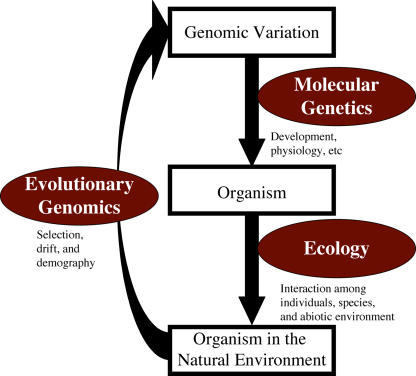
Why are some plants self-pollinating? What determines the timing of flowering and germination? Why do resistant and susceptible alleles of pathogen-resistant genes coexist in populations? These are just a few questions traditionally asked in the domain of ecology and evolutionary biology, and comprehensive answers to these and other questions are beginning to be addressed by molecular analyses. Increasingly, ecologists and evolutionists have been turning to Arabidopsis (Arabidopsis thaliana), the favored system for the study of plant molecular genetics and development (Mitchell-Olds, 2001; Pigliucci, 2002; Shimizu, 2002). Arabidopsis research has taken on an interdisciplinary focus, facilitating the integration of ecology, molecular genetics, and evolutionary genomics (Fig. 1). Genes, which were previously utilized in evolutionary and ecological research as mere molecular markers, are now being dissected to study the genetic basis of ecologically and evolutionarily important traits. Population genomic analyses of such genes can reveal molecular signatures associated with the pressures of natural selection, providing a glimpse of historical forces that may explain patterns of genetic and phenotypic variation observed in contemporary populations.
Figure 1.
Integration of three biological fields is required for comprehensive understanding of organismal biology. Molecular genetic studies are necessary to reveal which gene and the precise genetic variation responsible for developmental or physiological diversity. Ecological studies demonstrate how an organism interacts with conspecific individuals, other species, and abiotic environments to survive and reproduce. Evolutionary forces provide feedback mechanisms, including natural selection for adaptive traits, genetic drift, and demographic processes, which determines the genetic constitution of subsequent generations. Evolutionary genomics studies these forces by analyzing the genetics underlying phenotypic evolution in the context of genome-wide molecular information.
At the heart of these studies are attempts to understand traits in their broad ecological and evolutionary contexts. Ecology, which examines interactions among individuals, species, and their abiotic environment, requires knowledge of the response of plants to their natural environments. Evolutionary studies examine how selection, genetic drift, and evolutionary history have shaped patterns of variation at both the molecular and phenotypic levels. In this article, we will illustrate the power of Arabidopsis as a model plant for the study of ecology and evolutionary genomics, and the approaches that have led to key insights not only for these fields but also for plant biology in general (Mitchell-Olds, 2001; Shimizu, 2002).
EVOLUTIONARY HISTORY AND ECOLOGICAL PROFILE OF ARABIDOPSIS
Arabidopsis (L. Heynh.; family Brassicaceae) is a weedy annual plant, occupying disturbed habitats such as the margins of agricultural fields. It is a predominantly selfing species, with a reported out-crossing rate of approximately 1% (compiled by Hoffmann et al., 2003). Most natural populations adopt the winter annual life history strategy, which is characterized by plants germinating in the fall, overwintering as vegetative rosettes, and flowering in the spring. The spring annual strategy, with germination and flowering in spring, is also commonly observed (Pigliucci, 2002).
Arabidopsis diverged from other Arabidopsis species an estimated 5 to 6 million years ago, with its distribution and mating system apparently shaped by recent glacial-interglacial climate changes (Fig. 2). Its native range covers Eurasia and Northern Africa, and it is naturalized widely in the world, including in North America and Japan (Hoffmann, 2002). Evolutionary analysis of a set of genome-wide markers suggests that post-Pleistocene expansion in the current species range approximately 17,000 years ago occurred from two glacial refugia in the Iberian Peninsula and Asia (Sharbel et al., 2000). The evolutionary shift to self-pollination is thought to have facilitated the expansion in the geographic distribution of this species (Shimizu et al., 2004; see below).
Figure 2.
