Abstract
The gut microbiome has been recognised as a key component in the pathogenesis of inflammatory bowel diseases (IBD), and the wide range of metabolites produced by gut bacteria are an important mechanism by which the human microbiome interacts with host immunity or host metabolism. High-throughput metabolomic profiling and novel computational approaches now allow for comprehensive assessment of thousands of metabolites in diverse biomaterials, including faecal samples. Several groups of metabolites, including short-chain fatty acids, tryptophan metabolites and bile acids, have been associated with IBD. In this Recent Advances article, we describe the contribution of metabolomics research to the field of IBD, with a focus on faecal metabolomics. We discuss the latest findings on the significance of these metabolites for IBD prognosis and therapeutic interventions and offer insights into the future directions of metabolomics research.
Keywords: CROHN'S DISEASE, ULCERATIVE COLITIS, INFLAMMATORY BOWEL DISEASE, INTESTINAL MICROBIOLOGY
Key messages.
Advances in untargeted metabolomic technologies are expanding our understanding of metabolic alterations occurring in inflammatory bowel diseases (IBD).
Significant alterations in the faecal metabolome profiles of patients with ulcerative colitis (UC) and Crohn's disease (CD) have been described. The faecal metabolic signature of IBD includes changes in short-chain fatty acids, tryptophan metabolites, sphingolipids, and vitamin levels. While the metabolic signatures of the two IBD subtypes largely overlap, CD is primarily characterised by an enrichment of primary bile acids, whereas ulcerative colitis shows higher levels of proteolytic fermentation products.
Metabolomic profiling of IBD patients can provide complementary and mechanistic insights into the relationships between genetics, dysbiosis, and the disease.
Metabolomics is currently aiding in discovering novel therapeutic targets. In the near future, molecular profiling may facilitate patient stratification for personalised therapy.
Faecal metabolome profiles are emerging as a source of novel disease biomarkers, showing promise for disease diagnosis, behaviour, and treatment response prediction. However, these biomarkers still require validation in independent cohorts and clinical settings.
Introduction
Inflammatory bowel diseases (IBD) are chronic inflammatory disorders of the gastrointestinal tract that affect more than 7 million people worldwide.1 The two primary forms of IBD, Crohn’s disease (CD) and ulcerative colitis (UC), are characterised by intermittent inflammation and cumulative damage to the intestinal tract. The pathogenesis of IBD is multifactorial and involves an exaggerated intestinal immunological response in genetically predisposed individuals that is triggered by environmental and nutritional factors.2 Multiple lines of evidence from epidemiological, genomic, interventional and in vitro studies have revealed the important role of the gut microbiome in IBD pathogenesis.3
The gut microbiome—the trillions of micro-organisms, including bacteria, viruses, fungi and archaea living in the human gut—is an important factor in human health. This complex ecosystem impacts host digestion and nutrient absorption and helps modulate the host’s immune system. Knowledge of how the microbiome is involved in human health and disease is largely driven by a growing capacity to interrogate the gut microbiome using high-throughput technologies like whole-genome shotgun sequencing. These efforts have identified that the gut microbial composition of patients with IBD deviates from that of healthy individuals. The IBD gut microbiome is characterised by a decrease in bacterial richness and a reduction in beneficial species, for example, butyrate-producing bacteria, and an enrichment of opportunistic species, commonly referred to as pathobionts.4 These signatures seem to reflect more than just a state of chronic intestinal inflammation as gut microbiome changes have also been observed prior to the onset of CD and in healthy first-degree relatives, healthy individuals at high genetic risk for IBD and the non-affected twins of an IBD-affected twin.5,7 Furthermore, longitudinal studies on patients with IBD have shown that their gut microbiota undergoes temporal periods of ‘dysbiosis’ in which the loss of microbial diversity and blooming of pathobionts accentuates and co-occurs with metabolic and transcriptional changes in the gut.8,10
Despite the apparent involvement of the gut microbiota in the pathology of IBD, the mechanisms triggering intestinal inflammation are still unknown. Metabolites, that is, low-molecular-weight molecules including lipids, amino acids, small peptides, nucleic acids and organic acids, produced by the intestinal microbiota modulate host immunity and metabolism and have, therefore, been suggested to be critical factors in the development and progression of IBD.11 12 Because the composition and metabolic activity of the intestinal microbiota are contingent on nutrient intake, the interaction between dietary habits, microbiota and inflammation is becoming crucial for understanding the disease. A Westernised diet, characterised by increased consumption of simple carbohydrates, emulsifiers and lipids, along with reduced fibre intake, has been identified as a major risk factor for developing IBD.2 13 14 Experimental data have demonstrated that the accumulation of simple sugars and lipids in the intestinal lumen can induce inflammation in genetically susceptible rodent models and promote the expansion of pathobionts.15 16 However, whether dietary metabolites directly influence immune activation or whether effects are mediated by the microbiota remains unclear, and it is plausible that multiple mechanisms are involved.
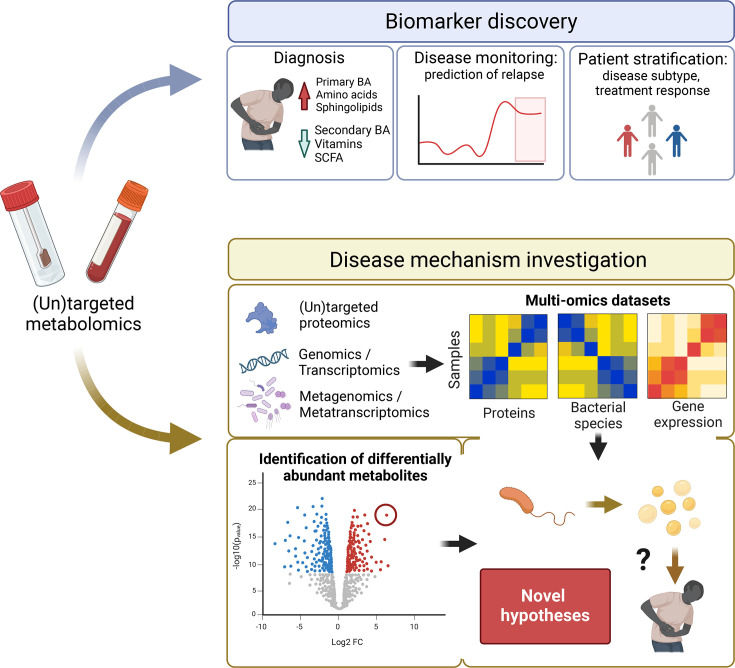
In the last decade, targeted and untargeted metabolomics analytical techniques, like mass spectrometry (MS) and nuclear magnetic resonance spectroscopy (NMR), have enabled high-throughput profiling of thousands of compounds.17 These technologies have been applied in multiple tissues, including blood,18 faeces19 and urine,20 and more recently, the luminal content of the gastrointestinal tract.21 The quantification and characterisation of metabolites in patients with IBD represent a promising strategy for the discovery of novel disease biomarkers and potential targets for therapy.22 Furthermore, combining metabolomics with genomics and metagenomics may help unravel the intricate molecular mechanisms underlying the disease (figure 1).
Figure 1. Applications of faecal metabolomics in IBD research. Overview of the potential of faecal metabolomics for the discovery of novel biomarkers and the metabolic pathways involved in disease development. Metabolite-based biomarkers could assist in the early diagnosis of disease and in monitoring disease activity to predict future relapses. Molecular profiles might also be useful for patient stratification and the design of personalised treatment strategies. Furthermore, metabolomics data provide complementary information to other omics datasets, which might be essential for understanding disease triggers. BA, bile acid; IBD, inflammatory bowel diseases; SCFA, short-chain fatty acid.
In this Recent Advances article, we highlight the contribution of high-throughput metabolomics to IBD research, discuss the latest findings regarding the significance of altered metabolites in IBD prognosis and therapeutic interventions and offer insights into the future directions of metabolomics research, with a focus on faecal metabolomics. Finally, we anticipate the potential breakthroughs and innovations we could expect in the coming years.
