Abstract
Paramyxovirus fusion proteins have two heptad repeat domains, HR1 and HR2, which have been implicated in the fusion activity of the protein. Peptides with sequences from these two domains form a six-stranded coiled coil, with the HR1 sequences forming a central trimer (K. A. Baker, R. E. Dutch, R. A. Lamb, and T. S. Jardetzky, Mol. Cell 3:309–319, 1999; X. Zhao, M. Singh, V. N. Malashkevich, and P. S. Kim, Proc. Natl. Acad. Sci. USA 97:14172–14177, 2000). We have extended our previous mutational analysis of the HR1 domain of the Newcastle disease virus fusion protein, focusing on the role of the amino acids forming the hydrophobic core of the trimer, amino acids in the “a” and “d” positions of the helix from amino acids 123 to 182. Both conservative and nonconservative point mutations were characterized for their effects on synthesis, stability, proteolytic cleavage, and surface expression. Mutant proteins expressed on the cell surface were characterized for fusion activity by measuring syncytium formation, content mixing, and lipid mixing. We found that all mutations in the “a” position interfered with proteolytic cleavage and surface expression of the protein, implicating the HR1 domain in the folding of the F protein. However, mutation of five of seven “d” position residues had little or no effect on surface expression but, with one exception at residue 175, did interfere to various extents with the fusion activity of the protein. One of these “d” mutations, at position 154, interfered with proteolytic cleavage, while the rest of the mutants were cleaved normally. That most “d” position residues do affect fusion activity argues that a stable HR1 trimer is required for formation of the six-stranded coiled coil and, therefore, optimal fusion activity. That most of the “d” position mutations do not block folding suggests that formation of the core trimer may not be required for folding of the prefusion form of the protein. We also found that mutations within the fusion peptide, at residue 128, can interfere with folding of the protein, implicating this region in folding of the molecule. No characterized mutation enhanced fusion.
Entry of enveloped viruses into susceptible cells requires fusion of viral and cellular membranes (12). This fusion is mediated by viral fusion proteins, which have recognizable sequence elements important for this activity. One of these sequences, the fusion peptide, inserts into target or cellular membranes attaching to these membranes and disordering the cellular lipid bilayer (12). Viral fusion proteins also often have heptad repeat regions (7). Results of studies in several systems indicate that heptad repeat domains are involved in conformational changes in the protein that take place upon activation of fusion. These conformational changes are proposed, in part, to pull viral and cellular membranes in close proximity required for subsequent fusion events (2, 6, 23). The heptad repeat domains may also be directly involved in these subsequent steps (15, 26).
Membrane fusion mediated by paramyxoviruses, such as Newcastle disease virus (NDV), is mediated by the fusion protein (F) (reviewed in reference 18). This protein is synthesized as a precursor (F0) which is activated upon proteolytic cleavage to produce disulfide-linked F1 and F2 polypeptides (reviewed in reference 18). Cleavage, which places the fusion peptide or fusion sequence at the new amino terminus of F1 (12), also results in a conformational shift indicated by increased hydrophobicity (14). Immediately adjacent to the fusion peptide is a heptad repeat sequence, heptad repeat 1 (HR1) (7). Mutational analysis of both regions has shown that both the fusion peptide and adjacent heptad repeat play a role in the fusion activity of the protein (13, 31). That peptides with sequences from the HR1 domain interfere with fusion activity of the intact protein additionally suggests a role of this domain in fusion (15, 42)
The paramyxovirus F proteins have, adjacent to the transmembrane region, HR2 domains, which have also been implicated in the fusion activity. Mutations in these regions abolish fusion, and peptides with sequences from these domains inhibit fusion (4, 11, 19, 27, 28, 39–41). Furthermore, these HR2 peptides interact with peptides from HR1 domains (2, 20, 22, 42), forming a six-stranded structure with a central core trimer of HR1 peptides with three HR2 peptides bound to the trimer (2, 20, 43). It is proposed that both HR1 and HR2 peptides mimic their respective domains in the intact protein interfering with interactions of the HR1 and HR2 domains necessary for fusion to proceed (2, 15, 42). The correlate to this hypothesis is that the two domains do not complex prior to activation of fusion and are therefore accessible to peptide binding.
These considerations suggest that the F protein is synthesized in a prefusion conformation which changes upon activation of fusion. Fusion activation clearly requires cleavage of the molecule. However, additional shifts may occur with the actual onset of fusion, analogous to changes detected in retroviral envelope proteins upon attachment of the SU-TM complex to receptors (8, 10, 32, 38) or upon acid activation of the influenza hemagglutinin protein (5, 6). To define properties of the prefusion and postfusion conformations of the NDV F protein, we have extended our previous mutational analysis of the HR1 domain (31), focusing on residues that form the hydrophobic core of the HR1 trimer in the peptide complexes (2, 43). We found that some of these mutations interfere with the proper folding of the molecule, likely perturbing structures required in the formation of the prefusion form of the protein. However, other mutations minimally alter folding and surface expression but impact the fusion activity of the protein. That mutation of some core residues had no effect on folding suggests that the HR1 trimer may not form during folding of F0. We also found that mutations at residue 154 interfere with the cleavage of the molecule without perturbing intracellular transport and may alter the conformation of the cleavage site in the F0 protein.
MATERIALS AND METHODS
Cells, vectors, and viruses.
Cos-7 cells, obtained from the American Type Culture Collection, were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with nonessential amino acids, vitamins, penicillin and streptomycin, and 10% fetal calf serum.
