Abstract
The tomato (Lycopersicon esculentum) Cf-9 resistance gene encodes the first characterized member of the plant receptor-like protein (RLP) family. Other RLPs such as CLAVATA2 and TOO MANY MOUTHS are known to regulate development. The domain structure of RLPs consists of extracellular leucine-rich repeats, a transmembrane helix, and a short cytoplasmic region. Here, we identify 90 RLPs in rice (Oryza sativa) and compare them with functionally characterized RLPs from different plant species and with 56 Arabidopsis (Arabidopsis thaliana) RLPs, including the downy mildew resistance protein RPP27. Many RLPs cluster into four distinct superclades, three of which include RLPs known to be involved in plant defense. Sequence comparisons reveal diagnostic amino acid residues that may specify different molecular functions in different RLP subtypes. This analysis of rice RLPs thus identified at least 73 candidate resistance genes and four genes potentially involved in development. Due to the synteny between rice and other Gramineae, this analysis should provide valuable tools for experimental studies in rice and other cereals.
Diverse pathogens cause plant disease, and plants have evolved a variety of defense mechanisms. Many plant resistance genes (R-genes) have been cloned and characterized (Dangl and Jones, 2001). A common structural unit in many R proteins is the Leu-rich repeat (LRR). LRRs are found throughout the tree of life and are thought to mediate protein-protein interactions (Kobe and Kajava, 2001). LRR-containing plant proteins have diverse overall structures and functions, including disease resistance (Jones and Jones, 1997). A 23- to 25-amino acid plant-specific extracellular LRR motif contains a conserved consensus sequence, LxxLxxLxLxxNxLt/sgxIpxxLG (Jones and Jones, 1997).
Several classes of plant proteins involved in defense signaling contain extracellular LRR-containing receptor-like proteins (RLPs), receptor-like kinases (RLKs), and polygalacturonase inhibiting proteins (PGIPs; Fig. 1). The first RLP identified was Cf-9 (Jones et al., 1994). Its sequence was originally divided into regions (called “domains”) A through G (Fig. 1). Domain A comprises a putative signal peptide. Domain B, which forms the N terminus of the mature protein, contains several Cys. Domain C, originally annotated as containing 27 LRRs, is separated by 28 amino acids (domain D) from the short acidic domain E. Domain F comprises the transmembrane (TM) helix; lastly, the short cytoplasmic region was designated domain G. Subsequently, domain C was divided into domains C1, C2, and C3 (Jones and Jones, 1997), with the non-LRR variable C2 region (the “loop-out” or “island” domain) dividing the 22 LRRs of C1 from the four LRRs of C3. All characterized RLPs (except TOO MANY MOUTHS [TMM; Nadeau and Sack, 2002], which does not have a C2 domain) have domain architecture similar to that of Cf-9. We refer to this domain organization as “Cf-9-like RLP” (Cf-RLP), to differentiate it from RLPs having other domain architectures. A multitude of plant proteins are known to be related structurally to the Cf-RLPs but do not match the canonical Cf-RLP domain structure (for review, see Shiu et al., 2004).
Figure 1.
Canonical Cf-9-like RLP, RLK, and PGIP domain structures. Domain A contains a putative signal peptide. Domain B contains one or two pairs of Cys that may play structural roles. Domain C contains multiple LRRs, and contains a variable “loop-out” C2 region in some RLPs and RLKs. Domain D is a linker domain between C and the acidic domain E. Domain F contains the TM helix, and domain G corresponds to the cytoplasmic region. M, Membrane.
RLPs are known to be involved both in defense and development. Characterized RLP genes involved in resistance include the Cf genes from tomato (Lycopersicon esculentum) that confer resistance against the fungal pathogen Cladosporium fulvum (Jones et al., 1994; Dixon et al., 1996, 1998; Thomas et al., 1997), tomato Ve genes (Kawchuk et al., 2001), apple (Malus domestica) HcrVf2 (Vinatzer et al., 2001), Arabidopsis (Arabidopsis thaliana) RPP27 (Tör et al., 2004), and tomato LeEIX1 and LeEIX2 (Ron and Avni, 2004). Developmental RLPs in Arabidopsis include TMM (Yang and Sack, 1995; Nadeau and Sack, 2002), which is involved in stomatal patterning, and CLAVATA2 (CLV2; Jeong et al., 1999) and its maize (Zea mays) ortholog FASCIATED EAR2 (FEA2; Taguchi-Shiobara et al., 2001), which are involved in meristem development.
The RLKs are a large gene family in plants involved in signaling (Shiu et al., 2004). The extracellular domain of the LRR-RLKs is similar to that of RLPs, but the RLKs differ from the RLPs in having a cytoplasmic kinase domain. To date, several RLKs have been characterized. Rice (Oryza sativa) Xa21 (Song et al., 1995) and Arabidopsis FLS2 (Gömez-Gömez and Boller, 2000) encode RLKs involved in defense. Arabidopsis CLAVATA1 (Clark et al., 1997) and RLK5/HAESA (Walker, 1993) are involved in development. Interestingly, Arabidopsis ERECTA has been reported to play a role in both development and defense (Torii et al., 1996; Godiard et al., 2003).
PGIPs are another group of plant proteins related to RLPs and RLKs. PGIPs are cell wall proteins that bind and inhibit polygalacturonases, cell wall-degrading enzymes thought to aid fungal penetration into host tissues (Albersheim and Anderson, 1971; De Lorenzo et al., 2001). These can be differentiated from RLPs and RLKs by their lack of a TM domain or kinase domain. Although no structure of an RLP or RLK has yet been reported, the structure of a bean (Phaseolus vulgaris) PGIP has recently been solved (Protein Data Bank ID 1OGQA), and provides a good basis for modeling RLPs and RLKs (Di Matteo et al., 2003).
