Abstract
The organization and dynamics of microtubules are regulated by microtubule-associated proteins, or MAPs. In Arabidopsis (Arabidopsis thaliana), nine genes encode proteins of the evolutionarily conserved MAP65 family. We proposed that different MAP65s might have distinct roles in the interaction with microtubules. In this study, two AtMAP65 proteins, AtMAP65-1 and AtMAP65-6, were chosen to test this hypothesis in vitro. Although both fusion proteins were able to cosediment with microtubules in vitro, different properties on tubulin polymerization and microtubule bundling were observed. AtMAP65-1 was able to promote tubulin polymerization, enhance microtubule nucleation, and decrease the critical concentration for tubulin polymerization. It also induced the formation of large microtubule bundles by forming cross-bridges between microtubules evenly along the whole length of microtubules. In the presence of AtMAP65-1, microtubule bundles were more resistant to cold and dilution treatments. AtMAP65-6, however, demonstrated no activity in promoting tubulin polymerization and stabilizing preformed microtubules. AtMAP65-6 induced microtubules to form a mesh-like network with individual microtubules. Cross-bridge-like interactions were only found at regional sites between microtubules. The microtubule network induced by AtMAP65-6 was more resistant to high concentration of NaCl than the bundles induced by AtMAP65-1. Purified monospecific anti-AtMAP65-6 antibodies revealed that AtMAP65-6 was associated with mitochondria in Arabidopsis cells. It was concluded that these two MAP65 proteins were targeted to distinct sites, thus performing distinct functions in Arabidopsis cells.
Microtubules, polymers of α- and β-tubulin dimers, are dynamic filaments that constantly undergo polymerization and/or depolymerization at both the plus end and the minus end. In higher plants, microtubules are organized into distinct arrays in differentiated cells (Cyr and Palevitz, 1995; Wasteneys and Galway, 2003). It is widely accepted that microtubule-associated proteins, or MAPs, play regulatory roles in microtubule organization (Lloyd and Hussey, 2001). However, it is still unclear how plant MAPs may contribute to the formation of different microtubule arrays.
Cortical microtubules are commonly found among interphase cells. Individual microtubules in the cortical array often have close contact with the plasma membrane and various organelles as revealed by electron microscopy (Gunning and Steer, 1996). It has been widely received that the close proximity is brought about by proteins that interact with both the plasma membrane/organelles and microtubules. The identity of such MAPs, however, is largely unknown.
Tireless quest of plant MAPs has recovered a class of MAPs of approximately 65 kD in both tobacco (Nicotiana tabacum) and carrot (Daucus carota) cells, thus called MAP65 (Jiang and Sonobe, 1993; Chan et al., 1996; Hussey et al., 2002). Biochemically purified tobacco MAP65 polypeptides are encoded by similar genes (Jiang and Sonobe, 1993; Smertenko et al., 2000). The Arabidopsis (Arabidopsis thaliana) genome contains nine genes encoding polypeptides similar to the tobacco MAP65 (Hussey et al., 2002). Sequence analysis indicates that plant MAP65 has significant sequence homology with the Ase1p protein from the budding yeast Saccharomyces cerevisiae and the PRC1 protein from human (Pellman et al., 1995; Jiang et al., 1998). Pan anti-MAP65 antibodies decorate microtubules in all arrays throughout the cell cycle (Jiang and Sonobe, 1993; Chan et al., 1996). Antibodies raised against a bacterially expressed MAP65, however, decorate only subpopulations of microtubules, especially in the mitotic spindle and the phragmoplast (Smertenko et al., 2000; Muller et al., 2004). Therefore, different MAP65s may participate in different cellular activities, despite the sequence similarity.
Evidence of functional divergence of different MAP65 proteins is emerging. In bean epicotyls, the MAP65 protein level positively correlates with the elongation activity, implying a potential role in cell elongation (Sawano et al., 2000). In carrot suspension cells, the expression level of a particular MAP65 protein is elevated when cells undergo rapid elongation (Chan et al., 2003). Thus, a certain MAP65 protein(s) is probably involved in establishing/maintaining the parallel transverse array of cortical microtubules. Genetic studies have also revealed a unique role of AtMAP65-3/PLEIADE as several mutant alleles of the gene cause failure in cytokinesis by altering microtubule organization in the phragmoplast (Muller et al., 2004).
To date, it has been concluded that MAP65 bundles microtubules by forming cross-bridges along neighboring microtubules (Jiang and Sonobe, 1993; Chan et al., 1996). Detailed analyses have been carried out in a particular form of MAP65, MAP65-1, and close relatives from Arabidopsis and tobacco (Smertenko et al., 2004; Van Damme et al., 2004; Wicker-Planquart et al., 2004). The conclusion from these studies is that MAP65-1 only contributes to the bundling activities, not tubulin polymerization. More importantly, it has been revealed that the C terminus of AtMAP65-1 contributes to microtubule binding and the N terminus to microtubule bundling (Smertenko et al., 2004).
Because nine forms of AtMAP65 have sequence identity from 28% to 79% (Hussey et al., 2002), different AtMAP65 proteins may be granted with distinct activities and different functions (Van Damme et al., 2004). While AtMAP65-1 has 78% sequence identity with AtMAP65-2, it only has approximately 44% sequence identity with AtMAP65-6 (Hussey et al., 2002). Because AtMAP65-6 shows significant sequence divergence with AtMAP65-1 at both N and C termini, we wondered whether the divergence would render different activities.
In this study, we have expressed AtMAP65-1 and AtMAP65-6 as fusion proteins in bacteria. Purified fusion proteins have been used to test their interaction with microtubules in vitro. We provide evidence that these two related proteins may regulate microtubule organization in different manners. We also demonstrate that AtMAP65-6 is targeted to mitochondria in Arabidopsis cells.