Challenges and opportunities of using metabolomics for mechanism discovery in IBD
High-throughput technologies, including MS and NMR, now allow the measurement of thousands of small compounds (<1.5 kDa) in single experiments. These technologies can target specific groups of metabolites, such as bile acids (BAs) or lipids, or they can take a wider untargeted approach.23 We have summarised the main metabolomic profiling technologies in box 1. However, despite the rapid advances in metabolite characterisation, only a small proportion of the chemical compounds detected in the human body can currently be assigned to known molecules. This represents one of the main limitations of untargeted techniques, posing a challenge to biological interpretability.24 25
Box 1. Main untargeted metabolomics techniques.
Nuclear MR (NMR) spectroscopy
NMR spectroscopy is a spectroscopic technique that relies on the interaction between electromagnetic radiation and atomic nuclei with spins, such as hydrogen (1H), carbon (13C) and nitrogen (15N). 1H NMR is widely used in metabolomics due to the natural abundance of hydrogen in biological samples. NMR spectroscopy can generate profiles of metabolites in complex mixtures with little sample preprocessing, including lipids, amino acids, nucleotides, organic acids and sugars. The advantages of NMR spectroscopy include robustness, high reproducibility and non-destructive characteristics. The main drawback is its relatively low sensitivity, resulting in the non-detection of less abundant metabolites.98
Mass spectrometry (MS)
MS is an analytical technique measuring the mass-to-charge ratio (m/z) of ions, allowing for the identification and quantification of chemicals by accurate measurement of mass spectra. It is often coupled with separation methods such as gas chromatography (GC) and liquid chromatography (LC) to allow for high-throughput measurement of many compounds. The complexity of biological samples is first reduced by separating constituents based on the interaction of different components with absorbent materials within the chromatographic column. After elution from the column, the m/z value of the separated molecule is detected by MS. The MS-based approach offers high sensitivity and selectivity but less robustness, and it requires complex sample ionisation processes.99
GC-MS uses inert gas, such as helium or nitrogen, as a mobile phase to transport samples through capillary columns for separation. It is used for naturally volatile substances or compounds that can be chemically modified into volatile derivatives, including short-chain fatty acids, amino acids, sugars, amines and organic acids. GC-MS has high sensitivity, selectivity and separation resolution, along with reproducible retention times, making it suitable for trace analysis. Well-developed mass spectral libraries make it a commonly used technique in metabolomics research. The inherent limitation is that GC-MS can only be used to separate and identify low molecular weight (ca. 50–600 Da) molecules and needs complex sample derivatisation.
LC-MS uses polar solvents as a mobile phase to distribute components, followed by mass analysis, and can work with different column chemistries, including reversed-phase LC for non-polar to moderately polar molecules and hydrophobic interaction liquid chromatography (HILIC) for ionic and polar molecules. LC-MS provides the broadest metabolite coverage without derivatisation, including proteins, nucleic acids, sugars, peptides and lipids and has become the most widely used technique in metabolomics. LC-MS can also provide high sensitivity and selectivity, but spectral libraries for LC-MS are currently less developed compared with GC-MS, leading to a higher number of unidentified spectra.100
A second important aspect to consider is that metabolites are the products of complex reactions occurring in the human body and are influenced by multiple factors, such as exposures (including diet), genetics and the gut microbiota. The pool of metabolites in a given tissue reflects different contributions from each of these factors. For instance, blood metabolites are strongly influenced by diet, whereas faecal metabolites predominantly reflect the metabolic activity of the intestinal microbiota.1926,28 In an Israeli cohort, dietary information derived from food frequency questionnaires accounted for the variation in over one-third of the measured serum metabolites (n=335), with the explained variation ranging from 4% to nearly 50% for individual metabolites. The microbiome accounted for the levels of 182 metabolites, whereas genetics influenced 83 serum molecules.27 A cohort study involving 1569 individuals from the USA found that, on average, genetics explained 4% and the microbiome 11% of the overall serum metabolome variation. Of the 595 metabolites associated with either genetics or the microbiome, approximately 410 were solely predicted by gut microbiota composition and 90 by genetics, though the impact of diet was not assessed in this cohort.29 In a Dutch cohort, long-term dietary patterns explained 9.3% of plasma metabolome variation. Compared with genetics or faecal microbiome composition, diet showed the largest number of associations with the serum metabolome, with 2854 associations between dietary habits and the levels of 769 circulating metabolites.26 Similarly, urine metabolite profiles have been shown to contain several biomarkers for food intake.30 31 In contrast, we demonstrated that only a few faecal metabolites, mainly related to coffee and tea intake, could be predicted using dietary data.19
These findings emphasise the importance of integrating diverse metabolomic measurements across different body sites with dietary and environmental data in the context of IBD. Moreover, since single metabolomic measurements only represent a snapshot of complex and dynamic processes, longitudinal measurments must be considered when investigating metabolic alterations in a disease context.
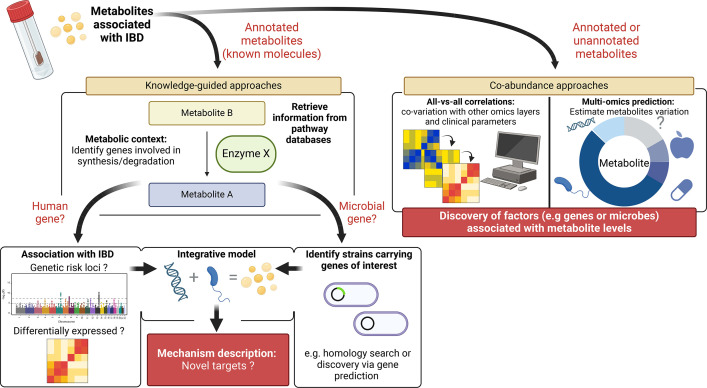
A third consideration is that the growing availability of multi-omics datasets generated from high-throughput technologies demands the development of effective methods for data integration. One of the current challenges in computational biology is designing robust, efficient algorithms that can handle large and complex datasets in order to facilitate the comprehension of biological events that trigger diseases. Two common approaches for integrating metabolomics data with, for example, genomic and metagenomics datasets rely either on knowledge of metabolic pathways (knowledge-guided approaches) or on statistical co-abundance analyses (agnostic approaches). Knowledge-guided approaches rely on the curated biological information in databases such as KEGG32 or METACYC.33 These frameworks use existing knowledge about metabolic pathways to associate genes with metabolites, facilitating the identification of precursor-product ratios between metabolites or the dysfunction of specific enzymatic reactions. Examples of such tools include MetaboAnalyst,34 IMPaLA,35 MIMOSA236 and Anansi.37 In contrast, agnostic approaches do not require prior knowledge of the biological relations between the different omics layers. A wide variety of agnostic techniques, ranging from correlations to advanced machine learning and AI algorithms, have recently been developed.38,41 While these approaches can be effective in discovering novel relations between features (genes, bacteria and metabolites), biological interpretation can often be challenging, and additional ex vivo or in vitro validations are therefore needed (figure 2).
Figure 2. Overview of two main strategies to integrate metabolomics in the context of IBD research. Differentially abundant metabolites in patients with IBD compared with non-IBD controls can be further investigated by integrating additional omics layers. For metabolites with a known identity, knowledge-guided approaches can be used to place metabolites in their metabolic context, allowing the identification of genes involved in their production and degradation. In the case of human genes, further investigation of its expression or the presence of polymorphisms in individuals with IBD can provide novel hypotheses of the pathomechanism of the disease. Similarly, faecal metagenomics and metatranscriptomics datasets can be used to explore the capacity of the gut microbiota to transform these molecules. When the information on the metabolic pathway is unavailable or the identity of the associated metabolites cannot be established, co-abundance analyses with other omics layers can help establish which environmental, host and microbial factors impact the levels of a specific metabolite. IBD, inflammatory bowel diseases.
Recently, we have witnessed not only a technological leap forward in the field of metabolomics and bioinformatics but also an increase in (untargeted) metabolomics profiling within large multi-omics cohorts of patients with IBD. The discovery of altered metabolites levels in patient samples, and their correlation with genetics, microbiota and clinical data, is providing new insights into the diversity and complexity of IBD. In the upcoming sections, we review recent findings from cohorts of patients with IBD where untargeted metabolomics has been employed.