Genes involved in development tend to have certain characteristics differentiating them from R-genes. R-genes are often found in multiple copies at genomic loci and are under strong diversifying selection, producing highly divergent sequences and structural variants with distinct recognition capacities (Bai et al., 2002; Meyers et al., 2003). By contrast, developmental genes are under evolutionary pressure to maintain a specific function, reducing sequence drift across orthologs. For this reason, while orthologs to genes involved in developmental processes can easily be identified, identification of orthologs to R-genes can be difficult. For example, the mean pairwise percent identity between rice and Arabidopsis RLPs is only 27%, and the RLP family as a whole exhibits a high degree of variability in the number of repeats. In comparison, the developmental genes FEA2 of maize and CLV2 of Arabidopsis have high sequence similarity (44% identity) and similar functional phenotypes (Taguchi-Shiobara et al., 2001). Hence, we anticipate that pairs of RLPs that are well conserved between two divergent species such as rice and Arabidopsis are likely be involved in development. Nevertheless, CLV2 proteins in three Arabidopsis accessions exhibit high polymorphism in the N-terminal portion of their LRRs (Jeong et al., 1999), perhaps indicating coevolution with another polymorphic signaling component.
In earlier work, we described 58 RLPs in Arabidopsis (Tör et al., 2004). Here, we identify 90 Cf-RLPs from rice (spp. japonica) and compare them with functionally characterized RLPs from other plants and a revised complement of 56 Arabidopsis RLPs. (Two of the original 58 genes were removed for different reasons. At4g13910.1 was merged with the At4g13900.1 locus by The Institute for Genomic Research [TIGR], and At4g13820.1 was determined to not contain a TM region, and is expected to encode a PGIP.) Our analysis has revealed four classes of Cf-RLPs, three of which include RLPs known to be involved in defense signaling. We have also identified potential rice homologs of functionally characterized developmental RLPs, as well as at least two putative novel developmental genes with orthologs in both rice and Arabidopsis. This work has also revealed conserved residues and motifs of RLPs, which may be of functional significance.
RESULTS
Identification and Genomic Organization of Rice RLPs
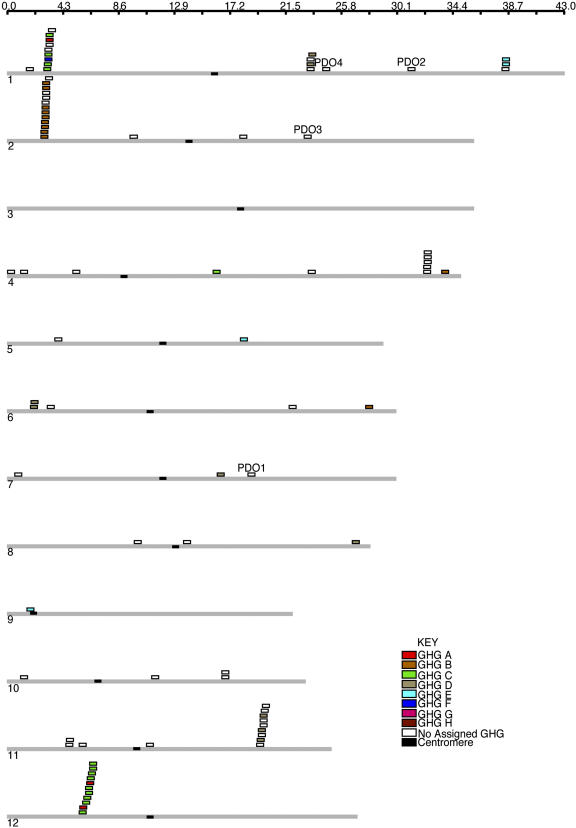
Since LRRs are often found in proteins with non-Cf-RLP folds, sequence-based methods of homolog detection can inadvertently include many non-Cf-RLPs in database searches. To discriminate canonical Cf-RLPs (referred to here as RLPs for simplicity) from proteins with different overall folds, we employed an intentionally stringent approach designed to include only those proteins sharing the canonical Cf-9-like RLP structure (described in “Materials and Methods”). This produced a set of 90 sequences in rice. For comparison, Cf-RLPs from other organisms as well as characterized RLK and PGIP sequences were identified using literature search and included in these analyses. A complete list of all Arabidopsis and rice RLP sequences, along with Cf-RLPs from other organisms and RLK and PGIP sequences, is available (Supplemental Table I). The rice sequences mapped to large clusters of genes on chromosomes 1, 2, 11, and 12, with smaller clusters and singletons scattered throughout the genome (Fig. 2). The rice genome has 38 loci containing RLPs, while Arabidopsis has 33 such loci. Thus, rice and Arabidopsis contain a similar number of RLP loci, yet rice has almost twice the number of genes predicted to encode RLPs. Furthermore, rice has an average of six RLPs at each locus containing more than one RLP, compared to 2.6 in Arabidopsis. This supports a hypothesis of recent tandem duplication contributing to the enlargement of the RLP family in rice. A similar conclusion was reached following analysis of the nucleotide-binding-LRR class of R-genes in rice (Bai et al., 2002).
Figure 2.