RESULTS
Fusion Proteins of AtMAP65-1 and AtMAP65-6 Bind to Microtubules in Vitro
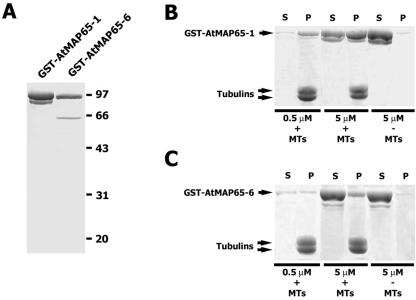
To obtain proteins for in vitro analysis, cDNA sequences of AtMAP65-1 and AtMAP65-6 were obtained in a cDNA clone (109M12) and by reverse transcription (RT)-PCR, respectively. Purified glutathione S-transferase (GST)-AtMAP65-1 fusion protein appeared in doublets (Fig. 1A). GST-AtMAP65-6 appeared in a major band with a minor band at a lower molecular mass (Fig. 1A). The lower band was a degradation product according to immunoblotting results (Fig. 6A).
Figure 1.
AtMAP65-1 and AtMAP65-6 cosediment with microtubules. A, Purification of AtMAP65-1 and AtMAP65-6. AtMAP65-1 fusion protein appeared in a doublet. AtMAP65-6 preparation had a minor band at 52 kD resulting from proteolysis. Positions of molecular mass markers were shown at right in kilodaltons. B, Cosedimentation of GST-MAP65-1 with microtubules. Cosedimentation experiments were performed with 0.5 and 5 μm fusion protein in the presence (+) or absence (−) of microtubules. Resulting proteins in the supernatants (S) and pellets (P) are shown. C, Cosedimentation of GST-MAP65-6 with microtubules. Similar to B, proteins in the supernatant (S) and pellets (P) were resolved by SDS-PAGE.
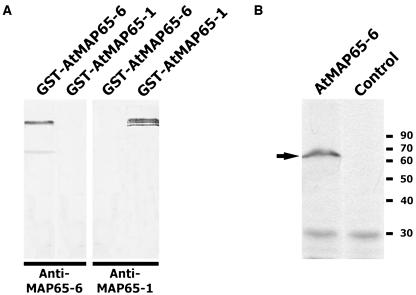
Figure 6.
Detection of the AtMAP65-6 protein. A, Monospecific anti-AtMAP65-6 antibodies. When both GST-AtMAP65-6 and GST-AtMAP65-1 were probed with affinity-purified anti-AtMAP65-6 antibodies, only GST-AtMAP65-6 was detected. The lower band was detected as well, indicating it was a degradation product. Purified anti-AtMAP65-1 antibodies only detected GST-AtMAP65-1, not GST-AtMAP65-6. B, Detection of native AtMAP65-6. Nitrocellulose membrane containing total proteins from Arabidopsis suspension cells was probed with anti-AtMAP65-6 and control IgG. A band between 60- and 70-kD markers was revealed by anti-AtMAP65-6. Positions of molecular mass markers are shown at right in kilodaltons.
When incubated with polymerized porcine brain microtubules, fusion proteins cosedimented with microtubules by centrifugation (Fig. 1, B and C). GST-AtMAP65-6 had lower cosedimentation efficiency than GST-AtMAP65-1. To verify that the cosedimentation was due to the interaction with microtubules, we tested whether the fusion proteins sedimented by themselves. Five micromolar AtMAP65 fusion proteins were centrifuged in the same conditions of cosedimentation experiments in the absence of microtubules. Both MAP65 fusion proteins remained predominantly in the supernatant (Fig. 1, B and C). Therefore, the fusion proteins were most likely active in terms of their binding to microtubules.
AtMAP65-6 Affects Microtubule Organization in a Different Manner Than AtMAP65-1
Because the fusion proteins bound to microtubules in vitro, we wanted to test whether they would affect microtubule organization. GST-AtMAP65-1 was used to test the microtubule-bundling activity according to earlier reports (Jiang and Sonobe, 1993; Chan et al., 1996). Large bundles of microtubules were formed (Fig. 2, A and B). As a matter of fact, microtubules were so overwhelmingly bundled that very few individual microtubules were found outside the bundles (Fig. 2B). This result again assured us that the GST fusion partner did not inhibit the bundling activity of AtMAP65-1.
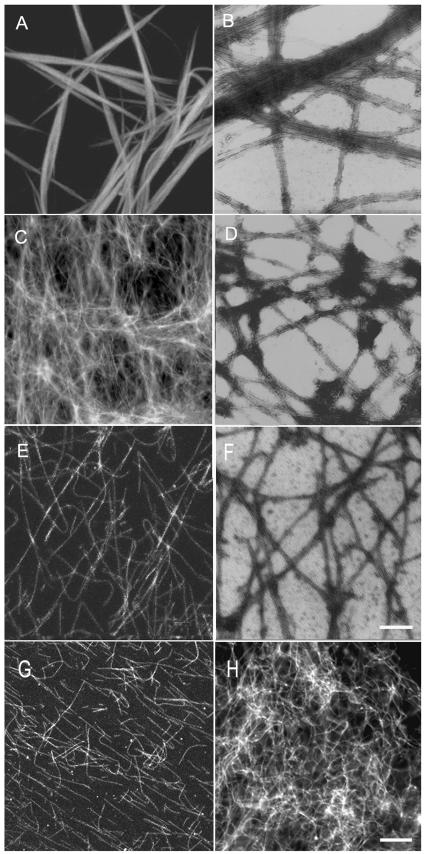
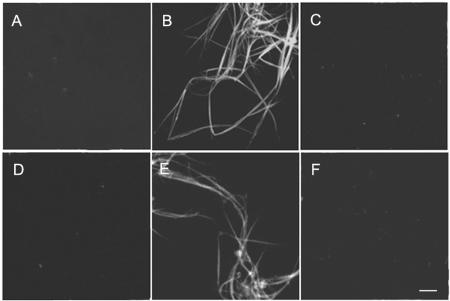
Figure 2.
Microtubule organization in the presence of AtMAP65-1 and AtMAP65-6. Fluorescent images are shown in A, C, and E, and electron micrographs are shown in B, D, and F. A and B, Microtubule bundles induced by AtMAP65-1. C and D, Microtubule mesh-like network induced by AtMAP65-6. E and F, Microtubules polymerized in the absence of MAP65. G, Microtubule bundles induced by AtMAP65-1 were sensitive to NaCl. Single microtubules were found upon 300 mm NaCl application. H, Microtubule mesh-like network induced by AtMAP65-6 was resistant to 500 mm NaCl treatment. Similar microtubule mesh was found as when there was no NaCl. Scale bar = 10 μm for fluorescent images and 0.25 μm for electron microscopic images.
