analyzed (A). The collateral sulcus label (CS) was not analyzed (B). “Appearance of …” indicates most anterior slice that contains landmark. “Last visualization of …” indicates most posterior slice that contains landmark. ASHS‐PMC diagram (B) is adapted from Xie et al. (2017). alERC = anterolateral entorhinal cortex, CS = collateral sulcus, DG = dentate gyrus, ERC = entorhinal cortex, FTJ = frontal temporal junction, PHC = parahippocampal cortex, PRC = perirhinal cortex, pmERC = posteromedial entorhinal cortex, SUB = subiculum.
2.4. Automated Segmentation
2.4.1. Automatic Segmentation of Hippocampal Subfields using the Olsen–Amaral–Palombo (ASHS‐OAP) Protocol
A custom atlas was built using the ASHS software (Yushkevich et al. 2015b) following the published procedures (https://sites.google.com/view/ashs‐dox/local‐ashs/building‐an‐atlas‐for‐t2‐mri), but not the slice heuristics procedure. We generated a custom atlas package (ASHS‐OAP) based on the 40 healthy older adults' manual segmentations from Olsen et al. (2017). Following atlas generation, automated segmentation using the ASHS‐OAP atlas was applied on all 40 older adult participants. The ASHS segmentation procedure contains an approach called multi‐atlas label fusion, which allows all participants in an atlas to “vote” on the label for each voxel, an approach known as multi‐atlas label fusion (Yushkevich et al. 2015b). As our atlas participants were also members of the target segmentation, we used leave‐one‐out cross‐validation to remove any possibility of bias in the target segmentation sample as described in the ASHS documentation (https://sites.google.com/view/ashs‐dox/local‐ashs/building‐an‐atlas‐for‐t2‐mri#h.p_8mP2fZnC5Wtu). More specifically, each atlas participants' vote was excluded from their own target segmentation during the labelling process. That is, to prevent inflating accuracy in their automated segmentation through bias, we did not use a participant's own manual segmentation when labelling their MR image (Schlichting et al. 2019). Atlas building in ASHS was performed on a Linux cluster with a Sun Grid Engine. The automated segmentation produced by ASHS‐OAP resulted in nine ROI, as also produced by the Manual‐OAP method. These regions were the tail and anterior head of the hippocampus, three subfields of the hippocampus (CA1, CA2/CA3/DG, SUB) and four MTL cortex subregions (alERC, pmERC, PRC, PHC) (Figure 1A). The tail and anterior head of the hippocampus were not analyzed. Therefore, in the analyses, which compared the ASHS‐OAP atlas to Manual‐OAP, seven ROI were assessed for each method (CA1, CA2/CA3/DG, SUB, alERC, pmERC, PRC, PHC). See https://doi.org/10.5061/dryad.6djh9w17h for more details.
2.4.2. Automatic Segmentation of Hippocampal Subfields using the Penn Memory Center (ASHS‐PMC) Protocol
Automated segmentation using the ASHS‐PMC atlas (Xie et al. 2017) (provided by ASHS team; Figure 1B) was also applied to the same group of 40 healthy older adults. The ASHS‐PMC atlas was trained on brain images of older adults with MCI or AD and thus can be used to segment hippocampal and extrahippocampal MTL subregions of older adults (55+ years of age). There are three atlases currently available for ASHS (ASHS‐PMC: Penn Memory Center T2‐weighted 3T MRI Atlas, ASHS‐PMC‐T1: Penn Memory Center T1‐Only Atlas for T1‐weighted 3T MRI, and ASHS‐Utrecht: Utrecht Medical Center 7T MRI Atlas); however, for ASHS to work effectively, the MRI scans provided by users must be sufficiently similar to the MRI scans used to form the atlas provided by the ASHS team. Therefore, the ASHS‐PMC atlas was chosen for the current study. It is important to note that the ASHS‐PMC atlas was trained on brain images of older adults with MCI or AD, whereas the ASHS‐OAP atlas was trained on brain images of healthy older adults.
Automated segmentation using the ASHS‐PMC atlas resulted in five subfield labels of the hippocampus (CA1, CA2, CA3, DG, SUB) and four MTL cortex subregion labels (ERC, BA35, BA36, PHC) (Figure 1B). Note that the ASHS‐PMC atlas labels the regions of the MTL differently than the Manual‐OAP/ASHS‐OAP methods (Xie et al. 2017). One of the goals in the current study was to compare the ASHS‐OAP atlas to the ASHS‐PMC atlas in terms of the relationship between their volumetric outputs and MoCA scores. Thus, in order to compare the volumetric outputs from the ASHS‐PMC atlas to the ASHS‐OAP atlas, we combined the volumes of BA35 and BA36 into one region (PRC) and CA2, CA3, and DG into another region (CA2/CA3/DG) from the volumetric outputs of ASHS‐PMC (Figure 1D). In addition, we combined alERC and pmERC volumes produced by ASHS‐OAP into one ERC region (Figure 1C). The ASHS‐PMC atlas segments the hippocampus into subfields along the entire longitudinal axis, so this protocol does not have an “anterior head” and “tail” label; therefore, these regions were not included in analyses comparing the ASHS‐OAP and the ASHS‐PMC atlas (Figure 1D). Thus, in the analyses which compared the ASHS‐PMC atlas to the ASHS‐OAP atlas, six ROI were assessed for each atlas: three subfields of the hippocampus (CA1, CA2/CA3/DG, SUB) and three MTL cortical regions (ERC, PRC, PHC) (Figure 1C,D).
2.4.3. Volume Extraction
The results of the automated segmentations (ASHS‐OAP and ASHS‐PMC) from the left and right hemispheres indicated three different segmentation variants depending on the segmentation algorithm used by ASHS (“Heur,” “NoGray,” and “UseGray”). We used the results from the segmentation algorithm “UseGray,” which is the preferred algorithm when the MRI intensity characteristics of the T2‐weighted image are similar to the intensity characteristics of the images in the atlas package (as stated in the ASHS published procedures: https://sites.google.com/view/ashs‐dox/local‐ashs/building‐an‐atlas‐for‐t2‐mri). In our case, the same T2‐weighted images were used in the atlas package and target sample for the ASHS‐OAP atlas. Similarly, we used the “UseGray” algorithm for extracting volumes from the ASHS‐PMC atlas.
Raw volumes obtained from automated and manual segmentation methods were adjusted for differences in overall head/brain size across all participants using an adapted version of the ANIMAL algorithm (Collins et al. 1995). Total brain volume was accounted for in each ROI using a linear regression where each ROI is regressed on total brain volume (Free et al. 1995). Each individual's estimated total brain volume (eTIV) was obtained from FreeSurfer (version 5.3) rather than ASHS, as Olsen et al. (2017) used FreeSurfer for their head size correction analyses (Buckner et al. 2004). This allowed for reliable comparison of the head/brain size corrected automated and manual volumes. Then the beta coefficient (i.e., regression slope, which represents the effect of brain size across participants on that region's volume), was obtained from the regression analysis. The residual value (i.e., the structure's actual size minus its predicted value based on the individual's brain volume) was accounted for in each ROI for each individual using the following formula (Free et al. 1995): NV = OV—Grad × (bVi−bVmean). In this formula, NV is the corrected MTL subregion volume, OV is the original MTL subregion volume, Grad is the gradient (i.e., beta coefficient) of the regression line between the MTL subregion volume and the total brain volume measure, bVi is the brain volume measurement for that participant (i.e., eTIV from FreeSurfer), and bVmean is the mean brain volume for all participants. For total volume analyses, we added the volumes of the left and right hemispheres for each participant's ROI for Manual‐OAP, ASHS‐OAP, and ASHS‐PMC methods.
Furthermore, all automated segmentations on the T2‐weighted images (voxel size = 0.43 × 0.43 × 3mm) were visually inspected for quality assurance. More specifically, visual inspection was applied using a pass or fail rating system for each ROI produced by ASHS‐OAP and ASHS‐PMC. A “fail” score was considered to be a ROI, which had more than 20 voxels under or over‐segmented on a given slice. All automated segmentations received a pass score for both ASHS‐OAP and ASHS‐PMC methods, and no participants were excluded from the current study. In addition, no manual corrections were applied to the automated segmentations.
2.5. Statistics
All analyses were performed using the R programming language (R Core Team 2022). In the analyses, which compared the ASHS‐OAP atlas to Manual‐OAP, seven ROI were assessed for each method (CA1, CA2/CA3/DG, SUB, alERC, pmERC, PRC, PHC).
In the analyses, which compared the ASHS‐PMC atlas to the ASHS‐OAP atlas, six ROI were assessed for each method (CA1, CA2/CA3/DG, SUB, ERC, PRC, PHC).
Variance‐stabilizing transformations (Draper and Smith 1998) were applied to all brain region volumes using the natural logarithm function “log()” in R. Log‐transformed volumes were then used in all statistical analyses to address heteroscedasticity and non‐normality in the data. Log‐transformation allowed for stabilization of the variance in brain regional volumes, reduced skewness, and allowed the data to more closely conform to the assumptions of parametric tests, resulting in more reliable and interpretable results. Note that all figures and tables report brain region volumes in their original state unless otherwise stated (i.e., corrected for total brain volume, but without the variance‐stabilizing transformation).
Initial exploration of the data assessed main effects and interactions with hemisphere using repeated‐measures or mixed‐design ANOVA models. When hemisphere did not interact with effects of interest, reported analyses were run on the summation of left and right hemisphere volumes.
