Abstract
The role of starch synthase (SS) III (SSIII) in the synthesis of transient starch in Arabidopsis (Arabidopsis thaliana) was investigated by characterizing the effects of two insertion mutations at the AtSS3 gene locus. Both mutations, termed Atss3-1 and Atss3-2, condition complete loss of SSIII activity and prevent normal gene expression at both the mRNA and protein levels. The mutations cause a starch excess phenotype in leaves during the light period of the growth cycle due to an apparent increase in the rate of starch synthesis. In addition, both mutations alter the physical structure of leaf starch. Significant increases were noted in the mutants in the frequency of linear chains in amylopectin with a degree of polymerization greater than approximately 60, and relatively small changes were observed in chains of degree of polymerization 4 to 50. Furthermore, starch in the Atss3-1 and Atss3-2 mutants has a higher phosphate content, approximately two times that of wild-type leaf starch. Total SS activity is increased in both Atss3 mutants and a specific SS activity appears to be up-regulated. The data indicate that, in addition to its expected direct role in starch assembly, SSIII also has a negative regulatory function in the biosynthesis of transient starch in Arabidopsis.
Starch serves a fundamental role in the life cycle of plants as the primary carbohydrate storage form for chemical energy. Transient starch is produced and degraded over short periods of time to satisfy the ongoing energy requirements of plant development (e.g. over the course of the diurnal cycle in the leaf). In contrast, storage starch accumulates and persists in heterotrophic tissues in anticipation of future plant energy needs, such as germination or sprouting. The anabolism and catabolism of both storage and transient starch forms requires precise control of the expression and/or activity of many different enzymes.
Starch consists of two homopolymers of α-d-glucosyl units joined in linear arrays by α-(1→4) glycosidic bonds, amylose (Am) and amylopectin (Ap). Ap is the more abundant polymer, and it contains α-(1→6) branch linkages at a frequency of approximately 5%. The organized architectural arrangement of linear and branched chains in Ap enables efficient packaging of large amounts of Glc into insoluble starch granules. The basic enzymatic steps responsible for Ap structure begin with the formation of the activated glucosyl donor ADP-Glc (ADPG), catalyzed by ADPG pyrophosphorylase. Reactions utilizing ADPG to build α-(1→4)-linked linear glucosyl chains are catalyzed by the starch synthases (SSs). Branch linkages are introduced by branching enzymes (BEs), which catalyze cleavage of an internal α-(1→4) linkage and creation of a new α-(1→6) linkage by transfer of the released reducing end to a C6 hydroxyl. Starch-debranching enzymes (DBEs) hydrolyze branch linkages and are thought to be involved in starch biosynthesis, based on evidence that DBE mutants produce less starch than normal and contain a highly branched polysaccharide not observed in wild type. Multiple SS, BE, and DBE isoforms exist in all plant species, and their strong evolutionary conservation suggests that each isoform class functions uniquely in starch metabolism.
Five distinct SS classes are known in all plants based on amino acid sequence similarities: granule-bound SS (GBSS), SSI, SSII, SSIII, and SSIV/V. All are highly similar in the C-terminal region, a span of approximately 450 amino acid residues comprising the catalytic and starch-binding domains, but differ significantly in the sequences of their N termini, with SSIII having the longest N-terminal arm. Genetic analyses in plants that accumulate storage starch indicate that at least three of the SS classes provide unique functions in starch biosynthesis: GBSS (Shure et al., 1983; Klösgen et al., 1986); SSIII (Gao et al., 1998; Cao et al., 1999); and SSII (Craig et al., 1998; Morell et al., 2003; Zhang et al., 2004). Mutations in these SS genes cause specific alterations in starch composition and/or starch structure, demonstrating that the function of each of these isoforms is not fully compensated by any other SS. A model to account for the roles of different SSs in determining the lengths of Ap chains proposes that SSI is primarily responsible for synthesis of the shortest chains, SSII lengthens these to provide most of the chains that crystallize, and SSIII produces longer length chains extending between clusters (Commuri and Keeling, 2001; James et al., 2003).
Although the SSs are thought to affect glucan structure primarily through their enzymatic extension of linear glucan chains, the SSIII isoform also is likely to influence the final molecular architecture of the starch indirectly through its association with other factors, particularly other starch-metabolizing enzymes. This idea is supported in part by previous biochemical analysis of enzyme activities in maize (Zea mays) kernels homozygous for a mutation in the dull1 (du1) gene, which codes for SSIII (Gao et al., 1998), in which the loss of SSIII activity is accompanied by the additional loss of a specific BEII activity (now known to be BEIIa; Preiss and Boyer, 1979). Furthermore, combinatorial genetic effects are produced in several maize double mutants involving du1, such that a more extreme kernel phenotype results than that exhibited by either single mutant. Such effects, which are observed with du1 in combination with sugary1 (su1), which codes for the DBE ISA1 (Cameron, 1947; Mangelsdorf, 1947), amylose extender (ae), which codes for BEIIb (Creech, 1965), or waxy (wx), which codes for GBSS (Creech, 1965; Shannon and Garwood, 1984), suggest either that physical interactions occur between the two gene products or that the mutations affect parallel or sequential steps in the starch synthesis pathway.
To date, investigations of the effects of altered SSIII activity have focused exclusively on storage starch metabolism. However, transient and storage starch forms are not synthesized by identical subsets of enzyme isoforms and can differ with respect to their Ap structures and Ap:Am ratios (Zeeman et al., 2002). In this study, changes in leaf starch synthesis were investigated in plants homozygous for mutations in the Arabidopsis (Arabidopsis thaliana) gene AtSS3, which codes for SSIII. Two independent null mutations in the AtSS3 locus were identified and found to cause changes in the structure of the Ap in leaf starch and in the degree of starch phosphorylation. In addition, both Atss3 mutations promote an overall increase in the amount of starch that is produced in the leaf during a single light phase of the diurnal cycle. The results indicate a unique function for SSIII in starch biosynthesis and, in addition, demonstrate a regulatory role for this protein in determining the overall rate of starch accumulation.
RESULTS
Identification of Null Mutations in the Gene Coding for SSIII
The Arabidopsis genomic locus At1g11720, referred to here as AtSS3, codes for a protein (GenBank accession no. NP_172637) that is highly similar in amino acid sequence to known SSIII proteins from several species (Li et al., 2000). A nearly full-length cDNA expressed from this locus was obtained by reverse transcription (RT)-PCR amplification of total RNA from Arabidopsis leaves, then cloned and sequenced (see “Materials and Methods”). This sequence matches a cDNA previously deposited in the GenBank database (accession no. NM_101044) with 100% identity, although in neither instance has the 5′ untranslated region of the transcript been characterized. Comparison of the cDNA sequence with that of the genomic DNA reveals a gene structure comprising 14 exons and 13 introns (Fig. 1A). The first 60 nucleotides of the available sequence from exon 1, starting at the presumed ATG initiation codon, encode a predicted plastid-targeting peptide of 20 amino acids according to analysis with the computational programs ChloroP and TargetP.