GST-AtMAP65-6 was then applied to microtubule suspensions under identical conditions. A different effect was observed. A very dense network was induced (Fig. 2C). Electron microscopic examination by negative staining indicated that microtubules were crisscrossed with each other, and very little if any bundling phenomenon was observed (Fig. 2D). These individual microtubules were brought together by the AtMAP65-6-induced crisscrosses to form a dense mesh-like network (Fig. 2C). To test whether such a dense microtubule network was indeed induced by AtMAP65-6, MAP-free microtubules were examined. In the absence of GST-MAP65-6, these microtubules appeared in single separated filaments (Fig. 2, E and F). Therefore, we concluded that the phenomena of microtubule bundling and microtubule crisscrossing, respectively, were caused by the addition of AtMAP65-1 and AtMAP65-6 fusion proteins.
To further test the effect of these MAPs on microtubule organization, we applied different concentrations of NaCl to strip the fusion proteins from the microtubules. Microtubule bundles induced by AtMAP65-1 became scattered in the presence of 100 or 200 mm NaCl (data not shown). When the NaCl concentration was brought to 300 mm or above, no more microtubule bundles were observed. Instead, all microtubules appeared in single individuals (Fig. 2G). The microtubule network induced by AtMAP65-6, however, was able to resist NaCl at concentrations as high as 500 mm (Fig. 2H). Thus, the microtubule network induced by AtMAP65-6 was more stable than the bundles induced by AtMAP65-1.
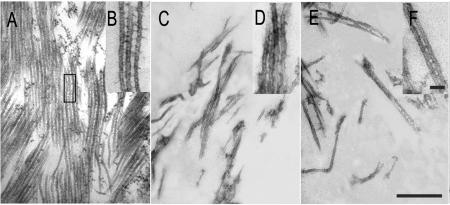
To reveal how these two MAP65 proteins affect microtubule organization differently, we examined microtubule bundles induced by GST-AtMAP65-1 and microtubule meshes induced by GST-AtMAP65-6 by thin sections. Microtubule bundles induced by GST-AtMAP65-1 were evenly spaced in parallel to each other, and the space between each other was approximately the diameter of microtubules (Fig. 3A). Microtubule bundles containing more than a dozen microtubules were frequently found, and bundles tended to merge with each other to form more complex super bundles (Fig. 3A). Spaced ladder-like cross-bridges with a distance close to the microtubule diameter of 25 nm were found between adjacent microtubules (Fig. 3B). These cross-bridges were identical to the data reported using native MAP65 proteins (Chan et al., 1999). Therefore, our data suggest that the GST tag did not interfere with the formation of cross-bridges between microtubules.
Figure 3.
Ultrastructural examination of microtubule bundles induced by AtMAP65 proteins. A and B, Large microtubule bundles induced by AtMAP65-1. Note that microtubules were evenly spaced out. The insert (B) showed ladder-like cross-bridges between adjacent microtubules. C, D, E, and F, Microtubule mesh-like network induced by AtMAP65-6. A few microtubules were found to be linked together, and microtubules were generally scattered (C and E). Inserts (D and F) show cross-bridges formed between microtubules. Scale bar = 0.5 μm in A, C, and E, and 0.05 μm in B, D, and F.
When the AtMAP65-6-induced microtubule meshes were sectioned, we did not observe a field densely occupied by microtubules (Fig. 3, C–F). Instead, a few microtubules occasionally jointed together at a given region, and most microtubules appeared to be scattered in a field (Fig. 3, C and E). When microtubules were jointed together, a space of <15 nm was observed between microtubules, and a few filamentous structures were occasionally present (Fig. 3, D and F). The jointed microtubules splayed out quickly.
AtMAP65-1, Not AtMAP65-6, Enhances Microtubule Polymerization and Promotes Nucleation
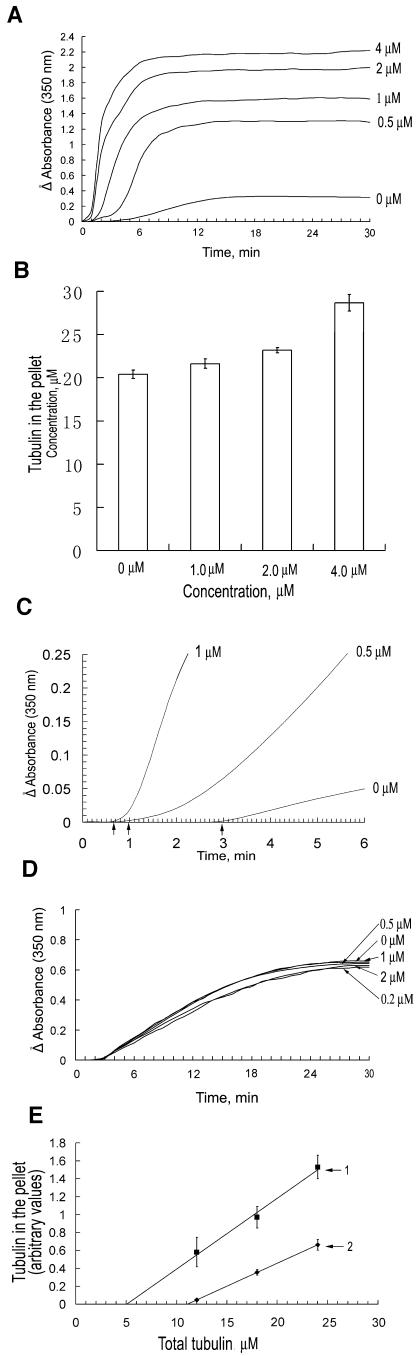
Because AtMAP65-1 and AtMAP65-6 fusion proteins induced microtubules into very different configurations, we wondered whether such different phenomena were caused by the effect of these two proteins on microtubule polymerization and nucleation. We examined microtubule polymerization and nucleation activities quantitatively in vitro, by monitoring changes of turbidity of tubulin suspension when the two fusion proteins were added. In a 30-min period, microtubule polymerization, reflected by the absorbance value at 350 nm, reached a steady state in approximately 12 min (Fig. 4A). When AtMAP65-1 fusion was added at 0.5 μm, the absorbance value jumped more than 4-fold at its steady state, indicating that more tubulins were polymerized into microtubules (Fig. 4A). The steady state was reached in approximately 8 min (Fig. 4A). When AtMAP65-1 was added at concentrations of 1, 2, and 4 μm, the steady state of turbidity increased even more, although not as dramatically as the difference between 0 and 0.5 μm (Fig. 4A). To test whether the increase of turbidity was not solely due to the bundling activity as suggested by an earlier report (Smertenko et al., 2004), we examined the amount of tubulin in the pellet of a high-speed centrifugation. Our results indicated that significantly more tubulins appeared in pellet in the presence of AtMAP65-1 than in the absence. In the presence of 4 μm, approximately 35% more tubulins appeared in the pellet than without AtMAP65-1 (Fig. 4B). Therefore, GST-AtMAP65-1 promoted microtubule polymerization in vitro.