Floral morphological diversity in the genus Arabidopsis. Left, Arabidopsis Col-0 accession. Right, Arabidopsis halleri subsp. gemmifera, collected in Osaka. This self-incompatible species has large petals to attract pollinator insects, and the anthers are separated from the stigma to avoid autopollination. Scale bar = 1 mm.
Arabidopsis displays a wide range of ecological relationships, including within- and between-species interactions and adaptations to abiotic environments. It responds physiologically and developmentally to a large variety of environmental cues, including light, daylength, vernalization, and nutrient and water levels (for review, see Pigliucci, 2002; Koornneef et al., 2004). Arabidopsis can be affected by bacterial and fungal pathogens and by insect herbivory. Despite being predominantly self-pollinating, flower visits by solitary bees, dipterans, and thrips have been observed in the field (Mitchell-Olds, 2001; Hoffmann et al., 2003). Furthermore, interactions with other individuals of Arabidopsis are ecologically important as mates and as competitors (Bates and Lynch, 2001; Shimizu et al., 2004; Köhler et al., 2005).
ECOLOGICAL AND EVOLUTIONARY GENOMICS: FINDING GENES ASSOCIATED WITH ECOLOGICAL TRAITS AND EVOLUTIONARY DIVERSIFICATION
The strength of Arabidopsis as a model system for ecological and evolutionary genetics is that it allows researchers to identify the genetic basis of a wide array of evolutionary and ecological phenomena. Several approaches to gene identification have been utilized to reveal the molecular genetic bases of various putative adaptive traits in this species, with the goal of determining the specific genetic polymorphism(s) responsible for ecologically and evolutionarily relevant phenotypic diversity.
Quantitative Trait Locus Mapping and the Isolation of Genes for Ecological and Evolutionary Traits
Many traits of interest to evolutionary biologists and ecologists, such as flowering time, water use efficiency, and trichome density, are quantitative in nature. It is no surprise that quantitative trait locus (QTL) mapping studies have been key components of Arabidopsis research, and this has been reviewed elsewhere (Koornneef et al., 2004). QTL mapping studies have been applied to examine the genetic basis of various traits, such as flowering time, inflorescence architecture, seed size, insect resistance, and light response (Koornneef et al., 2004; see below). Although only two accessions can be studied under standard QTL mapping techniques, the use of multiple populations can sample a wider range of natural variants, a point exemplified by a recent study of trichome density (Symonds et al., 2005). New genomic technologies, such as extreme array mapping using chip-based genomic arrays, are also beginning to provide more rapid mapping methods to identify QTL (Wolyn et al., 2004).
Linkage Disequilibrium Mapping and Candidate Gene Approaches as New Tools
A new genomics approach to trait locus mapping is linkage disequilibrium (LD) or association mapping, which may provide a new tool in the identification of genes underlying natural phenotypic (and perhaps adaptive) variation. In LD mapping, researchers exploit recombination and allele correlations that have occurred over evolutionary time to detect associations between particular genomic markers and specific phenotypes of interest. The use of LD mapping allows researchers to screen for alleles in a more diverse set of genotypes than is possible under standard QTL mapping studies. This procedure may, however, be complicated by nonindependence of individuals from each other in mapping populations due to population structure, but theoretical advances provide methods to take this into account. LD mapping in Arabidopsis has been used to detect correlations of flowering-time variation in CRY2 (Olsen et al., 2004) and FRI (Hagenblad et al., 2004). Estimates of LD in the Arabidopsis genome indicate that allele correlations can extend 50 to 250 kb, suggesting that whole-genome scanning by LD mapping in this species may be feasible (Nordborg et al., 2002). Whether such whole-genome LD mapping scans are possible remains to be seen; a recent study of LD mapping of known flowering-time genes shows that it may be difficult to detect genes underlying a trait if the genetic architecture is complex (e.g. involving epistasis or multiple alleles; Hagenblad et al., 2004).
