THE CHALLENGE OF DISSECTING GENE EXPRESSION IN MULTICELLULAR ORGANISMS
The complete genome sequences of two representative species of flowering plants, the monocot rice (Oryza sativa) and the eudicot Arabidopsis (Arabidopsis thaliana), provide us with a new opportunity to understand developmental and physiological events at the genome level (Arabidopsis Genome Initiative, 2000; Goff et al., 2002; Yu et al., 2002). Large numbers of genes in each organism (approximately 28,000 in Arabidopsis; Yamada et al., 2003; >40,000 in rice; Bennetzen et al., 2004) make the analysis of the regulation of every individual gene a considerable challenge. However, technological advances in quantifying RNA levels have made global gene analysis more feasible and reproducible. For Arabidopsis and rice, commercial oligonucleotide microarrays, which cover significant portions of genes in each genome, are available, as are custom-designed arrays (Zhu, 2003). For organisms whose genomes are not sequenced, spotted microarrays with expressed sequence tag (EST) clones are broadly available. In addition to hybridization-based measurements of gene expression, sequencing, or PCR-based gene expression analyses are being used. Available expression profiling technologies are well summarized by Breyne and Zabeau (2001).
One of the biggest challenges in studying global gene regulation in multicellular organisms is the heterogeneity of gene expression. Each organ is unique at the level of its tissues, cells, and gene expression profiles. In addition, there is growing evidence that responses to environmental stimuli or developmental signals occur differentially at the cell or tissue level. Thus, to better understand gene regulatory circuits, gene expression should be analyzed at a single cell or tissue type. Because the quantity of RNA, protein, or metabolites obtained from a single cell is very small, the development of sophisticated technologies is necessary. We need new techniques to efficiently isolate molecules from single cells and the equipment to detect and quantify small amounts of molecules.
SINGLE-CELL TRANSCRIPT PROFILING: BROADLY APPLICABLE TECHNIQUES
Improvements in RNA amplification techniques have made it possible to analyze small amounts of mRNA from very little starting material using either PCR amplification of primer-tagged cDNA (Hertzberg et al., 2001) or linear amplification through in vitro transcription on cDNA that is tagged with the promoter for RNA polymerase. In the latter case, optimized RNA amplification procedures have been shown to maintain the relative amounts of each RNA species (Van Gelder et al., 1990; Wang et al., 2000; Baugh et al., 2001). This technique is used for generating labeled RNA probes for hybridization to microarrays or for constructing cDNA libraries. Although the length of RNA decreases with amplification, it is sufficient for microarray hybridization or EST sequencing.
Another challenging problem of single-cell transcript profiling is the efficient and precise dissection of target cells from plant tissue. The presence of cell walls and large vacuoles of plant cells make the dissection of single cells difficult. However, several research groups have recently profiled gene expression at the single-cell level using various methods. Their approaches are summarized below.
Micropipetting
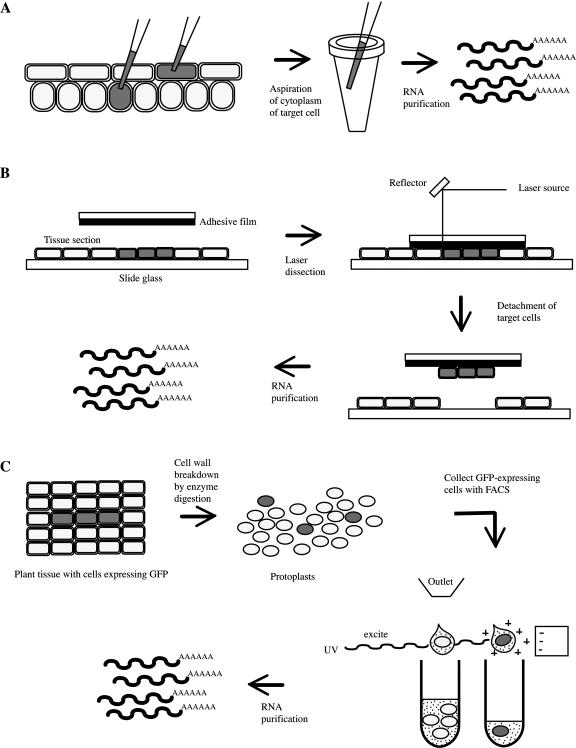
Micropipetting has been used to directly extract the contents of cells using microcapillaries or pipettes (Karrer et al., 1995). This method has been used to extract RNA from the epidermis, guard cells, and mesophyll of intact leaves (Fig. 1A). For global transcriptional profiling, extracts from 20 mesophyll and 50 epidermal cells were used for cDNA synthesis and microarray probe generation (Brandt et al., 2002). A total of 120 genes were differentially expressed in the two cell types; 98 genes increased in the mesophyll (enriched with genes involved in photosynthesis), and 22 genes were more highly expressed in the epidermis.