Representation of genomic loci of rice RLPs. The numbered gray bars represent the 12 rice chromosomes. The black bar on the chromosome represents the centromere; gene positions on chromosomes are indicated by the scale at top. RLPs are represented by rectangles colored according to their GHG classification. Positions of putative rice orthologs of the Arabidopsis genes CLV2 (OsPDO3) and TMM (OsPDO4) along with OsPDO1 and OsPDO2 are highlighted.
Classification into Global Homology Groups
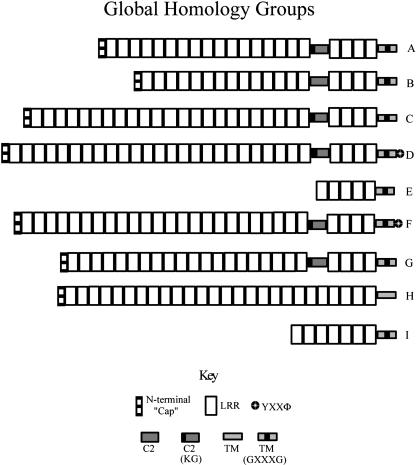
The Cf-RLPs of rice and Arabidopsis exhibit great variation in sequence and in the number of LRRs, making identification of the Cf-9-like domains difficult. To compensate for this, we clustered the full-length sequences into globally alignable subgroups (global homology groups, or GHGs), requiring all sequences in a GHG to have a length difference of no more than 15% and to share ≥30% sequence identity. Roughly 40% of Cf-RLPs from rice and Arabidopsis fell into nine GHGs containing three to 16 sequences (Fig. 3). We analyzed each GHG to determine the structural features relative to the canonical Cf-9 domain architecture.
Figure 3.
Features of GHGs. Shown in this figure are GHGs containing a minimum of three sequences. The striped rectangles represent the PGIP-like N-terminal regions.
Cf-RLPs of Rice and Arabidopsis Fall into Four Major Distinct Superclades
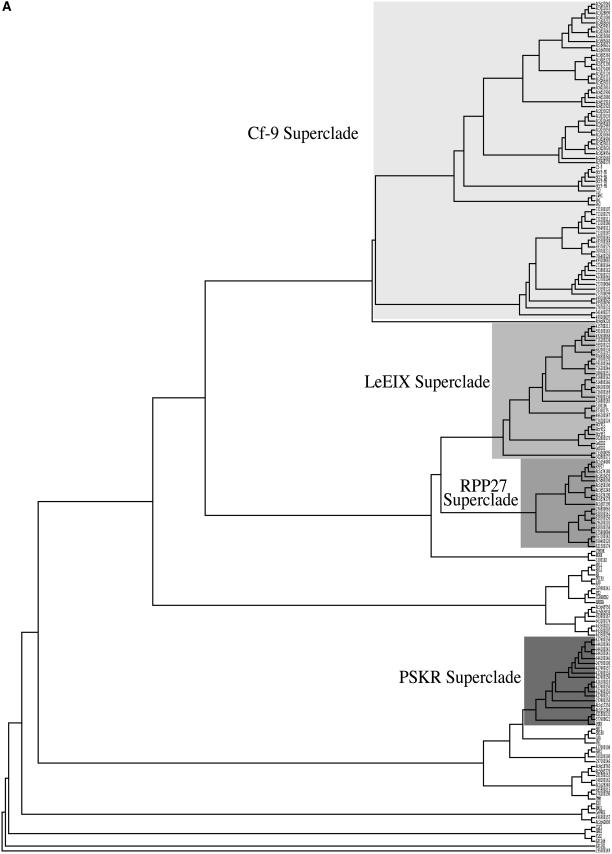
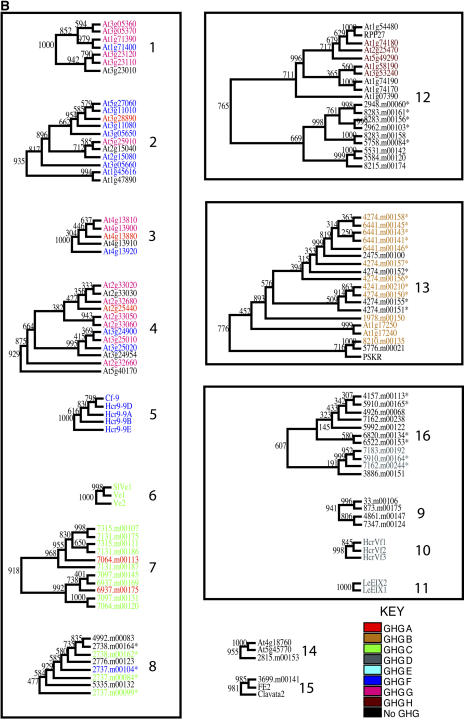
Although Cf-RLPs have low overall sequence similarity, domains C3 through F (the TM helix) are well conserved; we chose this region (C3-F) as the basis for phylogenetic tree construction (Fig. 4A). Phylogenetic analysis using several methods supports a set of 16 conserved clades (Fig. 4B; Table I). We defined clades as subtrees containing at least two sequences, greater than 60% bootstrap support, and no less than 50% average sequence identity. Of the 182 sequences in the tree, 123 are found in the 16 clades; we termed the remaining 59 phylogenetic singletons. Most clades segregate rice and Arabidopsis sequences, except for clades 12, 13, 14, and 15, which contain proteins from both species. Interestingly, among all the LRR-RLKs included in this analysis, only the carrot (Daucus carota) phytosulfokine receptor kinase (PSKR) groups with rice and Arabidopsis RLPs in clade 13. This is the only functionally characterized RLK that clusters with RLPs in our analyses.
Figure 4.
