Abstract
Phosphoinositides (PIs) are signaling molecules that regulate cellular events including vesicle targeting and interactions between membrane and cytoskeleton. Phosphatidylinositol (PtdIns)(4,5)P2 is one of the best characterized PIs; studies in which PtdIns(4,5)P2 localization or concentration is altered lead to defects in the actin cytoskeleton and exocytosis. PtdIns(4,5)P2 and its derivative Ins(1,4,5)P3 accumulate in salt, cold, and osmotically stressed plants. PtdIns(4,5)P2 signaling is terminated through the action of inositol polyphosphate phosphatases and PI phosphatases including supressor of actin mutation (SAC) domain phosphatases. In some cases, these phosphatases also act on Ins(1,4,5)P3. We have characterized the Arabidopsis (Arabidopsis thaliana) sac9 mutants. The SAC9 protein is different from other SAC domain proteins in several ways including the presence of a WW protein interaction domain within the SAC domain. The rice (Oryza sativa) and Arabidopsis SAC9 protein sequences are similar, but no apparent homologs are found in nonplant genomes. High-performance liquid chromatography studies show that unstressed sac9 mutants accumulate elevated levels of PtdIns(4,5)P2 and Ins(1,4,5)P3 as compared to wild-type plants. The sac9 mutants have characteristics of a constitutive stress response, including dwarfism, closed stomata, and anthocyanin accumulation, and they overexpress stress-induced genes and overaccumulate reactive-oxygen species. These results suggest that the SAC9 phosphatase is involved in modulating phosphoinsitide signals during the stress response.
Phosphoinositides (PIs) are a family of eight molecules in which the hydroxyl groups on the inositol moiety can be phosphorylated in a variety of combinations (Stevenson et al., 2000; Meijer and Munnik, 2003; van Leeuwen et al., 2004). PIs undergo cycles of phosphorylation and dephosphorylation through organelle-specific PI kinases and phosphatases, leading to distinct subcellular distributions of PI species (De Matteis and Godi, 2004). PIs control the timing and location of many cellular events including vesicle targeting, interactions between the membrane and the cytoskeleton, membrane budding and fusing, nuclear and cytoplasmic signal transduction, and activity of membrane channels (Hilgemann and Ball, 1996; Martin, 1998; Czech, 2000; Odorizzi et al., 2000; Stevenson et al., 2000; Simonsen et al., 2001; Hardie, 2003; Meijer and Munnik, 2003; Oliver et al., 2004; van Leeuwen et al., 2004). Specific PI-binding sites have been found on a variety of effector proteins including protein kinases, actin-binding proteins, GTPases, and membrane trafficking proteins, and it is thought that binding to PIs can target effector proteins to specific membrane locations (Martin, 1998; Hu et al., 1999; Yao et al., 1999; Dowler et al., 2000; Tall et al., 2000; Ellson et al., 2002; Itoh and Takenawa, 2002).
Unraveling the specific functions of the PI species and the enzymes that modify them is challenging for several reasons. Enzyme specificities do not always correlate between in vitro and in vivo assays; enzymes can display different in vitro specificities when purified from different heterologous expression systems, and different in vitro assay systems can lead to conflicting results (Mueller-Roeber and Pical, 2002; Ercetin and Gillaspy, 2004). It has been suggested that the conflicting results from in vitro assays may be due to the difficulty in presenting lipid substrates in a physiologically relevant manner in vitro (Drøbak and Heras, 2002). Both in vivo and in vitro, PI phosphatases can act upon more than one substrate; they are generally considered to have broader substrate specificities than the corresponding kinases (Mitchell et al., 2002; Roth, 2004; Zhong et al., 2004). Finally, functions of the different PIs and PI-modifying enzymes are not strictly conserved between eukaryotic kingdoms, making it difficult to extrapolate data obtained from yeast (Saccharomyces cerevisiae) and mammalian systems into the plant systems (Mueller-Roeber and Pical, 2002).
Phosphatidylinositol (PtdIns)(4,5)P2 has been studied extensively (Martin, 1998; Czech, 2003). Studies in which PtdIns(4,5)P2 localization or concentration is altered lead to defects in the actin cytoskeleton; PtdIns(4,5)P2 is known to bind the actin-binding protein profilin (Tall et al., 2000; Desrivieres et al., 2002; Yin and Janmey, 2003). Altering PtdIns(4,5)P2 distribution also affects exocytosis, perhaps via its effects on actin organization (Roth, 2004). PtdIns(4,5)P2 distribution has been studied though the use of chimeric proteins that specifically bind to PtdIns(4,5)P2, such as a fusion of green fluorescent protein (GFP) to the plekstrin homology domain of phospholipase C (GFP-PH-PLCδ; Varnai et al., 2002). In plants, GFP-PH-PLCδ showed that PtdIns(4,5)P2 accumulates at the tip of a growing pollen tube, where it can coordinate actin filament activity and exocytosis during polarized cell growth (Kost et al., 1999; Yao et al., 1999). Anti-PtdIns(4,5)P2 antibodies also showed a correlation between PtdIns(4,5)P2 localization and tip growth of root hairs (Braun et al., 1999).
There also is accumulating evidence that PI-derived signals are involved in plant stress response (Meijer et al., 2001; Munnik and Meijer, 2001; Wang, 2002; Park et al., 2004). Salt, cold, and osmotically stressed plants accumulate PtdIns(4,5)P2, synthesized mostly from PtdIns(4)P by a PtdIns(4)P 5-kinase (Smolenska-Sym and Kacperska, 1994; Pical et al., 1999; DeWald et al., 2001). A gene encoding a PtdIns(4)P 5-kinase is induced by drought, salt, and abscisic acid (ABA), suggesting a mechanism for increased levels of PtdIns(4,5)P2 in stressed plants (Mikami et al., 1998; Elge et al., 2001; Westergren et al., 2001; Mueller-Roeber and Pical, 2002). PtdIns(4,5)P2 signaling is terminated through the action of two types of phosphatases. The type II inositol polyphosphate 5-phosphatases (5PTases) act on both PIs and soluble inositol phosphates, while the Sac domain phosphatases are thought to act only on PIs (Mitchell et al., 2002; Whisstock et al., 2002).
PtdIns(4,5)P2 can serve as a direct precursor to other signaling molecules, including Ins(1,4,5)P3, generated through the action of phospholipase C (PLC). Ins(1,4,5)P3 has been well established as a component in the ABA and osmotic signaling pathways. Microinjection of Ins(1,4,5)P3 into guard cells is sufficient to induce stomatal closure (Blatt et al., 1990; Gilroy et al., 1990). Ins(1,4,5)P3 signaling is specifically terminated by the action of inositol polyphosphate phosphatases (PTases), of which different types have different specificities for each position on the inositol ring. Lowering or raising in vivo levels of Ins(1,4,5)P3 by overexpression or mutation of inositol polyphosphate 5PTase or 1-phosphatase genes also demonstrates the importance of Ins(1,4,5)P3 in osmotic and ABA signaling (Sanchez and Chua, 2001; Xiong et al., 2001; Perera et al., 2002; Burnette et al., 2003; Carland and Nelson, 2004). Type I 5PTases are generally thought to be specific for Ins(1,4,5)P3, but type II can also hydrolyze membrane-bound PIs (Zhong and Ye, 2004).
While both PtdIns(4,5)P2 and Ins(1,4,5)P3 accumulate in response to osmotic stress, the quantity of PtdIns(4,5)P2 can be 20- to 50-fold higher than Ins(1,4,5)P3 (DeWald et al., 2001). This observation is consistent with a model in which PtdIns(4,5)P2 itself has a specific role in the osmotic stress response. In animal cells, PtdIns(4,5)P2 has been extensively characterized as an important signal for several cellular processes including membrane trafficking (Czech, 2000, 2003). In plant cells and yeast, osmotic stress leads to alterations of membrane structure and trafficking, and several different proteins involved in vesicle formation, fusion, or targeting have been identified as being ABA or stress responsive or functionally important in stress response (Ristic and Ashworth, 1993; Leyman et al., 1999, 2000; Geelen et al., 2002; Levine, 2002; Mazel et al., 2004; Pratelli et al., 2004). A relationship between PIs and membrane responses to osmotic stress has been shown in yeast but not yet in plants (Homma et al., 1998; Delley and Hall, 1999; Audhya and Emr, 2004).
Elucidation of a specific role for PtdIns(4,5)P2 in plant signaling will be facilitated through in vivo studies. Plants in which genes encoding PI phosphatases and PTases are mutated or overexpressed help to shed light on their functions. The FRA3 gene encodes a Type II inositol polyphosphate phosphatase. In vitro and in vivo studies demonstrate that this enzyme recognizes both PIs and inositol phosphates as substrates; mutants lacking FRA3 accumulate both PtdIns(4,5)P2 and Ins(1,4,5)P3. The gene is predominantly expressed in developing fibers and vascular cells, and the mutant plants show a fragile fiber phenotype consistent with the abnormal actin organization and cell wall defects in these cells (Zhong et al., 2004).
