CHEMICALS AS BIOLOGICAL SWITCHES
Chemical genomics is a promising new technology for studying gene functions in the context of living organisms or cell systems. It complements existing molecular and genetics tools (e.g. mutagenesis, RNAi) by allowing fine-tunable in vivo modulations of protein functions and cellular processes (Blackwell and Zhao, 2003; Austin et al., 2004; Lipinski and Hopkins, 2004). This approach is feasible because of recent advances in the synthesis of large libraries of small chemicals. In chemical genomics experiments the libraries are used to identify in high-throughput screens interesting agonistic or antagonistic candidates that interfere with a biological process of interest. Typically, the libraries, used in these screens, consist of collections of diverse compounds with predicted drug-like properties (Lipinski et al., 1997; Oprea and Gottfries, 2001; Oprea, 2002; Baurin et al., 2004; Stockwell, 2004). In analogy to genetic screens, chemical genomic screens can utilize forward and reverse strategies (Schreiber, 1998; Haggarty et al., 2003). Forward chemical genomics screens probe modulations of complex biological processes rather than isolated targets. This is in contrast to small molecule discovery in the pharmaceutical and agricultural industry where the drug-able target is usually known and screened using in vitro systems. To fully understand the mode of action of isolated compounds with interesting biological activities, it is frequently necessary to identify their target(s) at a later stage of a screening project using biochemical and genetics gene or protein isolation techniques. In contrast to this forward strategy, reverse chemical genomics screens resemble, in their initial stage, drug discovery approaches by screening known targets (Drews, 2000). Subsequently, the isolated bioactive chemicals are used to study the molecular and biological functions of poorly characterized proteins in vivo. Both forward and reverse approaches utilize the identified chemicals as “research tools” for determining the functions, interactions, and architecture of cellular networks in living organisms. A potential pharmaceutical or agricultural application can be of interest but is not the central goal of this technology.
Chemical genomics has several outstanding advantages over classical genetics and molecular techniques for studying gene functions. Standard genetics approaches target one gene at a time and provide limited opportunity to control the extent of the downstream cellular effects. By contrast, chemicals can be targeted with spatiotemporal precision against a selected spectrum of proteins. They can be applied in defined dosages to distinct cells, organs, or developmental stages, often with rapid response times and reversible effects. Since chemical switches can act in a similar manner across a range of model or nonmodel organisms, their identification is of great interest for researchers working with different model systems. Finally, the chemicals can be used to inactivate a family of proteins with related sequences or structures in a single step. In the future, these “chemical family knock-downs” may be the method of choice for the functional characterization of paralogous genes with redundant functions.
AVAILABLE RESOURCES AND MISSING LINKS
In spite of the broad spectrum of new opportunities, chemical genomics has not yet evolved into a widely used strategy for biological systems analysis in academic research. This is due to several factors. One is the paucity of information resources, compound search and analysis services for annotated drugs, and agrochemicals in the public domain. An additional reason is the high cost of compound libraries and high-throughput equipment. This Update will provide a short outline of the existing open-access informatics resources that are relevant for chemical genomics-based research, and how ChemMine fills some of the missing links.
The critical software and database resources for bioactive chemical discovery projects are: tools for structure similarity comparisons, database searching, structure-activity comparisons, evaluations of the chemical descriptor (property) space, design of customized libraries (subsetting), lead optimization steps, and compound and screening databases. Despite the importance of these very basic enabling tools, most are not yet freely available. Recently, the first online services were established that give the public access to basic bioactivity information of drug-like compounds and virtual screening tools. The late start of such obvious and overdue information resources is particularly surprising since very similar resources are required in drug discovery, which is an established and well-funded research discipline (Strausberg and Schreiber, 2003; Savchuk et al., 2004). A strong commercialization trend may have contributed to the development of this “desert-like” landscape in academia with regard to freely available informatics tools and databases in this area.