Figure 1.
Schematic of isolation of single cell or tissue type for transcript profiling. A, Micropipetting. B, LCM. C, Protoplasting and sorting.
Laser-Capture Microdissection
Although micropipetting is a straightforward method for extracting RNA from intact tissues, its application is limited in that internal cells are not easily accessed with microcapillaries or pipettes, and there is no good way to visually supervise the extraction due to the limited depth of field of most microscopes. To isolate RNA from cells residing in anatomically complex tissues, a special dissecting tool for visualizing the target cells is essential.
Laser-capture microdissection (LCM) has been developed and used in gene expression profiling of animal cells (Emmert-Buck et al., 1996; Luzzi et al., 2003). Recently, it has also been applied to plant cells. A block of fixed tissue that is either frozen or embedded in paraffin is sectioned, and the region of interest is dissected out by a laser and immediately attached to adhesive film (Fig. 1B) or captured into the RNA extraction buffer in a tube cap by electrostatic action. Asano et al. (2002) constructed a cDNA library from 150 rice phloem cells using LCM and T7 polymerase RNA amplification. A paper by Kerk et al. (2003) describes technical details of LCM applied to a broad range of plant species and tissue types. For instance, LCM-based dissections of epidermal cells and vasculature from cryosections of fixed coleoptiles in maize (Zea mays) have been profiled on cDNA microarrays (Nakazono et al., 2003). About 1.5% of approximately 8,800 genes analyzed were found to be more than 2-fold enriched in epidermis versus vasculature and total coleoptile; a similar percentage of genes were also enriched in vasculature. Functional categories of genes enriched in each tissue showed differences, which partially reflect unique physiological roles of each tissue such as protection by epidermis and selective transport and support of plant body by vascular tissue. In the epidermis, genes involved in the shikimate pathway, secondary metabolism, and defense were enriched, whereas, in vasculatures, genes involved in transport, metal binding, lignin biosynthesis, and proteolysis were found to be enriched.
The LCM technique has at least two advantages: (1) it minimizes the extensive manipulation of tissues that could change the RNA profile, and (2) as tissues are fixed in large scale simultaneously, effects of collection time on experiments with important temporal components, such as the circadian clock, are reduced. However, in most cases, LCM is very labor intensive during the dissection step and is tricky for the isolation of small cells or tissues with few cell layers. Nakazono et al. (2003) collected more than 10,000 cells for RNA isolation, which likely needed hundreds of dissections. In particular, meristem cells are challenging for dissection as their size and number are small. Even with tissue sections of approximately 6 μm in thickness, the probability of contamination with neighboring cells could still be high.
