Abstract
Objectives
Systemic lupus erythematosus (SLE) shows a marked female bias in prevalence. X chromosome inactivation (XCI) is the mechanism which randomly silences one X chromosome to equalise gene expression between 46, XX females and 46, XY males. Though XCI is expected to result in a random pattern of mosaicism across tissues, some females display a significantly skewed ratio in immune cells, termed XCI-skew. We tested whether XCI was abnormal in females with SLE and hence contributes to sexual dimorphism.
Methods
We assayed XCI in whole blood DNA in 181 female SLE cases, 796 female healthy controls and 10 twin pairs discordant for SLE. Using regression modelling and intra-twin comparisons, we assessed the effect of SLE on XCI and combined clinical, cellular and genetic data via a polygenic score to explore underlying mechanisms.
Results
Accommodating the powerful confounder of age, XCI-skew was reduced in females with SLE compared with controls (p=1.3×10−5), with the greatest effect seen in those with more severe disease. Applying an XCI threshold of >80%, we observed XCI-skew in 6.6% of SLE cases compared with 22% of controls. This difference was not explained by differential white cell counts, medication or genetic susceptibility to SLE. Instead, XCI-skew correlated with a biomarker for type I interferon-regulated gene expression.
Conclusions
These results refute current views on XCI-skew in autoimmunity and suggest, in lupus, XCI patterns of immune cells reflect the impact of disease state, specifically interferon signalling, on the haematopoietic stem cells from which they derive.
Keywords: Lupus Erythematosus, Systemic; Autoimmune Diseases; Polymorphism, Genetic
WHAT IS ALREADY KNOWN ON THIS TOPIC
Increased prevalence of skewed X chromosome inactivation (XCI-skew) in females has been reported for numerous autoimmune diseases and it is hypothesised to contribute to disease development and sex biases in the prevalence. This hypothesis has not been robustly tested in systemic lupus erythematosus (SLE).
WHAT THIS STUDY ADDS
XCI-skew is reduced in SLE cases compared with healthy controls. This effect is driven by disease progression and likely reflects the impact of disease state, driven by chronic IFN-signalling, on haematopoietic stem and progenitor cells.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our work has implications for our understanding of immune system ageing in individuals with autoimmune diseases.
Introduction
Systemic lupus erythematosus (SLE) is a multisystem autoimmune disease, which presents with an incompletely understood sexual dimorphism. Females represent ~90% of cases and SLE is a leading cause of death in females aged under 34 years of age.1 Whereas hormonal factors were initially thought to explain the sex bias, attention has recently focused on the sex chromosomes as contributory factors.2 Strong epidemiological evidence supports X chromosome dosage as a substantial risk factor: males with Klinefelter’s syndrome (47, XXY) have a 14-fold increased prevalence of SLE compared with 46, XY males3; females with Turner’s syndrome (45, XO) have a lower risk, and 47, XXX females have a higher risk, compared with 46, XX females.4 5
There is complexity when interpreting the role of the X chromosome in disease. In mammals, X chromosome inactivation (XCI) ensures that only one X chromosome is active within each cell—any additional Xs are transcriptionally shut down, resulting in a functionally inactive chromosome (Xi). This process evolved to equalise the gene expression between 46, XX females and 46, XY males.6 In humans, during development the choice of which X is silenced in each cell is random and the Xi status is then clonally inherited by any daughter cells. Therefore, despite the karyotypic differences in sex chromosomes, every mammalian cell has only one active X chromosome.
Though XCI is expected to result in a random pattern of mosaicism across tissues, a great deal of variation has been observed across humans, with some females displaying a significantly unbalanced ratio, termed XCI-skew.7,9 The prevalence of XCI-skew in blood increases with age and represents a common age-acquired phenotype in females: one-third of females over 60 years have an XCI ratio in blood of 80:20 or greater.10 Age-acquired XCI-skew in blood is hypothesised to arise from long-term changes to the underlying haematopoietic stem and progenitor cells (HSPCs), such as stem cell exhaustion or clonal expansion, and is not thought to reflect short-term fluctuations in HSPC activity.11 12 Further, it has been hypothesised that XCI-skew of immune cells could play a causal role in the development of autoimmune disease.13 There is some evidence that the prevalence of XCI-skew in blood cells is increased in autoimmunity, including autoimmune thyroid disease,14,16 rheumatoid arthritis16 17 and systemic sclerosis.18 19 However, this is not consistent across autoimmune conditions.13 20 21
Conversely, autoimmune disease progression could also influence XCI-skew in blood cells. Given the selection of the Xi is stable across cell divisions, the XCI ratio of the peripheral immune cells must reflect that of the HSPCs from which they derive. HSPCs can be directly influenced by cytokine signalling,22 23 including interferon (IFN)-α which is a key cytokine in the pathogenesis of SLE.24 In a mouse model of SLE, inflammation resulted in significantly expanded HSPCs with increased self-renewal capacity.25 In humans, the dysfunction of immune cells in SLE can be traced back to HSPCs, where CD34+ HSPCs from SLE patients with severe disease showed enhanced proliferation and cell differentiation, together with a distinct gene expression signature.26 The impact of the cytokine environment of SLE on HSPCs could, therefore, be reflected in the XCI measures of the HSPC-derived immune cells.
Despite SLE having one of the most marked female sex predilections across all autoimmune conditions, and disease pathogenesis being driven by HSPC-derived immune cells, the role of XCI-skew of blood cells in SLE has yet to be established. We assayed XCI from whole blood in 181 female patients with SLE and 796 female controls from the TwinsUK population cohort, and combined clinical, cellular and genetic data to robustly investigate XCI in SLE.