The open National Cancer Institute (NCI) database was one of the first consolidated public efforts to change this situation by disseminating screening and bioactivity information for a larger compound set in a searchable database format for the cancer and HIV research community (Voigt et al., 2001; Ihlenfeldt et al., 2002; Couzin, 2003). ChemBank and PubChem are more recent implementations of compound and screening databases that are of relevance for a more general spectrum of users in basic and applied research areas (Strausberg and Schreiber, 2003; Austin et al., 2004). In addition, several online services have become available to provide noncommercial tools for virtual drug screening and compound similarity searching (e.g. Ligand.Info: von Grotthuss et al., 2004; ZINC: Irwin and Shoichet, 2005). Open-source and open-access projects, such as Open Babel (http://openbabel.sourceforge.net/), ChemPython.org (http://www.chempython.org/), CACTVS (Ihlenfeldt et al., 2002), and JOELib (http://www-ra.informatik.uni-tuebingen.de/software/joelib/index.html), are also very useful initiatives for promoting cheminformatics software development and circulating the available resources in the public domain.
THE CHEMMINE DATABASE
To further facilitate the incorporation of chemical genomics-based approaches in the discovery process of novel protein functions and gene networks, we have developed the ChemMine database (http://bioinfo.ucr.edu/projects/PlantChemBase/search.php). The first release of this public service provides access to an integrated suite of analysis and information retrieval tools for compound searching, structure-based clustering, descriptor generation (chemical properties), and retrieval of published bioactivity and target protein information (Fig. 1).
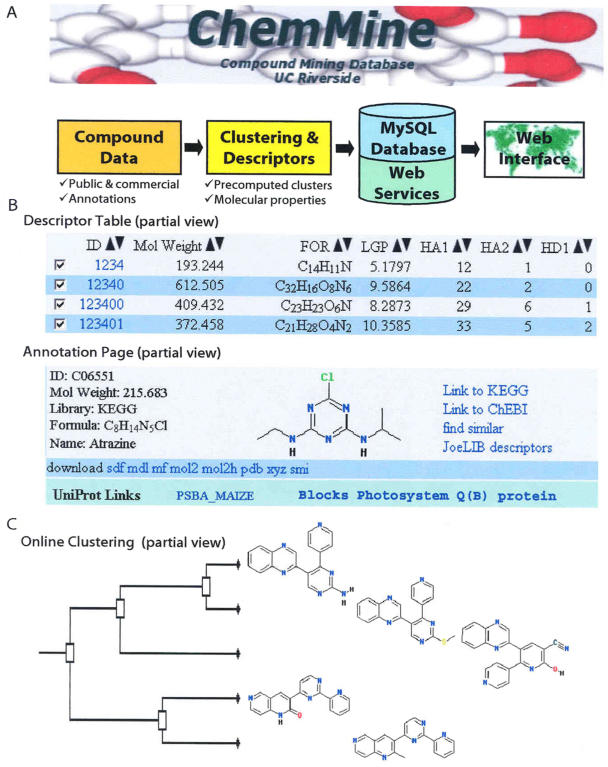
Figure 1.
Design overview of ChemMine. A, Outline of the available data and Web services. B, Selected examples of the result pages that are available through the ChemMine interface. C, Output sample of the hierarchical clustering tool.