Alternatively, researchers can exploit the high level of available molecular genetic information and use a candidate gene approach. With this method, genes that are known to affect a trait may be examined for further evidence that they are causally associated with a trait of interest in natural environments. Candidate gene approaches may also be used in conjunction with QTL and LD mapping strategies; this is most vividly illustrated by studies of the molecular genetic basis of ecological variation in flowering time (El-Assal et al., 2001). Knowledge of genetic networks has also been useful, as in a recent study in which researchers utilized information on the flowering-time pathway to identify a key epistatic regulatory interaction between the FRI and FLC genes that underlies a latitudinal cline in flowering time (Caicedo et al., 2004). It should be noted that all these association studies remain statistical associations until verified by genetic complementation (including transgenic complementation) techniques (see below).
Establishing Genetic Causality: Transgenic and Other Complementation Approaches
Transgenic methods provide important tools to prove that isolated genes (including candidate loci) actually underlie natural variation in the trait of interest. This is especially valuable in studying differences between species where genetic segregation analysis is impossible. For example, the transformation of SRK and SCR genes of Arabidopsis lyrata into Arabidopsis restored the self-incompatible response in the latter species, proving that mutations in these genes were responsible for the evolution of selfing (Nasrallah et al., 2002). Quantitative characters, however, may prove difficult to study by transgenic complementation, particularly if the alleles are of small to moderate effect. This is because independent transformants with the same transgenic genes very often display heterogeneous phenotypes, resulting from variability of transgene insertion locations and copy numbers. In certain cases, however, other methods may allow the analyses of quantitative characters by transgenic means (Tian et al., 2003).
Traditional genetic complementation tests are also routinely used to check whether a new laboratory-induced mutation is an allele of a known or novel gene; in principle, they could also be employed to identify genes underlying ecological traits or evolutionary changes. Maloof et al. (2001) used a complementation test to determine that a naturally occurring allele in the Lm-2 accession, which has reduced far-red sensitivity, is an allele of the PHYA gene.
Reverse Genomic Approaches: Adaptive Trait Locus Mapping
Finally, recent theoretical and experimental work suggests the utility of reverse genomic approaches collectively referred to either as adaptive trait locus or as hitchhiking mapping. These techniques rely on specific predictions of molecular evolutionary and population genetic theory on the levels and patterns of genetic variation expected for genes experiencing positive, or directional selection (selection that fixes an allele harboring an advantageous mutation that increases individual fitness; positive selection is often referred to as Darwinian selection) or balancing selection (selection that maintains variant alleles in a population, which may arise from heterozygote advantage, selection in variable environments, or fitness values that depend on allelic frequency; Luikart et al., 2003). For example, recent positive selection on a gene leads to reduced levels of nucleotide variation, while balancing selection is associated with increased levels of molecular diversity surrounding the selected mutation. Moreover, recurrent selection on protein sequence results in elevated levels of nonsynonymous (KA) nucleotide changes compared to synonymous (KS) substitutions. The latter approach has been used to identify genes associated with the divergence between Arabidopsis and its closely related congener A. lyrata (Barrier et al., 2003), and to examine the evolution of pollen genes between these two species (Schein et al., 2004). Reverse genetic approaches can then be used to determine the functions of identified adaptive trait genes and the phenotypic consequences associated with differential selection.
THE MOLECULAR BASIS OF ADAPTATION
Determining the genetic basis of adaptation is a central focus of evolutionary and ecological research. There have been concerted efforts in recent years to assess the genetics underlying putatively adaptive traits that vary within and between species. Using the approaches described above, we can identify the genes (and the specific polymorphisms within these genes) and determine the functional mechanisms underlying these adaptive traits and the evolutionary histories that gave rise to them (Mitchell-Olds, 2001; Shimizu, 2002; Luikart et al., 2003).
Successful studies along these lines have employed evolutionary genomic analyses to draw inferences on the evolutionary forces (including selection, drift, and population structure) that have shaped the history of these adaptive loci. A key concept of evolutionary genomics is that selection is a deterministic force that affects single genes, while population-level processes, such as population expansion and migration, are stochastic forces that affect all genes in the genome. Recent theoretical advances and the availability of genome-wide polymorphism data now permit researchers to discriminate between selective forces and population-level processes and thus identify genes underlying adaptive evolution (Luikart et al., 2003). Here, we discuss several examples that illustrate the utility of combining various approaches to understanding the diversification of adaptive traits, and an increasing number of such investigations using Arabidopsis are being employed to address evolutionary and ecological issues (Fig. 1).