NDV HN and F genes were expressed in Cos cells using pSVL (Pharmacia) as previously described (31).
Site-specific mutagenesis.
The F gene mutants were generated with a mutagenesis kit from Amersham Corporation using the appropriate oligomer for each mutation. Oligomers of 33 to 39 nucleotides were required to successfully isolate mutants of the fusion peptide and heptad repeat regions of the fusion protein gene. The entire gene of the mutant DNA was sequenced to verify that the rest of the gene remained unchanged by the mutagenesis reaction. The mutations isolated are shown in Fig. 1. Mutation names show the wild-type amino acid (in single-letter code), the position of the change, and the mutant amino acid.
FIG. 1.
Location of mutations. The sequence of the amino terminus of the F1 protein including both the fusion peptide and HR1 is shown. The overlapping dashed lines indicate that the boundary of the two domains is unclear. The positions of the hydrophobic residues in the heptad repeat of the HR1 domain are indicted by “a” and “d” above the sequence. Small arrows indicate the positions of mutations previously reported and partially characterized (31), and bold arrows indicate the new mutations described here. Upward and downward arrows indicate mutations that do not block surface expression and those that do block surface expression, respectively.
Transfections.
Transfections using Lipofectin or Lipofectamine (BRL/Gibco) were done as recommended by the manufacturer. Cos cells were plated at 3 × 105 per 35-mm plate and transfected 20 to 24 h later. For each 35-mm plate, a mix of DNA in 0.1 ml of OptiMem (BRL/Gibco) and 10 μl of transfection reagent in 0.2 ml of OptiMem was incubated at room temperature for 45 min and then diluted with 0.7 ml of OptiMem and added to a plate previously washed with OptiMem. Cells were incubated for 5 h, and then 2 ml of Cos cell medium was added.
Antibodies.
Antibodies used were anti-Ftail, anti-Fu1a, and anti-NDV. Anti-Ftail antibody was raised against a synthetic peptide with the sequence of the cytoplasmic tail of the fusion protein as described by Wang et al. (37) and prepared by the Peptide Core Facility of the University of Massachusetts Medical School. Anti-Fu1a is a monoclonal antibody obtained from Mark Peeples (30). Anti-NDV is a polyclonal antiserum raised in rabbits against UV-inactivated virions as previously described (30).
Immunofluorescence.
Cos cells were plated on 35-mm plates containing glass coverslips (Corning) and transfected as described above. The cells were washed twice with phosphate-buffered saline (PBS) and incubated at 4°C in PBS containing 3% bovine serum albumin (BSA), 0.02% sodium azide, and antibody (anti-NDV) for 1 h. Cells were washed three times with PBS containing BSA and azide and incubated with ice-cold PBS containing BSA, azide, and anti-rabbit immunoglobulin G (IgG) coupled to Alexa dye 488 (Molecular Probes) for 1 h. Cells were washed in ice-cold PBS containing azide and BSA. Pictures of the cells were taken immediately.
Flow cytometry.
Transfected cells were removed from plates with cell detachment buffer (Sigma Co.) after a 1-min pulse in trypsin (5 μg/ml), washed in PBS containing 1% BSA and 0.02% azide (fluorescence-activated cell sorting [FACS] buffer), and incubated with anti-NDV antibody for 1 h at 4°C. After three washes with FACS buffer, cells were incubated for 1 h at 4°C with goat anti-rabbit IgG coupled to Alexa dye 488. After three washes in FACS buffer, cells were resuspended in PBS containing 2% paraformaldehyde and subjected to flow cytometry (University of Massachusetts Medical School Flow Cytometry Facility). Cells transfected with vector alone and incubated with both primary and secondary antibody were used as controls.
Western analysis of mutant proteins.
Cell extracts were diluted in sample buffer and loaded onto 10% polyacrylamide gels without boiling. After electrophoresis, the gels were subsequently equilibrated in transfer buffer (25 mM Tris, 192 mM glycine, 5% methanol [pH 8.2]) and transferred to Immobilon-P (Millipore Corp.) membranes. The membrane was blocked in PBS containing 0.5% Tween 20 and 10% nonfat dried milk for 2 h at room temperature or overnight at 4°C. Membranes were washed in PBS-Tween 20 and incubated with primary antibody diluted in PBS-Tween 20 and 0.5% nonfat milk for 2 h at room temperature. Membranes were washed and then incubated in secondary antibody, anti-rabbit IgG coupled to horseradish peroxidase (Boehringer Mannheim) diluted in PBS-Tween and 0.5% nonfat milk, for 1 h at room temperature. Membranes were washed extensively, and bound antibody was detected using the ECL Western blotting detection reagent system (Amersham).
Radiolabeling and immunoprecipitation of protein.
Transfected cells were radiolabeled for 2 to 4 h at 37°C in DMEM lacking methionine but containing 100 μCi of [35S]methionine (Amersham) per ml. At the end of the labeling period, cells were washed in PBS and lysed in RSB buffer (0.01 M Tris-HCl [pH 7.4], 0.01 M NaCl) containing 1% Triton X-100, 0.5% sodium deoxycholate, 2 mg of iodoacetamide/ml, and 0.2 mg of DNase/ml as previously described (13, 14, 20). Immunoprecipitation of NDV proteins was accomplished as previously described (30).
Sucrose gradients.