Phylogenetic relationships of RLPs and selected RLKs and PGIPs. A, Neighbor-joining tree built using 1,000 bootstrap replicates, based on an alignment of the C3-F region of Arabidopsis, rice, and characterized RLPs, as well as LRR-RLKs and PGIPs. Superclades are shaded. B, Clades-superclades-GHG relationships. The 16 clades are divided into four major superclades (shown boxed in gray). Rice sequences found in large genomic clusters are indicated with an asterisk. See Table I for details.
Table I.
Phylogenetic clades with greater than 60% bootstrap support
The predicted function for sequences in each clade is based on functionally characterized sequences. Clade 14 does not contain a characterized RLP but does contain PDO2, for which we predict a role in plant development. The average percent identity (AVG % ID) is based on the conserved C3-F region used to construct the phylogenetic tree. Clades corresponding to genomic clusters at chromosomal loci are indicated in the last column. An asterisk denotes clades lacking genomic clusters. Chr, Chromosome. –, No rice sequences in the clade.
| Clade | No. of Sequences | No. from Rice | No. from Arabidopsis | AVG % ID | GHGs | Predicted Function | Rice Chr Locus |
|---|---|---|---|---|---|---|---|
| 1 | 7 | 0 | 7 | 69 | F, G | Unknown | – |
| 2 | 11 | 0 | 11 | 70 | A, F, G | Unknown | – |
| 3 | 5 | 0 | 5 | 86 | A, F, G | Unknown | – |
| 4 | 12 | 0 | 12 | 73 | A, F, G | Unknown | – |
| 5 | 5 | 0 | 0 | 94 | F | Resistance (Cf-9) | – |
| 6 | 3 | 0 | 0 | 83 | C | Resistance (Ve) | – |
| 7 | 11 | 11 | 0 | 77 | A, C | Unknown | 12 |
| 8 | 8 | 9 | 0 | 70 | C, F | Unknown | 1 |
| 9 | 4 | 4 | 0 | 64 | None | Unknown | – |
| 10 | 3 | 0 | 0 | 94 | D | Resistance (HcrVf) | – |
| 11 | 2 | 0 | 0 | 85 | D | Resistance (LeEIX) | – |
| 12 | 19 | 9 | 10 | 50 | H | Resistance (RPP27) | 4 |
| 13 | 19 | 16 | 2 | 55 | B | Unknown | 2 |
| 14 | 3 | 1 | 2 | 50 | I | PDO2 + homologs | – |
| 15 | 3 | 1 | 2 | 58 | None | Development (CLV2) | – |
| 16 | 11 | 11 | 0 | 56 | D | Unknown | 11 |
A coarser cut of the C3-F-region-based phylogenetic tree produced a higher-order classification, identifying four distinct superclades of RLPs with an average identity of at least 48% within the C3-F region (Fig. 4B; Table II). Eighty-three of the 90 rice RLPs and 47 of 56 Arabidopsis RLPs belong to a superclade. Each of these superclades contains at least one functionally characterized member, allowing us to infer possible functions for the proteins in each superclade. Although the phylogenetic tree is based on the C3-F region alone, the superclades defined by this tree correlate very well with phylogenetic distribution and genomic clusters and with domain architecture defined by the GHGs.
Table II.
Descriptions of superclades
The average and minimum percent IDs are with respect to the C3-F region only. Bootstrap support is based on 1,000 neighbor-joining bootstrap replicates. The number of rice and Arabidopsis sequences in each superclade is listed in the last column.
| Superclade | Avg % ID | Min % ID | Clades Contained | GHGs Contained | Bootstrap Support | Organisms |
|---|---|---|---|---|---|---|
| Cf-9 | 48 | 32 | 1–8 | A, C, F, G | 41% | Arabidopsis (35), rice (24), tomato |
| RPP27 | 50 | 35 | 12 | H | 77% | Rice (9), Arabidopsis (10) |
| LeEIX | 62 | 44 | 9–11, 17 | D | 12% | Rice (24), apple, tomato |
| PSKR | 52 | 35 | 13 | B | 78% | Arabidopsis (2), rice (16), carrot |
The Cf-9 superclade contains proteins from clades one through eight, including 24 rice and 35 Arabidopsis sequences and tomato Ve1 and homologs. While most superclades correspond to a single GHG, this superclade spans several GHGs due to variable numbers of LRRs in the C1 region. This is consistent with the observed variability of Cf-9 and Cf-2 homologs in the number of LRRs (Thomas et al., 1997; Dixon et al., 1998). The rice sequences in the Cf-9 superclade map to large genomic clusters on rice chromosomes 1 and 12.
The RPP27 superclade consists of sequences from clade 12 along with seven phylogenetic singletons from Arabidopsis and nine from rice. All sequences from the GHG H and the genomic cluster on rice chromosome 4 are represented in this class.
The LeEIX superclade contains HcrVf proteins and also includes clades 9 and 16, as well as six phylogenetic singletons from rice and no Arabidopsis sequences. This superclade includes sequences from the GHG D and the rice genomic cluster on chromosome 11.
The PSKR superclade consists of clade 13, including the carrot RLK PSKR and two phylogenetic singletons from Arabidopsis and 16 from rice. This superclade corresponds to GHG B, as well as the rice genomic cluster on chromosome 2.