At the current stage of this project, ChemMine centralizes compound structure and activity information from a growing number of public providers and vendors of chemical screening libraries. The incorporation of commercially available compounds provides access to their purchase information. This knowledge can be critical for follow-up studies and assembly of focused libraries in secondary screens when the resources for resynthesis of novel chemicals in larger quantities are limited or do not exist at all. It is expected that the current set of commercial compound collections in ChemMine (over 1 million) will quickly grow when more businesses realize the benefits of a public presence and express interest in participating in this project. In addition to commercial compounds, most collections from public initiatives are included in ChemMine. These highly annotated compound sets maximize access to bioactivity information, known target proteins, literature, and other useful annotation information, enabling the user to correlate screening results with available biological knowledge. Additional information will be included as it becomes available. Searches for analogs of metabolic compounds are available through the incorporation of the KEGG ligand database. Information about bioactive chemicals (e.g. known drugs, herbicides) and their functional characterization is provided through the data sets from ChEBI, ChemBank, NCI, PubChem, and other providers. The annotations from ChEBI illustrate the growing utility of these services (Fig. 1). This initiative was started to provide systematic target associations of small compounds that interfere with processes of living organisms. Via this linkage, ChemMine users can retrieve the target protein sequences, three-dimensional structures, and literature for annotated drugs or metabolic molecules that are available or hyperlinked in the UniProt database. Similar drug-to-target associations are available in the data sets from ChemBank and PubChem.
With regard to the specific needs of scientists working with proprietary or customized compound libraries, general purpose compound databases will remain incomplete no matter how many structures they contain. An additional reason for this limitation is that thousands of new compounds can be synthesized every day or their structures designed in silico. To counterbalance this inevitable incompleteness, the ChemMine project has a strong focus on online services. These features allow users to utilize most of ChemMine's analysis tools for external compound sets without being restricted to the compound coverage in the database. Since downstream analyses of compounds and their target proteins require the usage of various molecular modeling and computational chemistry programs, ChemMine supports interconversions of the most common structure formats (SDF, SMILES, PDB, etc.) for file exchange with other tools. The libraries from Open Babel (http://openbabel.sourceforge.net) are used for these reformatting steps.
The ChemMine interface allows queries in single or batch mode using one or many compound identifiers, compound names, or external annotations. The initial query results are displayed in a flexible table format that can be expanded and sorted by the chemical properties of the retrieved compounds. Annotations and structure images for each compound can be viewed on the next level for single or many entries. These pages contain links to additional information, such as available literature, target proteins, external annotation pages from different compound providers, and download options in different structure file formats. The structure images are generated with the batch rendering tool from the CACTVS package (Ihlenfeldt et al., 2002). In addition to textual search options, substructure and structure-based searches are one of the most important functions for exploring the chemical space. To perform similarity searches in ChemMine that are not dependent on the licensing restrictions of commercial software applications, we implemented an improved two-dimensional fragment-based similarity search technique, based on an algorithm suggested by Chen and Reynolds (2002). The current online version of this tool can use either atom pairs (Carhart et al., 1985) or atom sequences as structural descriptors, and uses the Tanimoto coefficient as similarity measure (Willett et al., 1998). According to the search studies from Chen and Reynolds (2002) and our own benchmark comparisons (data not shown), this search technique outperforms alternative tools with regard to the sensitivity and selectivity of identifying similar compounds. A disadvantage of this tool is its significantly lower speed compared to other search methods, such as fingerprint-based approaches. This tradeoff may become an issue for searching very large compound collections. The current search speed of the Web implementation is around 1 min per million compounds. The parallelization of the searches on available computer clusters and the implementation of an alternative high-speed search tool for querying large compound sets will be possible solutions when ChemMine grows beyond 3 million compounds. For substructure searches, ChemMine uses currently the corresponding tool from the Open Babel project in a speed-optimized implementation to increase the efficiency of this computationally expensive search type.
Structure-based clustering and descriptor space analyses are very useful strategies for both basic quantitative structure-relationship studies and lead optimization steps in compound screens. Structure-based clustering can be performed through the ChemMine interface using external or internal compounds or a combination of both. The similarity scores, generated by the fragment-based similarity search tool, are used for calculating the distance values required for clustering. The present set of clustering techniques consists of hierarchical clustering and a binning approach with variable similarity cutoffs. The open-source program Cluster 3.0 is used for the hierarchical clustering step (Eisen et al., 1998; de Hoon et al., 2004), while the resulting output is presented on the Web interface in form of interactive tree images that are generated by an internally developed tree viewing program (Girke et al., 2004). The resulting cluster tree pages provide hyperlinked compound identifiers at the terminal tree branches to guide users to the corresponding structure images of the clustered compounds. To increase the efficiency of this data visualization, the compound images are listed in the same order as the branches in the tree. In addition to the structure-based trees, all available molecular descriptors can be displayed in the corresponding order for constructing basic structure-activity tables in local spreadsheet programs.