Figure 1.
Characterization of Atss3 mutations. A, Gene map. The scaled linear map depicts the 14 exons as boxes and the 13 introns as lines between the boxes. The positions of the translational start and stop codons in exons 1 and 14, respectively, are noted. The locations of specific PCR primer sequences are noted, as well as the locations of the two T-DNA insertions in the gene (insertions are not drawn to scale). B, Analysis of AtSS3 5′-end transcripts. Control genomic DNA from wild-type leaves (lane 1) was amplified by PCR and total leaf RNA from Atss3-1 and Atss3-2 mutant plants (lanes 2 and 3, respectively) was amplified by RT-PCR using the gene-specific primers SS3-U2 and SS3-L1. C, Analysis of transcripts from the Atss3-1 mutant. Total RNA from leaves of wild-type plants (lane 1) and Atss3-1 plants (lanes 2 and 3) was amplified by RT-PCR. Primer pairs are SS3-TU1 and SS3-TL1 (lanes 1 and 3). Arabidopsis SSII gene-specific primers SS2-RP1 and SS2-LP1 served as a positive control (lane 2). D, Analysis of transcripts from the Atss3-2 mutant. Total RNA from leaves of wild-type plants (lane 1) and Atss3-2 plants (lanes 2 and 3) was amplified by RT-PCR. Primer pairs were SS3-RP2 and SS3-LP2 (lanes 1 and 3) or Arabidopsis SSII gene-specific primers SS2-RP2 and SS2-LP2 as a positive control (lane 2). E, Analysis of SSIII protein accumulation. Total soluble leaf extracts from wild-type, Atss3-1, and Atss3-2 plants were separated by SDS-PAGE and probed in immunoblot analysis with α-AtSSIII monoclonal antibody.
Two mutations in the AtSS3 locus are available in the Salk collection of T-DNA insertion lines (SALK_065732 and SALK_102605), designated Atss3-1 and Atss3-2, respectively. Gene-specific primers flanking the approximate insertion site for each mutation were designed so that PCR analysis of genomic DNA could identify the wild-type allele and each of the two different mutations. Using these primers, together with a primer from the T-DNA left border region, the mutations were confirmed to be located in exon 13 for Atss3-1 and exon 9 for Atss3-2 (Fig. 1A). PCR amplification distinguished the wild-type and mutant alleles, and homozygous mutant lines were identified and confirmed by segregation analysis for two successive generations.
Expression of the wild-type and mutant AtSS3 loci was examined at the level of mRNA and protein. RT-PCR amplification of leaf RNA revealed that, for both Atss3-1 and Atss3-2, transcripts accumulated corresponding to the 5′-region of the locus (Fig. 1B); however, no transcripts accumulated from the 3′-regions of the gene surrounding either insertion site (Fig. 1, C and D). Immunoblot analysis of total proteins extracted from leaves of wild-type, Atss3-1, or Atss3-2 plants was used to examine whether any SSIII protein accumulated in the mutants. A monoclonal antibody raised against a recombinant form of SSIII produced in Escherichia coli was used to detect the presence of the protein. A signal was detected in wild-type plants for a protein migrating slightly slower than the 150-kD molecular mass maker, in general agreement with the 116-kD molecular mass predicted from the AtSS3 cDNA sequence (Fig. 1E). This signal was completely absent from both the Atss3-1 and Atss3-2 plant extracts. Taken together, the data indicate that both Atss3 mutations produce abnormal transcripts that fail to direct expression of normal SSIII.
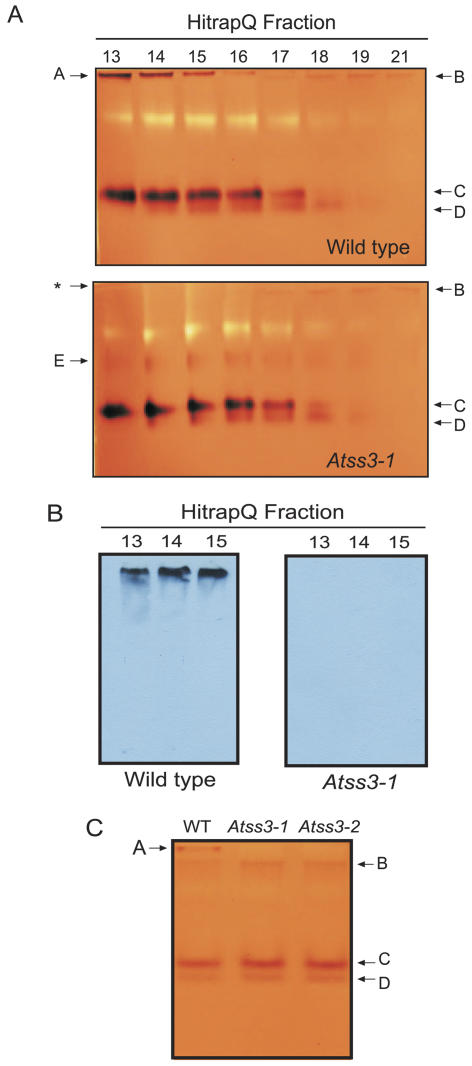
Native PAGE activity gel analysis (i.e. zymogram analysis) was employed to demonstrate that the two mutations result in loss of SSIII enzyme activity. Protein extracts from wild-type and mutant leaves were first separated by anion-exchange chromatography. The proteins in each column fraction were then separated in nondenaturing polyacrylamide gels impregnated with 0.3% glycogen. After electrophoresis, the gels were incubated in a buffer containing both glycogen and ADPG to establish conditions enabling the elongation of the exterior glycogen chains by SSs in the gel. Staining with iodine solution revealed each SS activity as a brown band on a tan background. Two major and two minor activity bands were observed in wild-type extracts (Fig. 2A). The most slowly migrating activity, one of the major bands, was completely missing in the extract from Atss3-1 leaves. Immunoblot analysis demonstrated that the SSIII protein comigrated with the most slowly migrating SS activity and that this signal was again missing from the Atss3-1 mutant plant (Fig. 2B). In a similar zymogram analysis using total leaf extract that had not been subjected to anion-exchange fractionation, the most slowly migrating SS activity also was found to be absent from Atss3-2 plants as well as the Atss3-1 mutant (Fig. 2C). Taken together, the data indicate that SSIII activity is completely missing in the two Atss3 mutants. These two homozygous lines were used in subsequent analyses to determine the effects of mutation of SSIII on starch biosynthesis.
Figure 2.