RLPs Conserved between Rice and Arabidopsis Represent Candidate Developmental Genes
As discussed earlier, developmental genes are less likely to be duplicated than R-genes, and are also more structurally and functionally conserved between distant species. Using these guidelines, we identified putative developmental orthologs (PDOs) between rice (OsPDO) and Arabidopsis (AtPDO). Candidate ortholog pairs were required to meet the following criteria: (1) global alignability (found in the same GHG), (2) support from bidirectional BLAST (i.e. each is the other's top-scoring hit in BLAST search of its genome), (3) placement in the same phylogenetic clade, (4) singleton at a genomic locus, and (5) significant sequence identity (at least two sds above the mean pairwise identity between rice and Arabidopsis Cf-RLPs). The first three criteria serve to ensure the two sequences are orthologous; the last two support a putative role in development. Only four rice RLPs (OsPDO1–OsPDO4) satisfied these stringent criteria.
OsPDO1 has approximately 60% sequence identity with its Arabidopsis ortholog AtPDO1 (At4g18760.1). OsPDO2 has approximately 45% sequence identity with AtPDO2 (At1g28340.1). Putative orthologs in other plant species are identifiable for both OsPDO1 and OsPDO2 in expressed sequence tag (EST) databases (Fig. 5) with an average of 70% and 55% identity with their top-scoring matches, respectively. In addition to the significant sequence conservation supporting a role in development for these proteins, EST data for PDO2 in other species reveal a consistent pattern of expression in seed-developing organs.
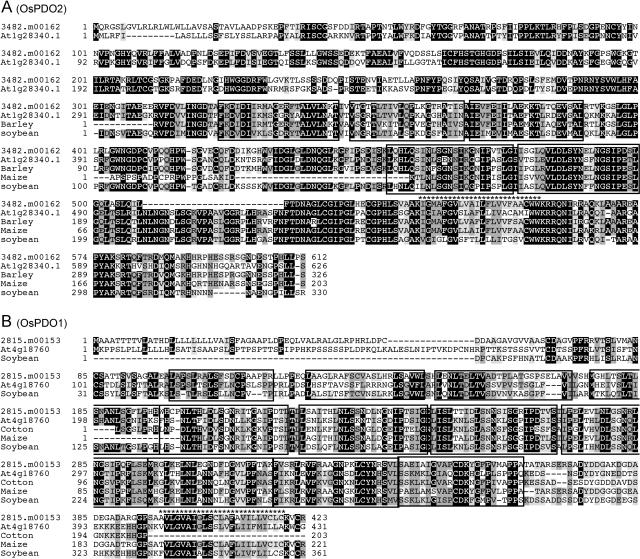
Figure 5.
Multiple sequence alignments of OsPDO2 (A) and OsPDO1 (B) and their homologs from other plants species. The protein sequences of the homologs are entire translated predicted open reading frames of ESTs from various plant species. LRRs are boxed, and the predicted TM domains are indicated with asterisks.
OsPDO3 and OsPDO4 are rice orthologs of CLV2 and TMM, respectively, with 45% and 48% identity. OsPDO3 also has 83% identity with the maize protein FEA2, which has a similar mutant phenotype to CLV2 (Taguchi-Shiobara et al., 2001). Finally, two Arabidopsis genes, At5g65830 and At3g49750, are closely related to each other and to three rice genes, and might represent an additional class involved in development. Interestingly, both of these Arabidopsis genes show increased expression in embryonic tissues, according to GENEVESTIGATOR, a Web-based interface used to mine publicly available Arabidopsis Affymetrix GeneChip data (Zimmermann et al., 2004). Since rice and Arabidopsis diverged about 150 million years ago (Chaw et al., 2004), it is conceivable that some of the genomic singleton RLPs that are not members of the superclades, such as At2g42800 or rice 1950.m00149, may be involved in a developmental function that is only maintained in the dicot or monocot clade.
Identification of Likely Functional Residues in Cf-RLPs
The clustering of sequences into global homology groups facilitated the identification of Cf-9-like domains and conserved motifs. Here, we describe conserved motifs in the Cf-RLPs that appear potentially important for maintaining the structure and/or function of these proteins.
The LRRs of Cf-9 are flanked by domains B and D, both of which contain conserved Cys. We found two structural variants in domain B of the Cf-RLPs. The first group has a single pair of conserved Cys and includes the RLPs known to be involved in development. The second group includes RLPs characterized in defense pathways (except Cf-2) and contains two pairs of conserved Cys. Sequence analysis suggests a homology between members of this second group and the amino-terminal 44 amino acids of PGIP (Protein Data Bank ID 1OGQA; average pairwise identity is 40% with very few gaps; Fig. 6A). The N terminus of the PGIP structure contains two pairs of conserved Cys, which form disulfide bridges capping its solenoid structure (Di Matteo et al., 2003). This apparent homology suggests a potential role for the corresponding Cys in the second group of RLPs. This group of RLPs includes 54 rice and 27 Arabidopsis sequences.
Figure 6.
Cf-RLP conserved motifs. A, Alignment of the B domain of Cf-RLPs to the N terminus of PGIP. 1OGQA is the Protein Data Bank identifier of the solved PGIP structure. Columns with highly conserved amino acids are highlighted in gray. Conserved Cys are marked with asterisks. B, Alignment of the C2 region of Cf-RLPs highlighting the Yx(n)KG motif. The conserved residues are indicated with asterisks and the columns are shaded as in A. C, Alignment of the TM regions of functionally characterized RLPs, RLKs, and PDOs that contain GXXXG-type motifs. The conserved Gly (or Ser) in the GXXXG-type motifs are highlighted in gray and are marked with asterisks.