Recent discoveries using untargeted metabolomics in combination with other omics layers
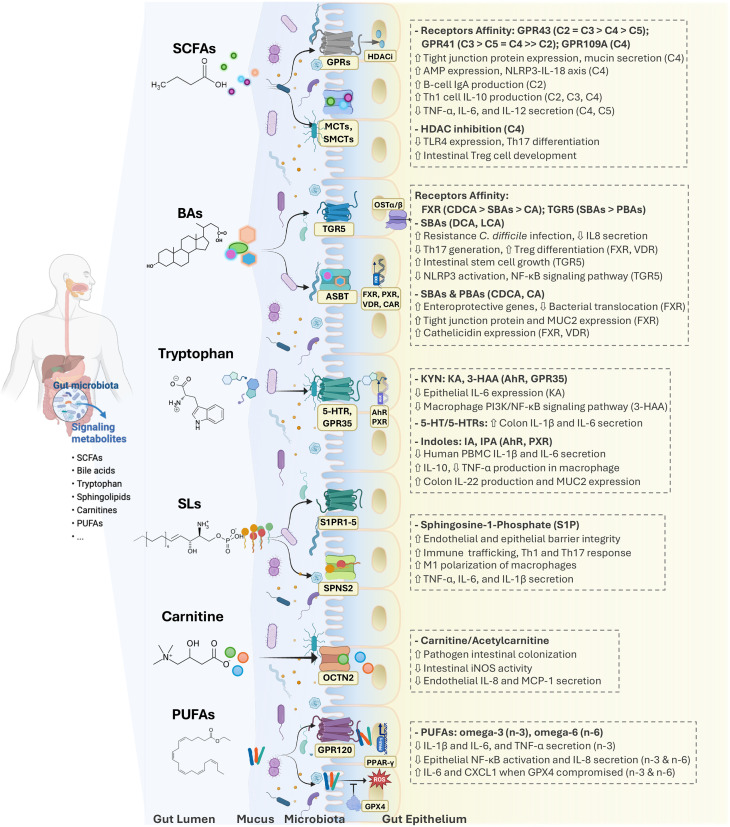
BAs and amino acids, particularly tryptophan, have been focal points in numerous studies. Current knowledge on these metabolites in the context of IBD was recently reviewed elsewhere.242,46 In box 2 and figure 3, we summarise metabolomic pathways that have been consistently associated with IBD and their suggested mechanism of action.
Box 2. Metabolite classes associated with inflammatory bowel diseases (IBD).
Other molecules associated with IBD
Other groups of molecules that have been associated with IBD include acylcarnitines, which are formed from fatty acids and L-carnitine, polyunsaturated fatty acids (PUFAs) and amino acids.
Acylcarnitines
Evidence from observational studies: In IBD, mitochondrial dysfunction and the subsequent reduction of fatty acid oxidation leads to elevated carnitine and acylcarnitine levels in the colon.117
Evidence from preclinical experimental studies: It has been recently demonstrated that increased levels of acylcarnitines promote the growth of pathobionts both in vitro and in the murine gut, providing evidence for metabolic host–microbiota interactions in the context of IBD.118
PUFAs
Evidence from observational studies: An imbalance in omega-6/omega-3 ratio of dietary PUFAs is associated with higher CD risk. Higher faecal PUFA levels have been observed in IBD.19
Evidence from preclinical experimental studies: Dietary PUFAs can trigger inflammatory response in genetically susceptible mice (Xbp1−/−IEC) via IL8 and TNF-α expression, significantly influenced by the compromised expression and enzymatic activity of glutathione peroxidase 4 (GPX4).119 Specifically, functional GPX4 mitigates the detrimental effects of dietary PUFAs on the intestinal epithelium by limiting the oxidation of membrane phospholipids.
Amino acids
Evidence from observational studies: Patients with IBD tend to present lower serum AA levels but increased faecal AA levels, potentially suggesting nutrient malabsorption and increased proteolytic fermentation in the gut.120
Evidence from preclinical experimental studies: Glutamine and arginine contribute to gut barrier integrity and modulate inflammation in colitis murine models.121
Figure 3. Intestinal metabolites have diverse regulatory effects on gut epithelium and mucosa immunity. The critical roles played by short-chain fatty acids (SCFAs), bile acids (BAs), tryptophan, sphingolipids (SLs), carnitines and polyunsaturated fatty acids (PUFAs) are highlighted here. 3-HAA, 3-hydroxyanthranilic acid; 5-HT, serotonin; ABST, apical sodium-dependent bile acid transporter; AhR, aryl hydrocarbon receptor; CA, cholic acid; CAR, constitutive androstane receptor; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; FXR, farnesoid X receptor; GPRs, G protein-coupled receptors; GPX4, glutathione peroxidase 4; HDAC, histone deacetylases; IA, indoleacrylic acid; IPA, indolepropionic acid; KA, kynurenic acid; KYN, kynurenine; LCA, lithocholic acid; MCT1, monocarboxylate transporter 1; OCTN2, organic cation/carnitine transporter 2; OST, organic solute transporter; PBAs, primary BAs; PPAR-γ, peroxisome proliferator-activated receptor gamma; PXR, pregnane X receptor; ROS, reactive oxygen species; SBAs, secondary BAs; SMCT1, sodium-coupled monocarboxylate transporter 1; S1PRs, sphingosine-1-phosphate receptors; SPNS2, spinster homolog 2; TGR5, transmembrane G protein-coupled receptor 5; VDR, vitamin D receptor.
The current eruption of studies employing untargeted techniques in large cohorts of patients with IBD provides a comprehensive overview of metabolic changes associated with the disease. For example, in a study using the Prospective Registry in IBD Study at MGH cohort (PRISM),47 which included 88 CD, 76 UC and 56 non-IBD controls, researchers identified 2729 metabolites associated with IBD. Notably, only 43% of the over 8000 metabolites detected in the faeces could be matched to a known compound in the Human Metabolome Database, highlighting the prevalence of unknown metabolites. Differential abundance analysis revealed 2456 metabolites associated with CD and 1049 associated with UC. In line with this, our study of 238 CD, 172 UC and 255 non-IBD controls also reported significant but comparable alterations in the faecal metabolomes of patients with CD and UC.19 Specifically, faecal levels of sphingolipids, primary BAs and ethanolamines were positively associated with IBD. Furthermore, both studies demonstrated a strong correlation between microbiota composition and faecal metabolite profiles, enabling the prediction of metabolite levels based on metagenomic data.
BAs, carnitines and propionate were identified as key alterations in the faecal metabolome of patients with IBD in the Integrative Human Microbiome Project (iHMP).10 These metabolites formed nodes in a large correlation network between faecal metabolomics, metagenomics, metatranscriptomics, metaproteomics and serological profiles. By examining the dynamics within each data layer throughout a 1-year progression of the disease, the authors found that changes in the BA profiles and enrichment of acylcarnitines were associated with periods of intestinal dysbiosis. While the directionality of these associations and their causal role in disease dynamics remain unclear, such analyses are helpful to prioritise targets and provide insights into the complex interaction between the host and their microbiota.
Faecal metabolomics was shown to be a strong predictor of disease activity in patients with UC in another cross-sectional multi-omics cohort.48 Phosphocholines, indoles and dipeptides were found to be the most enriched molecular classes in the faeces of this group of patients. Moreover, the authors showed that the enrichment of dipeptides was linked with increased protease activity of the gut commensal Bacteroides vulgatus. Remarkably, the authors were able to demonstrate that B. vulgatus protease activity induced colitis in IL10-deficient germ-free mice, providing evidence for novel therapeutic targets that aim to inhibit Bacteroides protease activity.
An enrichment of dipeptides was also observed in the stools of treatment-naïve paediatric UC patients with moderate or severe disease when compared with inactive disease in the Predicting Response to Standarized Colitis Therapy (PROTECT) cohort.49 Interestingly, the faecal metabolite profile variation was strongly associated with bacterial diversity, while the plasma metabolite variation was associated with disease activity. Participants with moderate or severe disease showed lower levels of secondary BAs and tryptophan metabolites, while phosphatidylcholines and sphingomyelins were among the most strongly enriched molecules. In plasma, long-chain triacylglycerols were the most significantly depleted metabolites, whereas acetylated polyamines were enriched.
In another cohort of 1313 individuals (484 UC, 464 CD and 365 non-IBD), an enrichment of sphingomyelins in both faeces and serum was associated with disease activity.18 Additionally, faecal secondary BAs were linked with inflammation extension in UC. Furthermore, integrating genomics and serum metabolomics through colocalisation and Mendelian randomisation analysis revealed two genetic loci influencing disease development via modulation of metabolite levels. These results suggested a protective effect for CD mediated by a polymorphism on chromosome 11 (rs4246215), resulting in lower fatty acid desaturase activity and a reduction in the conversion of linoleic acid into arachidonic acid.