Analysis of SSIII activity. A, Two-dimensional zymogram analysis. Proteins from equivalent fresh weights of leaves of wild-type (top) and Atss3-1 (bottom) plants were separated by anion-exchange chromatography. Proteins in equivalent volumes of the indicated HiTrapQ column fractions were separated in native polyacrylamide gels containing 0.3% glycogen, then stained with iodine solution. Identifiable SS activity bands are indicated. The asterisk in the Atss3-1 analysis indicates the position that corresponds to band A in the wild-type analysis. B, Immunoblot analysis. Duplicate gels to those shown in (A) were probed for the presence of SSIII with α-AtSSIII monoclonal antibody. C, One-dimensional zymogram analysis. Total soluble protein extracts from leaves of wild-type, Atss3-1, and Atss3-2 plants were analyzed for SS activity as described for (A). In this instance, there was no prior anion-exchange fractionation step.
Atss3 Mutations Condition Increased Starch Synthesis Rate
Homozygous mutant Atss3-1 and Atss3-2 plants appear to have normal seed germination properties, growth rates after germination, and flowering times. No obvious differences between mutant and wild type were observed in terms of plant size or morphology. However, investigation of transient starch metabolism in the leaves revealed that both mutants exhibit a starch excess phenotype. Plants grown under a photoperiod of 16-h light/8-h dark were initially examined for starch content by comparing the iodine-staining intensity of the leaf starch following the removal of chlorophyll. This qualitative analysis revealed an apparent excess of starch at the middle and end of the light cycle for both mutants (Fig. 3; data not shown). Quantitative data for starch content were then collected for homozygous wild-type and Atss3-1 plants subjected to the same light regime. For each time point, six whole plants were harvested and starch was quantified from each individual plant. This analysis revealed that the mutant accumulates 1.5 to 2 times more starch than wild type, and statistical analysis showed the difference to be highly significant (Fig. 4). The entire analysis of Figure 4 was repeated independently two times, several months apart, with essentially the same results, except that in one repetition the starch content increase was still evident at the end of the dark phase (data not shown).
Figure 3.
Qualitative analysis of leaf starch content. Wild-type, Atss3-1, and Atss3-2 Arabidopsis plants grown under long-day (16-h light/8-h dark) conditions (A–C) or short-day (12-h light/12-h dark) conditions (D and E) were decolorized and stained with iodine solution, then washed with water and photographed. A, Atss3-1 and wild-type leaves harvested at the end of the light phase. B, Atss3-1 and wild-type leaves harvested at the end of the dark phase. C, Atss3-2 and wild-type leaves harvested at the end of the light phase. D, Atss3-1 and wild-type leaves harvested at the end of the light phase. E, Atss3-1 and wild-type leaves harvested at the end of the dark phase.
Figure 4.
Starch accumulation in Arabidopsis leaves over the course of a diurnal cycle. Starch was harvested at 4-h intervals from wild-type plants (solid line) and Atss3-1 plants (dashed line) grown under a long-day photoperiod of 16-h light (white bar)/8-h dark (black bar). Starch quantification is in units of mg/g fresh weight of tissue. Each data point is the average of six independent plants and the se is indicated.
To discover whether the starch excess phenotype is specific to the imposed growth conditions of a long-day photoperiod, wild-type and mutant plants also were grown under a 12-h light/12-h dark regimen. For both Atss3-1 and Atss3-2, iodine staining of the leaf starch indicated an excess at the end of 12-h light period, but relatively little difference from wild type at the end of the dark period (Fig. 3; data not shown). Quantification of starch at the end of the 12-h light phase showed a small increase in starch content in the mutant, and equivalent amounts of remnant starch after 12 h of darkness (Table I).
Table I.
Leaf starch content
Values shown for starch content are in units of mg/g fresh weight ± se. Plants were grown under either long-day (LD; 16-h light/8-h dark) or short-day (SD; 12-h light/12-h dark) conditions. Plants were harvested either at the end of the light phase (EOL) or the end of the dark phase (EOD). For starch depletion, plants were grown in the LD regime, then grown for an extended period in the dark of 18 h, then returned to the light for 16 h.
| Growth Condition | Genotype | Collection Time | No. of Samples | Starch Content |
|---|---|---|---|---|
| Starch depleted | Wild type | EOL | 2 | 7.72 ± 0.69 |
| LD | Atss3-1 | EOL | 4 | 13.42 ± 0.95 |
| Wild type | EOD | 4 | 3.29 ± 0.32 | |
| Atss3-1 | EOD | 6 | 3.86 ± 0.30 | |
| Normal growth | Wild type | EOL | 5 | 8.39 ± 0.44 |
| SD | Atss3-1 | EOL | 5 | 9.43 ± 0.37 |
| Wild type | EOD | 7 | 2.37 ± 0.13 | |
| Atss3-1 | EOD | 7 | 2.43 ± 0.08 |
To test whether the increased starch accumulation occurs within a single photoperiod, the dark phase was extended to 18 h in order to completely deplete starch from the leaves. The starch-depleted plants then were allowed to grow in the light for 16 h, after which leaf starch was extracted and quantified. These data showed that, at the end of one light cycle, the starch content of the Atss3-1 leaves exceeded that of wild type by approximately 1.5-fold (Table I). There was little difference in the starch amounts in leaves after a successive dark cycle of 8 h (Table I). Together, the data suggest that the Atss3 starch excess phenotype can be attributed to an increased rate of starch biosynthesis and that the mutations have little or no effect on starch degradation. The effect was consistent in five repetitions of the analysis, either by iodine stain or by starch quantification, and for two independent alleles of AtSS3. Thus, the starch excess phenotype can be attributed specifically to the loss of SSIII.
Atss3 Mutations Alter the Structure of Leaf Ap
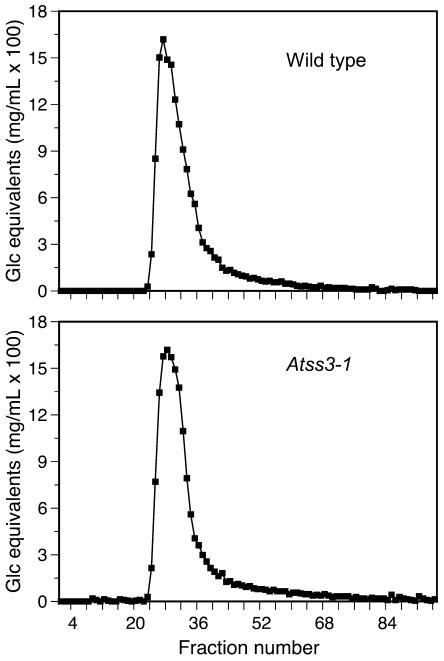
Compositional and structural changes in leaf starch that result from Atss3 mutations were investigated. To assess the relative amounts of component glucans, starches isolated from wild-type and Atss3-1 mutant leaves were separated according to molecular mass by gel permeation chromatography (GPC) on a CL-2B column. The profiles were very similar, indicating that Atss3-1 does not condition a significant change in the Ap:Am ratio (Fig. 5).
Figure 5.