Positive Selection on the Arabidopsis Selfing Locus
The evolutionary transition from out-crossing to selfing is one of the most prevalent trends in flowering plants. Charles Darwin (1876) proposed the reproductive assurance model to explain the prevalence of self-pollination in plants, suggesting that selfing can be evolutionarily advantageous when pollinators or mates are scarce in spite of inbreeding depression. Darwin's model also underlies Baker's rule, which states that colonizing species that disperse over long distances are generally self-compatible (Charlesworth, 2003; Shimizu et al., 2004, and refs. therein).
Self-incompatibility is a major mechanism to prevent selfing in plants. A. lyrata and many Brassicaceae species have a self-incompatible recognition system controlled by the Sterility (S)-locus, which harbors at least two functional genes, the female receptor gene SRK/Aly13 and the male ligand gene SCR/SP11. A number of S-haplotypes with divergent sequences are maintained by balancing selection in these species. Arabidopsis, however, has pseudogenes of SRK and SCR. Transgenic experiments showed that the loss of functional alleles at these genes is responsible for the emergence of selfing (Nasrallah et al., 2002).
The pseudoSCR1 gene in 21 Arabidopsis accessions has low levels of nucleotide diversity compared with neighboring genes in the pseudo S-locus and with genomic average. This low value is consistent with the hypothesis that the pseudogene allele of SCR1 was advantageous and recently spread to fixation in the species. Computer simulation based on coalescent theory (a mathematical theory to analyze the genealogy of DNA sequences, often used to derive inferences about demographic, population-level forces, and natural selection) demonstrates that this selection event most probably occurred very recently. The 95% confidence interval of the time estimate spans 0 to 320,000 years ago, when the planet experienced 100,000-year cycles of glacial-interglacial climate changes. Within this interval, the likelihood of the time estimate for the selective sweep was highest at T equals approximately 0 years, a time frame consistent with the expansion of the species range approximately 17,000 years ago after the last glacial retreats. If indeed selfing evolved during postglacial species expansion, it provides support for Darwin's reproductive assurance model, since rapid expansion would be accompanied by scarcities of mates and pollinators and thus selfing plants would have a selective advantage during long-distance dispersals (Shimizu et al., 2004). Also, the evolution of self-fertilization must have been followed by rapid evolution in floral morphological traits to facilitate selfing (Fig. 2). Those traits, including small flower size and autopollination, must have evolved after becoming self-compatible because they would have been deleterious if plants remained self-incompatible. These examples support the hypothesis that rapid adaptive evolution is a major response to climate change (for review, see Davis and Shaw, 2001).
Balancing Selection on the Disease Resistance Genes
Disease resistance genes are fascinating targets of selection, with their evolutionary dynamics driven by coevolution between the plant and the attacking pathogen (Bergelson et al., 2001). The Arabidopsis Col-0 accession is resistant to the bacterial pathogen Pseudomonas syringae avrRpm1, whereas the Nd-0 accession displays susceptibility. Mapping studies between Col-0 and Nd-0 showed resistance to this pathogen is conferred largely by one gene, resistance to Pseudomonas syringae pv maculicola (RPM1), which was isolated by map-based cloning (Grant et al., 1995).
Sequencing of the RPM1 gene in 26 Arabidopsis accessions revealed that resistant accessions had a functional allele while nonresistant accessions contained a large deletion spanning the gene. These two alleles showed high divergence in their flanking sequences, suggesting their long-term maintenance within the species, and the high divergence of disease resistance genes has previously been interpreted as evidence for an evolutionary arms race. This model posits that the dynamics of disease resistance genes drive selection in the plant to recognize rapidly evolving pathogens. Patterns of polymorphism at RPM1, however, indicate that balancing selection has maintained the resistant and nonresistant alleles for long evolutionary periods, inconsistent with a predicted high turnover of alleles as hypothesized by the arms-race model (Stahl et al., 1999). Instead, investigators proposed a trench warfare model in which advances and retreats of resistant allele frequency maintain variation for disease resistance as a dynamic polymorphism (Stahl et al., 1999).