Using the anova_test() function from the rstatix package in R (Kassambara 2023), a repeated‐measures ANOVA was run to test the significance of the interaction between brain region (within‐subjects factor) × segmentation method (within‐subjects‐factor; Manual‐OAP and ASHS‐OAP or ASHS‐OAP and ASHS‐PMC) on brain volume. Greenhouse–Geisser corrections (built into anova_test()) were applied to correct for violations of assumptions of sphericity (Mauchly's test of sphericity). Interaction effects were followed up by using paired t‐tests (two tailed) and corrected for multiple comparisons. The Holm‐Bonferroni method was used to control familywise error rate when performing multiple comparisons.
The accuracy of automated segmentation (ASHS‐OAP) relative to manual segmentation (Manual‐OAP) was assessed in terms of relative overlap by using the Dice similarity coefficient (DSC; Crum, Camara, and Hill 2006) for each MTL subregion in each hemisphere, which ASHS produced automatically as part of its software. The DSC produced a value between 0 and 1, where 0 indicated no overlap or similarity between two regions (complete dissimilarity) and 1 indicated perfect overlap or similarity between two regions (complete similarity) produced by Manual‐OAP and ASHS‐OAP. A higher DSC indicated that the automated segmentation method produced results that closely matched the manual segmentation (DSC > 0.70; high agreement), while a lower coefficient (0.5 > DSC > 0.7; moderate agreement, 0.5 > DSC; low agreement) indicated a greater divergence between the two methods. As ASHS‐OAP and ASHS‐PMC atlases are based on different segmentation protocols, DSC was not computed when these two atlases were compared.
The relationship between MTL subregion volumes and cognitive status was examined by treating MoCA performance as a categorical variable (healthy vs. at‐risk). Using the anova_test() function in R, MTL subregion volumes were entered into a single mixed‐design ANOVA to test for main effects of group (between‐subjects factor; healthy, at‐risk), brain region (within‐subjects factor), and group‐by‐region interactions on volume for each of the three segmentation methods separately. Greenhouse–Geisser corrections were applied to correct for violations of assumptions of sphericity. A second mixed‐design ANOVA, which included age as a covariate was also run to control for any age differences between groups for each segmentation method. Greenhouse–Geisser corrections were applied to correct for violations of assumptions of sphericity (Mauchly's test of sphericity). Interaction effects between group × brain region were followed up with independent t‐tests (one tailed) and corrected for multiple comparisons (Holm–Bonferroni method). Given the literature reporting volume reductions in MTL subregions as a function of AD severity (Adachi et al. 2003; Kerchner et al. 2010, 2012; La Joie et al. 2013), and our previous work on a similar group of individuals who demonstrated neural and behavioral impairments (D'Angelo et al. 2016; Newsome, Duarte, and Barense 2012; Newsome et al. 2013; Yeung et al. 2013), we had strong a priori hypotheses that brain volumes would be smaller in the at‐risk group; thus, one tailed tests were used.
Linear models were conducted to test the interaction between group (healthy, at‐risk) and segmentation method (Manual‐OAP and ASHS‐OAP or ASHS‐OAP and ASHS‐PMC) on regions that exhibited significant group differences (healthy, at‐risk).
3. Results
3.1. Manual‐OAP Versus ASHS‐OAP
3.1.1. Comparison of Manual Volumes (Manual‐OAP) and Automated Volumes (ASHS‐OAP)
Manual segmentation (Manual‐OAP) volumes were compared with automated segmentation volumes (ASHS‐OAP) of the same participant. Seven regions were assessed: three subfields of the hippocampus (CA1, CA2/CA3/DG, SUB) and four MTL cortical subregions (alERC, pmERC, PRC, PHC) (Figure 1). A repeated‐measures ANOVA revealed a nonsignificant protocol (Manual‐OAP and ASHS‐OAP) × brain region (natural log‐transformed volumes) × hemisphere interaction after correction for sphericity violations (p = 0.87); consequently, the reported analyses were run on the summation of left and right hemispheres. We present all main effects and interaction effects to provide a comprehensive account of our findings.
A repeated‐measures ANOVA was performed with brain region (seven ROI) and segmentation method (Manual‐OAP and ASHS‐OAP) as interaction terms and brain volume (natural log‐transformed volumes) as the dependent variable. This model was run in order to test whether the brain region × segmentation method was significant (indicating a significant difference between segmentation method on brain regional volume).
There was a significant interaction between segmentation method and brain region (F(6,234) = 2.87, p < 0.05), a significant main effect of segmentation method (F(1,39) = 5.90, p < 0.05), and a significant main effect of brain region (F(6,234) = 1361.55, p < 0.05). Mauchly's test for sphericity indicated violations for both brain region and the interaction term (p < 0.05). Following Greenhouse–Geisser corrections, the interaction term (p < 0.05) and main effect of brain region remained significant (p < 0.05). Thus, volumes produced by Manual‐OAP and ASHS‐OAP differed from one another significantly.
Mean volumes obtained from Manual‐OAP and ASHS‐OAP are shown in Table 1. Follow‐up paired t‐tests indicated that none of the ROI had significantly different volumes between Manual‐OAP and ASHS‐OAP. Note that the alERC (t(39) = 2.44, p < 0.05 uncorrected, two tailed) and pmERC (t(39) = 2.59, p < 0.05 uncorrected, two tailed) showed volumetric differences between Manual‐OAP and ASHS‐OAP; however, these effects did not survive correction for multiple comparisons.
TABLE 1.
Volumes (Mean ± SD) of manual (Manual‐OAP) and automated segmentation (ASHS‐OAP).
| Segmentation method | Manual‐OAP volume | ASHS‐OAP volume | ||||
|---|---|---|---|---|---|---|
| Hemisphere | Left | Right | Total | Left | Right | Total |
| CA1 | 625.87 ± 124.11 | 614.95 ± 121.63 | 1240.82 ± 128.50 | 624.13 ± 105.31 | 613.13 ± 94.55 | 1237.26 ± 113.47 |
| CA2/CA3/DG | 952.53 ± 151.92 | 990.24 ± 182.35 | 1942.77 ± 268.26 | 936.72 ± 108.62 | 967.45 ± 117.90 | 1904.16 ± 220.86 |
| SUB | 599.12 ± 195.26 | 540.45 ± 178.32 | 1139.57 ± 254.49 | 587.82 ± 141.02 | 538.73 ± 107.95 | 1126.55 ± 146.33 |
| alERC | 671.79 ± 176.31 | 700.59 ± 138.45 | 1372.38 ± 214.84 | 631.72 ± 120.39 | 664.70 ± 102.81 | 1296.42 ± 159.21 |
| pmERC | 219.38 ± 160.34 | 240.54 ± 138.74 | 459.93 ± 164.04 | 191.44 ± 133.98 | 219.48 ± 127.67 | 410.92 ± 140.74 |
| PRC | 2615.67 ± 505.95 | 2227.50 ± 636.84 | 4843.17 ± 1080.47 | 2524.22 ± 326.78 | 2098.59 ± 386.67 | 4622.81 ± 718.12 |
| PHC | 1894.22 ± 339.86 | 1774.50 ± 269.88 | 3668.72 ± 558.85 | 1918.20 ± 271.49 | 1756.55 ± 240.11 | 3674.76 ± 461.063 |
Note: Analyses were run on total volume; however, for completeness, we have reported left and right volumes for each MTL subregion. None of the regions of interest had significantly different volumes between Manual‐OAP and ASHS‐OAP when multiple comparisons were taken into account.
The DSC for Manual‐OAP versus ASHS‐OAP (Table 2) was the highest for CA2/CA3/DG in the hippocampal formation (Left: 0.81 ± 0.04, Right: 0.80 ± 0.05) and the lowest for the SUB (Left: 0.76 ± 0.06, Right: 0.75 ± 0.06). Moreover, the DSC of Manual‐OAP versus ASHS‐OAP (Table 2) was the highest for the PHC in the MTL cortex (Left: 0.78 ± 0.07, Right: 0.79 ± 0.06) and the lowest for pmERC (Left: 0.59 ± 0.12, Right: 0.62 ± 0.11).
TABLE 2.
Dice similarity coefficient (DSC) (mean ± SD) of volumes from Manual‐OAP and ASHS‐OAP.
| Hemisphere | Left | Right |
|---|---|---|
| CA1 | 0.78 ± 0.04 | 0.79 ± 0.04 |
| CA2/CA3/DG | 0.81 ± 0.04 | 0.80 ± 0.05 |
| SUB | 0.76 ± 0.06 | 0.75 ± 0.06 |
| alERC | 0.73 ± 0.06 | 0.74 ± 0.06 |
| pmERC | 0.59 ± 0.12 | 0.62 ± 0.11 |
| PRC | 0.74 ± 0.05 | 0.71 ± 0.08 |
| PHC | 0.78 ± 0.07 | 0.79 ± 0.06 |
Note: The DSC produced a value between 0 and 1, where 0 indicated no overlap or similarity between two regions (complete dissimilarity) and 1 indicated perfect overlap or similarity between two regions (complete similarity) produced by Manual‐OAP and ASHS‐OAP. (DSC > 0.70; high agreement, 0.5 > DSC > 0.7; moderate agreement, 0.5 > DSC; low agreement).