Size fractionation of glucan polymers from starch granules. Starch from wild-type and Atss3-1 plants was separated by GPC on a Sepharose CL-2B column, and the Glc equivalents in each column fraction were determined.
The frequency distribution of the linear glucan chain of each degree of polymerization (DP) from 3 to approximately 50 was determined in starch from Atss3-1, Atss3-2, and wild-type leaves by fluorophore-assisted capillary electrophoresis (FACE). For this analysis, the population of linear chains produced after complete hydrolysis of branch linkages by isoamylase treatment was labeled at the reducing ends with a fluorophore and separated based on DP. Subtraction of the normalized chain length distribution of wild-type starch from similar distributions of Atss3-1 and Atss3-2 mutant starches shows that the frequency of chains of DP 5 to 10 are increased in the mutants, chains of DP 14 to 20 are decreased, chains of DP 26 to 37 are increased, and chains of DP 43 to 46 are decreased (Fig. 6). In all cases, the degree of difference between the wild-type and the mutant starch profile is small, typically 0.5% or less. However, the difference pattern is highly repeatable in independent biological replicates, and the patterns for Atss3-1 and Atss3-2 starches are very similar. Thus, there is a small, but reproducible, change in the frequency in Ap of specific linear chain lengths less than DP 50 when SSIII is not present.
Figure 6.
FACE analysis of leaf starch chain length distributions. Starch from leaves of wild-type, Atss3-1, and Atss3-2 plants was debranched with commercial isoamylase. The reducing ends of the linear chains were fluorescently labeled and separated by FACE. The frequency of individual chain lengths in each starch sample was normalized to total peak area. In the two bottom images, the normalized value for each chain length from the wild-type starch was subtracted from that of either the Atss3-1 or the Atss3-2 mutant. Values are the average of three independent determinations and se is indicated.
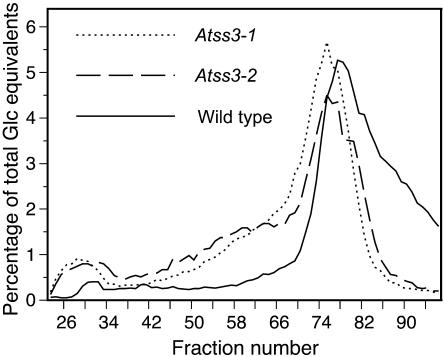
GPC also was used as a method to compare the frequency distribution of the longer linear chains in Ap, again after completely debranching the starch with isoamylase. The resolution of the method for separating linear glucan chains based on DP is far lower than the FACE method; however, GPC has the advantage of revealing the presence of longer chains that cannot be resolved by capillary electrophoresis (Fig. 7). Starch from both Atss3-1 and Atss3-2 mutants showed an obvious increase in the frequency of a population of longer length chains (fractions 48–63) compared to wild type. Analysis of the chain lengths present in selected GPC fractions indicates that the chains that are increased in abundance in the mutants are greater than DP 60 (data not shown).
Figure 7.
Size fractionation of linear glucan chains. Starch from leaves of wild-type (solid line), Atss3-1 (dotted line), and Atss3-2 (dashed line) plants was debranched to completion with commercial isoamylase and the resulting linear chains were separated by GPC on a Sepharose CL-2B column. The Glc equivalents in each column fraction were quantified and plotted as a percentage of total Glc equivalents.
Scanning electron microscopy was used to reveal whether there were significant changes in granule morphology in the absence of SSIII. The starch granules from wild-type, Atss3-1, and Atss3-2 leaves were all very similar in size and morphology (Fig. 8; data not shown). Thus, the Ap structural alterations caused by changes in chain length abundance in the plants lacking SSIII must not be of a nature that would cause gross changes to the structure of starch granules.
Figure 8.
Starch granule morphology. Starch granules from leaves of wild-type and Atss3-1 plants were coated with gold particles, visualized by scanning electron microscopy, and photographed.
Phosphate Content of the Starch Is Increased in Atss3 Mutants
Storage starches are known to contain monoesterified phosphate (Pi) groups at the C3 or the C6 position of some of the glucosyl units in Ap (Blennow et al., 2002). The extent of phosphorylation varies from a relatively high level in potato tuber starch to a minimal amount in the cereal starches (Blennow et al., 2000). To examine and compare the Pi content of starches in wild-type and Atss3 leaves of Arabidopsis, a modified Malachite Green procedure was employed that measures the staining intensity corresponding with the enzymatic release of Pi (Ekman and Jager, 1993). This method revealed increased phosphate in the leaf starches of both Atss3 mutants to a level approximately 2 to 3 times that of wild-type leaf starch (Table II). The same analysis was applied to starch from kernels of a maize mutant lacking SSIII activity, du1-M5, and again a large increase in the amount of phosphorylation was observed. This indicates that loss of SSIII activity directly or indirectly results in increased phosphorylation of transient starch as well as storage starch. This result is supportive of the previous finding that antisense suppression of SSIII in potato tubers (Solanum tuberosum) results in higher phosphate content of the tuber starch (Lloyd et al., 1999), although in the case of the Arabidopsis and maize mutants the relative increase is greater than was observed in potato.
Table II.
Phosphate content of leaf starch
The phosphate content of starch from Arabidopsis leaves or maize endosperm tissue was measured in units of μmol/mg starch. Starch isolated from Arabidopsis leaves at the end of the light phase or from imbibed mature maize kernels, was dried, then suspended in buffer. Phosphate content measurements were conducted on equivalent amounts of starch in duplicate or triplicate, as indicated.
| Organism | Genotype | Replications | Phosphate Content |
|---|---|---|---|
| Arabidopsis | Wild type | 3 | 4.333 ± 0.024 |
| Atss3-1 | 2 | 10.732 ± 0.031 | |
| Atss3-2 | 2 | 8.989 ± 0.001 | |
| Maize | Oh43 | 2 | 0.822 ± 0.019 |
| dul-M5 | 2 | 2.828 ± 0.144 |
Additional SS Activity in Leaves of Atss3 Mutants
The starch excess phenotype conditioned by Atss3 mutants suggests that SSIII activity loss is at least partially compensated by other SS isoforms, and that other types of starch biosynthetic enzyme activities also may be increased. To investigate specific effects of Atss3 mutations on starch-metabolizing enzymes, leaf protein extracts were subjected to starch zymogram analysis. Proteins were first separated by native PAGE, then transferred to another native gel containing starch and stained with iodine (Colleoni et al., 2003). This analysis, which enables visualization of the BE activities and the starch hydrolytic activities of the DBEs, α-amylases, and β-amylases, showed no differences between wild-type and mutant plants with respect to the migration, coloration, or band intensities of these activities (data not shown). Thus, loss of SSIII does not produce any alteration in these activities that is clearly observable by zymogram analysis.