Methods
SLE cohort
Archival DNA samples derived from whole blood (collected 2013–2019) were collected from 260 female patients with SLE at Guy’s and St Thomas’ Hospital, Birmingham Hospital, and Maidstone Hospital, and assayed for XCI. This resulted in 181 informative samples from unrelated individuals, with a median age of 50 years (table 1). All volunteers met the 1997 American College of Rheumatology criteria for SLE.27
Table 1. Descriptive of SLE cases and control cohorts.
| Controls | SLE cases | |
| Sample size | 796 | 181 |
| Age (median (range)) | 59.5 (20–74) | 50 (18–79) |
| Years of disease (median (range)) | – | 12 (1–47) |
| Age at diagnosis (median (range)) | – | 32 (5–75) |
| Hydroxychloroquine (n (%)) | – | 112 (61.9) |
| Methotrexate (n (%)) | – | 25 (13.8) |
| Biologics (n (%)) | – | 11 (9.1) |
| Azathioprine or mycophenolate (n (%)) | – | 42 (23.2) |
| With renal disease (n (%)) | – | 36 (20.4) |
| With genotype data (n (%)) | 750 (94.2) | 149 (82.3) |
| With full blood count data (n (%)) | 436 (54.8) | 42 (23.2) |
| XCI-skewing (mean) | 0.20 | 0.14 |
| XCI-skewing>80 (n (%)) | 175 (22.0) | 12 (6.6) |
SLEsystemic lupus erythematosusXCIX chromosome inactivation
Twins UK cohort
Archival DNA samples derived from whole blood (collected 1997–2017) were selected from individuals of the TwinsUK population cohort.28 2382 samples were assayed for XCI, which resulted in 1575 informative samples, as described previously.8 Individuals with self-reported SLE, as well as their co-twins or self-reported prior treatment with immunosuppressive medication, were excluded. Next, one individual from each twin pair was selected at random resulting in a cohort of 796 unrelated individuals, with a median age of 59.5 (table 1). During this sample selection process, we identified 10 pairs of SLE-discordant twins which were used in a follow-up twin analysis (see below).
The human androgen receptor assay
The human androgen receptor assay (HUMARA) method is a robust assay used extensively to measure XCI, which combines methylation-sensitive restriction enzyme digest and amplification of a highly polymorphic (CAG)n repeat in the first exon of the X-linked AR gene.29 The method used was exactly as described previously,8 using 625 ng of genomic DNA and processed on an ABI 3730xl. using the GeneScan 500 LIZ size standard.
Calculation of XCI
Data from the fragment analysis were analysed using the Microsatellite Analysis Software available on the ThermoFisher Cloud. The XCI status was calculated in each of the triplicates as follows:
Allele Ratio Mock Digestion (Rm)=allele 1 peak height/allele 2 peak height.
Allele Ratio HpaII Digestion (Rh)=allele 1 peak height/allele 2 peak height.
Normalised ratio (Rn)=Rh/Rm.
XCI percentage=[Rn/(Rn+1)]×100.
The SD and mean across the triplicates were used to calculate a coefficient of variation (CV) and samples with CV>0.15 were excluded from downstream analysis. The mean XCI percentage (0%–100%) was calculated for each sample, where 50% is a perfectly balanced XCI. The directionality of XCI away from 50% is uninformative (eg, both 0% and 100% are considered equal). Therefore, the XCI values are collapsed to a range of 50%–100% to create a continuous variable termed XCI-skew.
Whole blood count data
Whole blood count data obtained from standard Coulter-based clinical testing were date-matched to the XCI DNA sample, consisting of counts for white cell count (WCC), monocytes, lymphocytes and neutrophils. The proportion of lymphocytes was calculated by dividing the lymphocyte count by WCC, and monocyte-to-lymphocyte ratio and neutrophil-to-lymphocyte ratio were calculated by dividing the monocyte or neutrophil count, respectively, by lymphocyte count.
Medication use
Questionnaire data were used to match current medication use to the date of the blood sample for SLE patients. For each of hydroxychloroquine, methotrexate, biologics, azathioprine/mycophenolate, a categorical variable was created where healthy controls were coded as 0, SLE cases without medication use as 1, and SLE cases being treated with the medication as 2.
Renal disease
Questionnaire data were used to assess the history of renal disease (by ACR criteria) in SLE patients. A categorical variable was created where healthy controls were coded as 0, SLE cases without renal disease as 1 and SLE cases with renal disease as 2.
SLE polygenic score
A polygenic score (PGS) which captures SLE genetic susceptibility comprising 133 autosomal SNPs (MAF>1%) was used. The PGS model assumes an additive contribution of all SNPs, weighted by their effect sizes. However, skewed XCI will affect this additive assumption for X-linked SNPs, therefore, X-linked SNPs were excluded. Plink2 was used to calculate the SLE-PGS using genome-wide genotype data using the King’s College London CREATE system.30 94.2% and 82.3% of the TwinsUK controls and SLE samples had available genotype data, respectively (table 1). Samples were excluded if they were >3 s.d. away from the mean of heterozygosity across all SNPs.
Soluble SIGLEC-1 data
Soluble SIGLEC-1 (sSIGLEC-1) concentrations were measured using a non-isotopic time-resolved fluorescence assay based on the dissociation-enhanced lanthanide fluorescent immunoassay technology (PerkinElmer) in plasma samples from 304 SLE cases, as previously described.31 sSIGLEC-1 was measured in duplicate and individuals with a CV>0.3 were removed, together with individuals of non-European ancestry, resulting in a dataset of n=299. Patients were divided into groups based on sSIGLEC-1 serum level centiles (<50 th centile, 51st–74th centile, 75th–95th centile and >95 th centile). Of these, 41 individuals had matched XCI data and were used in the analyses.