Why, then, has the susceptible allele been maintained? One ecological hypothesis is that there is a cost associated with the disease resistance phenotype. To test this hypothesis, Tian et al. (2003) conducted a transgenic experiment to measure the fitness cost associated with plant defense by RPM1 in growing plants with and without the functional RPM1 gene in field conditions and in the absence of pathogen. They found a 9% decrease in seed number in plants with a functional RPM1 allele, demonstrating a high fitness cost associated with the maintenance of RPM1.
Other studies have continued to document the possible action of balancing selection on other disease resistance genes. Another noteworthy recent study revolves around the RPP13 gene, which confers resistance to the fungal pathogen Hyaloperonospora (Perenospora) parasitica associated with the expression of the ATR13 protein. High levels of amino acid polymorphisms of both the plant gene RPP13 and the fungal gene ATR13 suggest an interaction mediated by balancing selection (Allen et al., 2004). This study highlights the possibilities of exploring the molecular coevolutionary dynamics associated with biotic interactions between species.
The Ecological Genetics of Herbivory
Secondary metabolites play major roles in plant resistance to insect herbivores. QTL mapping studies in Arabidopsis have detected a major locus that underlies both the diversity of glucosinolate compounds and resistance to specialist (Plutella xylostella) and generalist (Trichoplusia ni) insect herbivores. Interestingly, high glucosinolate levels provide resistance to generalist, but not to specialist, herbivores (Kliebenstein et al., 2002), consistent with the ecological hypothesis that defense by secondary metabolites is effective against generalists but is overcome by specialist herbivores. Fine-mapping analysis for resistance genes showed that one of the major QTL for this trait harbors a small, tandem gene family encoding methylthioalkylmalate (MAM) enzymes, which determine the side-chain length of the aliphatic glucosinolates in the plant. Molecular population genetic analysis of this tandem family in 25 accessions reveals complex molecular variants, including evidence for frequent gene loss and gene conversion. These data also suggest the possible action of balancing selection in maintaining this variation at MAM (Kroymann et al., 2003), and may indicate that an ecological tradeoff between the cost of having these alleles and the resistance patterns to generalist and specialist insect herbivores is responsible for MAM genetic diversity.
Flowering Time
The onset of flowering is a major life history transition in flowering plants and is sensitive to various seasonal climatic signals, including photoperiod and vernalization (Koornneef et al., 2004). These cues vary systematically with latitude, and evolutionary adaptation to these ecological cues would be expected to lead to latitudinal clines in flowering times. Since Arabidopsis is a wild weed distributed over a wide latitudinal range in Eurasia and North Africa, latitude-dependent variation in flowering time might be expected to be associated with molecular polymorphisms in genes that regulate flowering time in response to environmental cues.
A number of genes have been identified that regulate flowering time in Arabidopsis. Polymorphisms in several of these, including the photoperiod pathway gene CRY2 (El-Assal et al., 2001; Olsen et al., 2004) and the vernalization gene FRI (Hagenblad et al., 2004), are known to underlie natural variation in flowering time in this species. Systematic variation in flowering time with latitude has been detected in common garden field experiments (Stinchcombe et al., 2004). This cline appears to be modulated by an epistatic interaction between FRI and the downstream gene FLC, which encodes a MADS box transcription factor (Caicedo et al., 2004).
Etiolation and Shade Avoidance
Light is a major ecological cue for plants. It affects diverse ecological phenomena, including etiolation, shade avoidance, and flowering time. Maloof et al. (2001) measured hypocotyl length of Arabidopsis seedlings in 141 accessions and found that this trait varied systematically with the latitude of origin of the accessions; those from lower latitudes generally had longer hypocotyls. The authors suggest that Arabidopsis has adapted to higher light intensities at lower latitudes by being less sensitive to light. A similar latitudinal cline in hypocotyl length has been detected using Norwegian populations, but in red and far-red light rather than white light (Stenøien et al., 2002).