Arabidopsis (Arabidopsis thaliana) encodes nine supressor of actin mutation (SAC) domain PI phosphatase-like proteins, falling into three different classes based on sequence (Zhong and Ye, 2003). Two of the classes show substantial sequence homology within them (SAC 1–5 and SAC 6–8), while SAC9 is unique. Mutations in the AtSAC1 gene cause abnormalities in cell wall synthesis and cellular morphology, suggesting a role for the AtSAC1 gene in vesicle trafficking or the actin cytoskeleton similar to that seen in yeast (Guo et al., 1999; Zhong and Ye, 2003). Preliminary results of the SAC 6-to-8 family suggests that loss of function of any one does not display a distinct mutant phenotype, consistent with functional redundancy (Despres et al., 2003).
We have identified and characterized mutants in the Arabidopsis SAC9 gene. The sac9 mutants constitutively express a systemic stressed phenotype, which may result from altered cellular signaling. Specific phenotypes displayed by the sac9 mutant plants include overexpression of stress-induced genes, elevated reactive oxygen species (ROS) accumulation, constitutively closed guard cells, and a corresponding dwarf, slow-growing phenotype. In the sac9 mutants, we measured elevated levels of both PtdIns(4,5)P2 and Ins(1,4,5)P3.
RESULTS
Identification of the SAC9 Gene
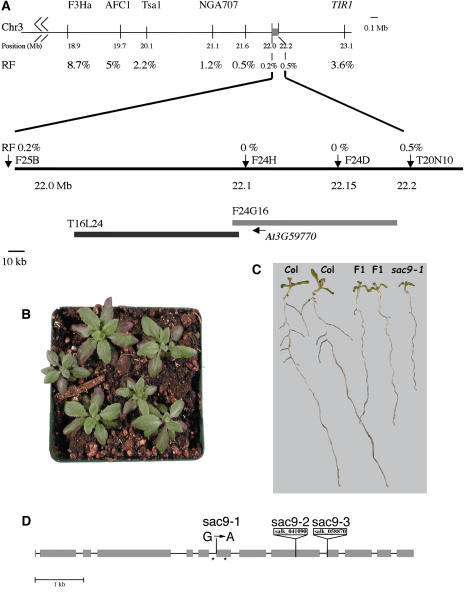
We identified an ethyl methanesulfonate (EMS)-generated recessive mutation, sac9-1, that causes a reduced growth rate, and hyponastic, deeply purple leaves. We used map-based cloning to identify the affected gene. We used a combination of cleaved amplified polymorphic sequences (CAPS) and simple sequence length polymorphisms (SSLPs) to narrow the interval containing the gene to a 200-kb region that is located on two bacterial artificial chromosomes (BACs; T16L24 and F24G16; Fig. 1A). Within this region, there are 52 putative genes. We obtained T-DNA lines containing inserts within the open reading frames of 14 of these genes and visually screened through these lines. We identified a plant homozygous for a T-DNA insertion in the gene At3g59770 (SALK_041090) that exactly resembles the sac9-1 mutant (Fig. 1B). The F1 progeny of a cross between this T-DNA insert mutant and the sac9-1 mutant also has the mutant phenotype, indicating that the two mutations are allelic (both alleles are recessive; Fig. 1C). We call this T-DNA insert allele sac9-2 and a second T-DNA insert allele subsequently obtained from the Salk Institute Genomic Analysis Laboratory (SIGnAL) database sac9-3 (SALK_058870).
Figure 1.
Identification of the SAC9 gene. A, Genetic mapping of SAC9 gene. We initially mapped the SAC9 gene to chromosome 3 between marker NGA707 (21.1 Mb) and the TIR1 gene (23.1 Mb). Three hundred plants were used for the fine-structure-mapping experiment. The interval was narrowed to a 200-kb region that was located on two BACs (T16L24 and F24G16) encoding 52 putative genes. We obtained T-DNA knockouts in 14 of these genes from the SIGnAL T-DNA database. B, T-DNA insertion mutant in the At3g59770 gene (SALK_041090) that exactly phenocopies the sac9-1 mutant in terms of slow growth and anthocyanin accumulation. C, Allelism test of sac9-1 and sac9-2. The F1 progeny of a cross between the SALK_041090 T-DNA insert mutant and the sac9-1 mutant has the mutant phenotype, indicating that the T-DNA insert allele is allelic to the EMS-generated sac9-1 allele. D, Structure of the SAC9 gene showing positions of mutations in sac9-1 (the original EMS mutant allele) and insertions in two T-DNA alleles, sac9-2 (SALK_041090) and sac9-3 (SALK_058870). The sac9-1 mutant has a G-to-A mutation at the last position of intron 6. cDNA isolated from sac9-1 either fail to splice out the intron, resulting in an in-frame stop codon within the intron, or splice out one additional nucleotide resulting in a frame shift mutation and a stop codon in exon 7 (stop codons indicated by *).
We sequenced the SAC9 genes from DNA isolated from plants carrying each of the three mutant alleles. In the EMS-generated allele of sac9-1, there is a G-to-A mutation in the last nucleotide of intron 6. To identify how this mutation affects the mRNA structure, we used reverse transcription (RT)-PCR to isolate and sequence cDNA from the sac9-1 mutant in the region around the mutation. PCR amplification of cDNA using primers flanking the mutation resulted in two bands, one corresponding in size to the fragment amplified from wild-type cDNA, and the other approximately 120 bp larger. Approximately 50% of the product is the larger band. Sequence analysis indicated that the larger band represents sac9-1 cDNA, in which intron 6 is not spliced out. As a result, there is an in-frame stop codon within this intron in the cDNA, which would result in a truncated protein (with a wild-type sequence until amino acid 749, then with 18 intron-encoded amino acids added before the stop codon; asterisk in Fig. 1D). The smaller band represents a transcript that has been improperly spliced, removing one nucleotide from the start of exon 7. As a result of this improper splicing, there is a reading frame shift leading to an in-frame stop codon within exon 7 (with a wild-type sequence until amino acid 749, then with 49 out-of-frame amino acids before the stop codon; asterisk in Fig. 1D). The position of the two T-DNA insertions in sac9-2 and sac9-3 are in exons 9 and 10, respectively. The three sac9 mutant alleles are most likely functionally null alleles. Using RT-PCR, we were unable to detect SAC9 transcript in RNA isolated from either of the T-DNA insert alleles (data not shown). Although the sac9-1 allele encodes a truncated protein including the catalytic domain, the phenotypes of the three mutants are identical suggesting that the truncated protein is nonfunctional or is degraded rapidly after synthesis.
The SAC9 Gene Encodes a Putative PI Phosphatase
The SAC9 gene encodes a 1,630-amino acid-long protein, and a closely related protein is encoded in the rice genome (Oryza sativa; GenBank accession no. BAB19411, The Institute for Genomic Research [TIGR] gene temp_id: 9629.t02350; Fig. 2A). The Arabidopsis gene has unusual splice sites, and the predicted splice sites of the rice gene are different in GenBank and the TIGR database (Zhong and Ye, 2003). We show an alignment with the TIGR predicted protein sequence, which is more similar to that encoded by the experimentally determined Arabidopsis cDNA sequence. A SAC domain (highlighted) is present from amino acids 146 to 640 (numbering relative to the Arabidopsis protein). The SAC domain was first identified in the yeast SAC genes (Novick et al., 1989). SAC domain proteins from yeast and humans have been shown to have PI phosphatase activity (Guo et al., 1999).
Figure 2.
The SAC9 gene encodes a putative PI phosphatase. A, Amino acid alignment of SAC9 from Arabidopsis (At) and rice (Os). The SAC domain is highlighted and extends from amino acids 146 to 640. The WW domain within the SAC domain is indicated, as are SAC domain motifs 6 and 7. Arrows above and below the alignment indicate positions of introns in the Arabidopsis and rice genes, respectively, and positions corresponding to the sites affected in each of the mutant alleles are indicated by arrowheads. The third rice exon (lowercase) is annotated as part of an intron by TIGR but may be part of the coding sequence as suggested by comparison to the experimentally determined Arabidopsis cDNA sequence. B, SAC domain family in Arabidopsis. SAC9 is unique among Arabidopsis SAC domain proteins but closely related to a protein encoded in the rice genome. The SAC domain is indicated by the black box, and the WW domain indicated by a gray box within SAC domain. C, The SAC9 catalytic domain has a substitution of a conserved Cys residue. The SAC catalytic domain and representative catalytic domains from Saccharomyces cerevisiae SAC1 and Arabidopsis SAC1 proteins are shown. SAC9 from Arabidopsis and rice have a Ser in place of the second Cys (highlighted), a feature that has been found in only one other protein, a SAC domain protein from G. lamblia.
The Arabidopsis genome encodes nine SAC domain proteins that fall into three distinct subgroups (Fig. 2B; Zhong and Ye, 2003). SAC 1 through 5 are of intermediate size and most similar to the yeast Fig4 protein sequence, SAC 6 through SAC 8 are the smallest and most similar to the yeast Sac1 protein sequence, and SAC9 is unique among the SAC family. The rice and Arabidopsis genes are similar, and the intron structure is identical for the eight introns in the downstream part of the coding sequence but differ in intron positions within the SAC domain (introns indicated by small arrows in Fig. 2A). The positions corresponding to those of the three mutations are also indicated as arrowheads in Figure 2A.