Figure 4.
AtMAP65-1, not AtMAP65-6, promoted microtubule polymerization. A, Polymerization of tubulins under 0, 0.5, 1, 2, and 4 μm AtMAP65-1 fusion protein for 30 min. The degree of polymerization was monitored by the A350 (y axis). B, Promotion of tubulin polymerization by AtMAP65-1 at different concentrations. Note that at the highest concentration (4 μm), approximately 35% more tubulins were present in the pellet than when there was no AtMAP65-1. C, AtMAP65-1 reduced lag time for tubulin polymerization. In the absence of AtMAP65-1, the lag time was about 3 min. In the presence of 1 or 0.5 μm AtMAP65-1, the lag time was estimated at about 40 or 60 s, respectively (see the arrows). D, Polymerization of tubulins in the presence of 0, 0.2, 0.5, 1, and 2 μm AtMAP65-6. Note there was no significant difference among them. E, The Cc for tubulin polymerization was lowered in the presence of AtMAP65-1. The Cc values of tubulin assembly were estimated at about 11 μm for polymerization of tubulins (line 2) and decreased to 5 μm when 1 μm of AtMAP65-1 was present (line 1).
We also examined the lag time of tubulin assembly, an indicator of microtubule nucleation at different concentrations of AtMAP65-1. In the absence of AtMAP65-1, the lag time was approximately 3 min (Fig. 4C). In the presence of 1 μm and 0.5 μm AtMAP65-1, however, the lag time was reduced to approximately 40 s and 60 s, respectively (Fig. 4C). Therefore, we concluded that the AtMAP65-1 fusion protein was able to promote the nucleation of microtubules.
When AtMAP65-6 was tested at identical concentrations added to the tubulin suspension, no significant changes in the polymerization curve were observed among different concentrations (0–2 μm) of the fusion protein (Fig. 4D). Thus, the results suggested that AtMAP65-6 did not bear the activity of promoting microtubule polymerization as AtMAP65-1 did.
We also examined the critical concentration (Cc) of tubulin assembly in the absence and presence of GST-AtMAP65-1. The Cc value of our purified tubulin preparation was 11 μm (Fig. 4E). In the presence of the AtMAP65-1 fusion protein at 1 μm, Cc dropped to 5 μm (Fig. 4E). We can conclude that tubulins are able to polymerize at much lower concentrations in the presence of AtMAP65-1 than the pure tubulins.
AtMAP65-1, Not AtMAP65-6, Stabilizes Microtubules against Cold Treatment and Dilution
We further tested whether these two MAP65 proteins were able to stabilize microtubules against depolymerizing challenges. When microtubules, previously polymerized by 20 μm rhodamine-labeled tubulins at 37°C for 30 min, were brought to 10°C for 30 min, these microtubules disassembled (Fig. 5A). In the presence of AtMAP65-1 at 0.5 μm, however, the identical microtubules resisted the cold temperature (Fig. 5B). When AtMAP65-6 was used in a similar test, all microtubules disassembled (Fig. 5C).
Figure 5.
Microtubule bundles under depolymerization challenges. A, Microtubules totally depolymerized after cold treatment. B, Microtubule bundles induced by AtMAP65-1 were resistant to cold treatment. C, In the presence of AtMAP65-6, microtubules were not detected after cold treatment. D, Microtubules were depolymerized completely upon dilution to 50× with warm buffer in 60 min. E, Microtubule bundles induced by AtMAP65-1 were resistant to the dilution treatment. F, Microtubule network induced by AtMAP65-6 was sensitive to the dilution treatment as the network disappeared from the treatment. Scale bar = 10 μm.
To further test the effect of these MAP65 proteins on microtubule stabilization, a dilution test was performed. In the test, microtubules were first assembled in the presence of MAP65 proteins and then diluted by 50× warm buffer solution. Similar to what we had observed in cold treatment, the presence of AtMAP65-1 made microtubules resistant to dilution treatment. Without the addition of MAP65s, most microtubules, if not all, were disassembled in 60 min (Fig. 5D). In the presence of AtMAP65-1, however, induced microtubules bundles resisted the dilution (Fig. 5E). On the contrary, AtMAP65-6 did not have such an effect. Microtubules were still disassembled with the dilution in the presence of AtMAP65-6 (Fig. 5F).
AtMAP65-6 Is Associated with Mitochondria
Earlier studies by immunofluorescence and green fluorescence protein tagging indicated that AtMAP65-1 is associated with cortical microtubules (Smertenko et al., 2004; Van Damme et al., 2004). We wondered whether AtMAP65-6 had a different intracellular localization pattern. To specifically detect AtMAP65-6, antibodies were raised against a specific peptide. Affinity-purified anti-AMAP65-6 only detected GST-AtMAP65-6, not GST-AtMAP65-1 (Fig. 6A). Conversely, anti-AtMAP65-1 antibodies only detected GST-AtMAP65-1, not GST-AtMAP65-6 (Fig. 6A). The monospecific anti-AtMAP65-6 antibodies were then used to detect the native AtMAP65-6 protein isolated from Arabidopsis suspension cells. A specific band of approximately 65 kD was revealed by anti-AtMAP65-6, not by control IgG (Fig. 6B). This molecular mass was consistent, with the calculated mass of 64.6 kD for AtMAP65-6.