To further examine how well volumes produced by Manual‐OAP matched with volumes produced by ASHS‐OAP, scatterplots for each ROI were created with data points representing each participant (using non‐log‐transformed volumes) (Figure 2; top panel). We found a strong correlation between volumes produced by Manual‐OAP and ASHS‐OAP for the PRC region (F(1,38) = 87.39, p < 0.001, R 2 = 0.70, R = 0.84). In addition, there was a moderate correlation between volumes produced by Manual‐OAP and ASHS‐OAP for CA1 (F(1,38) = 35.29, p < 0.001, R 2 = 0.48, R = 0.69), CA2/CA3/DG (F(1,38) = 42.58, p < 0.001, R 2 = 0.53, R = 0.73), SUB (F(1,38) = 51.08, p < 0.001, R 2 = 0.57, R = 0.75), alERC (F(1,38) = 42.33, p < 0.001, R 2 = 0.53, R = 0.73), and PHC (F(1,38) = 32.54, p < 0.001, R 2 = 0.46, R = 0.68). However, the relationship between volumes produced by Manual‐OAP and ASHS‐OAP for pmERC (F(1,38) = 8.48, p < 0.01, R 2 = 0.18, R = 0.42) was weak. In addition, Bland–Altman plots were used to assess the agreement of the log‐transformed volumes for each ROI between Manual‐OAP and ASHS‐OAP (Figure 2; bottom panel). Each point on the plot represents a single measurement, and most points are close to the central line (mean difference), indicating good agreement between the two methods. There are a few points near and outside the limits of agreement (e.g., pmERC), which indicates specific cases where the two methods differ. Overall, the majority of points fall within the limits of agreement, suggesting reasonable agreement between the manual and automated log‐transformed volume measurements, with some variation as indicated by the spread of points.
FIGURE 2.
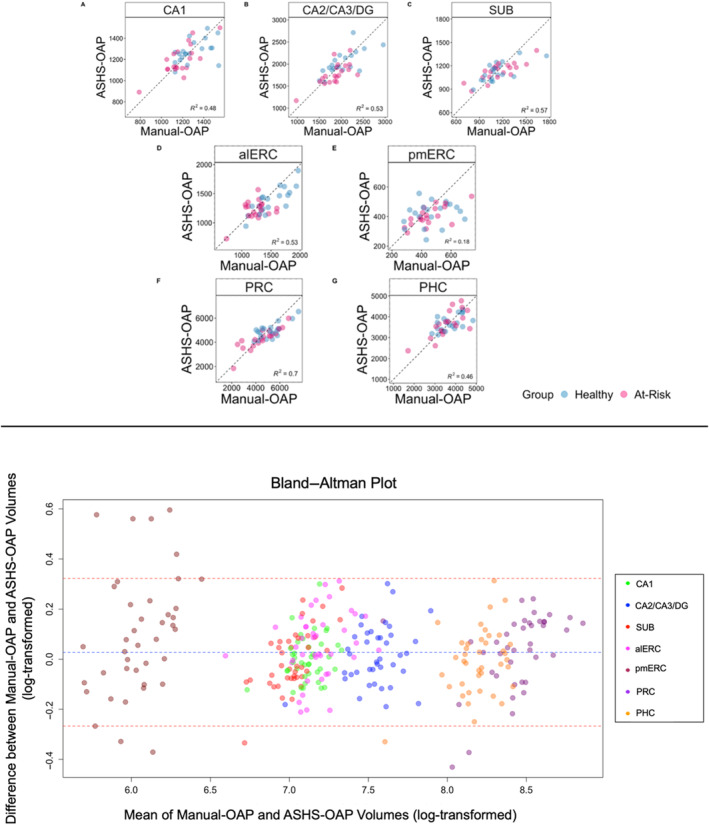
Top panel: Scatter plots depicting the variance explained between volumes produced by the Manual‐OAP and ASHS‐OAP atlas for each ROI (non‐log‐transformed volumes). Each data point represents one participant, categorised by healthy (blue) or at‐risk (pink). Dotted unity line represents equivalency between volumes produced by Manual‐OAP and ASHS‐OAP. Points that fall along the unity line indicate that the volumes of the two atlases are exactly the same. Deviation from the unity line indicates a discrepancy between the volumes of the two atlases. Volumes are measured in mm3. A: CA1, B: CA2/CA3/DG, C: Subiculum (SUB), D: Anterolateral entorhinal cortex (alERC), E: Posteromedial entorhinal cortex (pmERC), F: Perirhinal cortex (PRC), G: Parahippocampal cortex (PHC). Bottom panel: A Bland–Altman plot was used to assess the agreement between the Manual‐OAP and ASHS‐OAP methods (using log‐transformed volumes) by plotting the difference between the methods against their average. Each point is colored according to ROI. The central dashed line represents the mean difference between the log‐transformed volumes of Manual‐OAP and ASHS‐OAP. The line is close to 0, which suggests that there is no substantial systematic bias between the two methods. The other two dashed lines represent the limits of agreement, set at the mean difference ± 1.96 times the standard deviation of the differences.
3.1.2. Group Differences in MTL Subregion Volumes (Manual‐OAP, ASHS‐OAP)
A mixed‐design ANOVA was performed with brain region (seven ROI) and group (healthy, at‐risk) as interaction terms, and brain volume as the dependent variable (natural log‐transformed volumes) for Manual‐OAP and ASHS‐OAP methods separately. This test examined whether the brain region × group interaction was significant for a given segmentation method (indicating a significant difference between groups on brain regional volume). Initial exploration of the data using a mixed‐design ANOVA revealed no significant group (healthy, at‐risk) × brain region (natural log‐transformed volumes) × hemisphere interaction for Manual‐OAP (p = 0.53) and ASHS‐OAP (p = 0.63) after correction for sphericity violations; consequently, the reported analyses were run on the summation of left and right hemispheres.
For Manual‐OAP, a mixed‐design ANOVA revealed a significant main effect of group (F(1,38) = 5.33, p < 0.05) and brain region (F(6,228) = 842.72, p < 0.05). The group × brain region interaction effect was nonsignificant (F(6,228) = 1.76, p = 0.11). Mauchly's test for sphericity revealed violations for both brain region (p < 0.05) and the group × brain region interaction (p < 0.05). Following Greenhouse–Geisser corrections, the main effect of brain region remained significant (p < 0.05), while the group × brain region interaction remained nonsignificant (p = 0.15). When age was added as a covariate and after sphericity corrections were performed, there was still a significant main effect of brain region (p < 0.05) and a nonsignificant brain region × group interaction (p = 0.14). The effect of group was nonsignificant (p = 0.06).
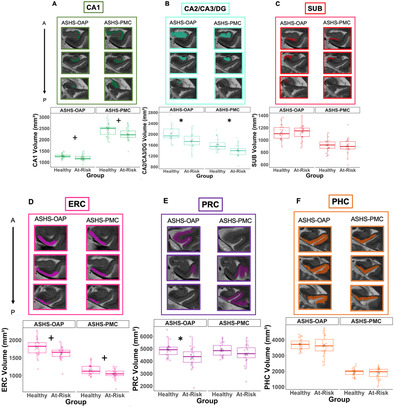
For Manual‐OAP, follow‐up independent samples t‐tests showed that only the alERC region was significantly larger in the healthy group versus the at‐risk group (t(38) = 3.27, p < 0.01, one tailed). The effect of alERC was still significant even when adding age into a follow‐up linear model with group and age as independent variables on volume. The CA1 subfield (t(36) = 2.42, p < 0.05, one tailed) and the PRC (t(31) = 2.19, p < 0.05, one tailed) also showed group differences; however, these effects did not survive correction for multiple comparisons. Boxplots for each region are plotted in Figure 3.
FIGURE 3.
Hippocampal subfields and MTL cortical volumes. Box plots, plotted separately for healthy and at‐risk participants and segmentation method (Manual‐OAP on the left and ASHS‐OAP on the right), *p < 0.05 after multiple comparison correction, +p < 0.05 before multiple comparison correction. Example of three segmentation slices produced for each ROI per segmentation method displayed for the same participant (healthy participant) (A–G). A = anterior, alERC = anterolateral entorhinal cortex; P = posterior; PHC = parahippocampal cortex; pmERC = posteromedial entorhinal cortex; PRC = perirhinal cortex; SUB = subiculum.
For ASHS‐OAP, a mixed‐design ANOVA revealed that there was a significant main effect of group (F(1,38) = 7.09, p < 0.05), brain region (F(6,228) = 156.13, p < 0.05), and group × brain region interaction effect (F(6,228) = 2.19, p < 0.05). Mauchly's test for sphericity indicated violations for both brain region (p < 0.05) and the group × brain region interaction (p < 0.05). Following Greenhouse–Geisser corrections, brain region (p < 0.05) remained significant, and the interaction effect became nonsignificant (p = 0.08). When age was added as a covariate, there was still a significant main effect of brain region (p < 0.05), and a significant main effect of group (p < 0.05) after corrections for sphericity. The brain region × group interaction (p = 0.09) remained nonsignificant.
For ASHS‐OAP, follow‐up independent samples t‐tests showed that the CA2/CA3/DG subfield (t(38) = 3.09, p < 0.05, one tailed) and the PRC (t(28) = 2.61, p < 0.05, one tailed) indicated group differences. Follow‐up linear models were conducted with age as a covariate. The effect of the CA2/CA3/DG subfield and PRC were still significant even when adding age into the linear models. The alERC (t(38) = 2.29, p < 0.05, uncorrected, one tailed) and CA1 region (t(34) = 2.10, p < 0.05, uncorrected, one tailed) showed group differences; however, these effects did not survive correction for multiple comparisons. Boxplots for each region are plotted in Figure 3.
Linear models were conducted to test the interaction between group (healthy, at‐risk) and segmentation method (Manual‐OAP and ASHS‐OAP) on regions that exhibited significant group differences for both Manual‐OAP and ASHS‐OAP methods (alERC, CA2/CA3/DG, PRC). The interaction between group and segmentation method was nonsignificant for alERC, CA2/CA3/DG, and PRC, which suggested that the observed volume differences between groups were not statistically different across Manual‐OAP and ASHS‐OAP methods.