At 48 h posttransfection, cells were lysed in RSB buffer, 1% Triton X-100, and 10 mM iodoacetamide and layered on top of 10-to-45% continuous sucrose gradients made in RSB buffer containing 0.1% Triton X-100. Gradients were spun in an SW41 rotor for 18 h at 38,000 rpm at 17°C and collected into 16 equal fractions. Proteins present were precipitated with trichloroacetic acid and resolved on polyacrylamide gels for Western analysis. Marker proteins used were ferritin (450 kDa), catalase (240 kDa), aldolase (158 kDa), and BSA (68 kDa) (Boehringer). The locations of the marker proteins in the gradients were determined by Coomassie blue staining of the gradient fractions.
Fusion assays. (i) Syncytium formation.
Cos cells were cotransfected with wild-type or mutant fusion protein genes and the wild-type HN protein gene using Lipofectin. At 24 and 48 h posttransfection, the numbers of nuclei in 40 fusion areas were counted to determine the average size of syncytia at each time point as previously described (30). Values obtained after transfection of the vector alone were subtracted.
(ii) Content mixing.
To measure content mixing, a plasmid encoding a tetracycline (TET)-responsive transcriptional activator, pTet-Off (Clontech), was transfected along with HN and F cDNAs. A separate population of cells was transfected with pTRE2 (Clontech) with a β-galactosidase gene inserted into the cloning cassette. This plasmid contains a TET-responsive element upstream from a cytomegalovirus promoter. After 30 h, Cos cells transfected with the β-galactosidase gene were trypsinized and then plated on Cos cells expressing HN and F protein as well as the TET-responsive transactivator. At 45 h posttransfection, when fusion was evident, the monolayers were lysed and extracts were assayed for β-galactosidase activity. Activity due to background fusion typical of Cos cells was measured after transfecting cells with comparable amounts of vector alone, and values obtained were subtracted from values obtained with cells expressing HN and wild-type F or mutant F proteins.
(iii) Lipid mixing.
The lipid mixing protocol used was similar to that previously described (16, 17). Avian red blood cells (RBC) (Crane Laboratories) were washed in PBS, resuspended in PBS, and incubated with 15 μg of R18 (Molecular Probes)/ml for 30 min at room temperature in the dark. Complete medium (3 volumes of DMEM with 10% fetal calf serum) was added, and incubation was continued for 30 min. The RBC were washed four times in ice-cold PBS, resuspended in PBS containing CaCl2, and added to transfected cells that had been washed in PBS with CaCl2. Cells were incubated for 1 h at 37°C, washed in cold PBS containing CaCl2, and visualized using a Nikon fluorescence microscope.
RESULTS
Mutagenesis of the amino terminus of F1.
Figure 1 shows the amino acid sequence of the amino terminus of the NDV F1 protein, including mutations previously made in this region (31) and new mutations described here. Mutations previously made in the heptad repeat region of the NDV F protein substituted charged residues for hydrophobic or polar amino acids and resulted in either transit-competent but fusion-negative proteins or proteins that were not properly folded (31). New mutations N147A and L154A introduced the uncharged amino acid alanine into two of these positions to determine if the introduction of a charge into these positions was responsible for the fusion-negative phenotype of the original set of mutants. In addition, mutations T161K, T161A, V168K, L175K, V179K, and M182K were made to define the role of these core amino acids in the HR1 domain.
Two new mutations were also made in the fusion peptide region, G123A and G128A. Previous mutation of these residues substituted lysine and leucine residues, respectively, into these positions, resulting in defective proteins. The G123K mutant, while expressed at the cell surface, was fusion negative. The G128L mutant failed to fold properly. In the very similar simian virus 5 (SV5) fusion protein, these residues have been changed individually to alanines, resulting in mutant proteins with enhanced fusion activity (13). Thus, these residues were changed to alanines to determine if the resulting NDV fusion proteins also had enhanced fusion activity.
Expression of mutant proteins.
The mutant proteins were expressed in Cos-7 cells using a simian virus 40-based vector and radioactively labeled with [35S]methionine for 2 h at 48 h posttransfection as previously described (29–31). The labeled proteins, precipitated with polyclonal antibody raised against a peptide with the sequence of the cytoplasmic tail (37), were electrophoresed in the presence (Fig. 2A) or absence (Fig. 2B) of reducing agent. In the presence of reducing agent, F0 and F1 were resolved and F2 was not detected (31). In the absence of reducing agent, the uncleaved F0 and the cleaved but disulfide-linked F1 and F2 comigrated as a single band with an approximate molecular weight of 66,000 (Fnr).
FIG. 2.
Characterization of mutant F proteins. (A and B) Precipitation of radiolabeled F proteins synthesized in cells transfected with plasmids encoding mutant F proteins (indicated above each lane). Cells transfected with 0.5 μg of DNA/35-mm plate of Cos cells were labeled with [35S]methionine for 2 h at 48 h posttransfection and subjected to a 4-h chase. Equivalent amounts of protein present in extracts (from 2 × 105 cells) were precipitated with anti-Ftail antibody, and the precipitated proteins were resolved on polyacrylamide gels in the absence (A) or presence (B) of reducing agent after boiling in sample buffer. (C and D) Western analysis of proteins present in 5 × 105 cells at 48 h posttransfection. Samples were not boiled prior to loading on polyacrylamide gels. Anti-Ftail was used to detect F protein. Fwt, wild-type F protein; M, marker proteins from infected cell extracts; V, vector.