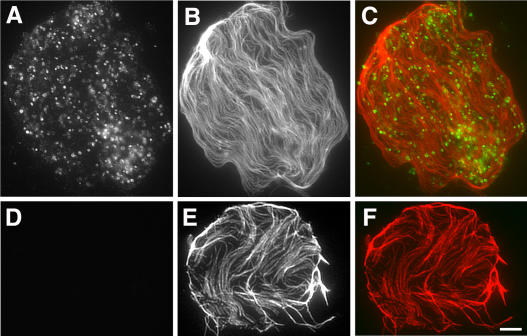
Because the monospecific antibodies detected the native protein, they were used to localize AtMAP65-6 in vivo. In protoplasts isolated from Arabidopsis suspension cells, numerous organelle-like particles were detected (Fig. 7A). These particles were in close contact with cortical microtubules attached to the plasma membrane (Fig. 7, B and C). When control rabbit IgG substituted anti-AtMAP65-6, no signal was detected, while cortical microtubules were still revealed (Fig. 7, D–F).
Figure 7.
Immunolocalization of AtMAP65-6. A to C, Dual localization of AtMAP65-6 and microtubules. AtMAP65-6 detected organelle-like structures (A) adjacent to cortical microtubules (B). Merged image (C) shows AtMAP65-6 in green and microtubules in red. D to F, Immunofluorescence with control IgG. No signal was detected with control rabbit IgG (D), while cortical microtubules were still visualized (E). Merged image is shown in F. Scale bar = 5 μm.
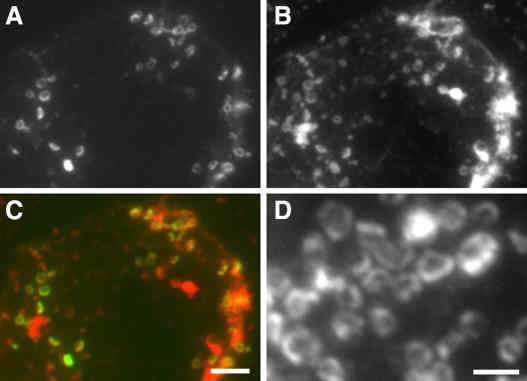
Because anti-AtMAP65-6 decorated an organelle-like structure, we wanted to determine the identity of the organelle. We tested markers of various organelles (data not shown). Anti-AtMAP65-6 signal showed colocalization with mitochondria, as revealed by the mitochondria-specific marker MitoTracker Red (Fig. 8, A–C). The AtMAP65-6 signal appeared in a ring surrounding the mitochondria (Fig. 7D). Thus, we concluded that AtMAP65-6 was targeted to the mitochondria in Arabidopsis cells.
Figure 8.
Colocalization of AtMAP65-6 with mitochondria. A to C, Dual localization of AtMAP65-6 and mitochondria. Anti-AtMAP65-6 signal (A) and MitoTracker Red-detected signal (B) were overlapping in merged image (C), in which AtMAP65-6 is shown in green and mitochondria in red. D, AtMAP65-6 formed a ring at the periphery of mitochondria. Enlarged images show that AtMAP65-6 decorated the peripheral region of the mitochondria (D). Scale bars = 5 μm (A–C) and 2 μm (D).
DISCUSSION
Our results indicated that two Arabidopsis MAP65 proteins, with 44% sequence identity (Hussey et al., 2002), acted differently on microtubules in vitro. In the presence of AtMAP65-1, microtubules formed large, bundles as reported by others. AtMAP65-6 induced a mesh-like network of crisscrosses. While AtMAP65-1 decorated cortical microtubules, AtMAP65-6 decorated mitochondria.
Interaction of Different MAP65 Proteins with Microtubules
Plant MAP65 proteins have been purified from tobacco BY-2 suspension-cultured cells and carrot suspension cells (Jiang and Sonobe, 1993; Chan et al., 1996). Three cDNA sequences have been isolated for three highly similar polypeptides, NtMAP65-1a, NtMAP65-1b, and NtMAP65-1c (Smertenko et al., 2000). Compared to the nine Arabidopsis MAP65 proteins, these three tobacco ones are closely related to the branch with AtMAP65-1 and AtMAP65-2 (Hussey et al., 2002). Biochemical data suggest that even these highly similar MAP65s exhibit different roles on microtubules, although they probably all have microtubule-bundling activities. NtMAP65-1a is capable of promoting microtubule polymerization, an activity found with MAP65 preparations directly purified from plant cells that contain multiple forms of MAP65 (Smertenko et al., 2000). NtMAP65-1b, however, does not promote polymerization (Wicker-Planquart et al., 2004). This is rather interesting as these two polypeptides have an amino acid sequence identity as high as 85% (Smertenko et al., 2000). Interestingly, AtMAP65-1 also bundles just microtubules without affecting polymerization (Smertenko et al., 2004). AtMAP65-2 has 78% sequence identity with AtMAP65-1 (Hussey et al., 2002), so it will be interesting to test whether they have different effects on microtubule polymerization.
Even closely related MAP65 proteins exhibit different activities; it was not surprising that we found AtMAP65-1 and AtMAP65-6 affected microtubule organization differently. It has been suggested that AtMAP65-1 homodimers make the cross-bridges between adjacent microtubules (Smertenko et al., 2004). The distance between adjacent microtubules in AtMAP65-1-induced bundles is close to the diameter of the microtubule. The distance between adjacent microtubules in the presence of AtMAP65-6, however, was much shorter than the microtubule diameter. Thus, this suggests to us that AtMAP65-6 might not form homodimers. The interaction between AtMAP65-1 dimers and microtubules allows parallel microtubules to be continuously associated with each other via the dimers. Because MAP65-1 has the microtubule-binding site at its C terminus and bundling activity at its N terminus (Smertenko et al., 2004), only an antiparallel configuration would form a bipolar dimer that cross-links microtubules. A bipolar dimer would bundle microtubules continuously. Because of the sequence divergence between AtMAP65-6 and AtMAP65-1, AtMAP65-6 may not form a bipolar dimer to grant a bundling activity.