Discordant twins
Questionnaire data were used to identify female twin pairs discordant for SLE (n=10 pairs; DZ=6; MZ=4) based on self-reported doctor’s diagnoses. DNA samples from twin pairs were date matched, and therefore, the XCI measures were perfectly matched for age.
Statistical analysis
For all linear and logistic regression models, XCI-skew was used as the dependent variable and age was included as a covariate. Results were quantified with effect sizes or ORs and 95% CIs. To assess the effects of each of the blood count variables on the disease associations in turn, linear regression models were constructed with XCI-skew as the dependent variable and the cell count as an independent variable (model 1). Residuals from model 1 were used as the dependent variable in a second model with SLE status as an independent variable and age as a covariate. For the sSIGLEC-1 analysis, an additional linear regression model was used with age as an interaction term. For discordant twin analyses, XCI-skew was compared using a one-sided paired sample Wilcoxon test. A p<0.05 was considered significant unless otherwise stated due to multiple testing correction using Bonferroni correction. All analyses were carried out using R V.4.1.1.
Results
XCI-skew is reduced in female SLE cases compared with female healthy controls
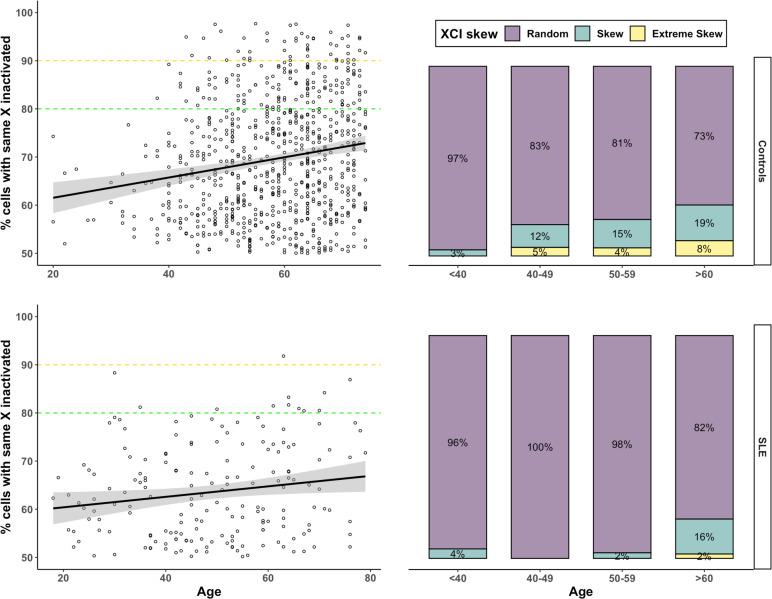
We quantified the degree of XCI-skew from 0 (representing 50:50 ratio) to 0.5 (representing a 100:0 ratio) across 181 SLE cases and 796 healthy controls from the TwinsUK population cohort (table 1). We found XCI-skew was positively correlated with age in the SLE cohort (p=0.027, r=0.14; figure 1), as previously described in healthy cohorts.8 10 32
Figure 1. XCI-skewing in SLE and controls The correlation between XCI (y-axis) and age (x-axis) is shown in panels on the left and the proportions of individuals (y-axis) with random (50%–79%), skewed (80%–89%) and extremely skewed (>90%) XCI across increasing age groups (x-axis) are shown in panels on the right. Controls (n=796) are in the upper panels and SLE cases (n=181) are in the lower panels. SLE, systemic lupus erythematosus; XCI, X chromosome inactivation.
However, using a linear regression model with the degree of XCI-skew as the dependent variable, and controlling for age as a covariate, we observed SLE status to be significantly and inversely correlated with XCI-skew (beta=−0.044; p=1.33×10−5). To ensure the case–control differences in XCI were not driven by the differences in the age distribution between the cohorts (table 1), we stratified the samples into four age groups, defined as under 40 years of age (yrs), 40–49 years, 50–59 years and over 60 years and applied the same linear regression model within each group (table 2). SLE status was significantly associated with reduced XCI-skew in 40–49 years (p=9.4×10−4) and 50–59 years (p=9.8×10−4), after Bonferroni correction (alpha=0.0125), and nominally significant in the over 60 years group (p=0.034; online supplemental figure S1). We saw no association in the under 40s group (p=0.61), which may reflect the low frequency of age-associated XCI-skew within this age group in both cases and controls.
Table 2. Case–control analysis within age groups.
| Controls (n (%)) | SLE cases (n (%)) | P value | Beta | |
| All samples | 796 (100) | 181 (100) | 1.33×10–5 | −0.044 |
| 40 and under | 31 (3.9) | 47 (26.0) | 0.61 | 0.010 |
| 40s | 133 (16.7) | 43 (23.8) | 0.00094* | −0.065 |
| 50s | 234 (29.4) | 40 (22.1) | 0.00098* | −0.064 |
| 60 and over | 398 (50.0) | 51 (28.2) | 0.034† | −0.040 |
nominally significantsignificant after Bonferroni correction.
significant after Bonferroni correction nominally significant.
SLEsystemic lupus erythematosus
We also defined XCI-skew (XCI≥80) and extreme XCI-skew (XCI≥90) as binary variables and used logistic regression models to assess their relationship with SLE. We confirmed that SLE status was associated with reduced odds of both XCI-skew, p=0.001; OR=0.90 (0.84–0.96) and extreme XCI-skew, p=0.024; OR=0.96 (0.92–0.99). In the SLE cohort, 6.6% have XCI-skew, and just one individual, equivalent to 0.55%, had extreme skew. A marked contrast with 22% and 6% of control samples, respectively, in these groups (figure 1; table 1).