Under reducing conditions, all mutant proteins were resolved and levels of expression were similar to the that of wild-type protein. The G128A, L154A, V168K, and V179K (Fig. 2A) and M182K (Fig. 2D) mutant DNAs expressed primarily the uncleaved fusion protein, F0, while cleavage of G123A, N147A, T161K, T161A, and L175K proteins was comparable to that of wild-type protein. In the absence of reducing agent, all mutant proteins were resolved, although very small amounts of uncleaved mutant proteins, G128A, L154K, V168K, V179K (Fig. 2B), and M182K (Fig. 2D), were detected. Decreased detection of these mutant proteins in the absence of reducing agent suggested that these proteins form disulfide linked aggregates that failed to enter the gel, a finding that suggests abnormal folding (3, 33).
The total amounts of cell-associated mutant proteins relative to wild-type protein were assessed by Western analysis (Fig. 2C and D). All mutant proteins were detected at levels comparable to those of the wild type.
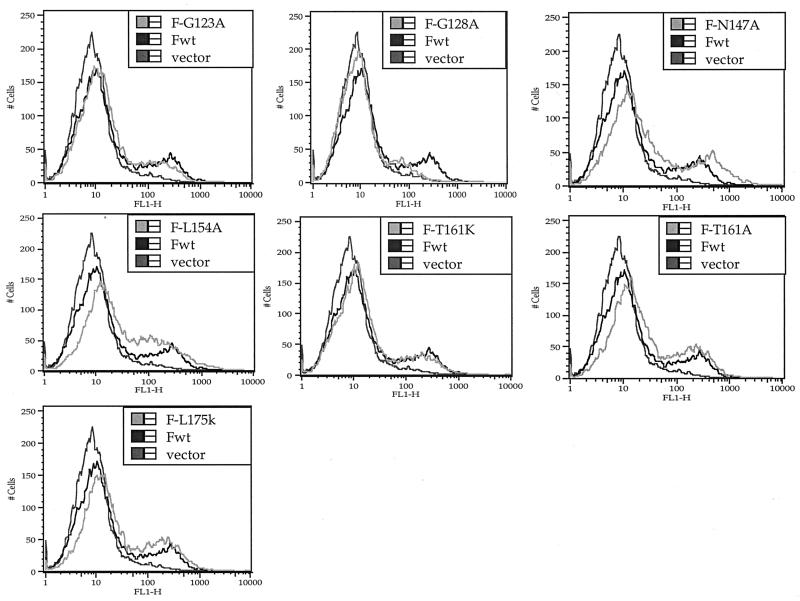
Surface expression of mutant proteins.
To determine which mutant proteins were transported to the cell surface, cell surface F protein was detected by immunofluorescence as previously described (31) (Fig. 3). The results were confirmed and quantitated by flow cytometry (Fig. 4 and 5). Cells were transfected with suboptimal levels of DNA in order to avoid effects due to overexpression of viral proteins. Figure 4 shows the percent positive cells detected relative to cells transfected with wild-type DNA. Figure 5 shows the intensity of surface expression for mutants detected at the surface. All mutant proteins that were cleaved could be detected at the cell surface (Fig. 3) and were expressed at densities comparable to that of the wild-type protein (Fig. 4 and 5). In addition, the L154A mutant was also expressed at the cell surface. However, the V168K, V179K, and M182K mutants were minimally detected at cell surfaces, and surface expression of the G128A protein was considerably reduced.
FIG. 3.
Immunofluorescence of cell surfaces expressing mutant proteins. Cells were transfected with 0.5 μg of DNA/35-mm plate of Cos cells and prepared for immunofluorescence as described in Materials and Methods. Pictures are of 4-s exposures of Kodak TMAXp3200 film using a Nikon inverted fluorescent microscope. Fwt, wild-type F protein.
FIG. 4.
Percent positive cells detected by flow cytometry analysis relative to wild type (Fwt; set at 100%) determined from data shown in Fig. 5 as well as duplicate experiments. The percentage of transfected Cos-7 cells expressing the wild-type F protein was 20% in the experiment shown.
FIG. 5.
Flow cytometry analysis of mutant-expressing cells. Cells transfected with 0.5 μg of DNA/35-mm plate (suboptimal levels) were processed for analysis by flow cytometry as described in Materials and Methods. Only data for mutant proteins expressed at the surface are shown. The primary antibody was anti-NDV. Each panel shows background (cells transfected with vector alone) and wild-type data (Fwt) as well as data for one mutant.
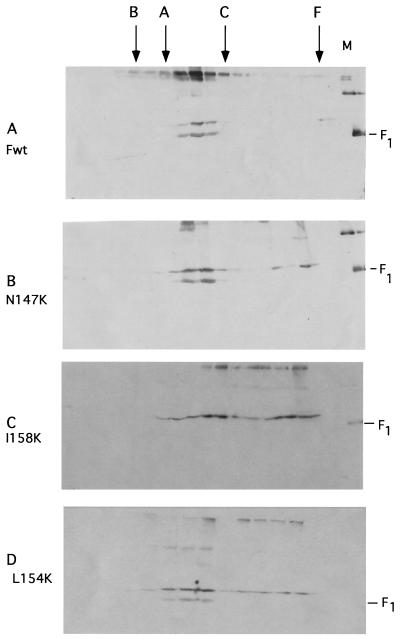
Sucrose gradient analysis of mutant proteins.