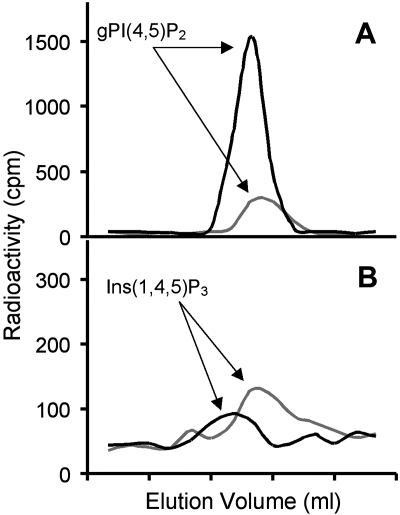
Figure 4.
PtdIns(4,5)P2 is elevated in roots of cold-stressed plants. Arabidopsis plants were metabolically labeled with 3H myoinositol. Control plants were removed from the media, roots and shoots were separated and treated as described in the text in order to extract radiolabeled phospholipids. Stressed plants were subjected to cold stress (0°C) for 1 h prior to root and shoot separation and phospholipid extraction. The peaks from the control plant samples are plotted with a gray line and the ones from the cold-stressed plant samples are plotted with a black line. Each tick on the x axis corresponds to 1 min. A, The gPI(4,5)P2 peaks from chromatography of control and cold-stressed plant glycerophosphoinositols are shown. B, The Ins(1,4,5)P3 peaks from chromatography of control and cold-stressed plant inositol phosphates are shown.
The SAC9 proteins from Arabidopsis and rice are unique among the plant SAC proteins in three ways. First, they contain a WW domain inserted between motifs 6 and 7 of the SAC domain (indicated in Fig. 2A). Second, within the SAC9 catalytic domain, the second conserved Cys is replaced by a Ser (Fig. 2C). Extensive searches of GenBank have revealed only one other protein, from Giardia lamblia (GenBank accession number EAA37060) that has this same Cys-to-Ser substitution in an otherwise functional-looking SAC domain. The first Cys of the catalytic domain is necessary for catalysis, but no such role for the second Cys has been reported. Whether or not the catalytic properties for SAC9 are affected by this change needs to be determined experimentally. Finally, the SAC9 proteins are much larger than other SAC domain proteins. In yeast and vertebrates, many SAC domain proteins have a second, 5PTase catalytic domain C terminal to the SAC domain (Hughes et al., 2000; Ha et al., 2003). We have been unable to identify any amino acid motifs or homologies to the SAC9 protein in the C-terminal 1,000-amino acid region, suggesting that this part of the protein does not contain any additional catalytic domains.
RT-PCR expression studies demonstrate that the different SAC domain proteins have different organ-specific mRNA expression patterns (Despres et al., 2003; Zhong and Ye, 2003). We found that SAC9 mRNA is most abundant in the roots, with lower expression levels throughout the leaves and shoot (data not shown), confirming the results of Zhong and Ye (2003) and results from the available microarray data.
PtdIns(4,5)P2 and Ins(1,4,5)P3 Accumulate in sac9-1 Mutant Roots
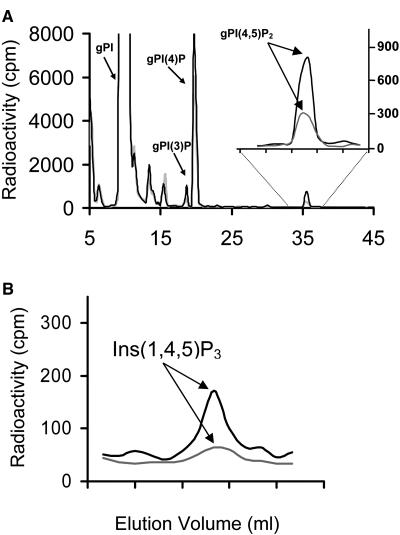
We hypothesized that the loss of function of the SAC9 gene would affect PI metabolism. To investigate this, we carried out a study in which 2-week-old hydroponically grown wild-type and sac9-1 Arabidopsis plants were labeled with [3H] myoinositol. After labeling, roots and shoots were separated, extracted independently, and PIs were deacylated and the corresponding glycerolphophoinositol head groups were analyzed by HPLC. No major differences were detected between wild-type and sac9-1 shoot extracts (Table I). By contrast, levels of PtdIns(4,5)P2 were approximately 4-fold higher in sac9-1 root extracts as compared to wild-type root extracts (Table I; Fig. 3A). In wild-type plants, PtdIns(4,5)P2 levels are consistently and reproducibly about 4-fold higher in shoots than roots. No significant differences were observed in PtdIns(3)P and PtdIns(4)P levels between sac9-1 and wild-type roots, although the average values between genotypes were more dissimilar than those found in shoots. A 3-fold increase in Ins(1,4,5)P3 was also detected in sac9-1 roots as compared to wild-type roots, but no difference in Ins(1,4,5)P3 was found in the shoots (Fig. 3B; data not shown). Prior studies have demonstrated that cold and osmotic stress leads to an increase in PtdIns(4,5)P2 levels in wild-type plants, but specific studies on roots have not been reported (Smolenska-Sym and Kacperska, 1994; Pical et al., 1999; DeWald et al., 2001). We examined wild-type Arabidopsis root tissue and found that 1 h at 0°C led to a 4-fold increase in PtdIns(4,5)P2 and no appreciable change in Ins(1,4,5)P3 levels (Fig. 4). These results show that cold treatment and the sac9-1 mutation both affect PI signaling in roots. It will be interesting to see whether cold and osmotic stress further affect PI levels in sac9 mutant roots.
Table I.
Relative abundance of various glycerophosphoinositol head groups (corresponding to different PI species) in terms of percent of the total glycerolphosphoinositols present in root or shoot tissue extracts from sac9-1 or wild-type plants, and a comparison of the relative abundance of each between sac9-1 and wild-type extracts
Values are averages of two to four separate experiments, number in parentheses are ses of the mean, and ratios are sac9-1 values divided by wild-type values.
| Glycerol-Phosphoinositol | Root sac9-1 | Root Wild Type | Root Ratio | Shoot sac9-1 | Shoot Wild Type | Shoot Ratio |
|---|---|---|---|---|---|---|
| gPI | 97.1 (0.3) | 97.2 (0.6) | 1.00 | 96.9 (0.15) | 96.5 (0.5) | 1.00 |
| gPI(3)P | 0.34 (0.04) | 0.20 (0.09) | 1.7 | 0.45 (0.15) | 0.50 (0.04) | 0.9 |
| gPI(4)P | 2.22 (0.5) | 3.25 (0.9) | 0.68 | 2.4 (0.2) | 2.12 (0.2) | 1.13 |
| gPI(4,5)P2 | 0.40 (0.1) | 0.11 (0.06) | 3.6 | 0.40 (0) | 0.47 (0.05) | 0.85 |
Figure 3.
PtdIns(4,5)P2 and Ins(1,4,5)P3 are elevated in roots of sac9-1 mutant plants. Wild-type or sac9-1 plants were metabolically labeled with 3H myoinositol. Roots and shoots were separated and treated as described in the text in order to extract radiolabeled phospholipids. The peaks from the wild-type plant samples are plotted with a gray line and the ones from the sac9-1 plant samples are plotted with a black line. Elution time in minutes is shown on the x axis, and each minor tick shown in the inset and in section B corresponds to 1 min. A, Chromatographs of sac9-1 (black) and wild-type (gray) glycerophosphoinositols are shown, with the gPI(4,5)P2 peaks expanded in an inset. In order to directly compare the relative PtdIns(4,5)P2 levels, equivalent glycerophosphoinositols counts per minute from sac9-1 and wild-type plants were loaded onto the HPLC column. B, The Ins(1,4,5)P3 peaks from chromatography of sac9-1 and wild-type plant inositol phosphates are shown. In order to directly compare the relative Ins(1,4,5)P3 levels, equivalent total inositol phosphates from sac9-1 and wild-type plants were loaded onto the column. Each HPLC run is representative of three independent analyses.