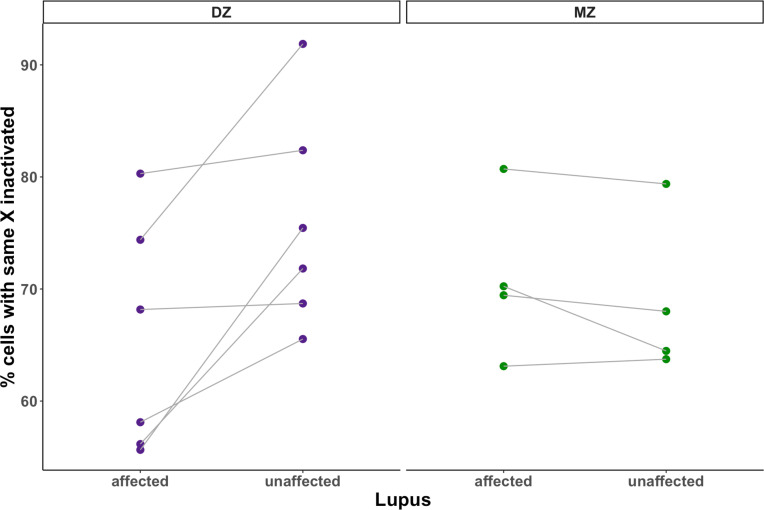
Replication using an intra-twin model of SLE discordant twins
Using twin pairs discordant for SLE, we next assessed whether XCI-skew was reduced in the affected twin compared with their unaffected cotwin and found a nominally significant association (p=0.080). Analysing the dizygotic (DZ) and monozygotic (MZ) twins separately, we observed the association was driven by differences between the discordant DZ twins (p=0.016, n=6; figure 2), whereas no effect was seen between the discordant MZ twins (p=0.94, n=4, figure 2), suggesting potential confounding genetic factors.
Figure 2. XCI-skewing in a discordant twin study using age-matched twin pairs discordant for SLE (Npairs =10), disease status is associated with decreased XCI skewing in the intratwin analysis of DZ twins (one-sided paired samples Wilcoxon test; p=0.016) but not MZ twins (one-sided paired samples Wilcoxon test; p=0.94). DZ, dizygotic; MZ, monozygotic; SLE, systemic lupus erythematosus; XCI, X chromosome inactivation.
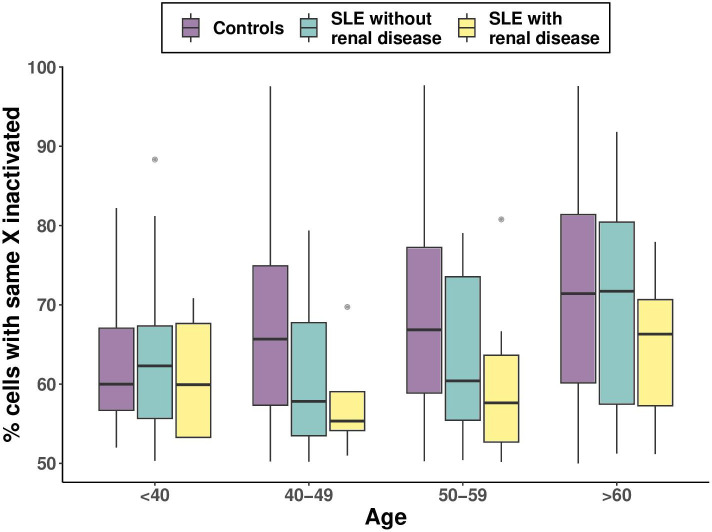
SLE severity further reduces XCI-skew
Given the association between SLE status and reduced XCI-skew, we hypothesised that a stronger effect would be observed in SLE cases with more severe disease, approximated by the presence of renal disease, which is associated with higher mortality and morbidity. To test this, we stratified the SLE cases based on a history of renal disease and compared each group (renal +ve and renal −ve) to the healthy controls. We observed a greater effect size on XCI-skew in those with renal disease (n=37; beta=−0.072; p=2.4×10−4) compared with those without renal disease (n=144; beta=−0.036; p=1.4×10−3; figure 3). Next, we compared which model was a better fit for the data. The first model included a binary variable which captured SLE status (controls/cases). The second model included a categorical variable which stratified the SLE cases by renal disease status (controls/renal −ve cases/renal +ve cases). We found the second model was nominally a better fit for the data (p=0.092). We also observed a nominally significant association between renal disease and reduced XCI-skew within the SLE samples only (beta=−0.031; p=0.091). Though the SLE patients with renal disease had a significantly higher mean age (56.7 years) compared with those without renal disease (47.3 years; Welch two sample t-test: p=6.5×10−4), only 1 individual (2.7%) with renal disease displayed a skewed XCI pattern (XCI≥80) compared with 11 individuals (7.6%) without renal disease (online supplemental figure S2).
Figure 3. The effect of disease severity on XCI-skewing boxplots representing the association of renal disease as a marker of SLE severity on XCI skewing. All boxplots display the median and IQR, with XCI-skewing on the y-axis and age category on the x axis. SLE, systemic lupus erythematosus; XCI, X chromosome inactivation.
Medication use does not explain the case–control differences in XCI
We hypothesised that disease treatment might be driving the differences between cases and controls. Using the same approach as carried out for renal disease, we stratified the SLE patients based on use of hydroxychloroquine, methotrexate, azathioprine/mycophenolate and biologics and observed no difference in effect size between groups compared with controls (online supplemental figure S3; online supplemental table 1), suggesting medication use does not explain the difference in XCI between cases and controls. We also saw no significant effect of mediation use on XCI-skew within the SLE samples only (online supplemental table 1).