The variable C2 region connects the C1 LRRs with the more conserved C3 LRRs. Various models have been proposed for the structural role of the C2 domain. The first hypothesis is that C2 is a flexible hinge-like region, which enables the two surrounding LRR regions to articulate relative to each other (Jones and Jones, 1997). Alternately, the C2 region could form a loop between the two LRR domains, minimally affecting the structure of the LRR scaffold. A third possibility is that this region folds into the regular LRR structure, forming an unbroken solenoid shape, much like the structure of PGIP. In total, 73 rice RLPs were identified that contain a C2 region varying nonintegrally in length from 30 to 80 amino acids. The variability in the C2 region belies its potential functional role. For example, mutation of conserved Gly in the C2 region of the Arabidopsis RLK BRI1 results in loss of brassinosteroid perception (Dievart and Clark, 2003), and this region has recently been proven to bind brassinolide (Kinoshita et al., 2005). Recent work has investigated conserved Cys and Trp in Cf-9 domain B, and putative glycosylation sites throughout the protein, as well as providing a model for domains B-D of Cf-9 (Van der Hoorn et al., 2005).
We also noted a novel conserved Yx(6-8)KG motif in the C2 region of 33 rice and 37 Arabidopsis RLPs (Fig. 6B); the function of this motif is unknown.
The GXXXG motif found in some TM regions has been shown to be involved in protein-protein interactions via intermolecular hydrogen bonds (Curran and Engelman, 2003). Eighty of the 90 rice Cf-RLPs and 55 of 56 Arabidopsis Cf-RLPs contain a (G/S/T)XXX(G/S/T) motif as do all functionally characterized RLPs, and LRR-RLKs (Fig. 6C). Considering the conservation of this motif across species, it is possible that they mediate intramolecular or intermolecular interactions.
The cytoplasmic tails of Cf-RLPs vary in length from one to 215 amino acids. A small fraction (nine of the 56 Arabidopsis Cf-RLPs and 20 of the 90 rice sequences) contained the YXXØ motif (where Ø represents any bulky, hydrophobic amino acid), shown to be required for rapid movement of TM proteins to lysosomes and lysosome-related organelles (Bonifacino and Traub, 2003). This motif is conserved within global homology group F containing the RLPs LeEIX1 and LeEIX2, in which the YXXØ motif has been shown to be necessary for function (Ron and Avni, 2004).
DISCUSSION
RLPs have been found in diverse plant species and are known to be involved in both defense and developmental pathways. R-gene families often expand by gene duplication and are under diversifying selection (Leister, 2004). Cf-RLPs in rice have been grouped by genomic locus, phylogenetic analysis, and global domain structure to form 16 clades and four superclades. The 16 clades contain more than 80% of the rice and Arabidopsis Cf-RLP sequences, of which 12 contain members from only one organism. This is in agreement with the findings of Shiu et al. (2004) showing 2-fold greater expansion of R-gene copies in rice. Superclade analysis shows three of the four include RLPs known to be involved in disease resistance. Most of the rice sequences in these superclades are clustered at genomic loci, indicating evolution by tandem gene duplication. We conclude therefore that the majority of Cf-RLPs are involved in resistance rather than development.
The PSKR superclade is the only one that includes an RLK. PSKR is a receptor for the plant peptide hormone phytosulfokine (Matsubayashi et al., 2002). Phytosulfokine is involved in cellular proliferation, dedifferentiation, and redifferentiation, along with plant hormones auxin and cytokinin (Matsubayashi, 2003). Though the closest RLP to PSKR is a genomic singleton, the other rice sequences in this superclade are clustered on chromosome 2. Given that R-genes are more likely to evolve by gene duplication than developmental genes, we speculate that the group of rice Cf-RLPs on chromosome 2 might function in recognizing PSK-like ligands secreted by a pathogen. Bacterial pathogens and symbionts are known to secrete phytohormones (Costacurta and Vanderleyden, 1995), and plants may have evolved receptors to recognize these signals.
Our analysis identifies four putative developmental genes in rice, OsPDOs 1 to 4. Attempts to determine a specific functional role and/or tissue-specific expression pattern based on comparisons with ESTs have not been fruitful for OsPDO1. However, EST data for close homologs for OsPDO2 allow us to infer that OsPDO2 may be expressed during seed production. Of the nine Arabidopsis RLPs that are not part of a superclade, six have been discussed here: the AtPDOs and At5g65830 and At3g49750. It is possible that the remaining three (At4g04220, At5g45720, and At2g42800) are involved in Arabidopsis-specific developmental processes.
Recent experimental investigation in functional characterization of Arabidopsis RLPs provides additional support for a developmental role for OsPDO1. To date, 20 homozygous T-DNA insertion lines have been obtained and challenged with a small range of microorganisms, including bacterial, oomycete, and fungal plant pathogens. Thus far, we have identified three RLPs involved in the defense response and one that has shown a developmental phenotype with slow growth, few leaf numbers, and late flowering (M. Tör, unpublished data). Interestingly, the gene for this mutant corresponds to AtPDO1 (At4g18760), supporting our inference that its rice ortholog, OsPDO1, may also play a role in development. Further characterization of this mutant is in progress.
Structural analyses of LRR proteins reveal a shared solenoid scaffold, with a β-sheet forming the concave side and stacked helices forming the convex side (Kobe and Kajava, 2001). While maintaining this common overall fold, Cf-RLPs exhibit variation in binding specificity, presumably mediated through variation in the solvent-exposed amino acids of the β-sheet and in the number of LRRs. In previous functional studies, the C1 region has been implicated in providing the recognition specificity of RLPs. Deletion of LRRs and introduction of point mutations in the C1 region of Cf-9 changes the specificity of Cf-9 to that of Cf-4 (Van der Hoorn et al., 2001; Wulff et al., 2001). Our analyses reveal the C1 region to be highly variable, particularly in the number of LRRs. This highlights the potential role of the N-terminal LRRs of the Cf-RLPs in determining specificity and function.