Protoplasting and Sorting
For the quick and accurate isolation of RNA from small meristematic cells, protoplasting and sorting techniques have been developed and then used to generate a global expression map for Arabidopsis roots (Fig. 1C; Birnbaum et al., 2003). Cells from transgenic lines expressing green fluorescent protein (GFP) in specific tissues or regions were isolated by protoplasting and sorting through a fluorescence-activated cell sorter (FACS), and the labeled RNA was hybridized to the Affymetrix ATH1 GeneChip (Santa Clara, CA). With this technique, about 107 cells were processed within 1.5 h. In addition to sorting GFP-expressing cells, longitudinal zones of the root were dissected into three stages, and RNA from each stage was hybridized to the ATH1 arrays. By combining the radial and longitudinal expression values, expression levels of approximately 22,000 genes in 15 root zones were generated. The expression profile of each gene in 15 root regions was termed a “digital in situ” and is available at the Arabidopsis Gene Expression database (AREX; http://www.arexdb.org).
Of the 10,492 genes that are detected in roots above a conservative threshold, 5,717 (approximately 54%) were differentially expressed in at least one subzone and had at least a 4-fold difference in expression level. This number is remarkably higher than in the other analyses discussed above, even considering the differences in analysis methods and types of microarrays used. Hierarchical clustering identified eight regions in which sets of genes are differentially expressed, and four regions (localized expression domains) had an enrichment of hormone-signaling genes (i.e. auxin, jasmonic acid, and gibberellic acid).
Although there were general concerns about the effect of protoplasting on transcriptional profiles, the number of genes whose expression changed was small (R2 > 0.9 for three replicates of the whole root versus the protoplasted root; Birnbaum et al., 2003).
By knowing the spatial expression patterns of most of the genes in the Arabidopsis genome, it is now possible to facilitate the positional cloning of root mutants according to their digital in situ patterns. Overlapping expression patterns of close homologs may also allow for more informed reverse genetic approaches to discover genes with potentially redundant functions.
In addition to these applications, the digital in situ can serve as a powerful tool to predict sets of tissue-specific genes. In order to identify cell type-specific transcription factors, we generated promoter::GFP transgenic plants for candidate genes based on the enrichment of transcripts in specific cells. In these experiments, GFP expression patterns showed a remarkable correlation to the digital in situ data, and we could generate new cell type-specific GFP lines for further transcriptional profiling (J.-Y. Lee, J. Colinas, J.Y. Wang, and P.N. Benfey, unpublished data).
One of the daunting tasks in profiling root cells is the characterization of transcripts in the quiescent center (QC). In Arabidopsis, there are only four to seven QC cells in the meristem. To understand the gene expression activity in root stem cells, transcripts of QC cells expressing the AGL42::GFP reporter were profiled as described above (Nawy et al., 2005). The first characterization of plant stem cell expression profiles illuminates how finely tuned the process of sorting and transcriptional profiling can get.
Although protoplasting and sorting is a very time-efficient and highly reproducible technique, it has only been used so far in roots. How well green parts of the plant may be sorted remains to be tested. Even though the generation of GFP-expressing lines is quite difficult for many plant species, Arabidopsis is easy to transform, and there are many publicly available collections of GFP lines (i.e. http://www.plantsci.cam.ac.uk/Haseloff/Home.html; http://enhancertraps.bio.upenn.edu).
SINGLE-CELL TRANSCRIPT PROFILING: CELL TYPE-DEPENDENT TECHNIQUES
The technologies introduced above may be applied, to a greater or lesser extent, to several species and various cell types. However, transcriptional profiling has been performed on single cells or tissue types that are easier to isolate, such as pollen, guard cells, xylem, and cambium.
Pollen Profiling
Mature pollen was physically separated from flowers (Honys and Twell, 2003) or only hydrated pollen was isolated by FACS (Becker et al., 2003), and the labeled RNA was hybridized to the Affymetrix approximately 8,300-gene Arabidopsis Genome Array (AGA). In both cases, the authors found that far fewer genes had presence calls when compared to the genes from other vegetative organs (21% versus 56%–64%; 13% versus 18%–32%, respectively). Many of the genes were uniquely expressed in pollen, and their functional categories were enriched for genes annotated as being involved in cell wall synthesis, cytoskeleton, or signal transduction. Hierarchical clustering analyses of genes in six gene families, including receptor-like kinases and glycoside hydrolases, found subsets of genes in those families that are expressed only in pollen and not in other vegetative organs, suggesting the acquisition of their exclusive roles for pollen development (Honys and Twell, 2003).