sac9 Mutant Phenotype
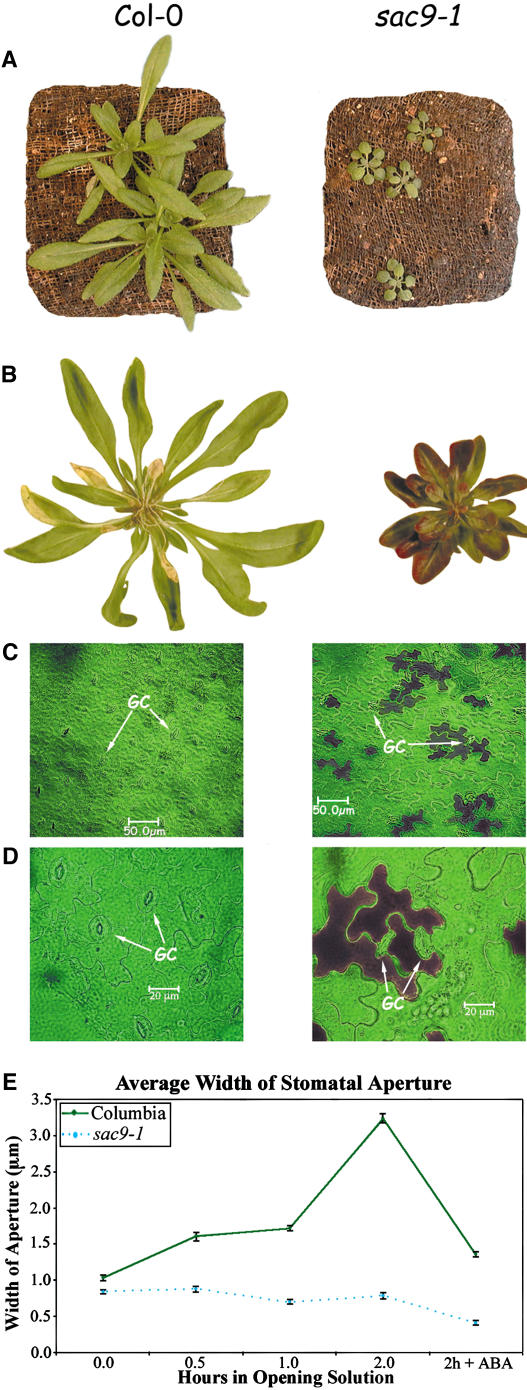
The Arabidopsis sac9-1 mutant is slow growing, makes small leaves with short petioles, and accumulates anthocyanin in the leaves (Fig. 5). Anthocyanin accumulates preferentially in the lower leaf epidermis (Fig. 5B) and specifically in cells adjacent to the guard cells (Fig. 5, C and D). While the anthocyanin-containing epidermal cells are of similar shape and size in the wild-type (Columbia [Col-0]) and mutant plants, the sac9-1 guard cells are noticeably smaller than wild-type guard cells. We quantified the guard cell size by measuring the stomatal aperture (Fig. 5E). Leaves were floated on stomatal-opening medium for varying lengths of time and then the lower epidermis peeled off and imaged (Kwak et al., 2002). In wild-type plants, stomatal apertures increased in size during this incubation, but in sac9-1 plants the apertures did not change. After 2 h on stomatal-opening solution, ABA was added to 1 μm, causing the wild-type stomata to close, but not affecting those in sac9-1 mutants. Therefore, the sac9-1 guard cells appear to be constitutively closed.
Figure 5.
Phenotypes of sac9-1 mutants. A, Four-week old wild-type (Col-0) and sac9-1 mutants grown in pots. The sac9-1 mutants grow much more slowly than wild-type plants and have short petioles and small leaves. B, Anthocyanin accumulates in the abaxial surface of sac9-1 leaves. C and D, Anthocyanin accumulates in the epidermal cells surrounding guard cells. Not all epidermal cells accumulate anthocyanin, but all of those that do are in direct contact with guard cells (GC). E, Stomata of sac9-1 plants are resistant to opening. Col-0 or sac9-1 leaves were floated on stomatal-opening solution for varying lengths of time and the stomatal apertures measured. After 2 h, ABA was added to 1 μm.
Reciprocal Grafting Experiments
Our biochemical studies did not show any differences in PI levels between sac9-1 and wild-type shoots, yet the mutation has a clear phenotypic effect in the shoot. This observation raises the possibility that the elevated PtdIns(4,5)P2 level in the root causes some sort of diffusible signal to move into the shoot. Ins(1,4,5)P3 potentially could be such a signal, although we do not record elevated Ins(1,4,5)P3 levels in the sac9 shoot. Alternatively, the sac9 mutation could lead to very localized changes in level or distribution of PtdIns(4,5)P2 in the shoot, which are not detected in our whole-shoot extracts. PtdIns(4,5)P2 distribution is at least as important as its quantity (Heilmann et al., 2001; Stefan et al., 2002). To differentiate between these possibilities, we carried out reciprocal grafting experiments between wild-type (Col) and sac9-1 mutant plants. A total of 120 successful grafts were obtained, including each of the four possible combinations (Table II). Grafted seedlings were transferred to pots and phenotypic observations of shoots were performed throughout development. We observed that shoots of Col self-grafts exhibited the wild-type Col phenotype, and shoots of sac9-1 self-grafts exhibited the sac9-1 phenotype. When Col shoots were grafted onto sac9-1 roots, 63% of the shoots exhibited the normal Col phenotype, while the remainder showed varying degrees of growth delay. However, none showed the extreme dwarfism, anthocyanin accumulation, and epinastic leaf habit of the sac9-1 shoots. This result suggests that the small and deficient sac9-1 root may have limited the shoot growth rate, perhaps through limitation of water or nutrient uptake, but that it was not sufficient to cause the wild-type shoot to phenocopy the sac9-1 shoot. Most importantly, sac9-1 shoots grafted onto Col roots all showed the typical sac9-1 phenotype, clearly demonstrating that the abnormal shoot phenotype of sac9 mutants is correlated with loss of function of SAC9 in the shoot. It is likely that there are cellular or subcellular alterations in PtdIns(4,5)P2 and/or Ins(1,4,5)P3 levels in the sac9-1 shoot that are not detectable by the bulk-labeling and extraction method we used. It will be important to carry out cell-specific expression and subcellular localization studies for SAC9, PtdIns(4,5)P2 and Ins(1,4,5)P3 to further understand the function of the SAC9 protein in the shoot.
Table II.
Results of grafting experiments between Col and sac9-1 mutant plants
Three- to 6-d-old seedlings grown on agar plates were grafted using a transverse-cut butt alignment. Three to 8 d after grafting, plants were transferred to soil and phenotypes assessed in terms of inflorescence height, leaf size, anthocyanin accumulation, and silique formation. In the Col/sac9-1 grafts, 63% of the shoots exhibited the normal Col phenotype, while the remainder showed varying degrees of growth delay; none showed the extreme dwarfism and anthocyanin accumulation characteristic of sac9-1 shoots.
| Scion/Stock | N | Shoot Phenotype |
|---|---|---|
| Col/Col | 25 | Col |
| sac9-1/sac9-1 | 21 | sac9-1 |
| Col/sac9-1 | 38 | Col and delayed Col |
| sac9-1/Col | 36 | sac9-1 |
sac9 Mutants Appear to be Constitutively Stressed
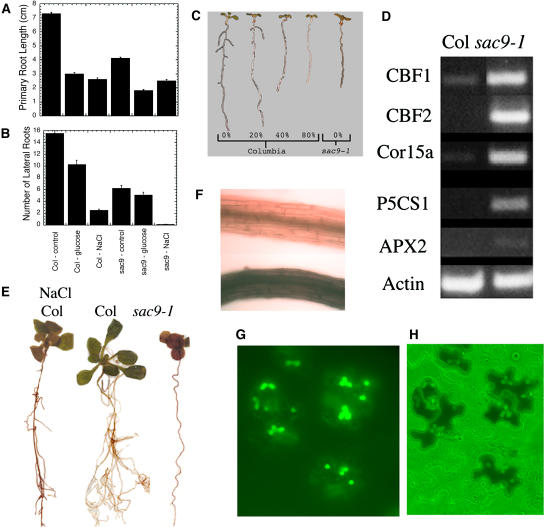
The sac9-1 mutants have shorter primary roots and fewer lateral roots than wild-type plants (Fig. 6, A and B). Lateral root production is disproportionately affected; when sac9-1 plants are grown until the primary root is the same length as wild-type plants, they underproduce lateral roots by about 50% (K. Parker, unpublished data). Defects in root growth, and specifically in lateral root production, have been observed in osmotically stressed plants. We examined the effects of osmotic stress on sac9-1 root growth using either 5% Glc or 60 mm NaCl, both of which hindered root growth in wild-type and sac9-1 plants. Strikingly, we observed that sac9-1 mutants grown on 60 mm NaCl essentially produced no lateral roots (Fig. 6B). Furthermore, sac9-1 but not wild-type plants grown on NaCl had bleached cotyledons, indicating that the mutants are hypersensitive to NaCl. Because both Glc and NaCl have additional effects besides affecting osmotic strength of the media, we used another procedure to affect only the osmotic potential of the plates (van der Weele et al., 2000). Wild-type plants grown on plates that had been equilibrated with high concentrations of polyethylene glycol (PEG) resemble sac9-1 mutants in their growth rate and pattern (Fig. 6C). Taken together, these results show that the sac9-1 plants phenotypically resemble osmotically stressed wild-type plants and are further hypersensitive to additional stress. Other experiments showed that sac9-1 plants are hypersensitive to both cold and high-light stress (data not shown).
Figure 6.
sac9-1 plants are constitutively stressed. A and B, Primary root length (A) and number of lateral roots (B) of Col-0 and sac9-1 plants grown for 4 d on control medium then transferred and grown for an additional 7 d on plates containing 5% Glc or 60 mm NaCl. C, Four-day-old plants were transferred to plates equilibrated with varying amounts of PEG-8000 and photographed after two additional days. D, Genes induced by stress are over-expressed in sac9-1 mutants. Col-0 or sac9-1 seedlings were grown for 4 weeks in pots at 22°C. RNA was isolated and reverse transcribed using oligo(dT) and Moloney murine leukemia virus-reverse transcriptase. PCR was carried out using primers specific for CBF1, CBF2, Cor15a, P5CS1, APX2, or actin, and product analyzed by agarose gel electrophoresis using ethidium bromide. E, Ten-day-old plants grown on plates with or without 50 mm NaCl were stained overnight with DAB. F, Col (top) and sac9-1 (bottom) roots stained with DAB. G and H, Epidermal peels of sac9-1 plants stained with 2′,7′-dichlorofluorescein diacetate showing colocalization of intense fluorescence staining with anthocyanin in epidermal cells adjacent to the guard cells.