To characterize the oligomeric structure of the mutant proteins, the sedimentation properties of mutant proteins were compared to those of the wild-type protein on 10-to-45% sucrose gradients as previously described (28). The proteins in each gradient fraction were detected by Western analysis using antibody raised against the cytoplasmic tail sequences. All mutants were characterized, and representative results are shown in Fig. 6. As previously reported (28), wild-type protein sedimented slightly faster than the marker protein aldolase (158 kDa) but slightly slower than catalase (240 kDa), a behavior consistent with a trimer (Fig. 6A). Figure 6B shows sedimentation characteristic of mutant proteins that were cleaved and surface expressed but fusion defective. These proteins sedimented as the wild-type protein, although some of the F0 sedimented in larger, more heterogeneously sized material. Figure 6C shows results characteristic of uncleaved mutants that were not expressed at the surface. These proteins sedimented very heterogeneously, and most of the protein formed very large heterogeneous material. The majority of the minimally cleaved 154-kDa protein sedimented as wild-type protein, although some material was also larger (Fig. 6D).
FIG. 6.
Sucrose gradient analysis of mutant proteins. At 48 h posttransfection, cells were lysed and extracts were layered onto a 10-to-45% sucrose gradient as described in Materials and Methods. Proteins present in each fraction were precipitated with trichloroacetic acid, incubated at 50°C for 10 min, and electrophoresed in the presence of reducing agent, and the F protein in each fraction was detected by Western analysis. Virion proteins (M) were electrophoresed in the last lane of each polyacrylamide gel and are shown in panels A to C. F0 migrates slightly slower than the marker F1. F, ferritin; C, catalase; A, aldolase; B, BSA; Fwt, wild type.
Fusion activities of mutant proteins.
The fusion activities of mutant proteins were determined by measuring syncytium formation, content mixing, and hemifusion. Syncytium formation directed by these mutant proteins was quantitated by syncytium size, as previously described (31), and the results are shown in Table 1. As expected, mutant proteins minimally detected at the cell surface (G128A, V168A, V179K, and M182K) did not direct syncytium formation, nor did the uncleaved L154A protein. However, G123A, N147A, T161A, and T161K mutants, which were found at the cell surface and proteolytically cleaved, had negligible syncytium-forming activity, results very similar to those obtained with G123K and N147K proteins (31). In contrast, L175K protein formed syncytia at nearly wild-type levels.
TABLE 1.
Syncytium formation by mutant proteins
| DNA | Size of syncytia ata:
|
% of wild typeb | |
|---|---|---|---|
| 48 h | 72 h | ||
| Wild type | 16 ± 2 | 21 ± 4 | 100 |
| G123A | 0.2 ± 0.2 | 0.4 ± 0.3 | 1.9 |
| G128A | 0.5 ± 0.2 | 0.4 ± 0.1 | 1.9 |
| N147A | 0.6 ± 0.2 | 0.5 ± 0.5 | 2.3 |
| L154A | 0.6 ± 0.2 | 0.1 ± 0.1 | 0.4 |
| T161K | 1.0 ± 0.7 | 0.4 ± 0.4 | 1.9 |
| T161A | 1.1 ± 1.0 | 0.4 ± 0.4 | 1.9 |
| V168A | 0.5 ± 0.2 | 0.25 ± 0.25 | 1.1 |
| L175K | 8 ± 3 | 17.4 ± 2 | 82.8 |
| V179K | 0.6 ± 0.5 | 0.1 ± 0.1 | 0.4 |
| M182K | 0.3 ± 0.1 | 0.6 ± 0.4 | 2 |
The average numbers of nuclei at 48 and 72 h posttransfection are shown. The background values obtained after transfection of the vector alone into Cos cells were subtracted, and the numbers are the averages of at least three separate determinations. The actual variation between all experiments is also shown.
The percentage of the wild-type value was calculated using the 72-h time points.
Content-mixing activities of these mutants were measured in a protocol similar, in principle, to previously reported protocols (25). Cells were transfected with a plasmid carrying the β-galactosidase gene driven by a promoter responsive to a TET-sensitive transactivator. These cells were mixed with cells transfected with plasmid DNAs containing HN and F genes as well as a plasmid encoding the TET-responsive transactivator of transcription. β-Galactosidase synthesis should be induced only when cells from the different populations fused. Indeed, significant activity was detected when both F and HN cDNAs were present (Fig. 7A). However, little enzyme activity was present when only F cDNA or only HN cDNA was present. Furthermore, there was little activity after transfection with HN cDNA and an F cDNA that encodes an uncleaved F protein (F115G) (21) (Fig. 7A). This assay is also dependent on the levels of expression of HN and F proteins, since there was a linear increase of activity as the amounts of DNA used were increased (24).
FIG. 7.
Fusion activity of mutant proteins measured by content mixing. The extent of content mixing of all mutants was measured as described in Materials and Methods and is expressed as a percentage of that observed for wild-type F protein (Fwt). (A) Control experiments. F-K115G is a cleavage mutant of F protein (21). (B) Content mixing of fusion peptide and HR1 mutants. The results are the averages of three separate experiments.
Content mixing directed by each of the F protein mutants in the presence of wild-type HN cDNA is shown in Fig. 7B. We have previously described other transport-competent mutations in this region (Fig. 1) which block syncytium formation (31). To determine their effects on content mixing as well as to compare effects of conservative and nonconservative mutations at similar positions, the content mixing directed by these mutant proteins was also determined. It is clear that the closer the mutation is to the amino terminus of F1, the greater the inhibition of content mixing. Furthermore, mutant proteins with alanines in positions 147 and 123 allowed more content mixing than mutant proteins with lysines at the same positions. The residue at position 161, however, made little difference in the activity of the protein.