3.2. ASHS‐OAP Versus ASHS‐PMC
3.2.1. Comparison of Automated Volumes Obtained From ASHS‐OAP and ASHS‐PMC
In the next sets of analyses, we were interested in comparing the ASHS‐OAP atlas to the ASHS‐PMC atlas. In order to compare the volumetric outputs from the ASHS‐PMC atlas to the ASHS‐OAP atlas, six regions were assessed for each atlas: three subfields of the hippocampus (CA1, CA2/CA3/DG, SUB) and three MTL cortical regions (ERC, PRC, PHC) (Figure 1C,D). Note that these sets of analyses are different than the previous section as we combined alERC and pmERC volumes produced by ASHS‐OAP into one ERC region. See Section 2 for further details. Initial exploration of the data using a repeated‐measures ANOVA revealed a significant protocol (ASHS‐OAP and ASHS‐PMC) × brain region (natural log‐transformed volumes) × hemisphere interaction (p < 0.05) after correction for sphericity violations. However, to align with our previous work (Olsen et al. 2017) and the prior Manual‐OAP versus ASHS‐OAP section of the current paper, the reported analyses were run on the summation of left and right hemispheres. We present all main effects and interaction effects to provide a comprehensive account of our findings.
A repeated‐measures ANOVA was performed with brain region (six ROI) and segmentation method (ASHS‐OAP and ASHS‐PMC) as interaction terms and brain volume (natural log‐transformed volumes) as the dependent variable. This model was run in order to test whether the brain region × segmentation method was significant. There was a significant interaction between brain region × segmentation method (F(5,195) = 894.61, p < 0.05), a significant main effect of segmentation method (F(1,39) = 303.60, p < 0.05), and a significant main effect of brain region (F(5,195) = 1139.22, p < 0.05). Mauchly's test for sphericity indicated violations for both brain region (p < 0.05) and the interaction effect (p < 0.05). Following Greenhouse–Geisser corrections, the main effect of brain region (p < 0.05) and the interaction effect (p < 0.05) remained significant. Therefore, volumes produced by ASHS‐OAP and ASHS‐PMC were significantly different from one another.
Mean volumes of six ROI (CA1, CA2/CA3/DG, SUB, ERC, PRC, PHC; Figure 1C,D) obtained from ASHS‐OAP and ASHS‐PMC are shown in Table 3. The follow‐up paired t‐tests indicated that PRC volumes generated by ASHS‐OAP and ASHS‐PMC were similar to one another, but ERC, PHC, CA2/CA3/DG, and SUB volumes produced by ASHS‐PMC were significantly smaller compared with ASHS‐OAP (p < 0.001, two tailed). CA1 volume produced by ASHS‐PMC was significantly greater than ASHS‐OAP (p < 0.001, two tailed).
TABLE 3.
Volumes (mean ± SD) of automated (ASHS‐OAP and ASHS‐PMC) segmentation methods.
| Segmentation Method | ASHS‐OAP volume | ASHS‐PMC volume | ||||
|---|---|---|---|---|---|---|
| Hemisphere | Left | Right | Total | Left | Right | Total |
| CA1 | 624.13 ± 105.31 | 613.13 ± 94.55 | 1237.26 ± 113.47 | 1156.03 ± 108.32 | 1180.15 ± 123.54 | 2336.19 ± 240.79 |
| CA2/CA3/DG | 936.72 ± 108.62 | 967.45 ± 117.90 | 1904.16 ± 220.8 | 753.41 ± 89.67 | 756.19 ± 96.32 | 1509.60 ± 187.30 |
| SUB | 587.82 ± 141.02 | 538.73 ± 107.95 | 1126.55 ± 146.33 | 469.07 ± 103.19 | 438.28 ± 92.19 | 907.35 ± 102.61 |
| ERC | 823.16 ± 133.81 | 884.1811 ± 133.01 | 1707.34 ± 113.96 | 564.04 ± 98.71 | 540.68 ± 85.63 | 1104.73 ± 112.88 |
| PRC | 2524.22 ± 326.78 | 2098.59 ± 386.67 | 4622.81 ± 718.12 | 2492.03 ± 278.44 | 2257.55 ± 282.58 | 4749.57 ± 594.96 |
| PHC | 1918.20 ± 271.49 | 1756.55 ± 240.11 | 3674.76 ± 461.06 | 924.19 ± 163.07 | 975.71 ± 159.50 | 1899.90 ± 263.74 |
Note: Bolded boxes indicate significantly different volumes between ASHS‐OAP and ASHS‐PMC (two tailed). Regions labeled by ASHS‐OAP that were significantly greater in volume compared to ASHS‐PMC included CA2/CA3/DG, SUB, ERC, and PHC. Regions labeled by ASHS‐PMC that were significantly greater in volume compared to ASHS‐OAP included CA 1 . Note that analyses were run on total volume; however, for completeness, we have reported left and right volumes for each MTL subregion.
To examine how well volumes produced by ASHS‐OAP matched with volumes produced by ASHS‐PMC, scatterplots for each ROI were created with data points representing each participant (using non‐log‐transformed volumes) (Figure 4; top panel). There was a strong correlation between automated volumes produced by ASHS‐PMC and ASHS‐OAP for the CA2/CA3/DG subfield (F(1,38) = 169.70, p < 0.001, R 2 = 0.82, R = 0.90). There was a moderate correlation between automated volumes produced by ASHS‐PMC and ASHS‐OAP for CA1 (F(1,38) = 86.88, p < 0.001, R 2 = 0.70, R = 0.83), SUB (F(1,38) = 102.20, p < 0.001, R 2 = 0.73, R = 0.85), and ERC (F(1,38) = 78.56, p < 0.001, R 2 = 0.67, R = 0.82). There was a weak relationship between automated volumes produced by ASHS‐OAP and ASHS‐PMC for PRC (F(1,38) = 26.58, p < 0.001, R 2 = 0.41, R = 0.64) and PHC (F(1,38) = 17.28, p < 0.001, R 2 = 0.31, R = 0.56). In addition, Bland–Altman plots were used to assess the agreement of the log‐transformed volumes between ASHS‐OAP and ASHS‐PMC for each ROI (Figure 4; bottom panel). Each point on the plot represents a single measurement, and most points are close to the central line (mean difference), indicating good agreement between the two methods. There are a few points near and outside the limits of agreement (e.g., CA1), which indicates specific cases where the two methods differ. Overall, the majority of points fall within the limits of agreement, suggesting reasonable agreement between the two methods, with some variation as indicated by the spread of points.
FIGURE 4.
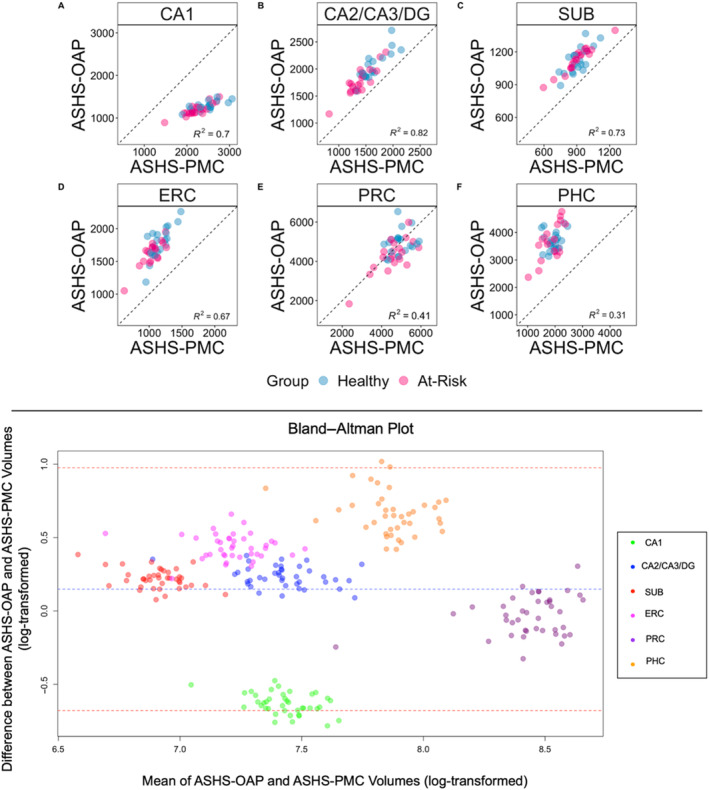
Top panel: Scatter plots depicting the variation explained between volumes produced by the ASHS‐OAP and ASHS‐PMC atlases for each region of interest. Each data point represents one participant, categorised by healthy (blue) or at‐risk (pink). Dotted unity line represents equivalency between volumes produced by ASHS‐OAP and ASHS‐PMC. Points that fall along the unity line indicate that the volumes of the two atlases are exactly the same. Deviation from the unity line indicates a discrepancy between the volumes of the two atlases. Volumes are measured in mm3. A: CA1, B: CA2/CA3/DG, C: Subiculum (SUB), D: Entorhinal cortex (ERC), E: Perirhinal cortex (PRC), F: Parahippocampal cortex (PHC). Bottom panel: A Bland–Altman plot was used to assess the agreement between the ASHS‐OAP and ASHS‐PMC methods (using log‐transformed volumes) by plotting the difference between the methods against their average. Each point is colored according to ROI. The central dashed line represents the mean difference between the log‐transformed volumes of ASHS‐OAP and ASHS‐PMC. The line is close to 0, which suggests that there is no substantial systematic bias between the two methods. The other two dashed lines represent the limits of agreement, set at the mean difference ± 1.96 times the standard deviation of the differences.