We investigated the stress response in sac9-1 mutants using RT-PCR analysis of several stress-induced genes. Transcript levels of a variety of genes correlated with stress response are elevated in the sac9-1 mutants. These genes include the CBF1 and CBF2 genes encoding cold-induced transcription factors, the cold response gene COR15a, P5CS1 (encoding the Pro biosynthetic enzyme Δ1-pyrroline-5-carboxylate synthase), and the ascorbate peroxidase-encoding gene APX2 (Fig. 6D; Karpinski et al., 1997).
Because ROS have been implicated in the stress response, and sac9-1 mutants seem to have a constitutive stress response, we speculated that they might also have elevated ROS levels. sac9-1 plants stain more than wild-type plants, particularly in the roots, with the ROS-stain 3,3′-diaminobenzidine (DAB; Fig. 6, E and F; Thordal-Christensen et al., 1997; Fryer et al., 2002, 2003). Enhanced DAB staining is correlated with osmotic stress as shown in wild-type plants grown on plates containing 50 mm NaCl (Fig. 6E). Soil-grown sac9-1 plants also stain more deeply than corresponding wild-type plants, with DAB staining apparent throughout the leaf including the trichomes (data not shown). We stained leaf epidermal peels of sac9-1 plants using the ROS indicator dye 2′,7′-dichlorofluorescein diacetate and observed a striking correlation of intense fluorescent staining in the epidermal cells that accumulate anthocyanin (Fig. 6, G and H; Cathcart et al., 1983). No similar pattern was found in wild-type plants. These results show that sac9-1 mutants have elevated levels of ROS throughout the plant.
DISCUSSION
PIs and Stress Signaling
Mutation of the SAC9 gene leads to dwarfed plants that accumulate ROS and up-regulate several stress-induced genes, and whose leaves accumulate anthocyanin, and have constitutively closed guard cells and altered PI signaling. Anthocyanin accumulation is a common response to environmental stress and may play a role in photoprotection (Steyn et al., 2002). Elevated ROS, also a response to stress, has profound effects on both gene expression and growth rate in plants. Our results support the importance of the PI-signaling pathways in stress response.
Plants activate multiple phospholipid-based signaling pathways in response to biotic and abiotic stress, and it is likely that there is significant cross talk, convergence, and divergence in the phospholipid-signaling pathways (Stevenson et al., 2000; Munnik, 2001; Wang, 2002, 2004; Wang et al., 2002; Meijer and Munnik, 2003). Phospholipid signaling includes both membrane-localized signals and the membrane-derived soluble signal Ins(1,4,5)P3, and it is likely that each plays distinct roles in osmotic stress response. Cleavage of PtdIns(4,5)P2 by PLC generates the lipid diacylglycerol (DAG) and the soluble Ins(1,4,5)P3 (Berridge and Irvine, 1984). In plants, the role of Ins(1,4,5)P3 in osmotic, ABA, and stress signaling is well established through numerous biochemical and genetic studies. Direct microinjection of Ins(1,4,5)P3 into guard cells is sufficient to cause them to close, while lowering the level of Ins(1,4,5)P3 through overexpression of PTases leads to decreased sensitivity to ABA (Blatt et al., 1990; Gilroy et al., 1990; Sanchez and Chua, 2001; Burnette et al., 2003).
Genetic loss-of-function studies investigating Ins(1,4,5)P3 function are complicated by the large number of genes encoding the PTases that can degrade it. The Arabidopsis genome encodes 11 putative type I 5PTases and four type II 5PTases (Berdy et al., 2001; Zhong and Ye, 2004). The only type I 5PTase gene with a known loss-of-function mutant phenotype is CVP2. Loss-of-function cvp2 mutants accumulate approximately three times more Ins(1,4,5)P3 than wild-type plants and are more sensitive to exogenous ABA, but they show no increased stress sensitivity and are morphologically normal other than having vein pattern defects in the cotyledons (Carland and Nelson, 2004). The CVP gene is expressed in most tissues, but functional redundancy may prevent phenotypic abnormalities elsewhere. Mutation of an inositol polyphosphate 1-phosphatase gene, FIERY1, results in a 10-fold increase in Ins(1,4,5)P3 in unstressed plants, and increased sensitivity to ABA, including decreased rates of germination and seedling growth in response to ABA and osmotic stress (Xiong et al., 2001). Mutants in the FRA3 gene encoding a type II 5PTase accumulate approximately twice as much Ins(1,4,5)P3 in inflorescence stems as compared to wild-type plants, but no difference in seedling levels, and show no change in ABA sensitivity (Zhong et al., 2004). Our results show that Ins(1,4,5)P3 levels are elevated in sac9 mutants approximately 3-fold over wild type, similar to the increase in the cvp2 and fra3 mutants but to a lesser extent than in fiery1 mutants. Although sac9 mutants are more sensitive to osmotic stress than wild-type plants, they are not hypersensitive to ABA in terms of germination or growth (data not shown). This observation may reflect differences in Ins(1,4,5)P3 levels between fiery1 and sac9 mutants, or may reflect differences in the cells and organelles in which the two genes function. We are in the process of identifying the cell type expression pattern of the SAC9 gene, which will aid us in interpreting the mutant phenotype. For example, a result showing that SAC9 is expressed in guard cells would be consistent with both our phenotypic data (flaccid guard cells) and our inability to detect significant changes in PI levels in whole-shoot extracts.
Besides sac9, the fra3 mutant is the only other Arabidopsis mutant known to accumulate PtdIns(4,5)P2. Unlike the sac9 mutant, the fra3 mutant is morphologically normal, except for the defect in secondary-wall thickening, which may reflect differences in gene expression patterns. The FRA3 gene is predominately expressed in the fiber cells that are affected in the mutant, and although the cell type expression pattern of SAC9 is not yet known, it is expressed in all organs with higher expression in the roots (Zhong and Ye, 2003). In fiber cells, the fra3 mutation alters actin organization, which is consistent with PtdIns(4,5)P2's ability to bind to profilin and other actin-binding proteins. It is likely that the defect in secondary-wall thickness in fra3 fiber cell is a consequence of the altered actin cytoskeleton (Zhong et al., 2004).
The altered levels of PI accumulation measured in the sac9 mutant root extracts are comparable to those measured in wild-type plants or cultured cells subject to stress. In wild-type plants treated with 0.25 m NaCl for 1 h, a 20-fold increase was observed in PtdIns(4,5)P2 and a 15-fold increase in Ins(1,4,5)P3 (DeWald et al., 2001). Using suspension-cultured cells, treatment with 0.8 m sorbitol or 0.4 m NaCl led to an 8- to 25-fold increase in PtdIns(4,5)P2 within 20 min (Pical et al., 1999). Previous studies also have found an increase in PtdIns(4,5)P2 in cold-stressed plants, which we confirmed in this study; after just 1 h at 0°C, Arabidopsis root extracts showed a 3-fold increase in PtdIns(4,5)P2 (Fig. 4; Smolenska-Sym and Kacperska, 1994). The 4-fold increase in PtdIns(4,5)P2 in sac9-1 roots is therefore within the range observed in stressed wild-type plants. Taken together, the accumulating data that stress leads to PtdIns(4,5)P2 accumulation, and the stressed phenotype of the sac9 mutant, which accumulates physiologically relevant levels of PtdIns(4,5)P2, suggest that PtdIns(4,5)P2 has a role in stress signaling or response.
It is likely that PtdIns(4,5)P2 contributes to the stress-induced responses of cell membranes and walls. Cold and osmotic stress lead to changes in cell turgor pressure, profoundly affecting membrane structure (Ristic and Ashworth, 1993; Kubitscheck et al., 2000). Stress also induces expression of numerous cell wall-localized proteins thought to aid in stress tolerance, including extensins, Gly- and Pro-rich proteins, and annexins (Kovacs et al., 1998; Garcia-Gomez et al., 2000; Wu and Cosgrove, 2000; Bray, 2002; Lee et al., 2004). PtdIns(4,5)P2 could play a role in stress response by reorganizing the actin cytoskeleton, affecting the targeting of vesicles or proteins to the plasma membrane or wall. It will be interesting to examine whether the actin cytoskeleton or membrane dynamics are affected in sac9 mutants.
Phosphatidic acid (PA) is a lipid-derived signal that is involved in numerous plant stress responses (Wang, 2002). PA increases in response to osmotic and cold stress, mediates ABA responses, and is involved in regulating guard cell aperture (Jacob et al., 1999; Frank et al., 2000; Munnik et al., 2000; Katagiri et al., 2001; Munnik, 2001; Sang et al., 2001b; Ruelland et al., 2002; Welti et al., 2002). PA is primarily the product of phospholipase D (PLD) cleavage of various lipid substrates, and PtdIns(4,5)P2 is an activator of some forms of PLD (Wang, 2002; McDermott et al., 2004). However, PtdIns(4,5)P2 can also contribute to PA synthesis via PLC activity. When PLC hydrolyzes PtdIns(4,5)P2, the resulting DAG can be phosphorylated to PA by DAG kinase (Munnik, 2001; Meijer and Munnik, 2003). Recent work has demonstrated that in some situations the extent of PLC-derived PA can be significant (Arisz et al., 2003; de Jong et al., 2004). PA is also involved in defense signaling and in the production of and protection from reactive oxygen (Sang et al., 2001a; Zhang et al., 2003). Thus, in light of our observations of ROS accumulation in the sac9 mutant, we are anxious to determine if the increased PtdIns(4,5)P2 results in elevated PA levels, and if so whether this is through PLD activation or PLC and DAG kinase activity. Genetic studies could then be used to distinguish between PtdIns(4,5)P2-specific responses, including effects on PLD activity and responses that are mediated by its cleavage by PLC to Ins(1,4,5)P3 and DAG/PA. Additional labeling studies are under way to further characterize the effect of the sac9 mutation on lipid signaling.