Our data also indicated that AtMAP65-1 and AtMAP65-6 affected microtubule stability differently. The difference further suggests that they may interact with microtubules in different manners. The continuous ladder-like interaction with microtubules allows microtubules to form a fortress-like structure by AtMAP65-1. Such a structure would certainly be more resistant to depolymerization challenges like cold and dilution treatments than mesh-like network of individual microtubules induced by AtMAP65-6.
Different MAP65 Proteins Have Different Intracellular Localization Patterns
Antibodies against tobacco and carrot MAP65 preparations decorate microtubules arrays evenly, which is indistinguishable from microtubule staining (Jiang and Sonobe, 1993; Chan et al., 1996). Antibodies raised against NtMAP65-1, AtMAP65-1, and AtMAP65-3, however, only detect subpopulations of microtubules in the mitotic spindle and the phragmoplast (Sawano et al., 2000; Muller et al., 2004; Smertenko et al., 2004). Unfortunately, it is not clear whether those antibodies recognize just a single form of MAP65s or multiple ones. Nevertheless, the localization pattern by these new antibodies in the spindle and phragmoplast midzone is very intriguing. It is worth noting that similar proteins in animal and fungal cells also demonstrate localization at similar regions (Pellman et al., 1995; Jiang et al., 1998; Schuyler et al., 2003). Functional analyses have also reached a common conclusion that such proteins play a critical role in cytokinesis (Jiang et al., 1998; Muller et al., 2004). The localization pattern in the spindle midzone raises speculation on how certain MAP65 proteins distinguish different sets of microtubules in a single cell.
Because AtMAP65-6 behaved differently than AtMAP65-1 in vitro, it was not surprising that AtMAP65-6 demonstrated a different intracellular localization pattern than AtMAP65-1. Our AtMAP65-6-specific antibodies revealed that AtMAP65-6 decorated mitochondria. Earlier scrutinizing pharmacological studies indicated that although plant mitochondria move along F-actin filaments, their positioning in the cortical cytoplasm relies on both F-actin and microtubules (Van Gestel et al., 2002). Thus, plant mitochondria may interact with cortical microtubules. Our results on AtMAP65-6 localization, therefore, provide direct molecular evidence showing that a MAP is targeted to this organelle. AtMAP65-6 may be one of the MAPs, if not the only one, that mediate the interaction between cortical microtubules and mitochondria.
The remaining task is to determine the localization patterns of six other members of the AtMAP65 family.
Potential Roles of AtMAP65s in Plant Growth
The different activities and localization patterns of AtMAP65-1 and AtMAP65-6 suggest that they play distinct roles in plant growth and development. Cortical microtubules are highly dynamic, and undergo reorganization upon receiving internal or external cues (Yuan et al., 1994). The cortical array consists of not only individual microtubules, but also microtubule bundles, even with mixed polarities (Shaw et al., 2003). These microtubule bundles could be induced and stabilized by proteins like AtMAP65-1, NtMAP65-1, and AtMAP65-5, as they all decorate cortical microtubules (Smertenko et al., 2000, 2004; Van Damme et al., 2004). But these proteins as stabilizing factors could have an inhibitory effect on microtubule reorganization. Transition from one orientation of cortical microtubules to another probably has an intermediate state when cortical microtubules appear more or less randomly. Random cortical microtubules can be found in some cells of the root epidermis (Sugimoto et al., 2000). It will be interesting to test whether these cells are in the transition from one orientation to another. It would also be interesting to learn the dynamics of these MAP65s in living cells.
While the budding yeast S. cerevisiae as a lower eukaryote has a single gene encoding a MAP65-like protein Ase1p, Arabidopsis as a higher plant contains multiple genes encoding MAP65 proteins. These proteins have been evolved to take on distinct tasks required for multifaceted cellular activities besides anaphase spindle elongation.
MATERIALS AND METHODS
AtMAP65-1 and AtMAP65-6 Fusion Protein Preparation
AtMAP65-1 and AtMAP65-6 are the protein products of the At5g55230 and At2g01910 genes, respectively. To express fusion proteins of AtMAP65-1 and AtMAP65-6, cDNA sequences were amplified from the EST clone 109M12 for AtMAP65-1, and by RT-PCR for AtMAP65-6. Arabidopsis (Arabidopsis thaliana) total RNA was purified from seedlings by the SV total RNA isolation kit (Promega, Madison, WI). The cDNA sequences, available from the GenBank (accession nos. NM-124905 and NM-126252, respectively), were referenced to design primers. Two primers of MAP65-1 (forward, 5′-GCGTCGACATGGCAGTTACAGATACTG-3′ [with the SalI site underlined], and reverse, 5′-TTGCGGCCGCTCATGGTGAAGCTGGAACTT-3′ [with the NotI site underlined]) were used to amplify AtMAP65-1 cDNA. The primers used for amplification of AtMAP65-6 cDNA were as follow: MAP65-6 forward, 5′-AACCCGGGAATGCTTATGGAATTAGA-3′ (with the SmaI site underlined), and MAP65-6 reverse, 5′-TAAACTCGAGTCAGCCTTGGAGAGG-3′ (with the XhoI site underlined). RT-PCR was carried out using Promega kit according to manufacturer's instruction. Amplified sequences were A-tailed and cloned into the pMD18-T vector at corresponding sites designed in the primers (TaKaRa Biotechnology, Dalian, China). cDNA sequences were verified by sequencing.
Full-length cDNAs of AtMAP65-1 and AtMAP65-6 were constructured into the vectors pGEX-4T2 and pGEX 4T1 (Amersham Biosciences, Piscataway, NJ) with SalI and NotI, and SmaI and NotI, respectively. Fusion proteins were expressed in the Escherichia coli strain BL21 (DE3) by induction with 1 mm isopropyl β-d-thiogalactopyranoside for 3 h at 30°C. The GST fusion protein was affinity purified with the glutathione-Sepharose resin according to the manufacturer's protocol (Amersham Biosciences). Protein samples were analyzed by SDS-PAGE and visualized by staining the gels with Coomassie Brilliant Blue R-250 (Sigma-Aldrich, St. Louis).
Microtubule Cosedimentation Assay
Porcine brain tubulins were purified according to a published method (Castoldia and Popov, 2003). The 5-(and 6-)carboxytetramethylrhodamine succinimidyl ester (NHS)-rhodamine was then conjugated to purified tubulin as reported before (Hyman, 1991). NHS-rhodamine-labeled tubulin then underwent another round of assembly/disassembly with 10% (v/v) dimethyl sulfoxide before being stored in liquid nitrogen as 10-μL aliquots, which were later used for polymerization and sedimentation assays.