Overall, these studies not only confirm the relevance of bile acids and tryptophan-derived metabolites in the IBD pathology but also point to other less studied molecules such as fatty acids and sphingolipids. Furthermore, the studies we have highlighted are elegant examples of how hypothesis-free approaches using multi-omics datasets in large cohorts, combined with statistical causal inference, can lead to novel mechanistic hypotheses in the context of IBD. Such discoveries are expected to increase significantly in the near future thanks to the growing availability of extensive molecular data in large population biobanks like the Finngen project,50 The UK Biobank,51 and LifeLines52 and disease-specific cohorts such as 1000IBD,53 iHMP,10 the Pediatrict Risk Stratification and Identification of Immunogenetic and Microbial Markers Study (RISK)54 and IBD Response.55 Importantly, mechanistic investigations will be needed to validate the relevance of novel metabolites or pathways associated with IBD, prioritise molecules for developing novel therapies and distinguish metabolites that trigger diseases from the by-products of inflammation.
Impact of disease subtype, location and intestinal resection on the faecal and serum metabolite profiles
Metabolomics studies have identified differences in the metabolomic profile between CD and UC patients. However, analyses in both the 1000IBD and PRISM cohorts revealed a substantial overlap between the UC and CD faecal metabolome signatures, although the enrichment of bile acids and ethanolamines was primarily observed in CD.19 47 In serum, metabolites related to lipid metabolism, TCA cycle-related molecules, and amino-acids were the main contributors distinguishing between CD and UC.56 57 58 Differences observed between CD and UC might be driven by disease location. Accumulating evidence indicates that ileal CD represents a distinct disease entity that differentiates itself from colonic CD. Consistently, the colonic-isolated and ileal-isolated CD subtypes also display clear differences in metabolomic profiles. Metabolites related to fatty acid biosynthesis, BA, and amino acids (tryptophan) exhibited a marked increase in faeces from ileal CD patients compared with those from colonic CD.59 60 Alterations in the bile acids profiles are also observed in the faeces of CD patients with resection in the ileum, with an increase on primary BA and a trend towards lower levels of secondary BAs.59 61 62 Considering the role of the ileum in bile acids reabsorption and nutrient absorption, inflammation or resection of this site is expected to have a large impact on the gut environment, and therefore, should be considered when studying the metabolome in IBD.
Identification of novel biomarkers for IBD based on faecal metabolites
The extent of the metabolic alterations observed in patients with IBD presents an opportunity to leverage metabolites as biomarkers, with combinations of metabolites suggested to be predictors of disease and treatment response. Below, we summarise recent proposed biomarkers identified using untargeted metabolomics approaches. Due to the semi-quantitative nature of these methods, biomarkers should be further assessed using targeted approaches, which are more sensitive and provide higher reproducibility compared to untargeted approaches.
Metabolomic markers for diagnosing and classifying IBD
Bacterial-associated metabolites, including short-chain fatty acids (SCFAs), medium-chain fatty acids, tryptophan-derivatives, BAs and sphingolipids, have been proposed as biomarkers for IBD diagnosis. For instance, in an Italian cohort of patients with IBD (n=132), 14 faecal metabolites enabled the separation of samples from patients with CD from those of non-IBD controls.63 Predictors included higher levels of biogenic amines, amino acids and lipids and lower levels of vitamins. In UC, 16 metabolites were found to be altered compared with controls, with 9 overlapping with CD.
Amino acid levels in faeces were also predictive of IBD in a study that included two cohorts from China (n=108 and n=70) and two derived from the US PRISM study (n=155 and n=65 from the Dutch replication cohort).64 A panel of 13 metabolites, more than half being derivates of amino acids, exhibited power in discriminating patients with IBD from controls, with an average area under the curve (AUC) of 0.916 in the Chinese cohorts and 0.885 in the US cohorts. Overall, this shows the robustness of these predictors across populations and two different ethnicities. Notably, none of the 13 metabolites were differentially abundant in colorectal cancer and type 1 diabetes. In another Chinese cohort comprising 158 UC, 130 CD and 138 healthy controls, targeted metabolomics in serum samples confirmed the potential of amino acids as biomarkers.65 Four and five amino acid levels were sufficient to distinguish UC and CD from healthy control serum samples, respectively (AUC=0.942 for UC, AUC=0.962 for CD), although none of the amino acids overlapped with those identified in the study mentioned above. Furthermore, a classification model using the levels of three amino acid metabolites (taurine, homocitrulline and kynurenine) accurately distinguished CD from UC (AUC=0.935). While these results are promising, replication in independent cohorts is still needed.
We recently used machine learning to identify the most discriminative faecal metabolomic features in patients with IBD.19 The ratio between two metabolites, sphingolipid lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) and L-urobilin, improved the accuracy of the calprotectin test in distinguishing samples from patients from those of non-IBD controls, reaching an AUC of 0.83 in the test dataset. Importantly, these findings were replicated in an independent cohort of Australian UC patients,66 and several additional studies have reported alterations in these two metabolites in faecal, plasma, mucosal and serum samples from patients with IBD, supporting the role of these metabolites as biomarkers.18 63 67 68
Taken together, and despite the diversity of methods and metabolites captured between studies, increased faecal levels of amino acid derivates and sphingolipids have been reported in multiple cohorts and are the most promising markers for IBD. Understanding the mechanism underlying the changes in these metabolites might provide useful leads for preventing and treating the disease. Alterations of other well-studied metabolites in IBD, for example, BAs and SCFAs,44 have also been reported in several other conditions.69 70 While this stresses that these metabolites play a critical role in gut health overall, they might be less suitable as unique IBD biomarkers. Furthermore, while the models we have described seem to perform well in discriminating IBD from non-IBD samples, subtype classification, that is, CD versus UC, has been shown to be less robust.19 47 To discover and validate subtype-specific metabolite signatures, large high-quality prospective cohorts are needed. In particular, it is important to distinguish the metabolic changes that are the product of intestinal inflammation from those that are subtype-specific.
Metabolomic markers for disease activity
Identifying changes in the level of metabolites preceding the development of flares can assist in disease monitoring and relapse prevention. Although multiple studies have found alterations of the faecal metabolome in relation to disease scores,48 71 whether these alterations can predict future disease relapse is still underexplored. In our recent study, we showed that the levels of two metabolites that distinguish IBD from non-IBD samples (lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) and L-urobilin) differed between individuals in long-term remission (no relapse registered 1 year before sample collection) and other patients with IBD.19 Validation in longitudinal cohorts will be needed to determine the value of tracking the levels of these two metabolites in faeces.
Promising biomarkers have been identified as well in blood samples from patients with IBD. In a prospective cohort study of 40 patients with UC, a combination of the levels of plasma metabolites exhibited an accuracy of 74% in predicting worsening postoperative endoscopic activity up to 7 months from sample collection.72 In addition, plasma histidine levels were found to be associated with an increased risk of relapse in patients with UC.73 Low histidine levels were also reported to be predictive of relapse within a 1-year period by Hisamatsu et al.73 In another study, patients in clinical remission presented higher levels of 3-hydroxybutyrate, acetoacetate and acetone in serum and of transaconitate in urine, whereas urinary acetamide and cystine levels decreased.74 Moreover, a prospective cohort study of 164 patients with IBD identified four serum metabolomic markers (sarcosine, carnitine, propionyl-l-carnitine and sorbitol) associated with clinical relapse within 2 years which showed a moderate performance in predicting relapse (AUC=0.70).75
Based on the currently available evidence, predictors for disease relapse are lacking due to the limited number of longitudinal studies and data heterogeneity across populations. Only plasma histidine levels have been reported to be reduced in active UC patients in multiple studies,72 73 76 but additional research is needed to assess its capacity to predict disease flares.
Metabolomic markers for response to treatment
Another promising avenue is the use of metabolomics profiles as predictors for treatment response. Given the suggested correlation between microbiome composition and treatment response to biologics,77,80 investigating the role of metabolites in this relationship is a logical next step for understanding the underlying mechanisms.