To identify clusters of structural similarity within entire libraries, ChemMine contains precomputed cluster tables for most of its compound sets. These tables summarize the number of similarity clusters using incremental similarity values as stringency cutoffs. The composition of each identified cluster is stored in the database and its members can be retrieved through the corresponding hyperlinks in each table. Since this data representation is particularly useful for evaluating the structural redundancy in customized compound sets (e.g. interlibrary comparisons), additional analyses will be uploaded to this site upon user request. Commercial libraries can only be included here after approval by their providers.
More than 40 different descriptors can be created in ChemMine for any set of externally provided compounds or for those represented in the database. They are generated with the open-source JOELib computational chemistry package. They include molecular properties, such as molecular weight, octanol/water partition coefficient, counts of hydrogen-bond donors/acceptors, rotable bonds, types of atoms, and reactive groups per molecule. The descriptors of the popular Lipinski's “rule of five” for drug-likeness prediction are included in this list (Lipinski et al., 1997).
FUTURE PERSPECTIVE
The ChemMine project is unique by providing several new online tools (e.g. clustering, descriptor generation) and integrating them with a wide variety of bioactive, natural, and screening compounds from public and commercial providers. Based on the experience from chemical genomics studies in plants and numerous discussions with colleagues (Zhao et al., 2003; Armstrong et al., 2004; Zouhar et al., 2004; Surpin et al., 2005), we anticipate a high demand for this resource because it closes many gaps in the collection of available Web services for utilizing compound knowledge in chemical genomics screens.
In the future, we will further develop ChemMine as an open-source project by implementing several new features. First, the database will be augmented with bioactivity information from internal and external screening programs using standardized and interchangeable formats that support screens from an unlimited number of organisms, in addition to those from plants. Second, an upload functionality for compound structures and screening data from external researchers will be integrated. Third, additional structure search tools will be implemented to increase the speed and functionality of the similarity searches. Fourth, complex query functions will be added to enable filtering on various descriptor fields and other criteria. Fifth, automated upload routines will be developed to easily expand compound collections in the database and to update their annotation and provider information in a timely manner. Sixth, the developed software tools will be released to the public via download options. Seventh, multicomponent clustering using variable sets of molecular descriptors and structural similarities will be implemented. Finally, we will work on the integration and interoperability of ChemMine with the ChemBank, NCI, PubChem, ChEBI, and other projects in this area. This effort will strongly support the vision that public activities in this area should have the common goal of developing an ultimate “meta-database” as a central depository and mining service for compound and screening data.
Acknowledgments
We thank Eric Brauner, Caroline Shamu, and Stephen Haggarty from the Institute for Chemistry and Cell Biology for their continuous support of this project.
This work was supported by the Center of Plant Cell Biology at the University California, Riverside, and by the Office of Biological and Physical Research of the National Aeronautics and Space Administration (grant no. NNA04CC73C).