The binding reaction was performed in 200 μL volume containing 1 μm microtubules and 0.5 and 5 μm MAP65-1 or MAP65-6, respectively, in the PEM buffer (1 mm MgCl2, 1 mm EGTA, and 100 mm PIPES-KOH, pH 6.9) containing 1 mm GTP and 20 μm taxol. After 30 min of incubation at room temperature, samples were centrifuged at 100,000g for 30 min. The supernatants and pellets were separated and brought to equal volumes in the SDS sample buffer. They were then analyzed by SDS-PAGE and visualized by staining the gels with Coomassie Brilliant Blue R-250. Gel scanning was performed on AlphaImager 2200 (Alpha Innotech, San Leandro, CA).
Assays of Microtubule Polymerization
A range of concentrations of AtMAP65-1 or AtMAP65-6 were added to 30 μm tubulin in the PEM buffer with 1 mm GTP. Microtubule polymerization was monitored turbidimetrically by the A350 with a 0.4-cm path quartz cell at 37°C in a DU-640 spectrophotometer (Beckman Coulter, Fullerton, CA).
The Cc for tubulin polymerization was determined according to a published method (Algaier and Himes, 1988). Briefly, 1 μm AtMAP65-1 was added to the PEM buffer with a range of concentrations of tubulin. After stabilization, the sample was centrifuged at 100,000g for 35 min at 37°C. The resulting pellet was resuspended with cold PEM buffer of the original volume. Protein concentration was determined by the Bio-Rad protein assay (Bio-Rad, Hercules, CA), using bovine serum albumin as the standard. Arbitrary values of tubulins in the pellet were plotted against the starting tubulin concentrations and were then used to determine the intercept with the x axis. As a control, the critical concentration of self-assembly of tubulin without MAP65-1 was measured.
Polymerized microtubules were challenged by the following treatments. For cold treatment, 20 μm NHS-rhodamine-tubulin was polymerized at 37°C for 30 min, with or without 0.5 μm AtMAP65-1 or AtMAP65-6. The preformed microtubules were then incubated at 10°C for 30 min before being fixed with 1% glutaraldehyde for observation. For the dilution challenge, microtubules were polymerized under identical conditions. But the samples were diluted by 50× (v/v) with prewarmed PEM buffer and incubated at 37°C for 30 min prior to being fixed with 1% glutaraldehyde for observation.
For the NaCl treatment, microtubules polymerized from 20 μm rhodamine-tubulin were used. Polymerized microtubules were first diluted 10× (v/v) with the PEM buffer plus 20 μm taxol and then centrifuged at 60,000g for 30 min at 37°C. The resulting pellet was resuspended with the PEM buffer of the same volume. AtMAP65-1 or AtMAP65-6 was added to a final concentration of 0.5 μm before the microtubule suspension was incubated at 37°C for 30 min. Final concentrations of NaCl were 100 mm, 200 mm, 300 mm, and 400 mm were for AtMAP65-1-treated microtubules, and 400 mm and 500 mm for AtMAP65-6-treated ones. Each sample was incubated at 37°C for 30 min prior to being fixed for observation.
Microtubule Observation by Confocal and Electron Microscopy
After being incubated at 37°C for 30 min, microtubule polymerization was terminated using 1% glutaraldehyde. The samples were then examined under a Leica DMR microscope (Wetzlar, Germany) equipped with a 100× oil objective (NA 1.3). Images were collected with an MRC1024 confocal laser scanning microscope equipped with argon-krypton laser (Bio-Rad).
Microtubules were either directly observed by negative staining or by thin sectioning. Negative staining was done with saturated uranyl acetate. Microtubules were collected by centrifugation at 100,000g. Microtubule pellets were fixed in 1% glutaraldehyde in PEM buffer, followed by 1% osmium tetroxide, and embedded in the LR White resin. Samples were sectioned and stained with uranyl acetate and lead citrate, then observed under a Hitachi 7500 electron microscope (Hitachi, Tokyo).
Polyclonal Antibody Production
The AtMAP65-1 peptide of amino acids 471 to 485, [H]-QEREEEKRRLREQKK-[OH], and the AtMAP65-6 peptide of amino acids 470 to 484, [H]-KPSPRRSSSFRKPNG-[OH], were chosen as antigens to raise specific antibodies. A Cys residue was added to the carboxyl terminus of each peptide for conjugation to the carrier Keyhole limpet hemocyanin. Antisera were produced at a commercial facility (GeneMed Synthesis, South San Francisco). Antibodies against each-peptide were affinity purified by the blot purification method (Tang, 1993). Briefly, GST-AtMAP65-1 and GST-AtMAP65-6 were separated on 12.5% SDS-PAGE gels and transferred to nitrocellulose membrane. The nitrocellulose membranes containing AtMAP65-1 or AtMAP65-6 were excised and blocked for 1 h in Tris-buffered saline (TBS) containing 3% (w/v) bovine serum albumin and 0.05% (v/v) Tween 20. Strips containing individual fusion proteins were incubated with corresponding serum diluted 1:10 with TBS containing 0.05% (v/v) Tween 20 at room temperature for at least 3 h. They were then washed three times with 10 mL TBS containing 0.05% (v/v) Tween 20. The antibodies were eluted with 2 mL elution buffer containing 0.1 m Gly-HCl, pH 3.0, 0.5 m NaCl, and 0.05% Tween 20 for 1 min with gentle shaking. Eluted antibodies were immediately neutralized with 0.25 mL of 1 m Tris, pH 8.0. Finally the antibodies were dialysed with PBS buffer, pH 8.0, and separated into aliquots and stored at −80°C. The antibodies were used at 1:100 (v/v) for immunoblotting experiments.
Immunoblotting and Immunofluorescence Experiments
Total proteins were extracted from Arabidopsis suspension cells according a published protocol (Preuss et al., 2003). Proteins were separated by a 12% SDS-PAGE gel and transferred to nitrocellulose membrane. Blots were probed with affinity-purified anti-AtMAP65-6 antibodies.