Specificity and Structure of PI Phosphatases
The presence of a SAC domain suggests that SAC9 encodes a PI phosphatase, and this interpretation is supported by the accumulation of PtdIns(4,5)P2 in sac9-1 mutants (Fig. 3; Table I). Although the Arabidopsis genome has eight other SAC domain-encoding genes, SAC9 is distinct from the rest, and we believe it may have a different function and may be plant specific (Zhong and Ye, 2003). Studies on the other Arabidopsis SAC genes suggest that they fall into two distinct classes, and there may be some functional redundancy between them (Despres et al., 2003). Only one other Arabidopsis SAC domain-encoding gene has been studied by loss-of-function analysis; mutations in the AtSAC1 gene cause abnormalities in cell wall synthesis and cellular morphology, suggesting a role for the AtSAC1 gene in vesicle trafficking or the actin cytoskeleton similar to that seen in yeast (Guo et al., 1999; Zhong and Ye, 2003). Levels of PI for other SAC domain mutants have not yet been reported.
It is intriguing to speculate that the SAC9 protein may have a unique function in plant cells. Unlike plants, yeast and animals have bifunctional synaptojanin or synaptojanin-like SAC domain proteins that contain a type II inositol PTase domain C terminal to the SAC domain (Guo et al., 1999). In plants, no homologs of bifunctional synaptojanin-like enzymes have been identified. Although we cannot find any sequences within SAC9 that suggest it is a plant homolog of a synaptojanin, we do note that its in vivo function more closely resembles that of a synaptojanin than other SAC domain proteins (Mitchell et al., 2002; Whisstock et al., 2002). In vitro expression and activity studies of full-length and truncated SAC9 proteins will determine if a cryptic catalytic activity can be detected in the C-terminal domain.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 EMS-mutagenized M2 seeds with the gl-1 marker were obtained from Lehle Seeds (Round Rock, TX). The sac9-1 mutant was selected based on its small size and anthocyanin accumulation. M3 seeds were collected, and mutant sac9-1 plants were backcrossed to Col-0 twice. After the first backcross, sac9-1 plants were crossed to Landsberg erecta for mapping. For growth on petri plates, seeds were surface sterilized in 15% bleach for 15 min, rinsed five times with sterile water, suspended in 0.1% agarose, and stored in the dark at 4°C for 5 to 7 d to induce germination. Seeds were plated on Murashige and Skoog (MS)/agar plates [0.7% (w/v) agar, 1% (w/v) Suc, 0.5 g/L 2-(N-morpholino) ethanesulfonic acid (MES; pH 5.7), 0.1 or 0.5× MS medium, 1× MS vitamins] and 5% Glc or 60 mm NaCl as indicated, and sealed with parafilm or surgical tape. The plates were placed in a Percival CU-32L (Boone, IA) tissue culture growth chamber set at a 24-h light period at 22°C and a photon flux of 75 μmol m−2 s−1. For osmotic stress studies, plates were equilibrated with PEG-8000 as described in van der Weele et al. (2000). Soil-grown plants were grown in Sunshine potting soil in a 22°C Percival AR60L growth chamber at a photon flux of 120 μmol m−2 s−1.
Positional Mapping
Genomic DNA was isolated from individual F2 plants from a mapping population showing the mutant phenotype. Primers for CAPS markers were purchased from Research Genetics (Huntsville, AL) or synthesized by Operon. CAPS and SSLPs markers were designed in the interval between At3g56930 and At3g62980 based on polymorphism data from the Cereon database (www.Arabidopsis.org). PCR amplification and digestion was performed using standard methods (Glazebrook et al., 1998). Details on CAPS and SSLPs markers designed for fine-structure mapping are available upon request. Once the interval was narrowed down to two BACs, we obtained SIGnAL T-DNA insert lines for genes on BACs T16L24 and F24G16 from the Arabidopsis Biological Resource Center (Ohio State University, Columbus; Alonso et al., 2003). Seeds were planted on plates and in pots, and the putative allelic lines were crossed to the sac9-1 mutant. After determining that the SALK_041090 and SALK_058870 lines were allelic to sac9-1, we confirmed the site of the insertion by amplifying and sequencing the expected region of the SAC9 gene from each line using cycle sequencing and an ABI sequencer provided by the Rancho Santa Ana Botanic Garden. The entire SAC9 gene from the sac9-1 mutant was sequenced to determine the site of the EMS-induced mutation.
Analysis of PIs and Inositol Phosphates
Plant Growth Conditions and Radiolabeling
A modified hydroponic system was used to grow wild-type and sac9-1 mutant Arabidopsis plants (Arteca and Arteca, 2000). Plants were grown in Magenta vessels (MAGENTA Corporation, Chicago) containing 0.5× MS medium under an 18-h d (24°C)/6-h night (22°C) cycle with irradiation of 100 to 200 μmol m−2 s−1. Roots of 3- to 4-week-old intact plants were bathed in 100 μCi of myo-[2-3H]inositol (Amersham-Pharmacia Biotech, Piscataway, NJ) per 1 mL of a 0.5× MS solution for 20 h.
Extraction of PIs and Inositol Phosphates
A detailed description of the lipid extraction from Arabidopsis plants has been already described (Hama et al., 2000; DeWald et al., 2001). Briefly, root and shoot tissues of the plants grown in the mini vessels were separately fixed in 5 to 7 mL of trichloroacetic acid (final concentration = 5%–6%) followed by incubation on ice for 1 h. Samples were washed three times with 10 mL H2O and then homogenized in a Dounce tissue grinder (VWR, West Chester, PA) by adding a mixture of 0.5 mL of 1 m HCl and 0.75 mL of a methanol:chloroform (1:1) solution. Homogenates were centrifuged and the bottom (organic) phase was recovered and aqueous phase reextracted once more with a chloroform:methanol (1:1) solvent. The aqueous phase containing soluble inositol phosphates were dried in vacuo, resuspended in H2O, and the radioactivity assayed using a Beckman LS 5801 liquid scintillation counter (Beckman, Fullerton, CA). The organic phases containing PIs were dried in vacuo and deacylated using a 40% methylamine/methanol/n-butanol/water (26.8%/ 45.7%/11.4%/16.1%) solution. The deacylated glycerophosphoinositol (gPI) head groups were extracted with n-butanol:petroleum ether:ethyl formate (20:4:1) three times and dried in vacuo. The glycerophosphoinositols were dried in vacuo, resuspended in H2O, and the radioactivity assayed using the liquid scintillation counter.
HPLC Analyses
Anion-exchange HPLC with Partisil 10 SAX (4.6×250 mm) columns (Whatman, Clifton, NJ) fitted with a SAX guard column (Phenomenex, Torrance, CA) was used to resolve the gPI head groups and Ins(1,4,5)P3. A portion of each sample (depending on each set of experiments) was mixed with approximately 80 nmol each of AMP, ADP, and ATP (which were used as internal controls monitoring the column performance) and applied to the column. DeWald et al. (2001) provides a detailed description of the gradients, reagents, and flow rate. Fractions were collected every 20 s, mixed with EcoLume scintillant (ICN Biomedicals, Irvine, CA), and counted in a liquid scintillation counter. As described earlier (Hama et al., 2000; DeWald et al., 2001), coelution of peaks with inositol phosphate or glycerophosphoinositol standards was used to identify various peaks in the chromatographs.
Cold Stress Protocol
Hydroponically grown radiolabeled Arabidopsis plants were submerged in 0.5× MS medium, similar to the protocol outlined by Yamaguchi-Shinozaki and Shinozaki (1994), and incubated at 0°C for 1 h. After the incubation period the plants were removed and their root and shoot tissue separated and placed in ice-cold trichloroacetic acid to stop cellular processes. PIs were extracted, deacylated, and quantified using HPLC.
Grafting Studies
Wild-type and mutant seeds were sterilized and plated on Millipore cellulose nitrate filters (type HA, pore diameter 0.45 μm; Millipore, Bedford, MA) on top of MS agar plates containing 1% Suc. Plates were placed vertically in the growth chamber for 3 to 6 d, at which time seedlings were cut horizontally mid hypocotyl using a 15° Sharpoint microdissecting knife (Fine Scientific Tools, Foster City, CA), and grafted together using a transverse-cut butt alignment (Turnbull et al., 2002). Plates were returned to the growth chamber in a horizontal position for 24 h and then restored to a vertical position. Grafts were assessed at 3 to 8 d post grafting by gently tugging on the shoot, and successful grafts were transferred to soil. Phenotypes of shoots were assessed in terms of inflorescence height, leaf size and orientation, anthocyanin accumulation, and silique formation.
