The field of biology has undergone several rounds of transformation in the approaches taken, ranging from theoretical to experimental perturbation to discovering molecular components. In the next decades to come, I believe it will take on another transformation to bioinformatical, where computational models of systems-wide properties could serve as the basis for experimentation and discovery. The ramifications of this will be not only the precise understanding of how organisms are built, but also the ability to engineer organisms to exhibit specified traits, to discover the causality of diseases, and to predict organisms' responses to changes in the environment. This could lead to prevention and targeted treatment of diseases, improved food production, and preservation of the environment.
Bioinformatics can be simply defined as an approach that uses computer science, engineering, and mathematical methodologies to manage, visualize, and analyze data to discover new patterns and build hypotheses and models. This includes database development, data management, software (algorithm) development, modeling (simulation), and quantitative analysis.
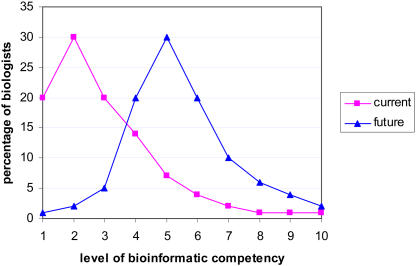
Currently, bioinformatics is conducted by a specialized group of individuals, such as database curators (Ph.D.-level biologists), database and software engineers, and computational biologists. On the fringe of this are the collaborative entities of biologists, mechanical or electric engineers (bioengineers), computer scientists, and mathematicians. The majority of the biologists, however, are on the other end of the spectrum in that they are users of the most basic bioinformatical tools. I see this as the major limitation of bioinformatics today. It is simply not as accessible to most biologists as it should be. In the future, I see that the distribution of people in this spectrum will change to a bell curve where the majority of biologists will have some basic skills such as programming, database development and management of large datasets, and quantitative and statistical analysis of data (Fig. 1). This change will not be unlike how molecular biology penetrated the field of biology some 30 years ago in changing how people thought about and conducted biological research. Recent publication of the Current Protocols in Bioinformatics series (http://www.does.org/cp/bioinfo.html) provides an example of this trend already in motion. The infiltration of bioinformatical biology may be more profound in shifting the biological research paradigm than molecular biology ever was. The richness and enormity of information, such as understanding the function of every gene in an organism, will shift research into more theoretical biology using bioinformatical approaches, with experiments carried out to find supporting or refuting evidence for the theories, models, and hypotheses. Biologists will generally have a much larger circumference of their domain of expertise and spend more of their time on the computer than at the bench. The concept of ownership of data will also change, and analyzing other people's data will be much more common place. This change will encourage, if not force, scientists to pay more attention to the quality of data annotation and actively participate in their improvement. Other problems in bioinformatics we are facing today include the heterogeneity of how data are analyzed, annotated, and displayed and the lack of connectivity among the available data. These problems arose partially because of the young age of the field of bioinformatics, with independent and disparate efforts carried out without the conventions and discipline associated with an established scientific community. Recent movements toward the creation of a scientific society for database curators (www.biocurator.org) and projects that bring together the efforts of different model organism databases (www.gmod.org) provide early hints to the development of bioinformatics into a more coherent discipline of biology.
Figure 1.
Desirable trend for bioinformatic competency in biologists. This graph depicts speculation about the changes in the proportion of biologists with varying degrees of bioinformatical competency currently and in the future. Pink squares represent the current situation, and blue triangles represent a desirable future trend. Bioinformatical competency is shown in a gradient of 1 to 10, where level 1 represents biologists whose extent of bioinformatical skills is to use PubMed and GenBank and level 10 signifies biologists who are self-sufficient in managing, mining, and analyzing all available data, including writing algorithms and creating databases.
Acknowledgments
I am grateful to Chris Somerville and Carolyn Lawrence for valuable discussion and helpful comments on the manuscript.