3.2.2. Group Differences in MTL Subregions (ASHS‐OAP Versus ASHS‐PMC)
A mixed‐design ANOVA was performed with brain region (six regions) and group (healthy, at‐risk) as interaction terms and brain volume as the dependent variable for ASHS‐OAP and ASHS‐PMC methods separately. Note that in order to compare ASHS‐OAP to ASHS‐PMC, we combined alERC and pmERC volumes produced by ASHS‐OAP into one ERC region. This model was run in order to test whether the brain region × group interaction was significant for a given segmentation method. Initial exploration of the data using a mixed‐design ANOVA revealed no significant group (healthy, at‐risk) × brain region (natural log‐transformed volumes) × hemisphere interaction for ASHS‐OAP (p = 0.34) and ASHS‐PMC (p = 0.75) after correction for sphericity violations; consequently, the reported analyses were run on the summation of left and right hemispheres.
For ASHS‐OAP, a mixed‐design ANOVA indicated that there was a significant brain region (natural log‐transformed volumes) × group interaction (F(5,190) = 3.0, p < 0.05), a significant main effect of group (F(1,38) = 7.34, p < 0.05), and a significant main effect of brain region (F(5,190) = 1016.66, p < 0.05). Mauchly's test for sphericity indicated violations for both brain region (p < 0.05) and the interaction effect (p < 0.05). Following Greenhouse–Geisser corrections, brain region (p < 0.05) and the interaction effect (p < 0.05) remained significant. When age was added to the statistical model as a covariate, there was still a significant brain region × group interaction (p < 0.05), a significant main effect of brain region (p < 0.05), and a significant main effect of group (p < 0.05) after sphericity corrections.
For ASHS‐OAP, follow‐up independent samples t‐tests showed that the CA2/CA3/DG subfield (t(38) = 3.09, p < 0.05, one tailed) and the PRC (t(28) = 2.61, p < 0.05, one tailed) indicated group differences. Follow‐up linear models were conducted with age as a covariate and the effect of CA2/CA3/DG subfield and PRC were still significant. Group differences were observed in the CA1 (t(34) = 2.10, p < 0.05, uncorrected, one‐tailed) and ERC (t(37) = 2.24, p < 0.05, uncorrected, one‐tailed). However, after applying corrections for multiple comparisons, the effects in the CA1 and ERC regions became marginally significant (p = 0.06, corrected). Boxplots for each region are plotted in Figure 5.
FIGURE 5.
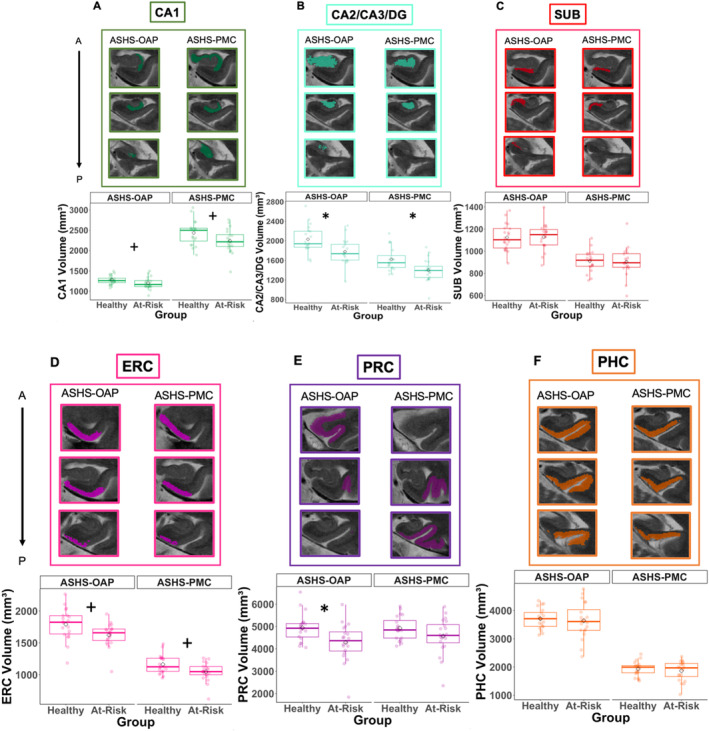
Hippocampal subfields and MTL cortical volumes. Box plots, plotted separately for healthy and at‐risk participants and segmentation method (ASHS‐OAP on the left and ASHS‐PMC on the right), *p < 0.05 after multiple comparison correction, +p < 0.05 before multiple comparison correction. Example of three segmentation slices produced for each ROI per segmentation method displayed for the same participant (healthy participant) (A–F). A = anterior, P = posterior; ERC = entorhinal cortex; PHC = parahippocampal cortex; PRC = perirhinal cortex; SUB = subiculum.
For ASHS‐PMC, a mixed‐design ANOVA indicated that the interaction between brain region and group was nonsignificant (F(5,190) = 2.12, p = 0.07), but there was a significant main effect of group (F(1,38) = 4.89, p < 0.05) and brain region (F(5,190) = 1284.52, p < 0.05). Mauchly's test for sphericity indicated violations for both brain region (p < 0.05) and the interaction effect (p < 0.05). Following Greenhouse–Geisser corrections, brain region (p < 0.05) remained significant, and the interaction effect remained nonsignificant (p = 0.09). When age was added to the statistical model as a covariate, there was still a nonsignificant brain region × group interaction (p = 0.12) and a significant main effect of brain region (p < 0.05). The main effect of group (p = 0.09) became nonsignificant.
For ASHS‐PMC, follow‐up independent samples t‐tests showed that the CA2/CA3/DG subfield (t(36) = 3.01, p < 0.05, one tailed) indicated group differences. Follow‐up linear models were conducted with age as a covariate. The effect of the CA2/CA3/DG subfield was still significant even when age was added into the linear model. Group differences were observed in the CA1 (t(37) = 2.05, p < 0.05, uncorrected, one‐tailed) and ERC (t(36) = 2.43, p < 0.05, uncorrected, one‐tailed) regions. However, after correcting for multiple comparisons, the effect in the CA1 region became nonsignificant (p = 0.10, corrected), while the effect in the ERC region became marginally significant (p = 0.05, corrected). Boxplots for each region are plotted in Figure 5.
Linear models were conducted to test the interaction between group (healthy, at‐risk) and segmentation method (ASHS‐OAP and ASHS‐PMC) on regions that exhibited significant group differences produced by the ASHS‐OAP atlas (CA1, CA2/CA3/DG, ERC, PRC) and ASHS‐PMC atlas (CA2/CA3/DG, ERC). The interaction between group and segmentation method was nonsignificant for CA1, CA2/CA3/DG, ERC, and PRC, which suggested that the observed volume differences between groups were not statistically different across ASHS‐OAP and ASHS‐PMC methods.
4. Discussion
Neuroscience research would benefit from the use of automated segmentation methods that efficiently and accurately delineate MTL subregions. We trained ASHS on the OAP manual segmentation protocol, as developed in our previous work (Olsen et al. 2017, 2013), to determine whether automated segmentation (ASHS‐OAP) would resemble manual segmentation (Manual‐OAP) and be consistent with volumes obtained through an existing automated ASHS atlas, ASHS‐PMC. Structural volumes from a group of older adults (Olsen et al. 2017) were compared across each of the methods, and were examined for their relationship with MoCA performance. The pattern of volumetric results was similar between the ASHS‐OAP atlas and manual segmentation for alERC and PRC (and to a lesser extent CA2/CA3/DG), suggesting that ASHS‐OAP is a viable alternative to manual segmentation methods in detecting early signs of neurodegeneration in MTL subregions. The two automated (ASHS‐OAP and ASHS‐PMC) methods produced significantly different volumes for most ROI (CA1, CA2/CA3/DG, SUB, ERC, PHC). However, both ASHS‐OAP and ASHS‐PMC identified early signs of neurodegeneration in CA2/CA3/DG and marginal differences in ERC. It is also important to note that ASHS‐OAP showed significant group differences in PRC and marginal differences in CA1, highlighting the differences between the segmentation protocols used by the two automated methods.
In general, brain volumes from ASHS‐OAP were similar to Manual‐OAP as assessed by DSC and nonsignificant differences in regional volumes between the two methods. Compared with the healthy group, there was a significant volume reduction in alERC volume in the at‐risk group produced by the Manual‐OAP method. In addition, compared with the healthy group, there was a significant volume reduction in the CA2/CA3/DG subfield and PRC volume in the at‐risk group produced by the ASHS‐OAP method. The nonsignificant interaction between group (healthy, at‐risk) and segmentation method for alERC, CA2/CA3/DG, and PRC suggests that there were no statistically significant differences in volumes produced by Manual‐OAP and ASHS‐OAP between the healthy and at‐risk groups.
In addition, although the ASHS‐OAP and ASHS‐PMC atlases were modified to label the same subregions of the MTL, there were still some discrepancies in the volumetric results produced as indicated by the significant interaction between segmentation method and brain region on volume. Different segmentation atlases/protocols may produce different volumetric results due to nuances in the segmentation rules that each protocol follows and the population with which the atlas was trained/developed for. Both ASHS‐OAP and ASHS‐PMC revealed significant group differences (healthy vs. at‐risk) in CA2/CA3/DG volumes and marginal differences in ERC. Additionally, ASHS‐OAP showed group differences in PRC volumes and marginal differences in CA1. Nonetheless, it can be inferred that ASHS‐OAP and ASHS‐PMC atlases did not statistically differ in volumes produced, as evidenced by the nonsignificant interaction between group (healthy vs. at‐risk) and segmentation method (ASHS‐OAP vs. ASHS‐PMC) for CA1, CA2/CA3/DG, ERC, and PRC volumes.