Because individual SS activities are more readily distinguished from one another after an initial separation by anion-exchange chromatography, partially purified proteins in HiTrapQ column fractions were analyzed in SS zymograms in which brown-colored, iodine-stained bands (Fig. 2A, bands A–E) are believed to represent SS activities. In control zymograms in which ADPG was omitted, band B was clearly visible, but bands A, C, D, and E were not observed (data not shown). This indicates that band B does not represent an SS activity, but bands A, C, D, and E do because the activities require the ADPG substrate. The zymogram analysis revealed not only the loss of SSIII activity in the mutants, but also an apparent increase in another SS activity, indicated by stronger iodine-staining intensity in the mutant (Fig. 2A, band E). Band E is only faintly discernible in wild-type extracts, and in the mutant extract this band appears to cofractionate with SS activity of band C. Based on the knowledge that a relatively fast migrating SS activity, such as that of band C, is missing in an Atss1 mutant (C. D'Hulst, personal communication), it is reasonable to predict that band C represents the SSI isoform. Thus, it is possible that the increased SS activity in band E in Atss3-1 leaves could be an alternative form of SSI. Another possibility is that band E could derive from increased activity of a GBSS, which is reported to further elongate long Ap chains in the context of a crystalline matrix (Dauvillee et al., 1999).
To quantify the effect of the loss of SSIII on total SS activity in the leaves, a biochemical assay was employed that measures incorporation of label from ADPG into elongated linear glucosyl chains in the presence of equivalent amounts of wild-type and Atss3 mutant leaf protein. Total SS activity was increased in both Atss3-1 and Atss3-2 mutants, from 1.2 to 1.5 times that of wild type (Table III). These data support the hypothesis that loss of SSIII is compensated by increases in the activity of one or more of the other SS isoforms.
Table III.
Total SS activity in leaves
SS activity was measured in units of μmol min−1 mg−1 protein ± se for equivalent amounts of protein from wild-type, Atss3-1, and Atss3-2 leaves. These values were determined by quantifying the incorporation of radiolabeled Glc from 14C-ADPG into acid-precipitable polysaccharide. Values were obtained from the indicated numbers of samples.
| Genotype | No. of Samples | Activity |
|---|---|---|
| Wild type | 5 | 129 ± 3.3 |
| Atss3-1 | 3 | 158 ± 1.6 |
| Atss3-2 | 3 | 188 ± 2.1 |
DISCUSSION
Characterization of Atss3 Alleles
This study demonstrates a unique collection of starch biosynthesis alterations that can be attributed to loss of the SS isoform, SSIII. The fact that mutation of AtSS3 is the causative agent of the observed phenotypes, as opposed to a secondary mutation in the T-DNA lines, is indicated by analysis of two independently derived mutant alleles. All of the observed phenotypic effects were very similar for both Atss3-1 and Atss3-2. The likelihood of the same unidentified mutation being present in independent lines, or of different unidentified mutations causing the same unique phenotype, is extremely small. We conclude, therefore, that all the observed effects on Arabidopsis starch biosynthesis arise as a result of mutations at the locus that codes for SSIII.
Both Atss3-1 and Atss3-2 clearly are null alleles in terms of SSIII expression at the level of enzyme activity (Fig. 2, A and C). The data also show definitively that SSIII protein expression is altered (Figs. 1E and 2B); however, the possibility that a remnant amino-terminal protein fragment of SSIII accumulates in the mutants cannot be ruled out. A monoclonal antibody was used to detect SSIII, and the epitope recognized by this antibody is not known because full-length recombinant SSIII protein was used as the antigen. Thus, if the epitope is located at the carboxyl terminus of SSIII, accumulation of an amino-terminal fragment lacking enzyme activity would not be detected. Nevertheless, it is clear that the C-terminal portion of SSIII is lacking in both mutants, as shown by analyses of transcript, protein, and enzyme activity.
A Starch Excess Phenotype Caused by Elimination of a Starch Biosynthetic Enzyme
An unexpected result of this study is that, when SSIII activity is eliminated from Arabidopsis leaves, there is an increase in the rate of starch accumulation during the light phase of a diurnal cycle. This result seemingly is at odds with the known function of SSs as positive factors for starch production. In this instance, however, one role of the biosynthetic enzyme SSIII apparently is to serve as a negative regulator of starch biosynthesis. The higher level of starch in Atss3 mutants cannot be attributed to a defect in starch degradation because excess accumulation was observed in a single light period after starch had been completely eliminated during extended growth in the dark. Previous studies of starch turnover show that little or no degradation occurs during the light in Arabidopsis leaves (Zeeman et al., 2002). Taken together, these data indicate that there is an increased rate of starch synthesis in the Atss3 mutants.
One possible explanation for increased starch synthesis could be the removal of an inhibitory effect of SSIII on the activities of one or more of the other starch biosynthetic enzymes, particularly the remaining SS isoforms. The data show that, in the Atss3 mutants, total SS activity is increased despite the loss of SSIII. This result is analogous to a previous observation in maize that mutation at the du1 locus, which codes for SSIII, causes increased total SS activity in developing kernels (Singletary et al., 1997). Furthermore, two-dimensional zymogram analysis of wild-type and mutant Arabidopsis leaf proteins showed increased activity of an SS (Fig. 2A, band E) that cofractionates with a strong, faster migrating SS (Fig. 2A, band C). This new activity could result from a modification of either SSI or SSII, perhaps owing to a change in the structure of the polypeptide or altered association with other factors.
An alternative explanation for the increase in the starch biosynthetic rate could be that changes in Ap structure caused by the loss of SSIII produce a branched glucan that provides a more efficient substrate for the activities of the remaining starch biosynthetic enzymes. Although the chain length differences in the mutant starches are not dramatically different from wild type, they could be such that they allow for increased SSI and/or SSII activities. The specific nature of some of the structural changes in Atss3 starch provides support for this hypothesis. Previous in vitro analysis of SSI activity suggests it functions primarily in the production of the shortest chains (Commuri and Keeling, 2001; C. D'Hulst, unpublished data), and the Atss3 mutant starch does have a slight excess of short chains of DP 10 or less that could result from increased SSI activity.
Arabidopsis SSIII has been implicated previously, although indirectly, in a starch excess phenotype (Sehnke et al., 2001). In that report, SSIII was shown to bind a trans-acting regulatory factor, one of the 14-3-3 proteins, and antisense inhibition of the 14-3-3 proteins was shown to cause excess starch accumulation in leaves. The starch excess phenotype was proposed to result from decreased inhibition of SSIII activity in response to reduced 14-3-3 binding in the mutant line. This study shows that loss of SSIII itself results in a starch excess phenotype, which is not entirely consistent with this model. However, if binding of the 14-3-3 factor to SSIII is required for SS activity, this could account for the observed phenotype. The question remains open as to whether the apparent negative regulatory effect of SSIII on starch biosynthetic rate is mediated in part by binding to a 14-3-3 protein.