Guard Cell Profiling and Abscisic Acid Signaling
Guard cells and mesophyll cells were isolated from mature leaves by protoplasting epidermal peels and chopped leaves, respectively. Highly pure populations of each cell type (99% purity for guard cells) were obtained by multiple iterations of filtering and washing the protoplasts through nylon mesh (with a pore size of 10 μm for guard cells and 30 μm for mesophyll cells). Their gene expression profiles were analyzed on the AGA microarray with and without treatment with abscisic acid (ABA; Leonhardt et al., 2004). Genes were identified that were differentially expressed in the two cell types, as well as differentially regulated by ABA in the two cell types. It is notable that the number of overlapping genes that showed changes in gene expression under ABA treatment between cell type-specific analysis and whole tissues was quite small. This suggests that plant hormone regulation can vary according to cell type, and the level of sensitivity differs greatly between isolated cells and whole tissues. As validation of their microarray results, the authors also identified a mutant that encodes for a protein phosphatase and is hypersensitive to ABA treatment. Considering the large number of genes in the protein phosphatase family (approximately 69 members) and the paucity of recessive phenotypes for these genes, the information about gene regulation at a single-cell level was indispensable in predicting the components of this signaling pathway. With the identification of genes regulated by ABA in each cell type, the authors could propose a working model of the ABA signaling pathway, which can provide a basis for future studies.
Xylem Cell Differentiation Profiling in Zinnia
Zinnia is an excellent system for studying xylem differentiation in vitro. Mesophyll cells isolated from leaves are placed in liquid culture and supplied with auxin and cytokinin. Differentiated mesophyll cells will undergo transdifferentiation into tracheary elements. Time-course expression profiling during transdifferentiation was reported by two research groups (Demura et al., 2002; Milioni et al., 2002). Despite the differences in the profiling methods (cDNA microarray by Demura et al., 2002; cDNA AFLP by Milioni et al., 2002), genes involved in auxin signaling and cell wall synthesis, as well as homeodomain transcription factors involved in xylem differentiation, were commonly found in both studies, demonstrating the sequential transcriptional regulation in the time course of transdifferentiation.
Cambium Profiling in Aspen
Due to technical difficulties, the molecular mechanisms of secondary growth have remained largely uncharacterized. However, EST sequencing and tools for global transcriptional profiling have recently shed light on the processes that are characteristic of trees. Schrader et al. (2004) reported high-resolution transcriptional profiling of vascular cambium. A series of approximately 20-μm-thin tangential sections around cambium was made, and mRNA was purified from each section for hybridization onto cDNA microarrays with probes representing approximately 13,000 genes. Cluster analysis revealed gene sets differentially expressed in three major regions: phloem mother cells, cambial stem cells, and xylem mother cells. Comparisons of cambial stem cell expression profiles to those of apical meristems showed that they shared genes that have been shown to be important in meristem maintenance, suggesting similar transcriptional pathways between primary and secondary meristems.
FUTURE APPLICATIONS OF HIGH-THROUGHPUT RNA ISOLATION TECHNOLOGIES
The three technologies described above are summarized in Table I. Each method has both advantages and disadvantages in terms of efficiency and applicability. Results of experimental technology, however, have added considerably to our understanding of gene regulation at the cellular level, and they will certainly become more widely used in the near future.
Table I.