Our analyses of Cf-RLPs identified several motifs that may facilitate intramolecular and intermolecular interactions. In LRR proteins, the hydrophobic amino acids of the LRR are protected from solvent exposure by their flanking regions (Kobe and Kajava, 2001). In the PGIP structure, two pairs of Cys at the N and C termini form disulfide bridges stabilizing an α-helical region that caps the LRRs. A motif similar to the N terminus of PGIP was seen in several Cf-RLPs, including RLPs known to be involved in defense (the developmental RLPs and the RLKs contain only one pair of Cys). Mutation of one of the N-terminal Cys in the RLK BRI1 leads to a weak brassinosteroid response phenotype (Dievart and Clark, 2003). Domain B is also required for proper function of Cf-4 (Van der Hoorn et al., 2001; Wulff et al., 2001). Thus, the conserved Cys are expected to be critical either for maintaining Cf-RLP structure or for mediating intermolecular interactions.
The N-terminal LRRs (domain C1) of many RLPs and RLKs are separated from the membrane-proximal four LRRs (domain C3) by an “island” or “loop-out” region of 30 to 80 amino acid sequences, referred to as the C2 domain. This domain in BRI1 has now been shown to be involved in direct binding to brassinolide (Kinoshita et al., 2005). On the other hand, the C2 domains of Cf-9 and Cf-4 are identical, and determinants of the distinct recognition achieved by these proteins reside in LRRs 10 to 16 of domain C1 (Van der Hoorn et al., 2001; Wulff et al., 2001).
Another conserved motif likely to be involved in protein-protein interactions is found in the TM region. The TM GXXXG motif is known to aid dimerization and activation of ErbB2, a mammalian receptor kinase (Bennasroune et al., 2004). This conserved motif in Cf-RLPs might mediate interaction between RLPs or between RLPs and other TM proteins, and play a critical role in RLP signal transduction from the extracellular, ligand-binding regions to the cytoplasmic space.
CONCLUSION
R-genes are known to be under diversifying selection to adapt to different pathogen challenges. Several gene families involved in defense response in plants, such as the nucleotide-binding-LRR proteins (Meyers et al., 2003), have been found in large genomic clusters, supporting a hypothesis of rapid evolution by tandem duplication and gene shuffling events. Our phylogenetic analysis identified four superclades of Cf-RLPs containing a majority of the rice and Arabidopsis Cf-RLPs (120 of the 146). All superclades contain sequences at multicopy tandem repeat-encoding loci, and three of the four also contain at least one functionally characterized R-gene. We hypothesize that the genes in these superclades are likely to function in disease resistance.
This analysis has identified 73 candidate R-genes in rice and at least four probable developmental genes for further experimental validation. Due to the close synteny between Gramineae (Paterson et al., 2003) this analysis should provide valuable tools for those seeking to locate R-genes in other cereals.
MATERIALS AND METHODS
Multiple Sequence Alignment
We used two methods for multiple sequence alignment. MUSCLE (Edgar, 2004) was used to align each global homology group. MAFFT (Katoh et al., 2002) was used to align the C3-F region for phylogenetic analysis.
Domain Identification
Hidden Markov models (HMMs) from the PFAM suite (http://pfam.wustl.edu) were used to identify the presence of LRRs and other structural domains. The TM prediction server TMHMM (version 2.0; Krogh et al., 2001) was used to detect the presence of putative TM domains. All HMMs used in our experiments were constructed using the UCSC HMM software suite (http://www.cse.ucsc.edu/research/compbio/sam.html). RLP-specific HMMs were constructed for individual domains (e.g. the N-terminal domain) using in-house tools to model subfamily-specific variants of these regions (Brown et al., 2005). The Cf-9-like RLPs from Arabidopsis (Arabidopsis thaliana), ecotype Col-0, were identified as described in Tör et al. (2004), and used to construct an HMM for the Cf-RLP C3 region.
Rice Cf-RLP Identification
To identify rice (Oryza sativa) proteins sharing the canonical Cf-RLP structure, we required sequences from release 2 of the TIGR Pseudomolecules (Yuan et al., 2003) to meet the following three criteria: (1) match the Arabidopsis C3 region HMM with an E value < 0.001, (2) have no matches to non-LRR domains in the PFAM library (using a permissive E value cutoff of 0.5), and (3) contain an identifiable TM domain at the C terminus.
Genomic Locus Identification
Unlike Arabidopsis, the genomic loci of rice proteins are not immediately available. To identify genomic locations of the rice RLPs, we used each sequence as a query in translated BLAST (Altschul et al., 1997) against the TIGR Rice Pseudomolecules database (version 2), requiring 100% identity between the query and the (translated) genomic sequences, allowing gaps for introns. This revealed chromosomal locations for 88 of the 90 rice RLPs (two sequences were not assigned a chromosomal location by TIGR). The location map for these sequences (Fig. 2) was created using GenomePixelizer (Kozik et al., 2002).
Global Homology Groups
We clustered rice and Arabidopsis RLPs, RLKs, and PGIPs into globally alignable sequence clusters, which we call GHGs, using a combination of BLAST, HMM, and heuristic methods. To enable us to assume the same overall architecture for all members of a GHG, we required all sequences in a GHG to have a minimum of 30% pairwise identity and a bidirectional alignment coverage (i.e. the percentage of a sequence's amino acids that are aligned to the other sequences in the cluster) of at least 85%.