4.1. Manual‐OAP and ASHS‐OAP
4.1.1. Volumes and DSC
To compare Manual‐OAP and ASHS‐OAP, the volumes of seven regions of the MTL were assessed in the same group of 40 healthy older adults: three subfields of the hippocampus (CA1, CA2/CA3/DG, SUB) and four MTL cortical subregions (alERC, pmERC, PRC, PHC). Based on the DSC results, it was evident that volumes produced by ASHS‐OAP was similar to volumes produced by Manual‐OAP. For both left and right hemispheres, the DSC for Manual‐OAP versus ASHS‐OAP was the highest in CA2/CA3/DG of the hippocampal formation and the lowest in SUB. Compared with the CA2/CA3/DG, the SUB can be a more difficult region to delineate as, according to the OAP protocol, there are two different rules for delineating the SUB and they depend on whether the SUB is on an anterior or posterior slice, which can lead to discrepancies in segmentation overlap between manual and automated methods. Moreover, ASHS tends to segment larger subfields of the hippocampus more accurately than smaller subfields (Wisse et al. 2016), which aligns well with the finding that the DSC was highest for CA2/CA3/DG compared with SUB.
In the MTL cortex, the DSC of Manual‐OAP versus ASHS‐OAP was the highest for the PHC and the lowest for pmERC in both hemispheres. The PHC is a one of the larger structures of the MTL and exhibits less variability in shape than other MTL cortical structures (as defined by the OAP protocol). By contrast, previous work by Wisse et al. (2016) noted that the anterior and posterior boundaries of the ERC were a large source of disagreement between manual segmentation and ASHS (ASHS‐Utrecht 7T atlas). In the current study, the alERC had a moderate DSC between ASHS‐OAP and Manual‐OAP, and pmERC had a low DSC between ASHS‐OAP and Manual‐OAP. Nonetheless, the majority of regions in the MTL cortex (as well as in the hippocampus) were in the moderate to high DSC range between ASHS‐OAP and Manual‐OAP methods for left and right hemispheres (DSC > 0.70). These patterns of results were also reflected in the Bland–Altman plots. In addition, there were no significant differences in volumes produced by Manual‐OAP and ASHS‐OAP, which provided further evidence that the ASHS‐OAP output was close to the “gold standard” of manually derived volumes.
4.1.2. Manual‐OAP, ASHS‐OAP, and MoCA
Given that Manual‐OAP and ASHS‐OAP were similar with respect to structural volumes, it is perhaps unsurprising that the methods were also alike with respect to the relationship between brain volumes and MoCA scores. Volumes of the SUB, pmERC, and PHC obtained from the Manual‐OAP and from the ASHS‐OAP methods had similar nonsignificant volumetric group differences between the healthy and at‐risk groups. In addition, the CA1 region showed group differences for both methods; however, these effects did not survive correction for multiple comparisons. Moreover, the majority of correlations between the volumes produced by Manual‐OAP and ASHS‐OAP were in the moderate to strong range for all MTL subregions. Finally, the effect of age on brain volume was nonsignificant for all regions that showed significant group differences for both Manual‐OAP and ASHS‐OAP methods.
Despite the similarities described above, there were some differences between Manual‐OAP and ASHS‐OAP for certain regions of the MTL and their relationship to MoCA performance. Using ASHS‐OAP, there were significant group differences in PRC volume and in CA2/CA3/DG volume. However, using Manual‐OAP, there were significant group differences in alERC volume only. Note the PRC showed group differences using Manual‐OAP; however, these effects did not survive correction for multiple comparisons. ASHS may bolster the relationship (e.g., capture more variance) between volume and cognitive performance for larger subfields of the hippocampus (e.g., CA2/CA3/DG) and subregions of the MTL (e.g., PRC). Previous work by Wisse et al. (2016) found that ASHS (ASHS‐Utrecht 7T atlas) segmented larger subfields (e.g., CA1, DG, and SUB) more accurately than smaller subfields (CA2, CA3) of the hippocampus. Importantly, for the alERC, a region that demonstrates some of the earliest signs of atrophy related to AD, group differences were exhibited for the ASHS‐OAP method; however, the effect did not reach statistical significance after correcting for multiple comparisons. Therefore, compared to automated methods, manual methods may be more sensitive in detecting subtle changes in grey matter volumes in more variable and smaller regions of the MTL that are sensitive to AD‐related atrophy.
4.2. ASHS‐OAP Versus ASHS‐PMC
4.2.1. Volumes
We compared a modified version of the ASHS‐OAP atlas to a modified version of the ASHS‐PMC atlas in order to assess whether atlases trained on different protocols would lead to discrepancies in volumetric results and in their relation to cognition in the same group of 40 healthy older adults. There was a significant interaction between brain region and protocol (ASHS‐OAP and ASHS‐PMC) on volume, which indicated that the two automated methods produced significantly different volumetric results. PRC volumes were similar across methods, but ERC, PHC, CA2/CA3/DG, and SUB volumes produced by ASHS‐PMC were smaller compared to volumes produced by ASHS‐OAP. Moreover, CA1 volume produced by ASHS‐PMC was significantly greater than CA1 volume produced by ASHS‐OAP. The correlations between the automated volumes generated by ASHS‐PMC and ASHS‐OAP ranged from weak (PRC, PHC) to moderate (CA1, SUB, ERC) to strong (CA2/CA3/DG) across all MTL subregions. These patterns of results were also reflected in the Bland–Altman plots.
The discrepancies in volumes produced by ASHS‐OAP and ASHS‐PMC may be explained by the fact that the atlases were, by definition, trained on different segmentation protocols that follow different rules for MTL segmentation. Indeed, research groups vary in their definitions based on which MTL structures are delineated and labeled (Yushkevich et al. 2015a). Although we matched the ROI segmented by the ASHS‐OAP atlas to those segmented by the ASHS‐PMC atlas, the volumetric results still differed among the majority of regions. For example, the ASHS‐PMC atlas segmented the CA1 subfield throughout the entire extent of the hippocampus, whereas the ASHS‐OAP atlas only segmented the CA1 in the anterior head and body of the hippocampus. Consequently, the CA1 volume produced by ASHS‐PMC was significantly greater than the CA1 volume produced by ASHS‐OAP. Another notable difference between the two atlases is related to the segmentation of PHC. The ASHS‐OAP atlas segmented the PHC more laterally than the ASHS‐PMC atlas and encompassed the entire CS. The ASHS‐PMC atlas defined the lateral boundary of the PHC as being only up to the medial bank of the CS (Xie et al. 2017). As a result, the PHC volume produced by ASHS‐PMC was significantly smaller than the PHC volume produced by ASHS‐OAP. Moreover, although PRC volumes obtained from ASHS‐OAP and ASHS‐PMC were not significantly different from one another, it is still important to note that each automated method labeled the anterior and lateral borders of PRC differently. More specifically, the ASHS‐PMC protocol defined the anterior most appearance of PRC as 2 mm anterior to the hippocampal head (Xie et al. 2017), but, for the ASHS‐OAP protocol, the most anterior appearance of PRC was defined as the slice that contained the most anterior appearance of the CS. In addition, unlike the ASHS‐OAP protocol, the ASHS‐PMC protocol defined the lateral border of PRC extending into the fundus of the occipito‐temporal sulcus (Xie et al. 2017).
In addition to differences in rules and landmarks used to define MTL subregions, different populations were used to train the ASHS‐PMC and ASHS‐OAP atlases. The ASHS‐PMC atlas was trained on brains of individuals with MCI and AD, whereas the ASHS‐OAP atlas was trained on healthy older adult brains who had no clinical diagnosis of MCI or AD. As the target sample in the current study comprised of healthy older adults, using the ASHS‐PMC atlas may have resulted in under‐segmented subregions of the MTL. The ASHS‐PMC atlas may have been “expecting” smaller and more atrophied regions of the MTL to segment, such as what is typically observed in individuals with MCI and/or AD. The ERC, PHC, CA2/CA3/DG, and SUB volumes produced by ASHS‐PMC were smaller compared with the volumes produced by ASHS‐OAP, and these subregions are typically smaller in AD (Pini et al. 2016). These findings highlight the importance of choosing an atlas that would best segment the MTL subregions of the target sample in order to avoid possible inaccurate segmentations (for more details, see Section 5).
4.2.2. ASHS‐OAP, ASHS‐PMC, and MoCA
ASHS‐OAP exhibited group differences as assessed by MoCA performance in CA2/CA3/DG, PRC, and marginal differences in ERC and CA1. The effect of CA2/CA3/DG subfield and the PRC were still significant even when adding age into the models. Using the ASHS‐PMC protocol, the two participant groups showed significant differences in the CA2/CA3/DG subfield and marginal differences in ERC. The effect of CA2/CA3/DG subfield was still significant even when adding age into the models. Despite some subtle differences in the specific regions that demonstrate group differences, it can be suggested that ASHS‐OAP and ASHS‐PMC atlases did not statistically differ in volumes produced based on the nonsignificant interaction between group and segmentation method (ASHS‐OAP and ASHS‐PMC) in CA1, CA2/CA3/DG, ERC, and PRC.
The ASHS‐OAP atlas and the ASHS‐PMC atlas both have their benefits and drawbacks in terms of which one is a “better” choice for detecting neurodegeneration. On the one hand, the ASHS‐OAP atlas is unique in the fact that it segments the anterior and posterior boundaries of the ERC, which may be of particular interest to researchers who want to study regions of the brain associated with early signs of AD‐related neurodegeneration. Moreover, the ASHS‐OAP atlas delineates the PRC more anteriorly than the ASHS‐PMC atlas; as a result, it captures greater volumetric variability in this region. On the other hand, the ASHS‐PMC protocol delineates the PRC into its substructures (BA35 and BA36), which may be of interest to researchers who want to study how these substructures change with neurodegeneration. In addition, the ASHS‐PMC atlas segments a wider range of hippocampal subfields (greater anterior extent) compared with the ASHS‐OAP atlas. Despite the benefits for each atlas, both of them exhibit a restricted range for certain subregions of the MTL. Thus, it is critical for researchers to understand the segmentation rules used to define MTL subregions in the atlas chosen to segment MTL subregions of their desired population.