References
- Armstrong JI, Yuan S, Dale JM, Tanner VN, Theologis A (2004) Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc Natl Acad Sci USA 101: 14978–14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, Brady LS, Insel TR, Collins FS (2004) NIH Molecular Libraries Initiative. Science 306: 1138–1139 [DOI] [PubMed] [Google Scholar]
- Baurin N, Baker R, Richardson C, Chen I, Foloppe N, Potter A, Jordan A, Roughley S, Parratt M, Greaney P, et al (2004) Drug-like annotation and duplicate analysis of a 23-supplier chemical database totalling 2.7 million compounds. J Chem Inf Comput Sci 44: 643–651 [DOI] [PubMed] [Google Scholar]
- Blackwell HE, Zhao Y (2003) Chemical genetic approaches to plant biology. Plant Physiol 133: 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carhart RE, Smith DH, Venkataraghavan R (1985) Atom pairs as molecular-features in structure activity studies—definition and applications. J Chem Inf Comput Sci 25: 64–73 [Google Scholar]
- Chen X, Reynolds CH (2002) Performance of similarity measures in 2D fragment-based similarity searching: comparison of structural descriptors and similarity coefficients. J Chem Inf Comput Sci 42: 1407–1414 [DOI] [PubMed] [Google Scholar]
- Couzin J (2003) NIH dives into drug discovery. Science 302: 218–221 [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Drews J (2000) Drug discovery: a historical perspective. Science 287: 1960–1964 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Lauricha J, Tran H, Keegstra K, Raikhel N (2004) The Cell Wall Navigator database. A systems-based approach to organism-unrestricted mining of protein families involved in cell wall metabolism. Plant Physiol 136: 3003–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL (2003) Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA 100: 4389–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihlenfeldt WD, Voigt JH, Bienfait B, Oellien F, Nicklaus MC (2002) Enhanced CACTVS browser of the Open NCI Database. J Chem Inf Comput Sci 42: 46–57 [DOI] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Comput Sci 45: 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C, Hopkins A (2004) Navigating chemical space for biology and medicine. Nature 432: 855–861 [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23: 3–25 [DOI] [PubMed] [Google Scholar]
- Oprea TI (2002) Chemical space navigation in lead discovery. Curr Opin Chem Biol 6: 384–389 [DOI] [PubMed] [Google Scholar]
- Oprea TI, Gottfries J (2001) Chemography: the art of navigating in chemical space. J Comb Chem 3: 157–166 [DOI] [PubMed] [Google Scholar]
- Savchuk NP, Balakin KV, Tkachenko SE (2004) Exploring the chemogenomic knowledge space with annotated chemical libraries. Curr Opin Chem Biol 8: 412–417 [DOI] [PubMed] [Google Scholar]
- Schreiber SL (1998) Chemical genetics resulting from a passion for synthetic organic chemistry. Bioorg Med Chem 6: 1127–1152 [DOI] [PubMed] [Google Scholar]
- Stockwell BR (2004) Exploring biology with small organic molecules. Nature 432: 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Schreiber SL (2003) From knowing to controlling: a path from genomics to drugs using small molecule probes. Science 300: 294–295 [DOI] [PubMed] [Google Scholar]
- Surpin M, Rojas-Pierce M, Carter C, Hicks G, Vasquez J, Raikhel N (2005) The power of chemical genomics to study the link between endomembrane system components and the gravitropic response. Proc Natl Acad Sci USA 102: 4902–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt JH, Bienfait B, Wang S, Nicklaus MC (2001) Comparison of the NCI open database with seven large chemical structural databases. J Chem Inf Comput Sci 41: 702–712 [DOI] [PubMed] [Google Scholar]
- von Grotthuss M, Koczyk G, Pas J, Wyrwicz LS, Rychlewski L (2004) Ligand. Info small-molecule Meta-Database. Comb Chem High Throughput Screen 7: 757–761 [DOI] [PubMed] [Google Scholar]
- Willett P, Barnard JM, Downs GM (1998) Chemical similarity searching. J Chem Inf Comput Sci 38: 983–996 [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J (2003) SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110 [DOI] [PubMed] [Google Scholar]
- Zouhar J, Hicks GR, Raikhel NV (2004) Sorting inhibitors (Sortins): chemical compounds to study vacuolar sorting in Arabidopsis. Proc Natl Acad Sci USA 101: 9497–9501 [DOI] [PMC free article] [PubMed] [Google Scholar]