Comparison of cell isolating technologies
| Technology | Micropipetting | LCM | Protoplasting-Sorting |
|---|---|---|---|
| Can any cell type be used? | No, only accessible cells by microcapillaries | Yes | No, only cells tagged with fluorescence |
| How long does it take to isolate target cells from living plants?a | Less than a day depending on the skills of using microcapillaries | Days due to the required time for fixation, mounting, and sectioning | Half a day for protoplasting and sorting |
| Can any plant species be used? | Yes | Yes | No, only species that are transformable |
| What is the main equipment required? | High-resolution dissecting microscope | Microdissection instrument for LCM | FACS |
| References | Brandt et al. (2002) | Kerk et al. (2003); Nakazono et al. (2003) | Birnbaum et al. (2003); Nawy et al. (2005) |
Once the target cells are isolated, RNA purification and the analysis by microarrays, PCR, or sequencing-based methods take equivalent times for all three techniques.
The importance of cell-level transcriptional profiling cannot be overemphasized, as the study of ABA responses in guard cells demonstrates (Leonhardt et al., 2004). Previous studies of whole-plant transcriptional profiles generated a very different picture from the one observed with single cell-type profiles. For instance, only 20% and 27% of genes induced by ABA in the profiling of mesophyll and guard cells, respectively, were also present in similar studies of whole plants. This suggests that regulatory networks that are inferred from whole-plant profiling may not be accurately projected onto the individual cell type, and vice versa. Thus, it would be more accurate to draw a big picture of gene regulation in response to hormones or other stimuli in the whole plant, based on an understanding of dynamic responses at the cellular level.
Root cell profiling has clearly demonstrated how differently gene expression may be regulated in each tissue and between developmental stages (Birnbaum et al., 2003). A total of 54% of genes expressed in roots are differentially expressed in root subzones, which is much higher than the proportion that is differentially expressed (37%) among seven organs in maize (Cho et al., 2002). More recent transcriptional profiling with more specific cell-type populations (i.e. protoxylem, pericycle, lateral root initials, etc.) in our lab is revealing an even greater number of differentially expressed genes. It is clear from these results that combining highly refined gene expression maps and molecular genetic approaches for physiological and developmental pathways will greatly facilitate understanding of root development and evolution in the coming years (Birnbaum and Benfey, 2004).
The accumulation of genome-level data makes the goal of building gene regulatory networks in plants much more attainable (for stress responses, see the review in Chen and Zhu, 2004). Currently, more than 1,500 Affymetrix chip data sets are deposited in The Arabidopsis Information Resource (http://www.arabidopsis.org), and the expression data for genes of interest can be searched or the entire data set can be downloaded for analysis. The program Genevestigator (Zimmermann et al., 2004) is a Web-based search engine for finding genes with specific expression patterns under particular physiological conditions, located in specific tissue types, and/or developmental stages from these microarray data sets. The AREX database (http://www.arexdb.org/index.jsp) also provides similar query tools for searching differentially expressed genes under user-defined criteria and for digitally visualizing individual gene expression profiles in the Arabidopsis root. These expression search tools can aid in determining gene functions and in designing experiments.
Combining the data sets from several cell and tissue types under well-designed experimental conditions, developmental and physiological pathways in the whole organism at a systems level can be identified (Pennisi, 2003; Levesque and Benfey, 2004). Obviously, for multicellular organisms, generating genomic data at the individual cell level for network building is an enormous amount of work. However, the accumulation of high-resolution and high-quality data is essential to fully understanding network connectivity (Kitano, 2002; Provart and McCourt, 2004). With advances in more reproducible cell-level transcriptional profiling, the highly complex map of plant development will achieve unprecedented resolution in the coming years.
Acknowledgments
We thank Juliette Colinas, Hongchang Cui, and Jean Wang for critical reading of the manuscript and comments.
This work was supported by the National Science Foundation AT2010 project (grant no. 0209754).