Another ecological response to light is the shade avoidance response, which allows plants to respond to the presence of overtopping neighbors and is mediated in part by phytochrome perception (Pigliucci, 2002). Callahan and Pigliucci (2002) demonstrated the shade avoidance response in other Scandinavian accessions but found no evidence of adaptation, raising the possibility that this response is a vestigial evolutionary characteristic (Pigliucci, 2002).
Conflict and Cooperation: Parental Imprinting and Pollen-Pistil Interactions
The competitive and cooperative strategies in pollen-pistil interaction and subsequent embryogenesis have been major themes in reproductive ecology. Intragenomic conflict theory proposes that mothers and fathers have conflicting interests in the allocation of nutrients from the mother to its embryos (Haig and Westoby, 1991). Since pollen from multiple fathers can succeed in fertilization in one mother, the paternal interest is to extract nutrients for his own offspring to the detriment of embryos sired by pollen from other plants. In contrast, the maternal interest is to distribute the nutrients equally among her embryos. Analysis of the MEDEA gene illustrates the molecular aspect of these conflicting interests. Genomic imprinting via methylation results in MEDEA being expressed only in the female side, which promotes the maternal interest by inhibiting the overgrowth of embryos due to the transcriptional repression of a MADS box gene PHERES1 (Köhler et al., 2005).
In contrast, cooperative interaction between female gametophytes is observed before fertilization. Relatedness among sibling female gametophytes has a mean of 0.5, since one-half of their genomes on average is shared after meiosis. Genetic analysis using maa gametophytic mutants showed that the female gametophyte was responsible for the prevention of polyspermy and facilitates fertilization of the sibling female gametophytes by making more male pollen tubes available. The strategy of female gametophytes bears striking similarities to those of altruistic worker bees, whose haplo-diploid social structure also results in shared genetic identity (Shimizu and Okada, 2000).
Male and female interactions also underlie speciation, since species are generated and maintained by reproductive isolation that inhibits hybridization. Rapid evolution is often the hallmark for genes responsible for reproductive traits and speciation in animals (Wu and Ting, 2004). It is thus noteworthy that the GRP gene family encoding pollen surface proteins also displays accelerated evolution, although the phenotypic consequences of this rapid diversification have yet to be established (Fiebig et al., 2004; Schein et al., 2004).
NATURAL, ECOLOGICALLY RELEVANT ENVIRONMENTS
Most phenotypic characterizations of Arabidopsis traits, whether by mutant analysis, natural variation among accessions, or QTL mapping studies, are generally undertaken in controlled growth conditions. Although some of these growth conditions are meant to mimic natural environments, they invariably are artificial. A biogeographic study by Hoffmann (2002), for example, showed that laboratory conditions (rather high temperatures and moisture levels) were not found in climatic conditions in the natural range of this species. While stable laboratory conditions are necessary to dissect plant phenotypes, it should be emphasized that controlled conditions may be a poor reflection of the complex, dynamically changing environments observed in nature.
It is increasingly clear that phenotypic traits and their underlying genetic bases may differ substantially between natural and artificial growth conditions. Weinig et al. (2003), for example, showed that the genetic loci for flowering-time variation in Col × Ler recombinant inbred lines differed in controlled photoperiod environments versus field settings in North Carolina and Rhode Island. Several of the QTL identified in laboratory environments were not detected in field experiments, suggesting that those genes may have little influence on flowering-time variation under these two field conditions. More importantly, several novel loci were identified only in field conditions and not in controlled environments. The importance of field conditions is further underscored by recent work that shows that a latitudinal cline in flowering time, associated with the epistatic interaction between FRI and FLC, can only be observed if the phenotypes are assayed in common garden field experiments (Caicedo et al., 2004; Stinchcombe et al., 2004).