In a longitudinal cohort study of 76 patients with CD, lipid and BA profiling from faeces, serum and urine showed distinct before-treatment signatures between those patients who responded to anti-TNF therapy and those who did not.81 Lipid profiling of serum unveiled alterations in the levels of four circulatory lipid markers (phosphocholines, ceramides, sphingomyelins and triglycerides) that accurately predicted the anti-TNF response. However, faecal lipid profile showed an even higher predictive accuracy than the serum lipid profile (faecal lipids: AUC=0.94±0.10, serum lipids: AUC=0.78±0.12). In addition, the authors demonstrated that BA profiles differed between responders and non-responders, with higher levels of primary BAs associated with non-response to treatment. Predictors built from the levels of three faecal BAs or five serum or five urine BAs showed a good performance in discriminating the two groups of patients (AUCs=0.81, 0.74 and 0.70, respectively).
The integration of stool metagenomics, serum metabolomics and proteomics allowed the prediction of response to anti-cytokine or anti-integrin therapy in 185 participants from the US PRISM cohort (AUC=0.963, 77 UC and 108 CD participants).82 Serum metabolomics alone showed moderate discriminative power (AUC=0.77 (95% CI 0.664 to 0.891)) but performed slightly worse than predictors based on proteomics (AUC=0.806) and metagenomics (AUC=0.849). Interestingly, responders at 14-week therapy presented higher levels of secondary BAs in their baseline serum sample, stressing the importance of microbial-produced metabolites in immunoregulatory functions.
In addition, we recently evaluated the capacity of the faecal metabolome and microbiome to predict response to ustekinumab and vedolizumab in a cohort of 100 patients. Our preliminary results suggest that faecal features have limited predictive power (AUC=0.71), similar to patient clinical characteristics. However, consistent with previous studies, we identified the levels of lithocholic acid, a secondary BA higher in responders than in non-responders, as a potential predictor of treatment success.83
Overall, while baseline faecal levels of BAs are promising predictors of treatment response, the mechanism behind this association is unclear. Considering the role of the intestinal microbiota in regulating the pool of primary and secondary BAs, these associations could represent a proxy of the microbiome alterations previously reported in non-responders. On the other hand, BAs are emerging as a key regulator of the immune system in the gut.84 Therefore, future research should elucidate the role of BAs in the response to biologicals.
It is important to stress that despite the many potential markers for diagnosis, monitoring and therapeutic responses, very few have been validated in independent cohorts. Estimations of accuracy have been primarily conducted in single cohorts using cross-validation approaches and predominantly in Caucasian cohorts. Consequently, the impact of different dietary habits, genetic backgrounds,85 and microbiome compositions86 among diverse human populations on metabolome-based biomarkers has been overlooked. Ideally, the robustness of these biomarkers should be tested across diverse and multiethnic patient cohorts and various stages of the disease. Moreover, integrating metabolome with other data layers that have also been proven as potential biomarkers, such as the proteome,87 88 microbiome4 47 64 or circulating antibodies,89 might increase diagnostic test accuracy and assist in understanding the disease heterogeneity, including subtypes and progression. Therefore, multiomics efforts to study the interaction between different data layers will likely yield better patient stratification and therapeutic strategies. Finally, any novel biomarkers should be validated using targeted metabolomics approaches and must exhibit superior accuracy to established markers such as faecal calprotectin, C-reactive protein or lactoferrin.
Future perspectives
Metabolite characterisation remains one of the main challenges in the field, as the identities of many molecules found in the gastrointestinal tract are still unknown. Understanding the chemical structure of metabolites, how they are produced, and their potential physiological impact on the gut environment will be crucial for developing novel therapeutic strategies for a wide range of health disorders.90 Therefore, efforts should be made to improve the analytical frameworks for quantifying and annotating metabolites. Equally important is the availability of metabolomic data in public repositories. Open-access metabolic data are critical for validating novel molecules and extrapolating their clinical relevance. For example, recently discovered microbial conjugated BAs (BAs that are reconjugated to amino acids by the gut microbiota) have been found to be elevated in faeces of patients with IBD upon reanalysing the MS data from the HMP2 cohort.91 In the same line, Gentry et al recently developed the so-called ‘reverse metabolomics’ strategy, an analytical pipeline that enables the systematic search of newly synthesised molecules in public repositories.92 Implementing this strategy at a large scale will certainly boost the discovery of relevant disease-associated molecules.
Despite the diversity of analytical approaches, in the context of IBD, sphingolipids, SCFAs, BAs, and tryptophan-derived metabolites have consistently been found as IBD-associated molecules. Therefore, targeted approaches to reveal the impact of these molecules and the molecular diversity on their derivatives should, in our opinion, centre the efforts of metabolomics research in IBD. Considering that these metabolites are partially regulated by the gut microbiota, either through synthesis or transformation, further understanding the metabolic capacity of the microbial communities in the gut will enable the identification of therapeutic strategies to modulate metabolites by targeting the microbiota. To this end, a deeper characterisation of gut microbes’ genetic and metabolic diversity will be necessary, as the function of a substantial proportion of bacterial genes remains unknown, and the impact of polymorphism and structural variants in bacterial genomes is still overlooked.93 94
Together with the benefits of technical improvements for discovering new metabolites, our understanding of the role of gut metabolism in health and diseases can be enhanced by sampling along the gastrointestinal tract using capsules. Compared with endoscopic sampling, gastrointestinal capsules are less invasive and allow sampling of different upper and lower gastrointestinal tract regions. A recent exploration using this technique to profile luminal metabolites of the upper intestine in 15 healthy controls showed that the chemical environment varied significantly along the gastrointestinal tract and revealed several molecules and microbially produced metabolites rarely detected in faeces.21 Comparing regional metabolic changes in patients with IBD during remission and periods of active disease or during clinical interventions will provide a better understanding of IBD triggers and host–microbiota crosstalk. Another promising strategy in the investigation of local host–microbiota interactions is spatial biology. Although still in its infancy, we expect that spatial technologies will lead to a better understanding of metabolic and immune interactions in the context of inflammation. Complementing measurements on gut metabolism with metabolomics measurements in blood and urine can lead to the discovery of novel mechanisms by which the microbial-produced molecules influence the host’s health beyond its impact on the gut environment.
Advances in our understanding of metabolic alterations in IBD are rapidly unveiling novel therapeutic approaches that target specific pathways. For instance, restoring the activity of aminoadipate aminotransferase, an enzyme crucial for converting tryptophan into xanthurenic and kynurenic acids, has shown promising results in promoting epithelial healing and modulating Th17 cell differentiation.95 Similarly, enhancing bacterial capacity to convert primary BAs into secondary BAs, namely lithocholic acid and deoxycholic acid, has been demonstrated to mitigate inflammation in murine colitis models.96 Although these findings illustrate exciting avenues in the treatment of IBD, it is important to keep in mind that, given the disease heterogeneity, it is improbable that strategies targeting a single metabolic pathway will serve as a ‘one-size-fits-all’ solution for IBD. In our view, patient stratification based on molecular profiles holds significant promise for tailoring treatments to individual patients. To this end, the identification of biomarkers will be essential. Beyond disease diagnosis, recent findings suggest that faecal metabolites can assist in predicting disease progression and drug response. In the near future, data from large IBD cohorts will allow the discovery and replication of metabolite-based biomarkers before these can be implemented in clinical practice.
Finally, data derived from (longitudinal) population cohorts or studies in first-degree relatives like the Genetic, Environmental, Microbial (GEM) study97 offer the opportunity to ascertain whether gut metabolic alterations precede disease onset. In combination with experimental data, these types of observation will be instrumental in establishing the causal role of dietary-derived or microbial-produced metabolites in the development of IBD.
Overall, we have highlighted the potential of untargeted metabolomics for investigating the molecular mechanisms involved in IBD and provided recent examples of how faecal metabolomics can help establish connections between genetic susceptibility, the microbiome and inflammation. Considering the recent findings presented above, we foresee that future technological innovations in metabolomics and bioinformatics will likely reveal new molecules suitable for developing new biomarkers and treatments for IBD.
Acknowledgements
RKW is supported by the Seerave Foundation and the EU Horizon Health consortium grant ID-DarkMatter-NCD (101136582) and the EU Horizon Europe Program grant miGut-Health: personalised blueprint of intestinal health (101095470).
Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council. Neither the European Union nor the granting authority can be held responsible for them. The funders had no role in study design, data collection and analysis, preparation of the manuscript or decision to publish.We like to thank Kate McIntyre for editorial support.
Footnotes
Funding: This study was funded by Seerave Foundation (n/a), HORIZON EUROPE European Innovation Council (101095470, 101136582). JWZ is supported by InnoHK, The Government of Hong Kong, Special Administrative Region of the People’s Republic of China, Research Grants Council-General Research Fund (RGC-GRF) Hong Kong (14121322) and National Nature Science Foundation of China (82100573).
Patient consent for publication: Not applicable.
Provenance and peer review: Commissioned; externally peer reviewed.
Contributor Information
Arnau Vich Vila, Email: a.vich.vila@umcg.nl.
Jingwan Zhang, Email: wendyzhang@magic-inno.hk.
Moting Liu, Email: m.liu@umcg.nl.
Klaas Nico Faber, Email: k.n.faber@umcg.nl.
Rinse K Weersma, Email: r.k.weersma@umcg.nl.
References
- 1.Collaborators GBDIBD The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol Hepatol. 2020;5:17–30. doi: 10.1016/S2468-1253(19)30333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adolph TE, Meyer M, Schwärzler J, et al. The metabolic nature of inflammatory bowel diseases. Nat Rev Gastroenterol Hepatol. 2022;19:753–67. doi: 10.1038/s41575-022-00658-y. [DOI] [PubMed] [Google Scholar]
- 3.Plichta DR, Graham DB, Subramanian S, et al. Therapeutic opportunities in inflammatory bowel disease: mechanistic dissection of host-microbiome relationships. Cell. 2019;178:1041–56. doi: 10.1016/j.cell.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vich Vila A, Imhann F, Collij V, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10:eaap8914. doi: 10.1126/scitranslmed.aap8914. [DOI] [PubMed] [Google Scholar]
- 5.Hedin CR, van der Gast CJ, Stagg AJ, et al. The gut microbiota of siblings offers insights into microbial pathogenesis of inflammatory bowel disease. Gut Microbes. 2017;8:359–65. doi: 10.1080/19490976.2017.1284733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand EC, Klaassen MAY, Gacesa R, et al. Healthy Cotwins share gut microbiome signatures with their inflammatory bowel disease twins and unrelated patients. Gastroenterol. 2021;160:1970–85. doi: 10.1053/j.gastro.2021.01.030. [DOI] [PubMed] [Google Scholar]
- 7.Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2008;2:716–27. doi: 10.1038/ismej.2008.37. [DOI] [PubMed] [Google Scholar]
- 8.Gilliland A, Chan JJ, De Wolfe TJ, et al. Pathobionts in inflammatory bowel disease: origins, underlying mechanisms, and implications for clinical care. Gastroenterol. 2024;166:44–58. doi: 10.1053/j.gastro.2023.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Levy M, Kolodziejczyk AA, Thaiss CA, et al. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219–32. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 10.Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nat New Biol. 2019;569:655–62. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 12.Adolph TE, Zhang J. Diet fuelling inflammatory bowel diseases: preclinical and clinical concepts. Gut. 2022;71:2574–86. doi: 10.1136/gutjnl-2021-326575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters V, Bolte L, Schuttert EM, et al. Western and carnivorous dietary patterns are associated with greater likelihood of IBD development in a large prospective population-based cohort. J Crohns Colitis. 2022;16:931–9. doi: 10.1093/ecco-jcc/jjab219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serrano-Moreno C, Brox-Torrecilla N, Arhip L, et al. Diets for inflammatory bowel disease: what do we know so far? Eur J Clin Nutr. 2022;76:1222–33. doi: 10.1038/s41430-021-01051-9. [DOI] [PubMed] [Google Scholar]
- 15.Tilg H, Zmora N, Adolph TE, et al. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol. 2020;20:40–54. doi: 10.1038/s41577-019-0198-4. [DOI] [PubMed] [Google Scholar]
- 16.Adolph TE, Meyer M, Jukic A, et al. Heavy arch: from inflammatory bowel diseases to metabolic disorders. Gut. 2024;73:1376–87. doi: 10.1136/gutjnl-2024-331914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirwan JA. Translating metabolomics into clinical practice. Nat Rev Bioeng. 2023;1:228–9. doi: 10.1038/s44222-023-00023-x. [DOI] [Google Scholar]
- 18.Di’Narzo AF, Houten SM, Kosoy R, et al. Integrative analysis of the inflammatory bowel disease serum metabolome improves our understanding of genetic etiology and points to novel putative therapeutic targets. Gastroenterol. 2022;162:828–43. doi: 10.1053/j.gastro.2021.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vich Vila A, Hu S, Andreu-Sánchez S, et al. Faecal metabolome and its determinants in inflammatory bowel disease. Gut. 2023;72:1472–85. doi: 10.1136/gutjnl-2022-328048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens NS, Siffledeen J, Su X, et al. Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J Crohn's Colitis. 2013;7:e42–8. doi: 10.1016/j.crohns.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 21.Folz J, Culver RN, Morales JM, et al. Human metabolome variation along the upper intestinal tract. Nat Metab. 2023;5:777–88. doi: 10.1038/s42255-023-00777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjerrum JT, Wang YL, Seidelin JB, et al. IBD metabonomics predicts phenotype, disease course, and treatment response. EBioMedicine. 2021;71:103551. doi: 10.1016/j.ebiom.2021.103551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alseekh S, Aharoni A, Brotman Y, et al. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat Methods. 2021;18:747–56. doi: 10.1038/s41592-021-01197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, et al. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol. 2022;20:143–60. doi: 10.1038/s41579-021-00621-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proc Natl Acad Sci U S A. 2015;112:12549–50. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Zhernakova DV, Kurilshikov A, et al. Influence of the microbiome, diet and genetics on inter-individual variation in the human plasma metabolome. Nat Med. 2022;28:2333–43. doi: 10.1038/s41591-022-02014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar N, Korem T, Weissbrod O, et al. A reference map of potential determinants for the human serum metabolome. Nat New Biol. 2020;588:135–40. doi: 10.1038/s41586-020-2896-2. [DOI] [PubMed] [Google Scholar]
- 28.Zierer J, Jackson MA, Kastenmüller G, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. 2018;50:790–5. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diener C, Dai CL, Wilmanski T, et al. Genome-microbiome interplay provides insight into the determinants of the human blood metabolome. Nat Metab. 2022;4:1560–72. doi: 10.1038/s42255-022-00670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Playdon MC, Sampson JN, Cross AJ, et al. Comparing metabolite profiles of habitual diet in serum and urine. Am J Clin Nutr. 2016;104:776–89. doi: 10.3945/ajcn.116.135301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Posma JM, Garcia-Perez I, Frost G, et al. Nutriome-metabolome relationships provide insights into dietary intake and metabolism. Nat Food. 2020;1:426–36. doi: 10.1038/s43016-020-0093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caspi R, Billington R, Ferrer L, et al. The metacyc database of metabolic pathways and enzymes and the biocyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–80. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang Z, Zhou G, Ewald J, et al. Using metaboanalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat Protoc. 2022;17:1735–61. doi: 10.1038/s41596-022-00710-w. [DOI] [PubMed] [Google Scholar]
- 35.Kamburov A, Cavill R, Ebbels TMD, et al. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–8. doi: 10.1093/bioinformatics/btr499. [DOI] [PubMed] [Google Scholar]
- 36.Noecker C, Eng A, Muller E, et al. Mimosa2: a metabolic network-based tool for inferring mechanism-supported relationships in microbiome-metabolome data. Bioinformatics. 2022;38:1615–23. doi: 10.1093/bioinformatics/btac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bastiaanssen TF, Quinn TP, Cryan JF. Knowledge-based integration of multi-omic datasets with anansi: annotation-based analysis of specific interactions. 2023;arXiv preprint:arXiv: 230510832. [Google Scholar]