5. Conclusion and Future Directions
ASHS is a promising alternative to current manual segmentation methods. In the same group of healthy older adults, volumes produced by the ASHS‐OAP atlas was similar to the volumes produced by the manual segmentation method that had been previously shown to be sensitive in detecting changes in grey matter associated with neurodegeneration (Olsen et al. 2017). Moreover, the ASHS‐PMC atlas was found to be sensitive in detecting early signs of neurodegeneration. However, delineation and definition of MTL subregions across different atlases and protocols need refinement. MTL segmentation protocols are highly discrepant across experts and employ different terminology and regional boundary definitions (Yushkevich et al. 2015a). As evidenced here, this can lead to different brain regional volume estimates even within the same group of participants.
The selection of atlas composition is important for accurate delineation of MTL subregions; however, the ideal parameters for atlas composition are yet to be defined. In the current paper, we applied the ASHS‐PMC atlas (trained on brain images of individuals with MCI and AD) to a set of healthy older adult brain images. This may have led to the underestimation of brain regional volumes as the ASHS‐PMC atlas may have been “expecting” smaller brain structure to segment due to neurodegeneration that is typically observed in MCI/AD. In addition, although not conducted in the current study, the ASHS‐OAP atlas (trained on healthy older adult brain images) may overestimate brain regional volumes if applied to a population of individuals with MCI and/or AD. Interestingly, Bender et al. (2018) suggested that using a more variable set of images (in their case, using a lifespan dataset over a sample‐specific dataset) to develop an atlas may lead to a more diverse set of neuroanatomical features in the atlas set, which would lead to better correspondence between manual and automated segmentations. Considering the findings by Bender et al. (2018), the current study suggests that future research should investigate whether training the ASHS‐OAP atlas on both healthy older adult brain images as well as brain images of older adults with MCI and AD may lead to better correspondence between manual and automated segmentations. Similarly, the ASHS‐PMC atlas may benefit from being trained on a subset of healthy older adult brain images in addition to the brain images of older adults with MCI and AD. Ultimately, having a more diverse set of atlas images would allow for both the ASHS‐OAP and ASHS‐PMC atlases to be reliably applied to either healthy or clinical populations, or a mix of the two groups.
Automated methods are promising but will only be as good as the protocol and data on which they are trained. Applying automated approaches to brains that do not fit certain expected “norms” (e.g., neurodegeneration in the MTL) may not produce results that are as accurate as a manual approach. A hybrid approach in which manual corrections are conducted after automated segmentation has been applied may help to alleviate segmentation discrepancies/errors. However, regardless of whether a manual or an automated approach is used for research, there is currently no single consensus MTL segmentation protocol; thus, volumetric results cannot be reliably compared across studies that use different segmentation methods. Fortunately, the Hippocampal Subfields Group (https://hippocampalsubfields.com) is currently developing a harmonized segmentation protocol that can be used by the scientific community to segment subregions of the MTL either manually and/or automatically (Wisse et al. 2017; Wuestefeld et al. 2023). These harmonization efforts will alleviate the discrepancies between protocols/atlases and will create a ground truth in MTL segmentation.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We would like to thank Dr. Malcolm Binns for his helpful guidance on the statistical analysis of this article.
Funding: This work was supported by Canadian Institutes of Health Research, PJT‐162274, PJT‐162292; National Institutes of Health, P30 AG066519, R01‐AG070592; James S. McDonnell Foundation; Canada Research Chair Program and a Max and Gianna Glassman Chair in Neuropsychology; Alzheimer Society of Canada.
Data Availability Statement
The data that support the findings of this study are openly available in Dryad at https://doi.org/10.5061/dryad.6djh9w17h.
References
- Adachi, M. , Kawakatsu S., Hosoya T., et al. 2003. “Morphology of the Inner Structure of the Hippocampal Formation in Alzheimer Disease.” American Journal of Neuroradiology 24: 1575–1581. [PMC free article] [PubMed] [Google Scholar]
- Bender, A. R. , Keresztes A., Bodammer N. C., et al. 2018. “Optimization and Validation of Automated Hippocampal Subfield Segmentation Across the Lifespan.” Human Brain Mapping 39, no. 2: 916–931. 10.1002/hbm.23891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, D. A. , Schneider J. A., Bienias J. L., Evans D. A., and Wilson R.. 2005. “Mild Cognitive Impairment is Related to Alzheimer Disease Pathology and Cerebral Infarctions.” Neurology 64, no. 5: 834–841. [DOI] [PubMed] [Google Scholar]
- Braak, H. , and Braak E.. 1995. “Staging of Alzheimer's Disease‐Related Neurofibrillary Changes.” Neurobiology of Aging 16, no. 3: 271–278. 10.1016/0197-4580(95)00021-6. [DOI] [PubMed] [Google Scholar]
- Braak, H. , and Del Tredici K.. 2020. “From the Entorhinal Region via the Prosubiculum to the Dentate Fascia: Alzheimer Disease‐Related Neurofibrillary Changes in the Temporal Allocortex.” Journal of Neuropathology & Experimental Neurology 79, no. 2: 2. 10.1093/jnen/nlz123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner, R. L., D. Head, J. Parker, et al. 2004. “A Unified Approach for Morphometric and Functional Data Analysis in Young, Old, and Demented Adults Using Automated Atlas‐Based Head Size Normalization: Reliability and Validation Against Manual Measurement of Total Intracranial Volume.” Neuroimage 23: 724–738. [DOI] [PubMed]
- Collins, D. L. , Holmes C. J., Peters T. M., and Evans A. C.. 1995. “Automatic 3‐D Model‐Based Neuroanatomical Segmentation.” Human Brain Mapping 3, no. 3: 190–208. 10.1002/hbm.460030304. [DOI] [Google Scholar]
- Crum, W. R. , Camara O., and Hill D. L.. 2006. “Generalized Overlap Measures for Evaluation and Validation in Medical Image Analysis.” IEEE Transactions on Medical Imaging 25: 1451–1461. [DOI] [PubMed] [Google Scholar]
- D'Angelo, M. C. , Smith V. M., Kacollja A., et al. 2016. “The Effectiveness of Unitization in Mitigating Age‐Related Relational Learning Impairments Depends on Existing Cognitive Status.” Aging Neuropsychology and Cognition 5585: 1e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damian, A. M. , Jacobson S. A., Hentz J. G., et al. 2011. “The Montreal Cognitive Assessment and the Mini‐Mental State Examination as Screening Instruments for Cognitive Impairment: Item Analyses and Threshold Scores.” Dementia and Geriatric Cognitive Disorders 31, no. 2: 126–131. 10.1159/000323867. [DOI] [PubMed] [Google Scholar]
- de Flores, R. , Wisse L. E. M., Das S. R., et al. 2020. “Contribution of Mixed Pathology to Medial Temporal Lobe Atrophy in Alzheimer's Disease.” Alzheimer's & Dementia: The Journal of the Alzheimer's Association 16, no. 6: 843–852. 10.1002/alz.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza, L. C. , Chupin M., Lamari F., et al. 2012. “CSF Tau Markers Are Correlated With Hippocampal Volume in Alzheimer's Disease.” Neurobiology of Aging 33, no. 7: 1253–1257. 10.1016/j.neurobiolaging.2011.02.022. [DOI] [PubMed] [Google Scholar]
- DeKraker, J. , Haast R. A., Yousif M. D., et al. 2022. “Automated Hippocampal Unfolding for Morphometry and Subfield Segmentation With HippUnfold.” eLife 11: e77945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, N. R. , and Smith H.. 1998. Applied Regression Analysis. Vol. 326. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- Free, S. L. , Bergin P. S., Fish D. R., Cook M. J., Shorvon S. D., and Stevens J. M.. 1995. “Methods for Normalization of Hippocampal Volumes Measured with MR.” American Journal of Neuroradiology 16, no. 4: 637–643. [PMC free article] [PubMed] [Google Scholar]
- Gilewski, M. J. , Zelinski E. M., and Schaie K. W.. 1990. “The Memory Functioning Questionnaire for Assessment of Memory Complaints in Adulthood and Old Age.” Psychology and Aging 5, no. 4: 482–490. [DOI] [PubMed] [Google Scholar]
- Hickling, A. L. , Clark I. A., Wu Y. I., and Maguire E. A.. 2024. “Automated Protocols for Delineating Human Hippocampal Subfields From 3 Tesla and 7 Tesla Magnetic Resonance Imaging Data.” Hippocampus 34: 302–308. [DOI] [PubMed] [Google Scholar]
- Iglesias, J. E. , Augustinack J. C., Nguyen K., et al. 2015. “A Computational Atlas of the Hippocampal Formation Using Ex Vivo, Ultra‐High Resolution MRI: Application to Adaptive Segmentation of In Vivo MRI.” NeuroImage 115: 117–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, C. R. , Dickson D. W., Parisi J. E., et al. 2002. “Antemortem MRI Findings Correlate With Hippocampal Neuropathology in Typical Aging and Dementia.” Neurology 58, no. 5: 5. 10.1212/WNL.58.5.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julayanont, P. , Brousseau M., Chertkow H., Phillips N., and Nasreddine Z. S.. 2014. “Montreal Cognitive Assessment Memory Index Score (MoCA‐MIS) as a Predictor of Conversion From Mild Cognitive Impairment to Alzheimer's Disease.” Journal of the American Geriatrics Society 62, no. 4: 679–684. 10.1111/jgs.12742. [DOI] [PubMed] [Google Scholar]
- Kassambara, A. 2023. “rstatix: Pipe‐Friendly Framework for Basic Statistical Tests_.” R package version 0.7.2. https://CRAN.R‐project.org/package=rstatix.