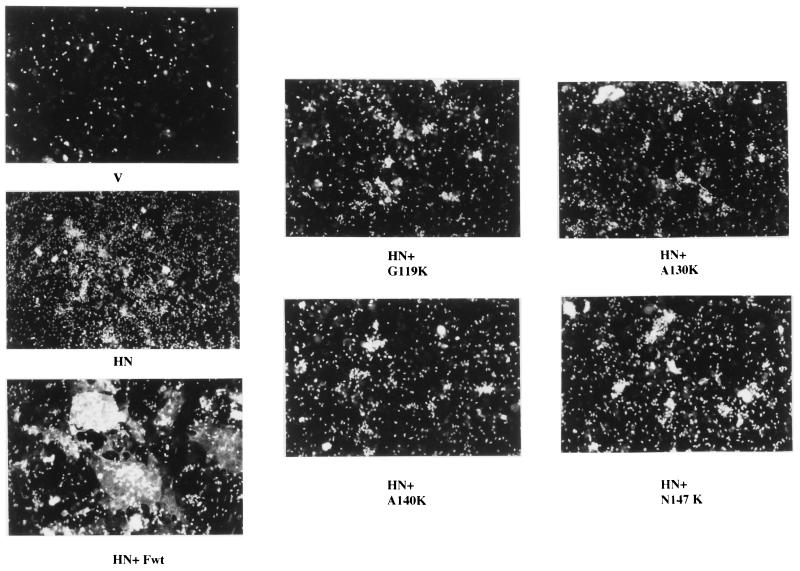
To determine if mutants which did not direct content mixing were capable of mediating lipid mixing, RBC fluorescently labeled with R18 were incubated with cells expressing these mutant proteins as previously described (16, 17). Figure 8 shows representative results. As expected, cells expressing the HN protein bound RBC, but there was no transfer of fluorescence from the RBC to the cells. In contrast, cells expressing both HN and F proteins became fluorescent as a result of the dye transfer. Not unexpectedly, cells expressing the HN protein as well as proteins with mutations in the fusion peptide (G119K and A130K) showed no evidence of lipid mixing. Furthermore, cells expressing HN protein as well as A140K or N147K protein showed little or no lipid mixing. Thus, these more amino-terminal mutations inhibit fusion at the earliest stages.
FIG. 8.
Lipid mixing directed by mutant proteins. Cells were transfected with the indicated DNAs and incubated with R18-labeled chick RBC as described in Materials and Methods.
DISCUSSION
Mutational analysis of the HR1 domain.
The published structure of a peptide with the SV5 HR1 sequence indicates that the region comparable to the NDV F protein from amino acids 136 to 199 has the potential to form a trimer (2). We have focused our present and previous mutational analyses on the hydrophobic or polar amino acids that form the interior of this trimer structure from amino acid 130 to 182 in order to determine the effect formation of this trimer may have on folding as well as fusion activity. The crystal structure of the NDV F protein was recently published (9), and the HR1 domain from amino acid 171 to 221 forms a trimer in this structure. However, the amino terminus of F1, from amino acid 117 to 170, was missing from the structure, and therefore the conformation of this region is unclear.
The trimer hydrophobic core in the SV5 peptide structure is formed by the “a” and “d” hydrophobic residues in the α helix formed by an HR1 monomer (2). In this study, we chose to replace hydrophobic or polar residues nonconservatively with the charged residue lysine, which can be incorporated into a monomer helix without its disruption (34). Some “d” residues were also replaced with alanine to assess differences between properties of proteins with conservative and nonconservative changes at the same position. Rather than giving rise to similar phenotypes, mutations of the core residues fall into four categories: mutations that block surface expression, mutations that allow surface expression and cleavage but inhibit fusion activity, mutations that allow surface expression but block proteolytic cleavage, and mutations at a single residue that have no effect on surface expression or fusion activity.
Recent studies of model peptides that form trimers have investigated the effect of different amino acids inserted into a central single site in either the “a” or “d” position (34–36). These experiments have shown that a single lysine residue in either an “a” or “d” position results in exclusively dimer formation. This result has been attributed to the significantly increased difficulty in burying the charge within the hydrophobic core of a trimer as opposed to a dimer. Substitution of an alanine in either an “a” or “d” position favors dimer formation but results in a mixed population of approximately 60% dimers and 30% trimers.
Studies with model peptides suggest that substitution of lysine at either “a” or “d” positions in the HR1 domain should have a similar disrupting effect on formation of the core trimer structure (34–36). If the core trimer must form during folding, then lysine residues in either “a” or “d” positions should block surface expression. While lysine residues in all “a” positions characterized were defective in folding, as determined by the expression of the protein at the cell surface and cleavage, five of seven “d” position substitutions had little detectable effect on folding and surface expression. That most of the “d” position substitutions allow cleavage and surface expression suggests that formation of an HR1 trimer characterized in peptide studies may not be absolutely required for F0 folding. Perhaps “a” position residues participate in a structure necessary for formation of the prefusion, precleavage form of the protein that is different than the core HR1 trimer defined by peptide studies. It follows, then, that formation of the core HR1 trimer may occur only upon proteolytic cleavage or activation of fusion.