Phylogenetic Tree Construction
Phylogenetic trees were estimated using the conserved C3-F region as input. We used several programs in these analyses, including Neighbor-Joining and Parsimony from the PHYLIP software suite (http://evolution.genetics.washington.edu/phylip.html), Mr Bayes (Ronquist and Huelsenbeck, 2003), SATCHMO (Edgar and Sjölander, 2003), and BETE (Sjölander, 1998). A total of 1,000 bootstrap replicates were performed using Neighbor-Joining, and clades having >60% bootstrap support were identified. Trees produced by other methods were examined for agreement with these clades. Phylogenetic trees are displayed with the TreeView software (Page, 1996).
Supplementary Material
Acknowledgments
J.D.G.J. thanks Joe Ecker at the Salk Institute for kindly hosting a sabbatical visit, during which this project was initiated.
This work was supported in part by the National Science Foundation (PECASE Award DBI–0238311 to K.V.S.) and in part by the Gatsby Charitable Foundation (to J.D.G.J.).
The online version of this article contains Web-only data.
References
- Albersheim P, Anderson AJ (1971) Proteins from plant cell walls inhibit polygalacturonases secreted by plant pathogens. Proc Natl Acad Sci USA 68: 1815–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, et al (2002) Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 12: 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasroune A, Fickova M, Gardin A, Dirrig-Grosch S, Aunis D, Cremel G, Hubert P (2004) Transmembrane peptides as inhibitors of ErbB receptor signaling. Mol Biol Cell 15: 3464–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72: 395–447 [DOI] [PubMed] [Google Scholar]
- Brown DK, Dale N, Christopher JW, Sjölander K (2005) Subfamily HMMs in structural and functional genomics. Pac Symp Biocomput 322–323 [PubMed]
- Chaw SM, Chang CC, Chen HL, Li WH (2004) Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58: 424–441 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Costacurta A, Vanderleyden J (1995) Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol 21: 1–18 [DOI] [PubMed] [Google Scholar]
- Curran AR, Engelman DM (2003) Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol 13: 412–417 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G, D'Ovidio R, Cervone F (2001) The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol 39: 313–335 [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, De Lorenzo G, Tsernoglou D, Cervone F (2003) The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci USA 100: 10124–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dievart A, Clark SE (2003) Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr Opin Plant Biol 6: 507–516 [DOI] [PubMed] [Google Scholar]
- Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JD (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10: 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84: 451–459 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Sjölander K (2003) SATCHMO: sequence alignment and tree construction using hidden Markov models. Bioinformatics 19: 1404–1411 [DOI] [PubMed] [Google Scholar]
- Godiard L, Sauviac L, Torii KU, Grenon O, Mangin B, Grimsley NH, Marco Y (2003) ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J 36: 353–365 [DOI] [PubMed] [Google Scholar]
- Gömez-Gömez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved on the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Jones JD (1997) The role of leucine-rich repeat proteins in plant defences. Adv Bot Res 24: 89–167 [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JD (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793 [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, et al (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 98: 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV (2001) The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol 11: 725–732 [DOI] [PubMed] [Google Scholar]
- Kozik A, Kochetkova E, Michelmore R (2002) GenomePixelizer—a visualization program for comparative genomics within and between species. Bioinformatics 18: 335–336 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20: 116–122 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y (2003) Ligand-receptor pairs in plant peptide signaling. J Cell Sci 116: 3863–3870 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y (2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Peterson DG, Estill JC, Chapman BA (2003) Structure and evolution of cereal genomes. Curr Opin Genet Dev 13: 644–650 [DOI] [PubMed] [Google Scholar]
- Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölander K (1998) Phylogenetic inference in protein superfamilies: analysis of SH2 domains. Proc Int Conf Intell Syst Mol Biol 6: 165–174 [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JD (1997) Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tör M, Brown D, Cooper A, Woods-Tör A, Sjölander K, Jones JD, Holub EB (2004) Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato Cf-9. Plant Physiol 135: 1100–1112 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn RA, Roth R, De Wit PJ (2001) Identification of distinct specificity determinants in resistance protein Cf-4 allows construction of a Cf-9 mutant that confers recognition of avirulence protein Avr4. Plant Cell 13: 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn R, Brande W, Rivas S, Durrant M, van der Ploeg A, de Wit P, Jones JDG Structure-function analysis of Cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17: 1000–1015 [DOI] [PMC free article] [PubMed]
- Vinatzer BA, Patocchi A, Gianfranceschi L, Tartarini S, Zhang HB, Gessler C, Sansavini S (2001) Apple contains receptor-like genes homologous to the Cladosporium fulvum resistance gene family of tomato with a cluster of genes cosegregating with Vf apple scab resistance. Mol Plant Microbe Interact 14: 508–515 [DOI] [PubMed] [Google Scholar]
- Walker JC (1993) Receptor-like protein kinase genes of Arabidopsis thaliana. Plant J 3: 451–456 [DOI] [PubMed] [Google Scholar]
- Wulff BB, Thomas CM, Smoker M, Grant M, Jones JD (2001) Domain swapping and gene shuffling identify sequences required for induction of an Avr-dependent hypersensitive response by the tomato Cf-4 and Cf-9 proteins. Plant Cell 13: 255–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Sack FD (1995) The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7: 2227–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Liu J, Suh B, Cheung F, Sultana R, Lee D, Quackenbush J, Buell CR (2003) The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res 31: 229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.