- 38.Morton JT, Aksenov AA, Nothias LF, et al. Learning representations of microbe-metabolite interactions. Nat Methods. 2019;16:1306–14. doi: 10.1038/s41592-019-0616-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodosthenous T, Shahrezaei V, Evangelou M. Integrating multi-OMICS data through sparse canonical correlation analysis for the prediction of complex traits: a comparison study. Bioinformatics. 2020;36:4616–25. doi: 10.1093/bioinformatics/btaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller E, Shiryan I, Borenstein E. Multi-omic integration of microbiome data for identifying disease-associated modules. Nat Commun. 2024;15:2621. doi: 10.1038/s41467-024-46888-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tataru CA, David MM. Decoding the language of microbiomes using word-embedding techniques, and applications in inflammatory bowel disease. PLoS Comput Biol. 2020;16:e1007859. doi: 10.1371/journal.pcbi.1007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michaudel C, Sokol H. The gut microbiota at the service of immunometabolism. Cell Metab. 2020;32:514–23. doi: 10.1016/j.cmet.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Duboc H, Rajca S, Rainteau D, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–9. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 44.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–37. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 45.Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugihara K, Morhardt TL, Kamada N. The role of dietary nutrients in inflammatory bowel disease. Front Immunol. 2018;9:3183. doi: 10.3389/fimmu.2018.03183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mills RH, Dulai PS, Vázquez-Baeza Y, et al. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides Vulgatus proteases with disease severity. Nat Microbiol. 2022;7:262–76. doi: 10.1038/s41564-021-01050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schirmer M, Stražar M, Avila-Pacheco J, et al. Linking microbial genes to plasma and stool metabolites uncovers host-microbial interactions underlying ulcerative colitis disease course. Cell Host Microbe. 2024;32:209–26. doi: 10.1016/j.chom.2023.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurki MI, Karjalainen J, Palta P, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–18. doi: 10.1038/s41586-022-05473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scholtens S, Smidt N, Swertz MA, et al. Cohort profile: lifelines, a three-generation cohort study and biobank. Int J Epidemiol. 2015;44:1172–80. doi: 10.1093/ije/dyu229. [DOI] [PubMed] [Google Scholar]
- 53.Imhann F, Van der Velde KJ, Barbieri R, et al. The 1000Ibd project: multi-omics data of 1000 inflammatory bowel disease patients; data release 1. BMC Gastroenterol. 2019;19:5. doi: 10.1186/s12876-018-0917-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kugathasan S, Denson LA, Walters TD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn's disease: a multicentre inception cohort study. Lancet. 2017;389:1710–8. doi: 10.1016/S0140-6736(17)30317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyatt NJ, Watson H, Anderson CA, et al. Defining predictors of responsiveness to advanced therapies in Crohn’s disease and ulcerative colitis: protocol for the IBD-RESPONSE and nested CD-metaRESPONSE prospective, multicentre, observational cohort study in precision medicine. BMJ Open. 2024;14:e073639. doi: 10.1136/bmjopen-2023-073639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scoville EA, Allaman MM, Brown CT, et al. Alterations in lipid, amino acid, and energy metabolism distinguish Crohn’s disease from ulcerative colitis and control subjects by serum metabolomic profiling. Metabolomics (Los Angel) 2018;14:17. doi: 10.1007/s11306-017-1311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams HRT, Willsmore JD, Cox IJ, et al. Serum metabolic profiling in inflammatory bowel disease. Dig Dis Sci. 2012;57:2157–65. doi: 10.1007/s10620-012-2127-2. [DOI] [PubMed] [Google Scholar]
- 58.Ooi M, Nishiumi S, Yoshie T, et al. GC/MS-based profiling of amino acids and TCA cycle-related molecules in ulcerative colitis. Inflamm Res. 2011;60:831–40. doi: 10.1007/s00011-011-0340-7. [DOI] [PubMed] [Google Scholar]
- 59.Jansson J, Willing B, Lucio M, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS ONE. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalez CG, Mills RH, Zhu Q, et al. Location-specific signatures of Crohn’s disease at a multi-omics scale. Microbiome. 2022;10:133. doi: 10.1186/s40168-022-01331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lapidus A, Akerlund J-E, Einarsson C. Gallbladder bile composition in patients with Crohn ’s disease. World J Gastroenterol. 2006;12:70–4. doi: 10.3748/wjg.v12.i1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang X, Vázquez-Baeza Y, Elijah E, et al. Gastrointestinal surgery for inflammatory bowel disease persistently lowers microbiome and metabolome diversity. Inflamm Bowel Dis. 2021;27:603–16. doi: 10.1093/ibd/izaa262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santoru ML, Piras C, Murgia A, et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep. 2017;7:9523. doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ning L, Zhou Y-L, Sun H, et al. Microbiome and metabolome features in inflammatory bowel disease via multi-omics integration analyses across cohorts. Nat Commun. 2023;14:7135. doi: 10.1038/s41467-023-42788-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou G, Liu H, Wei P, et al. Amino acids-targeted metabolomics reveals novel diagnostic biomarkers for ulcerative colitis and Crohn’s disease. Amino Acids. 2023;55:349–58. doi: 10.1007/s00726-023-03233-0. [DOI] [PubMed] [Google Scholar]
- 66.Chetwood JD, Paramsothy S, Haifer C, et al. Key metabolomic alterations are associated with ulcerative colitis disease state and activity: a validation analysis. Gut. 2024;73:1392–3. doi: 10.1136/gutjnl-2023-330196. [DOI] [PubMed] [Google Scholar]
- 67.Nyström N, Prast-Nielsen S, Correia M, et al. Mucosal and plasma metabolomes in new-onset paediatric inflammatory bowel disease: correlations with disease characteristics and plasma inflammation protein markers. J Crohn's Colitis. 2023;17:418–32. doi: 10.1093/ecco-jcc/jjac149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bushman FD, Conrad M, Ren Y, et al. Multi-omic analysis of the interaction between clostridioides difficile infection and pediatric inflammatory bowel disease. Cell Host Microbe. 2020;28:422–33. doi: 10.1016/j.chom.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong JMW, de Souza R, Kendall CWC, et al. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–43. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 70.Collins SL, Stine JG, Bisanz JE, et al. Bile acids and the gut microbiota: metabolic interactions and impacts on disease. Nat Rev Microbiol. 2023;21:236–47. doi: 10.1038/s41579-022-00805-x. [DOI] [PubMed] [Google Scholar]
- 71.Chen R, Zheng J, Li L, et al. Metabolomics facilitate the personalized management in inflammatory bowel disease. Therap Adv Gastroenterol. 2021;14:17562848211064489. doi: 10.1177/17562848211064489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Probert F, Walsh A, Jagielowicz M, et al. Plasma nuclear magnetic resonance metabolomics discriminates between high and low endoscopic activity and predicts progression in a prospective cohort of patients with ulcerative colitis. J Crohn's Colitis. 2018;12:1326–37. doi: 10.1093/ecco-jcc/jjy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hisamatsu T, Ono N, Imaizumi A, et al. Decreased plasma histidine level predicts risk of relapse in patients with ulcerative colitis in remission. PLoS ONE. 2015;10:e0140716. doi: 10.1371/journal.pone.0140716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keshteli AH, van den Brand FF, Madsen KL, et al. Dietary and metabolomic determinants of relapse in ulcerative colitis patients: a pilot prospective cohort study. World J Gastroenterol. 2017;23:3890–9. doi: 10.3748/wjg.v23.i21.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Borren NZ, Plichta D, Joshi AD, et al. Multi-‘-omics’ profiling in patients with quiescent inflammatory bowel disease identifies biomarkers predicting relapse. Inflamm Bowel Dis. 2020;26:1524–32. doi: 10.1093/ibd/izaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kohashi M, Nishiumi S, Ooi M, et al. A novel gas chromatography mass spectrometry-based serum diagnostic and assessment approach to ulcerative colitis. J Crohns Colitis. 2014;8:1010–21. doi: 10.1016/j.crohns.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 77.Ananthakrishnan AN, Luo C, Yajnik V, et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe. 2017;21:603–10. doi: 10.1016/j.chom.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caenepeel C, Falony G, Machiels K, et al. Dysbiosis and associated stool features improve prediction of response to biological therapy in inflammatory bowel disease. Gastroenterol. 2024;166:483–95. doi: 10.1053/j.gastro.2023.11.304. [DOI] [PubMed] [Google Scholar]