Protoplasts were prepared enzymatically from Arabidopsis suspension cells with 1% Cellulose RS and 0.1% Pectolyase Y23 (Karland, Cottonwood, AZ) in 0.3 m mannitol solution. Isolated protoplasts were attached to poly-l-lysine-coated slides and fixed with 4% paraformaldehyde as published before (Preuss et al., 2003). Microtubules were probed with the DM1A anti-α-tubulin antibody (Sigma). Secondary antibodies were Texas Red X-conjugated goat anti-mouse IgG and FITC-conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR). Mitochondria were detected with MitoTracker Red (Molecular Probes). All samples were observed under an Eclipse E600 microscope (Nikon, Melville, NY) equipped with epifluorescence optics. An Orca CCD camera (Hamamatsu Photonic Systems, Bridgewater, NJ) was used to capture images by the ImageProPlus 4.0 software package (Media Cybernetics, Silver Spring, MD).
All images presented in this article were assembled using the Photoshop 7.0 software package (Adobe Systems, San Jose, CA).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers NM-124905 for AtMAP65-1 and NM-126252 for AtMAP65-6.
Acknowledgments
We thank the Arabidopsis Biological Research Center for providing the cDNA clone 109M12.
This work was supported by the National Key Basic Research Project of China and the National Natural Science Foundation of China (grant no. 30128008 to B.L. and grant no. 30170057 to M.Y.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.052456.
References
- Algaier J, Himes RH (1988) The effects of dimethyl sulfoxide on the kinetics of tubulin assembly. Biochim Biophys Acta 954: 235–243 [DOI] [PubMed] [Google Scholar]
- Castoldia M, Popov A (2003) Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif 32: 83–88 [DOI] [PubMed] [Google Scholar]
- Chan J, Jensen CG, Jensen LCW, Bush M, Lloyd CW (1999) The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc Natl Acad Sci USA 96: 14931–14936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Mao G, Smertenko A, Hussey PJ, Naldrett M, Bottrill A, Lloyd CW (2003) Identification of a MAP65 isoform involved in directional expansion of plant cells. FEBS Lett 534: 161–163 [DOI] [PubMed] [Google Scholar]
- Chan J, Rutten T, Lloyd C (1996) Isolation of microtubule-associated proteins from carrot cytoskeletons: a 120 kDa map decorates all four microtubule arrays and the nucleus. Plant J 10: 251–259 [DOI] [PubMed] [Google Scholar]
- Cyr RJ, Palevitz BA (1995) Organization of cortical microtubules in plant cells. Curr Opin Cell Biol 7: 65–71 [DOI] [PubMed] [Google Scholar]
- Gunning BES, Steer MW (1996) Plant Cell Biology. Structure and Function. John and Bartlett Publishers, Sudbury, MA
- Hussey PJ, Hawkins TJ, Igarashi H, Kaloriti D, Smertenko A (2002) The plant cytoskeleton: recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol Biol 50: 915–924 [DOI] [PubMed] [Google Scholar]
- Hyman A (1991) Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence. J Cell Sci (Suppl) 14: 125–127 [DOI] [PubMed] [Google Scholar]
- Jiang C-J, Sonobe S (1993) Identification and preliminary characterization of a 65 kDa higher-plant microtubule-associated protein. J Cell Sci 105: 891–901 [DOI] [PubMed] [Google Scholar]
- Jiang W, Jimenez G, Wells N, Hope T, Wahl G, Hunter T, Fukunaga R (1998) PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell 2: 877–885 [DOI] [PubMed] [Google Scholar]
- Lloyd C, Hussey P (2001) Microtubule-associated proteins in plants—why we need a MAP. Nat Rev Mol Cell Biol 2: 40–47 [DOI] [PubMed] [Google Scholar]
- Muller S, Smertenko A, Wagner V, Heinrich M, Hussey P, Hauser M (2004) The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol 14: 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu Y, Fink G, Tu H (1995) Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J Cell Biol 130: 1373–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Delmer DP, Liu B (2003) The cotton kinesin-like calmodulin-binding protein associates with cortical microtubules in cotton fibers. Plant Physiol 132: 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano M, Shimmen T, Sonobe S (2000) Possible involvement of 65 kda MAP in elongation growth of azuki bean epicotyls. Plant Cell Physiol 41: 968–976 [DOI] [PubMed] [Google Scholar]
- Schuyler S, Liu J, Pellman D (2003) The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol 160: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S, Kamyar R, Ehrhardt D (2003) Sustained microtubules treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Smertenko A, Chang H, Wagner V, Kaloriti D, Fenyk S, Sonobe S, Lloyd C, Hauser M, Hussey P (2004) The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell 16: 2035–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A, Saleh N, Igarashi H, Mori H, Hauser-Hahn I, Jiang C, Sonobe S, Lloyd C, Hussey P (2000) A new class of microtubule-associated proteins in plants. Nat Cell Biol 2: 750–753 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Williamson R, Wasteneys G (2000) New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WJ (1993) Blot-affinity purification of antibodies. Methods Cell Biol 37: 95–104 [DOI] [PubMed] [Google Scholar]
- Van Damme D, Van Poucke K, Boutant E, Ritzenthaler C, Inzé D, Geelen D (2004) In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol 136: 3956–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gestel K, Kohler RH, Verbelen J-P (2002) Plant mitochondria move on F-actin, but their positioning in the cortical cytoplasm depends on both F-actin and microtubules. J Exp Bot 53: 659–667 [DOI] [PubMed] [Google Scholar]
- Wasteneys G, Galway M (2003) Remodeling the cytoskeleton for growth and form: an overview with some new views. Annu Rev Plant Biol 54: 691–722 [DOI] [PubMed] [Google Scholar]
- Wicker-Planquart C, Stoppin-Mellet V, Blanchoin L, Vantard M (2004) Interactions of tobacco microtubule-associated protein MAP65-1b with microtubules. Plant J 39: 126–134 [DOI] [PubMed] [Google Scholar]
- Yuan M, Shaw P, Warn R, Lloyd C (1994) Dynamic reorientation of cortical microtubules, from transverse to longitudinal, in living plant cells. Proc Natl Acad Sci USA 91: 6050–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]