Although attempting to analyze responses to natural environments may seem daunting, ecologists have developed experimental and statistical methods to dissect plant responses to complex environments. Engelmann and Schlichting (2005), for example, examined the effect of water stress on Arabidopsis morphology and life history, varying not only the amount, but also the grain of the water treatments (constant versus unstable). Unstable water treatments are more reflective of natural conditions given the idiosyncratic nature of rainfall patterns in the wild. Indeed, unstable water treatments generated responses that were distinct from those under constant water regimes, including differences in which specific morphological or phenological traits display plasticity to water regimes. They also demonstrated interactions between treatment regimes; for example, that a low-level, constant water treatment resulted in early flowering, reminiscent of a drought escape response, and that mortality was highest with a nonconstant application rate of low water amounts. Studies like this allow molecular geneticists to design novel experiments such as mutant screening and microarray analysis in conditions that better reflect the natural environment that plants experience.
PERSPECTIVES
Although there has been tremendous progress in the last decade, a gap still exists between ecological studies, molecular genetics, and evolutionary genomics. Each field tends to employ a different vocabulary and interaction among these disparate fields remains limited. There is a pressing need to synthesize these somewhat distinct fields for two reasons. First, further understanding of some evolutionary and ecological processes clearly requires us to probe the molecular nature underlying organismal phenotypes and responses. It is only by isolating relevant genes, examining the nature of variation at these loci, and determining the resultant functional consequences of such variation that we can begin to gain insight into the nature and history of ecological and evolutionary diversification. Molecular analysis can study signatures of adaptive change, including those that have occurred in the evolutionary past. Molecular dissection has allowed us to examine, for example, the ecology of disease resistance or the adaptive evolution of selfing during past climate change. Molecular tools are also being developed in other species of Arabidopsis, which display a variety of ecological phenotypes and allow us to extend our studies to other ecological and evolutionary phenomena (Mitchell-Olds, 2001).
Second, understanding the ecological and evolutionary setting in which genes operate allows us to gain a deeper appreciation of their functions. Functions of genes are generally described in biochemical, developmental, or physiological terms. They can also be viewed, however, in light of the ecological context in which they are expressed or in their associations with evolutionary adaptations. Indeed, the power of ecological and evolutionary investigation may lead us to find new genes underlying long-studied traits, such as flowering time, which may escape notice under standard laboratory conditions. This, in turn, provides a foundation for a more comprehensive annotation of genome function. It is in both these areas that Arabidopsis offers unique opportunities: to harness the power of genetics and genomics to further our understanding of ecology and evolution, on one hand, and of development and physiology, on the other.
Acknowledgments
We thank Johanna Schmitt, Tanaka Kenta, Hiroshi Kudoh, and the Purugganan laboratory members for helpful discussions and/or critical reading of the manuscript.
This work was supported in part by a fellowship from the Japan Society for the Promotion of Science (to K.K.S.) and by grants from the National Science Foundation (to M.D.P.).
References
- Allen RL, Bittner-Eddy PD, Grenville-Briggs LJ, Meitz JC, Rehmany AP, Rose LE, Beynon JL (2004) Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306: 1957–1960 [DOI] [PubMed] [Google Scholar]
- Barrier M, Bustamante CD, Yu J, Purugganan MD (2003) Selection on rapidly evolving proteins in the Arabidopsis genome. Genetics 163: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP (2001) Root hairs confer a competitive advantage under low phosphate availability. Plant Soil 236: 243–250 [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D (2001) Evolutionary dynamics of plant R-genes. Science 292: 2281–2285 [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD (2004) Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA 101: 15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan HS, Pigliucci M (2002) Shade-induced plasticity and its ecological significance in wild populations of Arabidopsis thaliana. Ecology 83: 1965–1980 [Google Scholar]
- Charlesworth D (2003) Effects of inbreeding on the genetic diversity of populations. Philos Trans R Soc Lond B Biol Sci 358: 1051–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C (1876) The Effects of Cross and Self Fertilization in the Vegetable Kingdom. John Murray, London