Structural Changes in Ap Resulting from Loss of SSIII
Mutations in the Arabidopsis gene coding for SSIII affect both the physical and chemical structure of leaf starch, i.e. chain length distribution and phosphate content, respectively. Most of the Atss3 chain length differences are relatively small in terms of the percentage change from wild type and they do not result in gross morphological changes in starch granules. The effects of Atss3-1 and Atss3-2 on starch physical structure include more Ap short chains of DP 10 or less, which most likely represent unbranched external A chains, and fewer Ap chains of approximately DP 11 to 22, which are thought to comprise the crystalline lamellae of the cluster unit (Umemoto et al., 2002). The explanations for these complex changes cannot yet be known, although possibilities include both altered substrate availability and enzyme regulation. As mentioned previously in this discussion, enhanced SSI activity might account for the increase in short chains in the range of DP 6 to 10. However, the decrease of chains in the range of DP 11 to 20 in the Atss3 mutants could be the result of less activity of SSII. Studies in maize, pea (Pisum sativum), potato, rice (Oryza sativa), and Chlamydomonas indicate that SSII functions to elongate very short chains of DP 6 to 10, producing intermediate chains of DP approximately 11 to 24 for the crystalline lamellae (Fontaine et al., 1993; Craig et al., 1998; Edwards et al., 1999; Umemoto et al., 2002; Zhang et al., 2004). Because Atss3 leaf starch has fewer chains of these intermediate lengths, a potential down-regulation of SSII is suggested. Thus, the starch structural changes produced by SSIII loss are likely to be due to the combined effects of both positive and negative alterations in other starch biosynthetic enzyme activities. Furthermore, SS activity may not be the only factor contributing to the chain length distribution because BE activity also alters this physical characteristic of the polymer.
Perhaps the most significant structural difference observed in the Atss3 mutants is with respect to very long chains, estimated to be greater than DP 60, which are more prevalent in the mutant starch. This result is consistent with previous analysis of endosperm starch in maize plants homozygous for mutations in the du1 gene, which showed that this SSIII mutant starch contained the longest chains of any starch form that has been characterized (Wang et al., 1993). Longer chains appearing when SSIII activity is eliminated might result from a compensating soluble SS, although the known properties of SSI and SSII are not consistent with the idea that these isoforms produce chains of more than DP 25. It is possible that SSIV may be involved in long-chain production or that BE activity is reduced in the SSIII mutants, so that many long chains remain without being cleaved into shorter lengths during branch formation. No change in BE activity was observed in vitro in the Atss3 mutants; however, zymogram analysis is not quantitative and may not have the sensitivity to detect small activity changes. Another explanation for the very long chains in Atss mutant starch could be an increased or redirected activity of GBSS, which has been shown to function in the elongation of long Ap chains in Chlamydomonas in addition to its major role in the synthesis of Am (Delrue et al., 1992; Dauvillee et al., 1999). The Am:Ap ratio is unchanged by the Atss3 mutations, but it is possible that the structural changes in Ap molecules in the mutant could provide a better substrate for GBSS activity.
The production of a starch with more long chains is likely to account for the observed increase in starch phosphate content in the Atss3 mutants. This is in line with studies in transgenic plants that indicate starch phosphorylation depends on the structure of the starch. The specific suppression of SSIII in transgenic potato causes a 70% increase in the phosphorylation frequency, most likely due to increases in the lengths of the longest Ap chains (Abel et al., 1996; Lloyd et al., 1999). Antisense suppression of BE activity, which causes formation of longer length Ap chains, also resulted in significantly increased levels of starch-bound phosphate (Schwall et al., 2000). In this study, the phosphate content of the Atss3-1 and Atss3-2 leaf starches in Arabidopsis and du1 mutant endosperm starch was 2 to 3 times that of wild-type starch, an even greater effect than has been observed in the transgenic potato studies. The biochemical reason that excess frequency of very long chains correlates with higher phosphate level remains to be discovered.
Continued use of the Arabidopsis reverse genetic resources, such as in this study, is likely to provide in the near future significant new insights into questions regarding the complex direct and indirect functions of each starch biosynthetic isoform. Single-mutant characterizations have been completed for SSIII (this study), SSII (X. Zhang, unpublished data), and SSI (C. D'Hulst, unpublished data), and analysis of double-mutant combinations is under way.
MATERIALS AND METHODS
Plant Materials
Wild-type Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) and Atss3 mutant lines in the same genetic background were sown in Sunshine Soil mix. Sown seeds were incubated at 4°C for 2 to 3 d, then grown at 21°C, 60% relative humidity, in a 12-h light/12-h dark photoperiod, or a 16-h light/8-h dark photoperiod. Two Atss3 mutant lines were identified by PCR screening of Arabidopsis plants from the Salk collection that contain random T-DNA insertions. PCR primers were derived from the T-DNA left border (LBa1, TGG TTCACGTAGTGGGCCATCG) and from the SSIII gene sequence (SS3-TU1, AGGTTGTCTTATTAGGTTCAGC; SS3-TL1, GTGCAGAGTGATAGAGCTCAAG; SS3-RP2, GCCCCAGCATAAATCTGGAGAG; SS3-LP2, CCTCTTCTCTGAAGCCCTTCCC; Fig. 1A). Nucleotide sequence analysis of PCR products generated by LBa1, together with a gene-specific primer (SS3-TU1 or SS3-LP2), confirmed the locations of the T-DNA insertion sites for both alleles, termed Atss3-1 and Atss3-2. Control amplifications were conducted for each mutant allele using primers from the Arabidopsis gene coding for SSII (AT3G01180; SS2-RP1, GCTACCAATATCACATTCATGAC; SS2-LP1, CTTACCATGATTTGCCTTCTG; LP2, CCTCTTCTCTGAAGCCCTTCCC; RP2, AGTGGTGGAAAATTAGGGGCG).
Expression of Recombinant SSIII in Escherichia coli and Monoclonal Antibody Production
Nearly full-length SSIII cDNA was amplified from total RNA isolated from Arabidopsis leaves by RT-PCR using the following gene-specific primers: SS3-U2, GCTTCAGGACCAAAAAGCTC and SS3-L1, AAGAAGAAAGGATCAAACTCTC (Fig. 1A). The coding region of the SSIII cDNA minus the 60-nucleotide sequence coding for a predicted transit peptide was cloned into a pDONR plasmid vector using Gateway cloning technology (Invitrogen, Carlsbad, CA) to generate plasmid pESS3 (entry clone). The SSIII cDNA sequence was transferred to the pDEST15 expression vector by means of the PCR-based Gateway homologous recombination system, during which the Arabidopsis SSIII was fused to a sequence coding for a glutathione S-transferase (GST) tag. The resulting plasmid pDSS3 was introduced into E. coli cells by electroporation and expressed as a fusion protein with the N-terminal GST tag according to standard Gateway system protocols. Recombinant SSIII protein was affinity purified using a GST-agarose column, from which it was eluted by competition with free glutathione. The purified protein was analyzed by SDS-PAGE. Purified recombinant SSIII protein was sliced from the gel for use as antigen for the production of monoclonal antibodies. Inoculation of mice and screening of sera were conducted at the Iowa State University hybridoma facility. The hybridoma culture fluid designated as α-AtSSIII was used undiluted for immunoblot analysis.