- Kerchner, G. A. , Hess C. P., Hammond‐Rosenbluth K. E., et al. 2010. “Hippocampal CA1 Apical Neuropil Atrophy in Mild Alzheimer Disease Visualized With 7‐T MRI.” Neurology 75: 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchner, G. A., G. K. Deutsch, M. Zeineh, R. F. Dougherty, M. Saranathan, and B. K. Rutt. 2012. “Hippocampal CA1 Apical Neuropil Atrophy and Memory Performance in Alzheimer’s Disease.” Neuroimage 63: 194–202. [DOI] [PMC free article] [PubMed]
- Khan, U. A. , Liu L., Provenzano F. A., et al. 2014. “Molecular Drivers and Cortical Spread of Lateral Entorhinal Cortex Dysfunction in Preclinical Alzheimer's Disease.” Nature Neuroscience 17, no. 2: 304–311. 10.1038/nn.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Joie, R. , Perrotin A., de La Sayette V., et al. 2013. “Hippocampal Subfield Volumetry in Mild Cognitive Impairment, Alzheimer's Disease and Semantic Dementia.” Neuroimage Clinics 3: 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, S. M. , Lockhart S. N., Baker S. L., and Jagust W. J.. 2017. “Tau and β‐Amyloid Are Associated With Medial Temporal Lobe Structure, Function, and Memory Encoding in Normal Aging.” Journal of Neuroscience 37, no. 12: 3201. 10.1523/JNEUROSCI.3769-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwick, A. , Zamboni G., and de Jager C. A.. 2012. “Profiles of Cognitive Subtest Impairment in the Montreal Cognitive Assessment (MoCA) in a Research Cohort With Normal Mini‐Mental State Examination (MMSE) Scores.” Journal of Clinical and Experimental Neuropsychology 34, no. 7: 750–757. 10.1080/13803395.2012.672966. [DOI] [PubMed] [Google Scholar]
- Mazloum‐Farzaghi, N. , Barense M., Ryan J., et al. 2024. “Data From: ASHS‐OAP Atlas for Automatic Entorhinal Cortex Segmentation [Dataset].” Dryad. 10.5061/dryad.6djh9w17h. [DOI] [Google Scholar]
- Nasreddine, Z. S. , Phillips N. A., Bédirian V., et al. 2005. “The Montreal Cognitive Assessment, MoCA: A Brief Screening Tool for Mild Cognitive Impairment.” Journal of the American Geriatrics Society 53, no. 4: 695–699. [DOI] [PubMed] [Google Scholar]
- Nelson, P. T. , Alafuzoff I., Bigio E. H., et al. 2012. “Correlation of Alzheimer Disease Neuropathologic Changes With Cognitive Status: A Review of the Literature.” Journal of Neuropathology & Experimental Neurology 71, no. 5: 362–381. 10.1097/NEN.0b013e31825018f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome, R. N. , Duarte A., and Barense M. D.. 2012. “Reducing Perceptual Interference Improves Visual Discrimination in Mild Cognitive Impairment: Implications for a Model of Perirhinal Cortex Function.” Hippocampus 22: 1990–1999. [DOI] [PubMed] [Google Scholar]
- Newsome, R. N., C. Pun, V. M. Smith, S. Ferber, and M. D. Barense. 2013. “Neural Correlates of Cognitive Decline in Older Adults At‐Risk for Developing MCI: Evidence From the CDA and P300.” Cognitive Neuroscience 4: 152–162. [DOI] [PubMed]
- Olsen, R. K. , Nichols E. A., Chen J., et al. 2009. “Performance‐Related Sustained and Anticipatory Activity in Human Medial Temporal Lobe During Delayed Match‐to‐Sample.” Journal of Neuroscience 29, no. 38: 11880–11890. 10.1523/JNEUROSCI.2245-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, R. K. , Palombo D. J., Rabin J. S., Levine B., Ryan J. D., and Rosenbaum R. S.. 2013. “Volumetric Analysis of Medial Temporal Lobe Subregions in Developmental Amnesia Using High‐Resolution Magnetic Resonance Imaging.” Hippocampus 23, no. 10: 855–860. 10.1002/hipo.22153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, R. K. , Yeung L.‐K., Noly‐Gandon A., et al. 2017. “Human Anterolateral Entorhinal Cortex Volumes Are Associated With Cognitive Decline in Aging Prior to Clinical Diagnosis.” Neurobiology of Aging 57: 195–205. 10.1016/j.neurobiolaging.2017.04.025. [DOI] [PubMed] [Google Scholar]
- Palombo, D. J. , Amaral R. S. C., Olsen R. K., et al. 2013. “KIBRA Polymorphism Is Associated With Individual Differences in Hippocampal Subregions: Evidence From Anatomical Segmentation Using High‐Resolution MRI.” Journal of Neuroscience 33, no. 32: 13088–13093. 10.1523/JNEUROSCI.1406-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, R. C. 2004. “Mild Cognitive Impairment as a Diagnostic Entity.” Journal of Internal Medicine 256, no. 3: 183–194. 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- Pini, L. , Pievani M., Bocchetta M., et al. 2016. “Brain Atrophy in Alzheimer's Disease and Aging.” Ageing Research Reviews 30: 25–48. [DOI] [PubMed] [Google Scholar]
- Pipitone, J. , Park M. T. M., Winterburn J., et al. 2014. “Multi‐Atlas Segmentation of the Whole Hippocampus and Subfields Using Multiple Automatically Generated Templates.” NeuroImage 101: 494–512. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2022. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Schlichting, M. L. , Mack M. L., Guarino K. F., and Preston A. R.. 2019. “Performance of Semi‐Automated Hippocampal Subfield Segmentation Methods Across Ages in a Pediatric Sample.” NeuroImage 191: 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling, R. A. , Aisen P. S., Beckett L. A., et al. 2011. “Toward Defining the Preclinical Stages of Alzheimer's Disease: Recommendations From the National Institute on Aging‐Alzheimer's Association Workgroups on Diagnostic Guidelines for Alzheimer's Disease.” Alzheimer's & Dementia 7, no. 3: 280–292. 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse, L. E. M. , Chételat G., Daugherty A. M., et al. 2021. “Hippocampal Subfield Volumetry From Structural Isotropic 1 mm3 MRI Scans: A Note of Caution.” Human Brain Mapping 42, no. 2: 539–550. 10.1002/hbm.25234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse, L. E. , Daugherty A. M., Olsen R. K., et al. 2017. “A Harmonized Segmentation Protocol for Hippocampal and Parahippocampal Subregions: Why Do We Need One and What Are the Key Goals?” Hippocampus 27, no. 1: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse, L. E. M. , Gerritsen L., Zwanenburg J. J., et al. 2012. “Subfields of the Hippocampal Formation at 7 T MRI: In Vivo Volumetric Assessment.” NeuroImage 61, no. 4: 1043–1049. [DOI] [PubMed] [Google Scholar]
- Wisse, L. E. M. , Kuijf H. J., Honingh A. M., et al. 2016. “Automated Hippocampal Subfield Segmentation at 7T MRI.” American Journal of Neuroradiology 37, no. 6: 1050–1057. 10.3174/ajnr.A4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuestefeld, A. , Baumeister H., Adams J. N., et al. 2023. “Comparison of Histological Delineations of Medial Temporal Lobe Cortices by Four Independent Neuroanatomy Laboratories.” Hippocampus 34, no. 5: 241–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, L. , Pluta J. B., Das S. R., et al. 2017. “Multi‐Template Analysis of Human Perirhinal Cortex in Brain MRI: Explicitly Accounting for Anatomical Variability.” NeuroImage 144: 183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, L. , Wisse L. E., Pluta J., et al. 2019. “Automated Segmentation of Medial Temporal Lobe Subregions on In Vivo T1‐Weighted MRI in Early Stages of Alzheimer's Disease.” Human Brain Mapping 40, no. 12: 3431–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, L. K. , Olsen R. K., Bild‐Enkin H. E., et al. 2017. “Anterolateral Entorhinal Cortex Volume Predicted by Altered Intra‐Item Configural Processing.” Journal of Neuroscience 37, no. 22: 5527–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, L. K. , Olsen R. K., Hong B., et al. 2019. “Object‐In‐Place Memory Predicted by Anterolateral Entorhinal Cortex and Parahippocampal Cortex Volume in Older Adults.” Journal of Cognitive Neuroscience 31, no. 5: 711–729. [DOI] [PubMed] [Google Scholar]
- Yeung, L.‐K. , Ryan J. D., Cowell R. A., and Barense M. D.. 2013. “Recognition Memory Impairments Caused by False Recognition of Novel Objects.” Journal of Experimental Psychology: General 142, no. 4: 1384–1397. 10.1037/a0034021. [DOI] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Amaral R. S., Augustinack J. C., et al. 2015a. “Quantitative Comparison of 21 Protocols for Labeling Hippocampal Subfields and Parahippocampal Subregions in In Vivo MRI: Towards a Harmonized Segmentation Protocol.” NeuroImage 111: 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Pluta J. B., Wang H., et al. 2015b. “Automated Volumetry and Regional Thickness Analysis of Hippocampal Subfields and Medial Temporal Cortical Structures in Mild Cognitive Impairment.” Human Brain Mapping 36, no. 1: 258–287. 10.1002/hbm.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in Dryad at https://doi.org/10.5061/dryad.6djh9w17h.