- Davis MB, Shaw RG (2001) Range shifts and adaptive responses to quaternary climate change. Science 292: 673–679 [DOI] [PubMed] [Google Scholar]
- El-Assal SE-D, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29: 435–440 [DOI] [PubMed] [Google Scholar]
- Engelmann KE, Schlichting CD (2005) Coarse- versus fine-grained water stress in Arabidopsis thaliana (Brassicaceae). Am J Bot 92: 101–106 [DOI] [PubMed] [Google Scholar]
- Fiebig A, Kimport R, Preuss D (2004) Comparisons of pollen coat genes across Brassicaceae species reveal rapid evolution by repeat expansion and diversification. Proc Natl Acad Sci USA 101: 3286–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Hagenblad J, Tang C, Molitor J, Werner J, Zhao K, Zheng H, Marjoram P, Weigel D, Nordborg M (2004) Haplotype structure and phenotypic associations in the chromosomal regions surrounding two Arabidopsis thaliana flowering time loci. Genetics 168: 1627–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D, Westoby M (1991) Genomic imprinting in endosperm: its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos Trans R Soc Lond B Biol Sci 333: 1–13 [Google Scholar]
- Hoffmann MH, (2002) Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae). J Biogeogr 29: 125–134 [Google Scholar]
- Hoffmann MH, Bremer M, Schneider K, Burger F, Stolle E, Moritz G (2003) Flower visitors in a natural population of Arabidopsis thaliana. Plant Biol 5: 491–494 [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T (2002) Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Page DR, Gagliardini V, Grossniklaus U (2005) The Arabidopsis thaliana MEDEA polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37: 28–30 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T (2003) Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci USA (Suppl 2) 100: 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4: 981–994 [DOI] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T (2001) Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol Evol 16: 693–700 [Google Scholar]
- Nasrallah ME, Liu P, Nasrallah JB (2002) Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297: 247–249 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, Oefner PJ, et al (2002) The extent of linkage disequilibrium in Arabidopsis thaliana. Nat Genet 30: 190–193 [DOI] [PubMed] [Google Scholar]
- Olsen KM, Halldorsdottir SS, Stinchcombe JR, Weinig C, Schmitt J, Purugganan MD (2004) Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167: 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M (2002) Ecology and evolutionary biology of Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi 10.1199/tab.0003, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Schein M, Yang Z, Mitchell-Olds T, Schmid KJ (2004) Rapid evolution of a pollen-specific oleosin-like gene family from Arabidopsis thaliana and closely related species. Mol Biol Evol 21: 659–669 [DOI] [PubMed] [Google Scholar]
- Sharbel TF, Haubold B, Mitchell-Olds T (2000) Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol Ecol 9: 2109–2118 [DOI] [PubMed] [Google Scholar]
- Shimizu KK (2002) Ecology meets molecular genetics in Arabidopsis. Popul Ecol 44: 221–233 [Google Scholar]
- Shimizu KK, Cork JM, Caicedo AL, Mays CA, Moore RC, Olsen KM, Ruzsa S, Coop G, Bustamante CD, Awadalla P, et al (2004) Darwinian selection on a selfing locus. Science 306: 2081–2084 [DOI] [PubMed] [Google Scholar]
- Shimizu KK, Okada K (2000) Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127: 4511–4518 [DOI] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671 [DOI] [PubMed] [Google Scholar]
- Stenøien HK, Fenster CB, Kuittinen H, Savolainen O (2002) Quantifying latitudinal clines to light responses in natural populations of Arabidopsis thaliana (Brassicaceae). Am J Bot 89: 1604–1608 [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J (2004) A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci USA 101: 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds VV, Godoy AV, Alconada T, Botto JF, Juenger TE, Casal JJ, Lloyd AM (2005) Mapping quantitative trait loci in multiple populations of Arabidopsis thaliana identified natural allelic variation for trichome density. Genetics 169: 1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Weinig C, Dorn LA, Kane NC, German ZM, Halldorsdottir SS, Ungerer MC, Toyonaga Y, Mackay TF, Purugganan MD, Schmitt J (2003) Heterogeneous selection at specific loci in natural environments in Arabidopsis thaliana. Genetics 165: 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyn DJ, Borevitz JO, Loudet O, Schwartz C, Maloof J, Ecker JR, Berry CC, Chory J (2004) Light-response quantitative trait loci identified with composite interval and eXtreme array mapping in Arabidopsis thaliana. Genetics 167: 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Ting CT (2004) Genes and speciation. Nat Rev Genet 5: 114–122 [DOI] [PubMed] [Google Scholar]