Immune cell type composition does not explain the case–control differences in XCI
SLE manifests with significant changes to the abundance of cell populations in the peripheral blood and we postulated that cell composition could explain differences in XCI between cases and controls. As expected, we observed stark differences in full blood count data between cases and controls, with lower levels of lymphocytes and higher levels of monocytes and neutrophils in the SLE patients (online supplemental table 2). Given the strong correlation between SLE status and immune cell counts, it was not possible to add cell counts as covariates directly to the model. Instead, for each cell type in turn (n=7), we first regressed out their effects on XCI-skew (see the ‘Methods’ section) and found SLE status was still significantly associated with the residuals of XCI-skew following the removal of the effects of each cell type (online supplemental table 3). Of note, controlling for monocyte count, which we have previously reported to be positively associated with XCI-skew in a population cohort,8 augmented the effect of SLE status on XCI-skew (online supplemental figure S4). Therefore, the differences in cell proportions between cases and controls do not explain the XCI association.
Genetic susceptibility to SLE is not associated with XCI-skew
Genome-wide association studies (GWASs) have identified many common genetic variants which increase the risk of developing SLE and the additive effect of the disease-associated variants can be captured using a PGS.33 We calculated the SLE-PGS across the cases and controls with available date (n=899) and demonstrated it is significantly associated with SLE status (online supplemental figure S5) p=0.001, OR=1.35 (95% CI 1.13 to 1.62)). However, we see no association between the SLE-PGS and XCI-skew (p=0.37, beta=0.0036), suggesting the inherited genetic susceptibility to SLE so far identified through GWAS does not influence XCI (online supplemental figure S6).
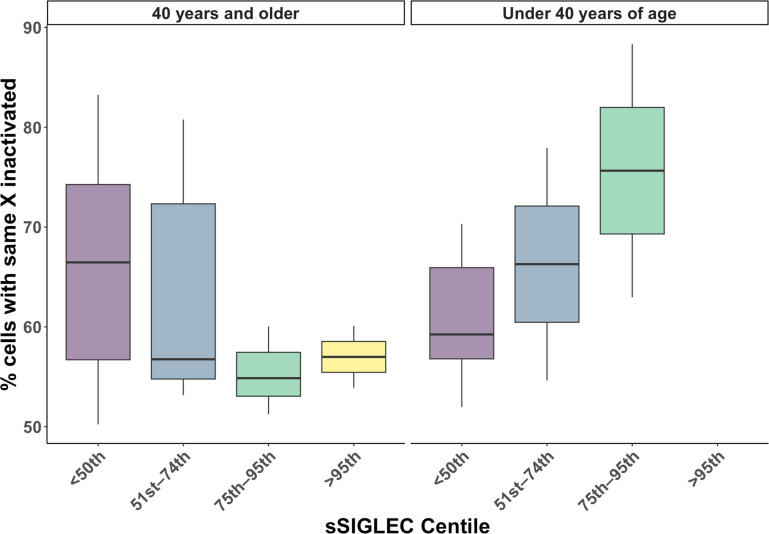
Interferon signalling is associated with reduced XCI-skew in an age-dependent manner
We postulated that the effects of chronic interferon (IFN) signalling, a key hallmark of lupus pathology, could be the mechanism underpinning the disease effects on XCI-skew. We tested this hypothesis using measures of soluble SIGLEC-1 (sSIGLEC-1), a plasma biomarker for type I interferon-regulated gene expression.31 We observed a non-linear relationship between age and sSIGLEC-1 (online supplemental figure S7), and therefore, assessed the association between sSIGLEC-1 and XCI-skew using age as an interaction term (XCI-skew−sSIGLEC-1×age). We found this model was a better fit for the data (p=0.004) compared with the model with age as a covariate (XCI-skew−sSIGLEC-1+age). We observed an age-dependent association between sSIGLEC-1 and XCI-skew (n=41; p=0.009), with a significant interaction with age (p=0.004; figure 4).
Figure 4. The age-dependent effects of IFN-signalling on XCI-skewing boxplots representing the association of non-linear sSIGLEC-1 percentile groups on XCI-skewing. All boxplots display the median and IQR, with XCI-skewing on the y-axis, and percentile groups on the x-axis. XCI, X chromosome inactivation.
Discussion
The prevalence of XCI-skew in HSPC-derived immune cells increases with age and represents a common age-acquired phenotype in females.10 32 It has been hypothesised that XCI-skew of immune cells may play a causal role in the development and sex biases of autoimmune disease, with inadequate thymic deletion being the proposed mechanistic driver.13 Here, to the best of our knowledge, we present the largest study of XCI in SLE to date and demonstrate reduced XCI-skew in SLE cases compared with healthy controls, thus refuting the hypothesis that XCI-skew contributes to the sex bias of SLE.13
Instead, our results, which demonstrate that disease severity impacts XCI-skew, whereas differential WCC, medication or genetic susceptibility to SLE do not, suggest that the disease state itself is affecting XCI-skew. Further, we demonstrated an age-dependent association between XCI-skew and sSIGLEC-1, a plasma biomarker for type I interferon (IFN-I)-regulated gene expression, a key hallmark of lupus pathogenesis. We propose a mechanism in which the persistent IFN-I signature common in SLE impacts the HSPCs, which results in the XCI-skew differences compared with a control population. Both stem cell exhaustion and clonal expansion have been hypothesised as mechanisms underlying the XCI-skew signature commonly observed in ageing females.10 Though the role of interferons on the homeostasis of HSPCs is complex,23 24 34 the IFN-I signature in SLE has been shown not to cause stem cell exhaustion.35 36 Indeed, mice with active lupus have been shown to have HSCs with enhanced self-renewal capacity.25 Such an effect in SLE patients, driven by chronic IFN-I stimulation, could prevent stem cell exhaustion and maintain balanced XCI ratios in peripheral blood cells. These results reveal important insights into the ageing immune system in individuals with lupus and warrant follow-up studies across other autoimmune diseases and interferonopathies.