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K-I (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 32: 401–408 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Brown EL, Hunter CP (2001) Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res 29: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Coleman C, Liu R, Ma J, Ramakrishna W (2004) Consistent over-estimation of gene number in complex plant genomes. Curr Opin Plant Biol 7: 732–736 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Benfey PN (2004) Network building: transcriptional circuits in the root. Curr Opin Plant Biol 7: 582–588 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Sasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brandt S, Kloska S, Altman T, Kehr J (2002) Using array hybridization to monitor gene expression at the single cell level. J Exp Bot 53: 2315–2323 [DOI] [PubMed] [Google Scholar]
- Breyne P, Zabeau M (2001) Genome-wide expression analysis of plant cell cycle modulated genes. Curr Opin Plant Biol 4: 136–142 [DOI] [PubMed] [Google Scholar]
- Chen WJ, Zhu T (2004) Networks of transcriptional factors with roles in environmental stress response. Trends Plant Sci 9: 591–596 [DOI] [PubMed] [Google Scholar]
- Cho Y, Fernandes J, Kim S-H, Walbot V (2002) Gene-expression profile comparisons distinguish seven organs of maize. Genome Biol 3: research0045 [DOI] [PMC free article] [PubMed]
- Demura T, Tashiro G, Horiguchi G, Kishimoto N, Kubo M, Matsuoka N, Minami A, Nagata-Hiwatashi M, Nakamura K, Okamura Y, et al (2002) Visualization by comprehensive microarray analysis of gene expression program during transdifferentiation of mesophyll cells into xylem cells. Proc Natl Acad Sci USA 99: 15794–15799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA (1996) Laser capture microdissection. Science 274: 998–1001 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Hertzberg M, Sievertzon M, Aspeborg H, Nilsson P, Sandberg G, Lundeberg J (2001) cDNA microarray analysis of small plant tissue samples using cDNA tag target amplification protocol. Plant J 25: 585–591 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer EE, Lincoln JE, Hogenhout S, Bennett AB, Bostock RM, Martineau B, Lucas WJ, Gilchrist DG, Alexander D (1995) In situ isolation of mRNA from individual plant cells: creation of cell-specific cDNA libraries. Proc Natl Acad Sci USA 92: 3814–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM (2003) Laser capture microdissection of cells from plant tissues. Plant Physiol 132: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H (2002) Looking beyond the details: a rise in system-oriented approaches in genetic and molecular biology. Curr Genet 41: 1–10 [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analysis of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2c mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque M, Benfey PN (2004) Systems biology. Curr Biol 14: R179–R180 [DOI] [PubMed] [Google Scholar]
- Luzzi V, Mahadevappa M, Raja R, Warrington JA, Watson MA (2003) Accurate and reproducible gene expression profiles from laser capture microdissection, transcript amplification, and high density oligonucleotide microarray analysis. J Mol Diagn 5: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milioni D, Sado P-E, Stacey NJ, Roberts K, McCann MC (2002) Early gene expression associated with the commitment and differentiation of a plant tracheary element is revealed by cDNA-amplified fragment length polymorphism analysis. Plant Cell 14: 2813–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono M, Qui F, Borsuk LA, Schnable PS (2003) Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissue of maize. Plant Cell 15: 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee J-Y, Colinas J, Wang JY, Thongrad SC, Malamy JE, Birnbaum K, Benfey PN (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- Pennisi E (2003) Tracing life's circuitry. Science 302: 1646–1649 [DOI] [PubMed] [Google Scholar]
- Provart NJ, McCourt P (2004) Systems approaches to understanding cell signaling and gene regulation. Curr Opin Plant Biol 7: 605–609 [DOI] [PubMed] [Google Scholar]
- Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandburg G (2004) A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH (1990) Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA 87: 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM (2000) High-fidelity mRNA amplification for gene profiling. Nat Biotechnol 18: 457–459 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Zhu T (2003) Global analysis of gene expression using GeneChip microarrays. Curr Opin Plant Biol 6: 418–425 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]