Stomatal Aperture Assay and Reactive Oxygen Imaging
For stomatal aperture measurements, leaves were floated in bright light on stomatal-opening solution (5 mm KCl, 50 μm CaCl2, and 10 mm MES/Tris pH 6.15) for 2 h, at which time ABA was added to 1 μm. Apertures were measured in the focal plane of the outer edges of guard cells in epidermal strips (Kwak et al., 2002). Leaves or seedlings were stained in an acidic solution of DAB overnight (Thordal-Christensen, 1997; Fryer et al., 2002). For fluorescence staining, abaxial strips of leaf epidermis were prepared by pressing leaves onto double-sided tape and then scraping away the upper cell layers (Falbel et al., 2003). Epidermal peels were incubated in 30 μm 2′,7′-dichlorofluorescein diacetate in 30 mm KCl and 10 mm MES-KOH (pH 6.15) then photographed using a Zeiss fluorescence microscope (Jena, Germany; Murata et al., 2001).
RT-PCR Studies
RT-PCR studies were carried out on RNA purified from seedlings, roots, or shoots using the TRI reagent (Sigma, St. Louis). Five micrograms of RNA was DNAse treated, then reverse transcribed using Moloney murine leukemia virus (Promega) and oligo(dT) as a primer. Three microliters of this product was used as a template for PCR. For quantification of the SAC9 transcript, we used primers located near the 3′ end of the transcript SAC9UP6 (5′-GGTTTGCAGAATATCGACC) and SAC9LO6 (5′-GGATCGAACCAAGCTACG). Actin primers were used as a control Act1 (5′-ATGAAGATTAAGGTCGTGGCAC) and Act2 (5′-GTTTTTATCCGAGTTTGAAGAGGC; Berdy et al., 2001). To identify the structure of the sac9-1 cDNA in the region around the mutation, we carried out RT-PCR using exonic primers flanking this region, SAC9UP4 (5′-ACCATATCACACGGTGCG) and SAC9LO7 (5′-TAATCTTCCTCCAGTACGTTC). For gene expression studies of stress-regulated genes, the following primers were used: CBF1-F (5′-GAGATTATTGTCCGACGTTG), CBF1-R (5′-GATTCGTGGTCGTCGTAT), CBF2-F (5′-ATGTTTGGCTCCGATTACG), CBF2-R (5′-GACCATGAGCATCCGTCG), Cor15aF (5′-CAGAAAACTCAGTTCGTCG), COR15aR (5′-TGCTTTACCCTCCGCGAA), P5CS1F (5′-CCAGCAGCCTGTAATGCG), and P5CS1R (5′-GGAACACAGCAGCGCTAT). APX2 primers were as described (Panchuk et al., 2002).
Note Added in Proof
While this paper was under revision, Zhong et al. (2005) published the results of a phosphoinositide analysis of the Arabidopsis mutants in the SAC1 gene.
Zhong R, Burk DH, Nairn CJ, Wood-Jones A, Morrison WH III, Ye ZH (2005) Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 17: 1449–1466
Acknowledgments
We are grateful to the Arabidopsis Biological Resource Center and SIGnAL for providing BAC DNA and mutant seeds, and Syngenta for access to the Ler sequence polymorphism database for marker design. We thank Elaine Guerra and Harvey Mudd College students Mark Cameron, Kristine Funkhouser Nowak, Justin Pava, and Vivi Nguyen for contributions to the research, the Rancho Santa Ana Botanic Garden for DNA sequencing, Lew Feldman and Keni Jiang at University of California, Berkeley for sabbatical support for M.E.W., and Almut Vollmer for critical reading of the manuscript. We appreciate the insightful comments of two anonymous reviewers.
This work was supported by the National Science Foundation (grant no. IBN–9722191), by the Harvey Mudd College Biology Department, and by the Utah Agricultural Experiment Station (paper no. 7655).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.061317.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arisz SA, Valianpour F, van Gennip AH, Munnik T (2003) Substrate preference of stress-activated phospholipase D in Chlamydomonas and its contribution to PA formation. Plant J 34: 595–604 [DOI] [PubMed] [Google Scholar]
- Arteca RN, Arteca JM (2000) A novel method for growing Arabidopsis thaliana plants hydroponically. Physiol Plant 108: 188–193 [Google Scholar]
- Audhya A, Emr SD (2004) Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J 23: 3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdy SE, Kudla J, Gruissem W, Gillaspy GE (2001) Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating inositol trisphosphate signaling. Plant Physiol 126: 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Irvine RF (1984) Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature 312: 315–321 [DOI] [PubMed] [Google Scholar]
- Blatt MR, Thiel G, Trentham DR (1990) Reversible inactivation of K+ channels of Vicia stomatal guard cells following the photolysis of caged inositol 1,4,5-trisphosphate. Nature 346: 766–769 [DOI] [PubMed] [Google Scholar]
- Braun M, Baluska F, von Witsch M, Menzel D (1999) Redistribution of actin, profilin and phosphatidylinositol-4,5-bisphosphate in growing and maturing root hairs. Planta 209: 435–443 [DOI] [PubMed] [Google Scholar]
- Bray EA (2002) Classification of genes differentially expressed during water-deficit stress in Arabidopsis thaliana: an analysis using microarray and differential expression data. Ann Bot (Lond) 89: 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette RN, Gunesekera BM, Gillaspy GE (2003) An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiol 132: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland FM, Nelson T (2004) COTYLEDON VASCULAR PATTERN2-mediated inositol (1,4,5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. Plant Cell 16: 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathcart R, Schwiers E, Ames BN (1983) Detection of picomole levels of hydroperoxides using a fluorescent dichlorofluorescein assay. Anal Biochem 134: 111–116 [DOI] [PubMed] [Google Scholar]
- Czech MP (2000) PIP2 and PIP3: complex roles at the cell surface. Cell 100: 603–606 [DOI] [PubMed] [Google Scholar]
- Czech MP (2003) Dynamics of phophoinositides in membrane retrieval and insertion. Annu Rev Physiol 65: 791–815 [DOI] [PubMed] [Google Scholar]
- de Jong CF, Laxalt AM, Bargmann BOR, de Wit PJGM, Joosten MHAJ, Munnik T (2004) Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J 39: 1–12 [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Godi A (2004) PI-loting membrane traffic. Nat Cell Biol 6: 487–492 [DOI] [PubMed] [Google Scholar]
- Delley PA, Hall MN (1999) Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J Cell Biol 147: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres B, Bouissonnie F, Wu H-J, Gomord V, Guilleminot J, Grellet F, Berger F, Delseny M, Devic M (2003) Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J 34: 293–306 [DOI] [PubMed] [Google Scholar]
- Desrivieres S, Cooke FT, Morales-Johansson H, Parker PJ, Hall MN (2002) Calmodulin controls organization of the actin cytoskeleton via regulation of phosphatidylinositol (4,5)-bisphosphate synthesis in Saccharomyces cerevisiae. Biochem J 366: 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H (2001) Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol 126: 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR (2000) Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J 351: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drøbak BK, Heras B (2002) Nuclear phosphoinositides could bring FYVE alive. Trends Plant Sci 7: 132–138 [DOI] [PubMed] [Google Scholar]
- Elge S, Brearley C, Xia HJ, Kehr J, Xue HW, Mueller-Roeber B (2001) An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: synthesis of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in insect cells by 5-phosphorylation of precursors. Plant J 26: 561–571 [DOI] [PubMed] [Google Scholar]
- Ellson CD, Andrews S, Stephens LR, Hawkins PT (2002) The PX domain: a new phosphoinositide-binding module. J Cell Sci 115: 1099–1105 [DOI] [PubMed] [Google Scholar]
- Ercetin ME, Gillaspy GE (2004) Molecular characterization of an Arabidopsis gene encoding a phospholipid-specific inositol polyphosphate 5-phosphatase. Plant Physiol 135: 938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbel TG, Koch LM, Nadeau JA, Segui-Simarro JM, Sack FD, Bednarek SY (2003) SCD1 is required for cell cytokinesis and polarized cell expansion in Arabidopsis thaliana. Development 130: 4011–4024 [DOI] [PubMed] [Google Scholar]
- Frank W, Munnik T, Kerkmann K, Salamini F, Bartels D (2000) Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell 12: 111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR (2003) Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J 33: 691–705 [DOI] [PubMed] [Google Scholar]
- Fryer MJ, Oxborough K, Mullineaux PM, Baker NR (2002) Imaging of photo-oxidative stress responses in leaves. J Exp Bot 53: 1249–1254 [PubMed] [Google Scholar]
- Garcia-Gomez BI, Campos F, Hernandez M, Covarrubias AA (2000) Two bean cell wall proteins more abundant during water deficit are high in proline and interact with a plasma membrane protein. Plant J 22: 277–288 [DOI] [PubMed] [Google Scholar]
- Geelen D, Leyman B, Batoko H, Di Sansabastiano G-P, Moore I, Blatt MR (2002) The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: evidence from competition with its cytosolic domain. Plant Cell 14: 387–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Read ND, Trewavas AJ (1990) Elevation of cytoplasmic calcium by caged calcium or caged inositol trisphosphate initiates stomatal closure. Nature 346: 769–771 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Drenkard E, Preuss D, Ausubel FM (1998) Use of cleaved amplified polymorphic sequences (CAPS) as genetic markers in Arabidopsis thaliana. In J Martinez-Zapater, J Salinas, eds, Methods in Molecular Biology: Arabidopsis Protocols, Vol 82. Humana Press, Totowa, NJ, pp 173–182 [DOI] [PubMed]