The age-dependent effects of sSIGLEC-1 on XCI-skew raise important considerations, which warrant further investigation. Specifically, older females are more likely to have had lupus for longer, and therefore, the effects of chronic IFN signalling could be more pronounced. Notably, it is at older ages that we expect to see a higher prevalence of XCI-skewing, and indeed we observed no difference in XCI-skew between cases and controls in the under 40s, therefore, this also could be an issue of power. It is important to note we have very limited numbers of younger patients with higher sSIGLEC scores.
Our work contrasts with earlier studies which also measured XCI using the HUMARA method and reported increased XCI-skew in autoimmunity, including rheumatoid arthritis,16 17 systemic sclerosis18 19 and autoimmune thyroid disease.14,16 Given these discrepant findings across autoimmune disease, it will be of great interest to establish whether XCI points to mechanistic differences in the development or effect of specific diseases, or indeed whether analytical methods underpin the different results. With this latter point in mind, it is of paramount importance that age is fully controlled for in case–control analyses due to the strong effect of ageing on XCI. Of note, some studies which report case–control differences in XCI did not observe the expected positive correlation between XCI-skew and age in the healthy control samples.15 16 19 In addition to controlling for age in every regression model, we ensured that our findings were not spuriously driven by differences in the age distribution of the cases and controls in two important ways. First, we validated the association within age strata (<40 years, 40–49 years, 50–59 years and ≥60 years). Second, we replicated our study using a small, independent cohort of SLE-discordant twin pairs who are perfectly matched for age.
As well as replicating our findings, the discordant twin study revealed an intriguing result: the discordant DZ twins had significant differences in XCI, whereas the MZ twins did not. This difference in DZ and MZ twins is in line with two previous studies. First, a small study in MZ twins discordant for SLE found no difference in XCI skew.37 Second, a twin study of XCI and serum levels of autoantibodies to thyroid peroxidase, a measure of subclinical thyroid disease, found differences in XCI between DZ twin pairs but not MZ twin pairs.38 The authors hypothesised that the findings could suggest XCI is not a causative factor in levels of autoantibodies to thyroid peroxidase, but instead that the two phenotypes could be driven by the same genetic confounders.38 Likewise, our findings suggest that XCI and SLE may be affected by the same underlying genetic factors. However, we observe no effect of the PGS for SLE on XCI. Of note, a limitation of PGS, and indeed GWAS more broadly, is that they typically only capture the effects of common single nucleotide variants. It is plausible that other sources of genetic variation, such as copy number variation or rare variants, could underpin unexplained genetic effects.
XCI is initiated by the long non-coding RNA XIST which acts in cis to recruit protein complexes and epigenetic changes to silence the inactivated X (Xi). Functional studies have demonstrated a dynamic role of XIST during the development and maturation of T and B cells, where XIST is no longer localised to the Xi in naïve cells.39 40 Further, this dynamic role of XIST has been demonstrated to be disrupted in both a mouse model of SLE and human SLE patients, suggestive of impaired transcriptional regulation of the X chromosome in lymphocytes as a feature of SLE.39 41 Crucially, DNA methylation, which is required for the epigenetic memory of the Xi to be maintained throughout cell divisions, is not thought to be disrupted during loss of XIST RNA localisation.40 42 The HUMARA assay is dependent on the methylation of the AR locus on the Xi, and therefore, would not be impacted by the dysregulation of XIST in SLE.
Our study does have limitations. The discordant twin study was of limited sample size. However, such discordant twin models are powerful. For some phenotypes, notably whole blood count data and the sSIGLEC-1 data, we had a high percentage of samples with missing data. This prevented us from carrying out further analyses, such as controlling for the effects of cell composition when testing differences between those with and without renal disease. Likewise, we lacked sufficient data to assess molecular and cellular markers of disease activity, which may have informed our interpretation. Of note, work by our group has previously reported no correlation between C reactive protein and XCI-skew.8
In summary, we have demonstrated that XCI-skew is reduced in SLE cases compared with healthy controls drawn from a population cohort. We postulate that chronic IFN-I signalling impacts HSPCs, and this is reflected in the XCI patterns of HSPC-derived immune cells. Further work is needed to confirm this mechanism, which could reveal important insights into the ageing immune system in individuals with autoimmunity.
supplementary material
Acknowledgements
We thank Prof. Frances M. K. Williams for her constructive feedback on the manuscript. We also thank all volunteers who participated in the study.
The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Footnotes
Funding: KSS acknowledges funding from the Medical Research Council (MR/M004422/1 and MR/R023131/1). CCYW acknowledges funding from the NIHR Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. This paper represents independent research (part) funded by the NIHR Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, Chronic Disease Research Foundation (CDRF), Zoe Globald and the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London.
Provenance and peer review: Not commissioned; externally peer reviewed.
Handling editor: Josef S Smolen
Patient consent for publication: Not applicable.
Ethics approval: This study involves human participants and was approved by London Brent Research Ethics Committee 12/LO/1273. TwinsUK has received ethical approval associated with TwinsUK Biobank (19/NW/0187), TwinsUK (EC04/015) or Healthy Ageing Twin Study (HATS) (07 /H0802/84) studies from NHS Research Ethics Service Committees London—Westminster. Participants gave informed consent to participate in the study before taking part.