While the role of “a” position residues in fusion cannot be assessed because of the absence of surface expression of these mutant proteins, all surface-expressed mutants with “d” position residues altered to lysine, with the exception of one with a mutation at amino acid 175, did not direct fusion. That most “d” position residues do affect fusion activity argues, not surprisingly, that a stable HR1 trimer is required for optimal fusion activity. The substitution of alanine also inhibited fusion, although not as stringently as the lysine substitutions, as might be expected since alanine should only decrease the frequency of trimer formation in the population (34) and should have a less destabilizing effect than lysine (34). Interestingly, fusion inhibition by these mutants was more stringent the closer the mutations were to the amino terminus of F1. This result may indicate that stability of the core timer at its amino-terminal end may be more critical to the formation of a functional HR1-HR2 complex.
The third phenotype of HR1 mutations was that of L154A mutant protein. This mutation, in a “d” position, had little detectable effect on folding of F0. Expression at the surface was comparable to that of the wild-type protein, yet this protein remained uncleaved. We have previously reported that substitution of lysine at this position had an identical effect on the protein (31). In addition, L154K mutant protein reacted to a conformationally sensitive monoclonal antibody (31), also indicating no gross conformational abnormalities. These results suggest that mutation at this site in the HR1 sequence alters the conformation or accessibility of the cleavage site region of the molecule, rendering it resistant to host cell proteases. Since this region of the F protein was missing in the crystal structure (9), the effect of this residue on the cleavage site is unknown.
A surprising finding was that one mutation, a change of a leucine to a lysine residue at position 175, had no apparent effect on folding or fusion. This residue is at the amino terminus of the region visualized in the F protein crystal structure (9) and forms part of the core of the trimer resolved in the structure, a trimer that extends from amino acid 171 to 221. However, if this residue is at the boundary of the trimer, then mutations may have minimal effects on folding, in contrast to mutations at residues 179 and 182, which fall within the trimer and do block folding. Chen et al. (9) proposed that upon fusion activation, the entire HR1 domain forms an extended trimer similar to the SV5 peptide structure. In such a structure, residue 175 would be in the middle of the extended trimer. The mutation at residue 175 may not affect fusion because, as suggested above, the stability of the amino-terminal end of the trimer may be more important for fusion than stability of more carboxyl-terminal regions.
Mutational analysis of the fusion peptide.
Here we describe two mutants with changes in the fusion peptide sequence, G123A and G128A. We have previously reported properties of mutants with nonconservative changes of these residues (31). G123K allowed surface expression but not fusion, while G128L blocked surface expression. However, conservative changes at the same residues in the very similar SV5 F protein actually enhanced fusion (13). A conservative G-to-A change at amino acid 123 in the NDV F sequence, however, still resulted in a fusion-defective protein. A conservative G-to-A change at position 128 still resulted in a folding-defective protein, although the surface expression of this mutant was slightly increased over that of the protein with the G-to-L change previously reported. These observations are interesting in two ways. First, the different phenotypes of similar mutants in the SV5 and NDV fusion proteins underscore the differences between these two proteins, the most significant of which is the difference in requirement of HN protein for fusion (1). Second, that a mutation in the fusion peptide interferes with surface expression argues that this domain participates in a conformation critical to the overall folding of the molecule. However, no mutations in a potential “a” or “d” position, relative to the HR1 domain, influenced the folding of the molecule.
In summary, mutational analysis of hydrophobic core residues in the HR1 trimer suggests that the amino-terminal half of the HR1 core trimer may not form during folding of the F protein but may form only upon conformational shifts in the molecule upon cleavage or initiation of fusion.
ACKNOWLEDGMENTS
This work was supported by grants GM 37745 and AI 30572 from the National Institutes of Health.
REFERENCES
- 1.Bagai S, Lamb R A. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol. 1995;69:6712–6719. doi: 10.1128/jvi.69.11.6712-6719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Structural basis for paramyxovirus-mediated membrane fusion Mol. Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 3.Braakman I, Helenius J, Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckland R, Malvoisin E, Beauverger P, Wild F. A leucine zipper structure present in the measles virus fusion protein is not required for its tetramerization but is essential for fusion. J Gen Virol. 1992;73:1703–1707. doi: 10.1099/0022-1317-73-7-1703. [DOI] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Carr, C. M., and P. S. Kim. 1993. A spring loaded mechanism for the conformational change of influenza hemagglutinin. Cell 823–832. [DOI] [PubMed]
- 7.Chambers P, Pringle C R, Easton J J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 8.Chan D C, Kim P S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Gorman J J, McKimm-Breschkin J, Lawrence L J, Tullock P A, Smith B J, Colman P M, Lawrence M C. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 10.Furuta R A, Wild C T, Weng Y, Weiss C D. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh J K, Peisajavich S G, Ovadia M, Shai Y. Structure-function study of a heptad repeat positioned near the transmembrane domain of Sendai virus fusion protein which blocks virus-cell fusion. J Biol Chem. 1998;273:27182–27190. doi: 10.1074/jbc.273.42.27182. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 13.Horvath C M, Lamb R A. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J Virol. 1992;66:2443–2455. doi: 10.1128/jvi.66.4.2443-2455.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu M, Scheid A, Choppin P. Activation of the Sendai virus fusion protein (F) involves a conformational change with exposure of a new hydrophobic region. J Biol Chem. 1981;256:3557–3563. [PubMed] [Google Scholar]