Leaf Protein Extraction
Proteins were extracted from Arabidopsis leaves by grinding 200 to 400 mg of leaf tissue (fresh weight) under liquid nitrogen with a mortar and pestle. The tissue was homogenized in 2 volumes of extraction buffer (0.1 m HEPES-KOH, pH 7.0, 20 mm 2-mercaptoethanol, 0.1 mg/mL PMSF, 0.1% [v/v] Triton X-100, 1 mm EDTA, and 20% [v/v] glycerol). The homogenate was centrifuged at 13,000 rpm for 20 min at 4°C, and the supernatant was frozen immediately with liquid nitrogen, then stored at −80°C. Protein content of the supernatant was determined by standard methods (Bradford, 1976).
Qualitative Comparison of Leaf Starch Content and Starch Quantification
Leaves of individual plants (100–400 mg fresh weight) were boiled in 50 mL 80% (v/v) ethanol. The decolored leaves were then ground with a mortar and pestle in 80% ethanol, centrifuged for 10 min at 13,000 rpm, and the pellet was washed twice with 80% ethanol. After centrifugation, the insoluble material was suspended in 1 mL of distilled water and boiled for 30 min. Total starch in each leaf sample was quantified using a commercial Glc assay kit (catalog no. E0207748; R-Biopharm, Darmstadt, Germany), according to instructions in the kit protocol. The assay employs amyloglucosidase to achieve complete digestion of the starch sample to Glc, and measurement of the Glc by application of a Glc oxidase reagent.
For qualitative comparison of leaf starch content, whole plants were harvested at specific times in the diurnal cycle and decolorized as described in the previous paragraph. Plants were stained with fresh iodine solution (I2/KI [5 g KI, 0.5 g I2 in 500 mL water]) for 5 min, destained in water for 1–to 2 h, and photographed immediately. In all instances, pairs of wild-type and mutant plants were stained and destained identically.
Starch Granule Analysis
Arabidopsis leaves were harvested in the middle or end of the light phase of the 16-h light/8-h dark photoperiod. Leaves were washed in water, then homogenized in 5 volumes of isolation buffer (100 mm 3-[N-morpholino] propanesulfonic acid [MOPS], pH 7.2, 5 mm EDTA) using a mortar and pestle. The homogenate was filtered through two layers of Miracloth and centrifuged at 3,000g for 10 min at 4°C. The starch granules in the pellet were purified by means of two centrifugations through 90% Percoll/10% isolation buffer, then washed repeatedly with water and dried at room temperature. Scanning electron photomicrographs of gold-coated starch granules were produced by the Iowa State University microscopy facility.
Partial Purification of Leaf Proteins and SS Activity Analysis
For partial purification of leaf proteins, 10 g of leaf tissue harvested at the middle of the light phase of the photoperiod were ground to a fine powder in liquid nitrogen with a mortar and pestle. The powdered tissue was suspended in 8 mL of buffer (50 mm Tris-acetate, pH 7.5, 10 mm DTT, and 1× protease inhibitor cocktail [catalog no. P-8465; Sigma, St. Louis]), then centrifuged at 13,000 rpm for 20 min at 4°C. The supernatant was passed through a 0.22-μm filter and proteins were separated by anion-exchange chromatography on a HiTrapQ column (catalog no. 17-1164-01; Amersham-Pharmacia Biotech, Uppsala) using AKTA FPLC instrumentation (Amersham-Pharmacia). After the bound proteins were eluted with a linear NaCl gradient (0%–45%), they were separated and equivalent fraction volumes were analyzed in native activity gels (i.e. SS zymograms), as described previously (Zhang et al., 2004). Proteins in duplicate gels were transferred to a nitrocellulose membrane for hybridization with α-AtSSIII monoclonal antibody.
Total soluble SS activity in leaves was assayed according to the methanol-KCl precipitation method described by Cao et al. (1999). Fresh leaves (200 mg) were ground into fine powder in liquid nitrogen and suspended in 400 μL of extraction buffer. Protein concentration was determined by the Bradford method, and SS activity assay was then conducted with different amounts of protein. Control experiments demonstrated that the amount of 14C incorporated into methanol-precipitable product is linear with respect to the amount of protein in the assay (data not shown).
Characterization of Starch Structure and Properties
Relative Ap and Am content was determined as follows. Solubilized starches from mutant and wild-type leaves were size fractionated on a Sepharose CL-2B column (Amersham-Pharmacia), according to a previously described method (Dinges et al., 2001). The Am percentage is defined as (Glc equivalents in Am-containing fractions/total Glc equivalents) × 100. The Glc equivalents in each fraction were measured following complete hydrolysis of the sample with amyloglucosidase (catalog no. 80610; Megazyme, Bray, Ireland), using a colorimetric Glc oxidase/peroxidase method (catalog no. 510-A; Sigma). In a separate analysis, starch was debranched to completion with Pseudomonas isoamylase prior to separation on the CL-2B column, as described previously (Dinges et al., 2001) and the Glc equivalents in each column fraction were measured.
The distribution of the linear chain lengths in starch samples was determined by FACE of the debranched starch, as described previously (O'Shea et al., 1998; Dinges et al., 2003). Measurements were performed on three independent starch samples for both wild-type and each Atss3 mutant.
Determination of Starch Phosphate Content
Total phosphate in the starch was determined using the Malachite Green method (Ekman and Jager, 1993). Three milligrams of dry starch from wild-type, Atss3-1, and Atss3-2 mutant leaves of Arabidopsis or 10 mg of dry starch from maize wild-type inbred Oh43 or du1-M5 mutant endosperm (Gao et al., 1998) were suspended in 0.3 mL concentrated H2SO4 and charred over a gas burner. The solution was clarified by dropwise addition of H2O2 (30%, w/v). The volume was increased by the addition of 1.032 mL of water and the sequential addition of 1.068 mL 10.19 n NaOH, and the samples were capped and boiled for 10 min. A 100-μL aliquot of the digested sample was transferred to a cuvette, together with 100 μL of Clark and Lubs buffer (0.054 m KCl, 0.146 n HCl), followed by 40 μL 6 n HCl. Sixty microliters of dH2O were added to balance the 6 n HCl to 100 μL. Phosphate reagent [1 vol 10% (NH4)6Mo7O24, 3 vol 0.2% Malachite Green] was added (100 μL), and the mixture was incubated at room temperature for 20 min. The phosphate content was determined by measurement of the A660 and comparisons with standards with known KH2PO4 contents.