Data availability free text: All data relating to SLE samples are available on reasonable request, with the exception of clinical phenotype data. All data relating to TwinsUK samples have been deposited to the TwinsUK BioResource data management team and are available by application to the Twin Research Executive Access committee (TREC) at King’s College London. The TwinsUK BioResource is managed by TREC, which provides governance of access to TwinsUK data and samples. TwinsUK data users are bound by data sharing agreement set out in the data access application form (https://twinsuk.ac.uk/wpcontent/uploads/2018/11/DTR_DataAccess_Policy_0318.pdf). This includes responsibilities with respect to third party data sharing and maintaining participant privacy. Further responsibilities include a responsibility to acknowledge data sharing.
Patient and public involvement: Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Contributor Information
Amy L Roberts, Email: amy.roberts@kcl.ac.uk.
Alessandro Morea, Email: alessandro.morea@ifom.eu.
Ariella Amar, Email: ariella.amar@kcl.ac.uk.
Magdalena West, Email: magdalena.west@kcl.ac.uk.
Sarah Karrar, Email: sarah.karrar@kcl.ac.uk.
Rhiannon Lehane, Email: rhilehane@gmail.com.
Philip Tombleson, Email: philip.tombleson@kcl.ac.uk.
Deborah Cunningham Grahman, Email: deborah.cunninghame-graham@kcl.ac.uk.
John A Reynolds, Email: john.reynolds5@nhs.net.
Chloe C Y Wong, Email: chloe.wong@kcl.ac.uk.
David L Morris, Email: david.l.morris@kcl.ac.uk.
Kerrin S Small, Email: kerrin.small@kcl.ac.uk.
Timothy J Vyse, Email: timothy.vyse@kcl.ac.uk.
Data availability statement
Data are available on reasonable request.
References
- 1.Yen EY, Singh RR. Brief report: lupus—an unrecognized leading cause of death in young females: A population-based study using nationwide death certificates, 2000–2015. Arthritis Rheumatol . 2018;70:1251–5. doi: 10.1002/art.40512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libert C, Dejager L, Pinheiro I. The X Chromosome in immune functions: when a Chromosome makes the difference. Nat Rev Immunol. 2010;10:594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 3.Scofield RH, Bruner GR, Namjou B, et al. Klinefelter’s syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X Chromosome. Arthritis Rheum. 2008;58:2511–7. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooney CM, Bruner GR, Aberle T, et al. 46,X,Del(X)(Q13) Turner’s syndrome women with systemic lupus erythematosus in a pedigree Multiplex for SLE. Genes Immun. 2009;10:478–81. doi: 10.1038/gene.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu K, Kurien BT, Zimmerman SL, et al. X Chromosome dose and sex bias in autoimmune diseases: increased 47,XXX in systemic lupus erythematosus and Sjögren’s syndrome. Arthritis & Rheumatology. 2016;68:1290–300. doi: 10.1002/art.39560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LYON MF. Gene action in the X-Chromosome of the Mouse. Nature. 1961;190:372–3. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 7.Bolduc V, Chagnon P, Provost S, et al. No evidence that Skewing of X Chromosome inactivation patterns is transmitted to offspring in humans. J Clin Invest. 2008;118:333–41. doi: 10.1172/JCI33166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts AL, Morea A, Amar A, et al. Age acquired SKEWED X Chromosome inactivation is associated with adverse health outcomes in humans. Elife. 2022;11:e78263. doi: 10.7554/eLife.78263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amos-Landgraf JM, Cottle A, Plenge RM, et al. X Chromosome–inactivation patterns of 1,005 Phenotypically unaffected females. Am J Hum Genet. 2006;79:493–9. doi: 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busque L, Mio R, Mattioli J, et al. Nonrandom X-inactivation patterns in normal females: Lyonization ratios vary with age. Blood. 1996;88:59–65. [PubMed] [Google Scholar]
- 11.Tonon L, Bergamaschi G, Dellavecchia C, et al. Unbalanced X-Chromosome inactivation in Haemopoietic cells from normal women. Br J Haematol. 1998;102:996–1003. doi: 10.1046/j.1365-2141.1998.00867.x. [DOI] [PubMed] [Google Scholar]
- 12.van Dijk JP, Heuver L, Stevens-Linders E, et al. Acquired Skewing of Lyonization remains stable for a prolonged period in healthy blood donors. Leukemia. 2002;16:362–7. doi: 10.1038/sj.leu.2402379. [DOI] [PubMed] [Google Scholar]
- 13.Chitnis S, Monteiro J, Glass D, et al. The role of X-Chromosome inactivation in female predisposition to Autoimmunity. Arthritis Res Ther . 2 doi: 10.1186/ar118. n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmonds MJ, Kavvoura FK, Brand OJ, et al. Skewed X Chromosome inactivation and female preponderance in autoimmune thyroid disease: an association study and meta-analysis. J Clin Endocrinol Metab. 2014;99:E127–31. doi: 10.1210/jc.2013-2667. [DOI] [PubMed] [Google Scholar]
- 15.Yin X, Latif R, Tomer Y, et al. Thyroid epigenetics: X chromosome inactivation in patients with autoimmune thyroid disease. Ann N Y Acad Sci. 2007;1110:193–200. doi: 10.1196/annals.1423.021. [DOI] [PubMed] [Google Scholar]
- 16.Chabchoub G, Uz E, Maalej A, et al. Analysis of SKEWED X-Chromosome inactivation in females with rheumatoid arthritis and autoimmune thyroid diseases. Arthritis Res Ther. 2009;11:R106. doi: 10.1186/ar2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zito A, Davies MN, Tsai P-C, et al. Heritability of SKEWED X-inactivation in female twins is tissue-specific and associated with age. Nat Commun. 2019;10:5339. doi: 10.1038/s41467-019-13340-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozbalkan Z, Bagişlar S, Kiraz S, et al. Skewed X Chromosome inactivation in blood cells of women with scleroderma. Arthritis Rheum. 2005;52:1564–70. doi: 10.1002/art.21026. [DOI] [PubMed] [Google Scholar]