- 79.Doherty MK, Ding T, Koumpouras C, et al. Fecal microbiota signatures are associated with response to ustekinumab therapy among Crohn’s disease patients. MBio. 2018;9:e02120-17. doi: 10.1128/mBio.02120-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kolho K-L, Korpela K, Jaakkola T, et al. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am J Gastroenterol. 2015;110:921–30. doi: 10.1038/ajg.2015.149. [DOI] [PubMed] [Google Scholar]
- 81.Ding NS, McDonald JAK, Perdones-Montero A, et al. Metabonomics and the gut microbiome associated with primary response to anti-TNF therapy in Crohn’s disease. J Crohns Colitis. 2020;14:1090–102. doi: 10.1093/ecco-jcc/jjaa039. [DOI] [PubMed] [Google Scholar]
- 82.Lee JWJ, Plichta D, Hogstrom L, et al. Multi-omics reveal microbial determinants Impacting responses to biologic therapies in inflammatory bowel disease. Cell Host Microbe. 2021;29:1294–304. doi: 10.1016/j.chom.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prins FM, Hidding IJ, Klaassen MAY, et al. Limited predictive value of the gut microbiome and metabolome for response to biological therapy in inflammatory bowel disease. medRxiv. 2024 doi: 10.1101/2024.05.10.24307195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guzior DV, Quinn RA. Review: microbial transformations of human bile acids. Microbiome. 2021;9:140. doi: 10.1186/s40168-021-01101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shanahan F, Ghosh TS, O’Toole PW. Human microbiome variance is underestimated. Curr Opin Microbiol. 2023;73:102288. doi: 10.1016/j.mib.2023.102288. [DOI] [PubMed] [Google Scholar]
- 87.Hatsugai M, Kurokawa MS, Kouro T, et al. Protein profiles of peripheral blood mononuclear cells are useful for differential diagnosis of ulcerative colitis and Crohn’s disease. J Gastroenterol. 2010;45:488–500. doi: 10.1007/s00535-009-0183-y. [DOI] [PubMed] [Google Scholar]
- 88.Bourgonje AR, Hu S, Spekhorst LM, et al. The effect of phenotype and genotype on the plasma proteome in patients with inflammatory bowel disease. J Crohns Colitis. 2022;16:414–29. doi: 10.1093/ecco-jcc/jjab157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bourgonje AR, Andreu-Sánchez S, Vogl T, et al. Phage-display immunoprecipitation sequencing of the antibody epitope repertoire in inflammatory bowel disease reveals distinct antibody signatures. Immunity. 2023;56:1393–409. doi: 10.1016/j.immuni.2023.04.017. [DOI] [PubMed] [Google Scholar]
- 90.Arifuzzaman M, Collins N, Guo C-J, et al. Nutritional regulation of microbiota-derived metabolites: implications for immunity and inflammation. Immunity. 2024;57:14–27. doi: 10.1016/j.immuni.2023.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quinn RA, Melnik AV, Vrbanac A, et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature. 2020;579:123–9. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gentry EC, Collins SL, Panitchpakdi M, et al. Reverse metabolomics for the discovery of chemical structures from humans. Nature. 2024;626:419–26. doi: 10.1038/s41586-023-06906-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhernakova DV, Wang D, Liu L, et al. Host genetic regulation of human gut microbial structural variation. Nature. 2024;625:813–21. doi: 10.1038/s41586-023-06893-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schloissnig S, Arumugam M, Sunagawa S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Michaudel C, Danne C, Agus A, et al. Rewiring the altered tryptophan metabolism as a novel therapeutic strategy in inflammatory bowel diseases. Gut. 2023;72:1296–307. doi: 10.1136/gutjnl-2022-327337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sinha SR, Haileselassie Y, Nguyen LP, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27:659–70. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raygoza Garay JA, Turpin W, Lee S-H, et al. Gut microbiome composition is associated with future onset of Crohn’s disease in healthy first-degree relatives. Gastroenterology. 2023;165:670–81. doi: 10.1053/j.gastro.2023.05.032. [DOI] [PubMed] [Google Scholar]
- 98.Moco S. Studying metabolism by NMR-based metabolomics. Front Mol Biosci. 2022;9:882487. doi: 10.3389/fmolb.2022.882487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beale DJ, Pinu FR, Kouremenos KA, et al. Review of recent developments in GC-MS approaches to metabolomics-based research. Metabolomics (Los Angel) 2018;14:152. doi: 10.1007/s11306-018-1449-2. [DOI] [PubMed] [Google Scholar]
- 100.Emwas A-HM. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol Biol. 2015;1277:161–93. doi: 10.1007/978-1-4939-2377-9_13. [DOI] [PubMed] [Google Scholar]
- 101.Parada Venegas D, De la Fuente MK, Landskron G, et al. Short chain fatty acids (SCFaS)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peng L, Li Z-R, Green RS, et al. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–25. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun M, Wu W, Chen L, et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun. 2018;9:3555. doi: 10.1038/s41467-018-05901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee JG, Lee J, Lee A-R, et al. Impact of short-chain fatty acid supplementation on gut inflammation and microbiota composition in a murine colitis model. J Nutr Biochem. 2022;101:108926. doi: 10.1016/j.jnutbio.2021.108926. [DOI] [PubMed] [Google Scholar]
- 105.Schaap FG, Trauner M, Jansen PLM. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11:55–67. doi: 10.1038/nrgastro.2013.151. [DOI] [PubMed] [Google Scholar]
- 106.Inagaki T, Moschetta A, Lee Y-K, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–5. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.D’Aldebert E, Biyeyeme Bi Mve M-J, Mergey M, et al. Bile salts control the antimicrobial peptide cathelicidin through nuclear receptors in the human biliary epithelium. Gastroenterol. 2009;136:1435–43. doi: 10.1053/j.gastro.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 108.Mawe GM, Hoffman JM. Serotonin signalling in the gut--functions, dysfunctions and therapeutic targets. Nat Rev Gastroenterol Hepatol. 2013;10:473–86. doi: 10.1038/nrgastro.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alexeev EE, Lanis JM, Kao DJ, et al. Microbiota-derived Indole metabolites promote human and murine intestinal homeostasis through regulation of Interleukin-10 receptor. Am J Pathol. 2018;188:1183–94. doi: 10.1016/j.ajpath.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Quintana FJ, Basso AS, Iglesias AH, et al. Control of T(Reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature New Biol. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 111.Venkatesh M, Mukherjee S, Wang H, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–24. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 113.Wang S, van Schooten F-J, Jin H, et al. The involvement of intestinal tryptophan metabolism in inflammatory bowel disease identified by a meta-analysis of the transcriptome and a systematic review of the metabolome. Nutrients. 2023;15:2886. doi: 10.3390/nu15132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature New Biol. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Verstockt B, Vetrano S, Salas A, et al. Sphingosine 1-phosphate modulation and immune cell trafficking in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2022;19:351–66. doi: 10.1038/s41575-021-00574-7. [DOI] [PubMed] [Google Scholar]
- 116.Brown EM, Ke X, Hitchcock D, et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe. 2019;25:668–80. doi: 10.1016/j.chom.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith SA, Ogawa SA, Chau L, et al. Mitochondrial dysfunction in inflammatory bowel disease alters intestinal epithelial metabolism of hepatic acylcarnitines. J Clin Invest. 2021;131:e133371. doi: 10.1172/JCI133371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lemons JMS, Conrad M, Tanes C, et al. Enterobacteriaceae growth promotion by intestinal acylcarnitines, a biomarker of dysbiosis in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2024;17:131–48. doi: 10.1016/j.jcmgh.2023.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schwärzler J, Mayr L, Vich Vila A, et al. PUFA-induced metabolic enteritis as a fuel for Crohn’s disease. Gastroenterol. 2022;162:1690–704. doi: 10.1053/j.gastro.2022.01.004. [DOI] [PubMed] [Google Scholar]
- 120.Massironi S, Viganò C, Palermo A, et al. Inflammation and malnutrition in inflammatory bowel disease. Lancet Gastroenterol Hepatol. 2023;8:579–90. doi: 10.1016/S2468-1253(23)00011-0. [DOI] [PubMed] [Google Scholar]
- 121.Coburn LA, Gong X, Singh K, et al. L-arginine supplementation improves responses to injury and inflammation in dextran sulfate sodium colitis. PLoS ONE. 2012;7:e33546. doi: 10.1371/journal.pone.0033546. [DOI] [PMC free article] [PubMed] [Google Scholar]