Acknowledgments
The authors thank the Arabidopsis Biological Resource Center and the Salk Institute Genomic Analysis Laboratory for providing T-DNA insertion mutants. We also thank the staff of the Iowa State University microscopy facility, Ms. Tracie Bierwagen for help with FACE analysis and stock maintenance, and Dr. Akiko Kubo for helpful discussions. We also are grateful to Dr. Christophe D'Hulst for sharing unpublished data.
This work was supported by an award from the National Science Foundation (grant no. DBI 0209789 to A.M.M. and M.G.J.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060319.
References
- Abel GJW, Springer F, Willmitzer L, Kossmann J (1996) Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L.). Plant J 10: 981–991 [DOI] [PubMed] [Google Scholar]
- Blennow A, Engelsen SB, Munck L, Moller BL (2000) Starch molecular structure and phosphorylation investigated by a combined chromatographic and chemometric approach. Carbohydr Polym 41: 163–174 [Google Scholar]
- Blennow A, Nielsen TH, Baunsgaard L, Mikkelsen R, Engelsen SB (2002) Starch phosphorylation: a new front line in starch research. Trends Plant Sci 7: 445–450 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cameron JW (1947) Chemico-genetic bases for the reserve carbohydrates in maize endosperm. Genetics 32: 459–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Imparl-Radosevich J, Guan HP, Keeling PL, James MG, Myers AM (1999) Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol 119: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni C, Myers AM, James MG (2003) One- and two-dimensional native PAGE activity gel analyses of maize endosperm proteins reveal functional interactions between specific starch metabolizing enzymes. J Appl Glycosci (1999) 50: 207–212 [Google Scholar]
- Commuri PD, Keeling PL (2001) Chain-length specificities of maize starch synthase I enzyme: studies of glucan affinity and catalytic properties. Plant J 25: 475–486 [DOI] [PubMed] [Google Scholar]
- Craig J, Lloyd JR, Tomlinson K, Barber L, Edwards A, Wang TL, Martin C, Hedley CL, Smith AM (1998) Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. Plant Cell 10: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creech RG (1965) Genetic control of carbohydrate synthesis in maize endosperm. Genetics 52: 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillee D, Colleoni C, Shaw E, Mouille G, D'Hulst C, Morell M, Samuel MS, Bouchet B, Gallant DJ, Sinskey A, et al (1999) Novel, starch-like polysaccharides are synthesized by an unbound form of granule-bound starch synthase in glycogen-accumulating mutants of Chlamydomonas reinhardtii. Plant Physiol 119: 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski JM, Van Den Koornhuyse N, Maddelein ML, Fournet B, Ball S (1992) Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J Bacteriol 174: 3612–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, James MG, Myers AM (2003) Mutational analysis of the pullulanase-type debranching enzyme in maize indicates multiple functions in starch metabolism. Plant Cell 15: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, Myers AM, James MG (2001) Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol 125: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A, Fulton DC, Hylton CM, Jobling SA, Gidley M, Rossner U, Martin C, Smith AM (1999) A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J 17: 251–261 [Google Scholar]
- Ekman P, Jager O (1993) Quantification of subnanomolar amounts of phosphate bound seryl and theronyl residues in phosphoproteins using alkaline hydrolysis and malachite green. Anal Biochem 214: 138–141 [DOI] [PubMed] [Google Scholar]
- Fontaine T, Hulst CD, Maddelein M-L, Routier F, Pepin TM, Decq A, Wieruszeski J-M, Delrue B, Van den Koornhuyse N, Bossu J-P, et al (1993) Toward an understanding of the biogenesis of the starch granule. J Biol Chem 268: 16223–16230 [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM (1998) Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell 10: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James MG, Denyer K, Myers AM (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6: 215–222 [DOI] [PubMed] [Google Scholar]
- Klösgen RB, Gierl A, Schwarz-Sommer Z, Saedler H (1986) Molecular analysis of the waxy locus of Zea mays. Mol Gen Genet 203: 237–244 [Google Scholar]
- Li Z, Mouille G, Kosar-Hashemi B, Rahman S, Clarke B, Gale KR, Appels R, Morell MK (2000) The structure and expression of the wheat starch synthase III gene. Motifs in the expressed gene define the lineage of the starch synthase III gene family. Plant Physiol 123: 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Landschutze V, Kossmann J (1999) Simultaneous antisense inhibition of two starch-synthase isoforms in potato tubers leads to accumulation of grossly modified amylopectin. Biochem J 338: 515–521 [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf PC (1947) The inheritance of amylaceous sugary endosperm and its derivatives in maize. Genetics 32: 448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li Z (2003) Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J 34: 173–185 [DOI] [PubMed] [Google Scholar]
- O'Shea MG, Samuel MS, Konik CM, Morell MK (1998) Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labeling and high-resolution separation. Carbohydr Res 307: 1–12 [Google Scholar]
- Preiss J, Boyer CD (1979) Evidence for independent genetic control of the multiple forms of maize endosperm branching enzymes and starch synthases. In JJ Marshall, ed, Mechanisms of Polysaccharide Polymerizaton and Depolymerization. Academic Press, NY, pp 161–174 [DOI] [PMC free article] [PubMed]
- Schwall GP, Safford R, Westcott RJ, Jeffcoat R, Tayal A, Shi YC, Gidley MJ, Jobling SA (2000) Production of very-high-amylose potato starch by inhibition of SBE A and B. Nat Biotechnol 18: 551–554 [DOI] [PubMed] [Google Scholar]
- Sehnke PC, Chung HJ, Wu K, Ferl RJ (2001) Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. Proc Natl Acad Sci USA 98: 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Garwood DL (1984) Genetics and physiology of starch development. In EF Paschall, ed, Starch: Chemistry and Technology. Academic Press, Orlando, FL, pp 25–86
- Shure M, Wessler S, Federoff N (1983) Molecular identification and isolation of the Waxy locus in maize. Cell 35: 225–233 [DOI] [PubMed] [Google Scholar]
- Singletary GW, Banisadr R, Keeling PL (1997) Influence of gene dosage on carbohydrate synthesis and enzymatic activities in endosperm of starch-deficient mutants of maize. Plant Physiol 113: 293–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y (2002) Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor Appl Genet 104: 1–8 [DOI] [PubMed] [Google Scholar]
- Wang Y-J, White P, Pollak L, Jane J (1993) Amylopectin and intermediate materials in starches from mutant genotypes of the Oh43 inbred line. Cereal Chem 70: 521–525 [Google Scholar]
- Zeeman SC, Tiessen A, Pilling E, Kato KL, Donald AM, Smith AM (2002) Starch synthesis in Arabidopsis. Granule synthesis, composition, and structure. Plant Physiol 129: 516–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Colleoni C, Ratushna V, Sirghie-Colleoni M, James MG, Myers AM (2004) Molecular characterization of the Zea mays gene sugary2, a determinant of starch structure and functionality. Plant Mol Biol 54: 865–879 [DOI] [PubMed] [Google Scholar]