- 19.Kanaan SB, Onat OE, Balandraud N, et al. Evaluation of X Chromosome inactivation with respect to HLA genetic susceptibility in rheumatoid arthritis and systemic sclerosis. PLoS One. 2016;11:e0158550. doi: 10.1371/journal.pone.0158550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brix TH, Hansen PS, Bennedbak FN, et al. X Chromosome inactivation pattern is not associated with Interindividual variations in thyroid volume: A study of euthyroid Danish female twins. Twin Res Hum Genet. 2009;12:502–6. doi: 10.1375/twin.12.5.502. [DOI] [PubMed] [Google Scholar]
- 21.Brix TH, Hansen PS, Kyvik KO, et al. The pituitary-thyroid axis set point in women is uninfluenced by X Chromosome inactivation pattern? A twin study. Clin Endocrinol (Oxf) 2010;73:666–70. doi: 10.1111/j.1365-2265.2010.03848.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang CC, Lodish HF. Cytokines regulating hematopoietic stem cell function. Curr Opin Hematol. 2008;15:307–11. doi: 10.1097/MOH.0b013e3283007db5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King KY, Goodell MA. Inflammatory modulation of Hscs: viewing the HSC as a foundation for the immune response. Nat Rev Immunol. 2011;11:685–92. doi: 10.1038/nri3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Essers MAG, Offner S, Blanco-Bose WE, et al. IFNα activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–8. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 25.Niu H, Fang G, Tang Y, et al. The function of hematopoietic stem cells is altered by both genetic and inflammatory factors in lupus mice. Blood. 2013;121:1986–94. doi: 10.1182/blood-2012-05-433755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grigoriou M, Banos A, Filia A, et al. Transcriptome Reprogramming and myeloid Skewing in haematopoietic stem and progenitor cells in systemic lupus erythematosus. Ann Rheum Dis . 2020;79:242–53. doi: 10.1136/annrheumdis-2019-215782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arthritis rheumatism - 2005 - Hochberg - updating the American college of rheumatology revised criteria for the. doi: 10.1002/art.1780400928. [DOI] [PubMed]
- 28.Verdi S, Abbasian G, Bowyer RCE, et al. Twinsuk: the UK adult twin Registry update. Twin Res Hum Genet. 2019;22:523–9. doi: 10.1017/thg.2019.65. [DOI] [PubMed] [Google Scholar]
- 29.Cutler Allen R, Zoghbi HY, Annemarie Moseley IB, et al. Methylation of Hpall and Hhal sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X Chromosome inactivation. Am J Hum Genet. 1992;51:1229–39. [PMC free article] [PubMed] [Google Scholar]
- 30.King’s College London . King’s Computational Research, Engineering and Technology Environment (CREATE) 2022. [Google Scholar]
- 31.Oliveira JJ, Karrar S, Rainbow DB, et al. The plasma biomarker soluble SIGLEC-1 is associated with the type I interferon transcriptional signature, ethnic background and renal disease in systemic lupus erythematosus. Arthritis Res Ther. 2018;20:152. doi: 10.1186/s13075-018-1649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mengel-From J, Thinggaard M, Christiansen L, et al. Skewed X inactivation and survival: A 13-year follow-up study of elderly twins and Singletons. Eur J Hum Genet. 2012;20:361–4. doi: 10.1038/ejhg.2011.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Wang Y-F, Liu L, et al. Genome-wide assessment of genetic risk for systemic lupus erythematosus and disease severity. Hum Mol Genet. 2020;29:1745–56. doi: 10.1093/hmg/ddaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demerdash Y, Kain B, Essers MAG, et al. Yin and Yang: the dual effects of Interferons on Hematopoiesis. Exp Hematol. 2021;96:1–12. doi: 10.1016/j.exphem.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodero MP, Crow YJ. Type I Interferonâ-mediated Monogenic Autoinflammation: the type I Interferonopathies, a conceptual overview. J Exp Med. 2016;213:2527–38. doi: 10.1084/jem.20161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietras EM, Lakshminarasimhan R, Techner J-M, et al. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I Interferons. J Exp Med. 2014;211:245–62. doi: 10.1084/jem.20131043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Q, Parfitt A, Grennan DM, et al. X-Chromosome inactivation in Monozygotic twins with systemic lupus erythematosus. Autoimmunity. 1997;26:85–93. doi: 10.3109/08916939709003851. [DOI] [PubMed] [Google Scholar]
- 38.Brix TH, Hansen PS, Kyvik KO, et al. Preliminary evidence of a Noncausal association between the X-Chromosome inactivation pattern and thyroid Autoimmunity: A twin study. Eur J Hum Genet. 2010;18:254–7. doi: 10.1038/ejhg.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Syrett CM, Kramer MC, et al. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc Natl Acad Sci USA. 2016;113:E2029–38. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savarese F, Flahndorfer K, Jaenisch R, et al. Hematopoietic precursor cells transiently reestablish Permissiveness for X inactivation. Mol Cell Biol. 2006;26:7167–77. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Syrett CM, Paneru B, Sandoval-Heglund D, et al. Altered X-Chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight. 2019;4:e126751. doi: 10.1172/jci.insight.126751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syrett CM, Sindhava V, Hodawadekar S, et al. Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic Yy1-dependent two-step process in activated B cells. PLoS Genet. 2017;13:e1007050. doi: 10.1371/journal.pgen.1007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on reasonable request.