- 15.Joshi S B, Dutch R E, Lamb R A. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 16.Kemble G W, Danieli T, White J W. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 17.Kemble G W, Henis Y I, White J M. GPI- and transmembrane anchored influenza hemagglutinin differ in structure and receptor binding activity. J Cell Biol. 1993;122:1253–1265. doi: 10.1083/jcb.122.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1177–1206. [Google Scholar]
- 19.Lambert D M, Barney S, Lambert A L, Guthrie K, Medinas R, Davis D E, Bucy T, Erickson J, Merutka G, Petteway S R. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion Proc. Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawless-Delmedico M K, Sista P, Sen R, Moore N C, Antczak J B, White J M, Greene R J, Leanza K C, Matthews T J, Lambert P M. Heptad repeat regions of respiratory syncytial virus F(1) protein form a six membered coiled-coil complex. Biochemistry. 2000;39:11684–11695. doi: 10.1021/bi000471y. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Sergel T, Razvi E, Morrison T. Effect of cleavage mutants on syncytium formation directed by the wild-type fusion protein of Newcastle disease virus. J Virol. 1998;72:3789–3795. doi: 10.1128/jvi.72.5.3789-3795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathews J M, Young T F, Tucker S P, MacKay J P. The core of the respiratory syncytial virus fusion protein is a trimeric, coiled coil. J Virol. 2000;74:5911–5920. doi: 10.1128/jvi.74.13.5911-5920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews T J, Wild C, Chen C-H, Bolognesi D P, Greenberg M L. Structural rearrangements in the transmembrane glycoprotein after receptor binding. Immunol Rev. 1994;140:93–104. doi: 10.1111/j.1600-065x.1994.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 24.McGinnes L, Sergel T, Reitter J, Morrison T. Carbohydrate modifications of the NDV fusion protein heptad repeat domains influence maturation and fusion activity. Virology. 2001;238:332–342. doi: 10.1006/viro.2001.0899. [DOI] [PubMed] [Google Scholar]
- 25.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peisajovich S G, Samuel O, Shai Y. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J Mol Biol. 2000;296:1353–1365. doi: 10.1006/jmbi.2000.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rapaport D, Ovadia M, Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reitter J, Sergel T, Morrison T. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J Virol. 1995;69:5995–6004. doi: 10.1128/jvi.69.10.5995-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sergel T, McGinnes L W, Morrison T G. The fusion promotion activity of the NDV HN protein does not correlate with neuraminidase activity. Virology. 1993;196:831–834. doi: 10.1006/viro.1993.1541. [DOI] [PubMed] [Google Scholar]
- 30.Sergel T, McGinnes L W, Peeples M E, Morrison T G. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology. 1993;193:717–726. doi: 10.1006/viro.1993.1180. [DOI] [PubMed] [Google Scholar]
- 31.Sergel-Germano T, McQuain C, Morrison T. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J Virol. 1994;68:7654–7658. doi: 10.1128/jvi.68.11.7654-7658.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan K, Liu J-H, Wang J-H, Shen S, Lu M. Atomic structure of a thermostable subdomain of the HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatu U, Braakman I, Helenius A. Membrane glycoprotein folding, oligomerization, and intracellular transport: effects of dithiothreitol in living cells. EMBO J. 1993;12:2151–2157. doi: 10.1002/j.1460-2075.1993.tb05863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tripet B, Wagschal K, Lavigne P, Mant C T, Hodges R S. Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled-coil: 20 amino acid substitutions in position “d.”. J Mol Biol. 2000;300:377–402. doi: 10.1006/jmbi.2000.3866. [DOI] [PubMed] [Google Scholar]
- 35.Wagschal K, Tripet B, Hodges R S. De novo design of a model peptide sequence to examine the effects of single amino acid substitutions in the hydrophobic core on both stability and oligomerization state of coiled-coils. J Mol Biol. 1999;285:785–803. doi: 10.1006/jmbi.1998.2284. [DOI] [PubMed] [Google Scholar]
- 36.Wagschal K, Tripet B, Lavigne P, Mant C T, Hodges R S. The role of position “a” in determining the stability and oligomerization state of alpha-helical coiled coils: 20 amino acid stability coefficients in the hydrophobic core of proteins. Protein Sci. 1999;8:2312–2329. doi: 10.1110/ps.8.11.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Raghu G, Morrison T, Peeples M E. Intracellular processing of the paramyxovirus F protein: critical role of the predicted amphipathic alpha helix adjacent to the fusion domain. J Virol. 1992;66:4161–4169. doi: 10.1128/jvi.66.7.4161-4169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiessenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp-41. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 39.Wild T F, Buckland R. Inhibition of measles virus infection and fusion with peptides corresponding to the leucine zipper region of the fusion protein. J Gen Virol. 1997;78:107–111. doi: 10.1099/0022-1317-78-1-107. [DOI] [PubMed] [Google Scholar]
- 40.Yao Q, Compans R W. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]
- 41.Young J K, Hicks R P, Wright G E, Morrison T G. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology. 1997;238:291–304. doi: 10.1006/viro.1997.8834. [DOI] [PubMed] [Google Scholar]
- 42.Young J K, Li D, Abramowitz M C, Morrison T G. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J Virol. 1999;73:5945–5956. doi: 10.1128/jvi.73.7.5945-5956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao X, Singh M, Malashkevich V N, Kim P S. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc Natl Acad Sci USA. 2000;97:14172–14177. doi: 10.1073/pnas.260499197. [DOI] [PMC free article] [PubMed] [Google Scholar]