- Guo S, Stolz LE, Lemrow SM, York JD (1999) SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem 274: 12990–12995 [DOI] [PubMed] [Google Scholar]
- Ha S-A, Torabinejad J, DeWald DB, Wenk MR, Lucast L, De Camilli P, Newitt RA, Aebersold R, Nothwehr SF (2003) The synaptojanin-like protein Inp53/Sjl3 functions with clathrin in a yeast TGN-to-endosome pathway distinct from the GGA protein-dependent pathway. Mol Biol Cell 14: 1319–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Takemoto JY, DeWald DB (2000) Analysis of phosphoinositides in protein trafficking. Methods 20: 465–473 [DOI] [PubMed] [Google Scholar]
- Hardie RC (2003) Regulation of TRP channels via lipid second messengers. Annu Rev Physiol 65: 735–759 [DOI] [PubMed] [Google Scholar]
- Heilmann I, Perera IY, Gross W, Boss WF (2001) Plasma membrane phosphatidylinositol 4,5-bisphosphate levels decrease with time in culture. Plant Physiol 126: 1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgemann DW, Ball R (1996) Regulation of cardiac Na+, Ca2+ exchange and KATP potassium channels by PIP2. Science 273: 956–959 [DOI] [PubMed] [Google Scholar]
- Homma K, Terui S, Minemura M, Qadota H, Anraku Y, Kanaho Y, Ohya Y (1998) Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem 273: 15779–15786 [DOI] [PubMed] [Google Scholar]
- Hu MH, Bauman EM, Roll RL, Yeilding N, Abrams CS (1999) Pleckstrin 2, a widely expressed paralog of pleckstrin involved in actin rearrangement. J Biol Chem 274: 21515–21518 [DOI] [PubMed] [Google Scholar]
- Hughes WE, Cooke FT, Parker PJ (2000) Sac phosphatase domain proteins. Biochem J 350: 337–352 [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Takenawa T (2002) Phosphoinositide-binding domains: functional units for temporal and spatial regulation of intracellular signalling. Cell Signal 14: 733–743 [DOI] [PubMed] [Google Scholar]
- Jacob T, Ritchie S, Assmann SM, Gilroy S (1999) Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc Natl Acad Sci USA 96: 12192–12197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM (1997) Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9: 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Takahashi S, Shinozaki K (2001) Involvement of a novel Arabidopsis phospholipase D, AtPLDdelta, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J 26: 595–605 [DOI] [PubMed] [Google Scholar]
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua N-H (1999) Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol 145: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs I, Ayaydin F, Oberschall A, Ipacs I, Bottka S, Ponger S, Dudits D, Toth EC (1998) Immunolocalization of a novel annexin-like protein encoded by a stress and abscisic acid responsive gene in alfalfa. Plant J 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Kubitscheck U, Homann U, Thiel G (2000) Osmotically evoked shrinking of guard-cell protoplasts causes vesicular retrieval of plasma membrane into the cytoplasm. Planta 210: 423–431 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Moon J-H, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK (2004) Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 16: 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A (2002) Regulation of stress responses by intracellular vesicle trafficking? Plant Physiol Biochem 40: 531–535 [Google Scholar]
- Leyman B, Geelen D, Blatt MR (2000) Localization and control of expression of Nt-Syr1, a tobacco snare protein. Plant J 24: 369–382 [DOI] [PubMed] [Google Scholar]
- Leyman B, Geelen D, Quintero FJ, Blatt MR (1999) A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science 283: 537–540 [DOI] [PubMed] [Google Scholar]
- Martin TF (1998) Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu Rev Cell Dev Biol 14: 231–264 [DOI] [PubMed] [Google Scholar]
- Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol 134: 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott M, Wakelam MJO, Morris AJ (2004) Phospholipase D. Biochem Cell Biol 82: 225–253 [DOI] [PubMed] [Google Scholar]
- Meijer HJG, Berrie CP, Iurisci C, Divecha N, Musgrave A, Munnik T (2001) Identification of a new polyphosphoinositide in plants, phosphatidylinositol 5-monophosphate (PtdIns5P), and its accumulation upon osmotic stress. Biochem J 360: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HJG, Munnik T (2003) Phospholipid-based signaling in plants. Annu Rev Plant Biol 54: 265–306 [DOI] [PubMed] [Google Scholar]
- Mikami K, Katagiri T, Iuchi S, Yamaguchi-Shinozaki K, Shinozaki K (1998) A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J 15: 563–568 [DOI] [PubMed] [Google Scholar]
- Mitchell CA, Gurung R, Kong AM, Dyson JM, Tan A, Ooms LM (2002) Inositol polyphosphate 5-phosphatases: lipid phosphatases with flair. IUBMB Life 53: 25–36 [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C (2002) Inositol phospholipid metabolism in Arabidopsis: characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130: 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T (2001) Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci 6: 227–273 [DOI] [PubMed] [Google Scholar]
- Munnik T, Meijer HJG (2001) Osmotic stress activates distinct lipid and MAPK signalling pathways in plants. FEBS Lett 498: 172–178 [DOI] [PubMed] [Google Scholar]
- Munnik T, Meijer HJG, ter Riet B, Hirt H, Frank W, Bartels D, Musgrave A (2000) Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J 22: 147–154 [DOI] [PubMed] [Google Scholar]
- Murata Y, Pei Z-M, Mori IC, Schroeder J (2001) Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13: 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Osmond BC, Botstein D (1989) Suppressors of yeast actin mutations. Genetics 121: 659–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD (2000) Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci 25: 229–235 [DOI] [PubMed] [Google Scholar]
- Oliver D, Lien C-C, Soom M, Baukrowitz T, Jonas P, Fakler B (2004) Functional conversion between A-type and delayed rectifier K+ channels by membrane lipids. Science 304: 265–270 [DOI] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schoffl F (2002) Heat stress and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Gu Y, Lee Y, Yang Z, Lee Y (2004) Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol 134: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Love J, Heilmann I, Thompson WF, Boss WF (2002) Up-regulation of phosphoinositide metabolism in tobacco cells constitutively expressing the human type I inositol polyphosphate 5-phosphatase. Plant Physiol 129: 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pical C, Westergren T, Dove SK, Larsson C, Sommarin M (1999) Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thaliana cells. J Biol Chem 274: 38232–38240 [DOI] [PubMed] [Google Scholar]
- Pratelli R, Sutter J-U, Blatt MR (2004) A new catch in the SNARE. Trends Plant Sci 9: 187–195 [DOI] [PubMed] [Google Scholar]
- Ristic Z, Ashworth EN (1993) Changes in leaf ultrastructure and carbohydrates in Arabidopsis thaliana L. (Heyn) cv. Columbia during rapid cold acclimation. Protoplasma 172: 111–123 [Google Scholar]
- Roth MG (2004) Phosphoinositides in constitutive membrane traffic. Physiol Rev 84: 699–730 [DOI] [PubMed] [Google Scholar]
- Ruelland E, Cantrel C, Gawer M, Kader J-C, Zachowski A (2002) Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol 130: 999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J-P, Chua N-H (2001) Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13: 1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Cui D, Wang X (2001) Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol 126: 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Zheng S, Li W, Huang B, Wang X (2001. a) Regulation of plant water loss by manipulating the expression of phospholipase D. Plant J 28: 135–144 [DOI] [PubMed] [Google Scholar]
- Simonsen A, Wurmser AE, Emr SD, Stenmark H (2001. b) The role of phosphoinositides in membrane transport. Curr Opin Cell Biol 13: 485–492 [DOI] [PubMed] [Google Scholar]
- Smolenska-Sym G, Kacperska A (1994) Phosphatidylinositol metabolism in low temperature-affected winter oilseed rape leaves. Physiol Plant 91: 1–8 [Google Scholar]
- Stefan CJ, Audhya A, Emr SD (2002) The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol Biol Cell 13: 542–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Heilmann I, Persson S, Boss WF (2000) Inositol signaling and plant growth. Trends Plant Sci 5: 252–258 [DOI] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155: 349–361 [DOI] [PubMed] [Google Scholar]
- Tall EG, Spector I, Pentyala SN, Bitter I, Rebecchi MJ (2000) Dynamics of phosphatidylinositol 4,5-bisphosphate in actin-rich structures. Curr Biol 10: 743–746 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- van der Weele CM, Spollen WG, Sharp RE, Baskin TI (2000) Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J Exp Bot 51: 1555–1562 [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Okresz L, Bogre L, Munnik T (2004) Learning the lipid language of plant signalling. Trends Plant Sci 9: 378–384 [DOI] [PubMed] [Google Scholar]
- Varnai P, Lin X, Lee SB, Tuymetova G, Bondeva T, Spat A, Rhee SG, Hajnoczky G, Balla T (2002) Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains: studies on the PH domains of phospholipase C delta 1 and p130. J Biol Chem 277: 27412–27422 [DOI] [PubMed] [Google Scholar]
- Wang X (2002) Phospholipase D in hormonal and stress signaling. Curr Opin Plant Biol 5: 408–414 [DOI] [PubMed] [Google Scholar]
- Wang X (2004) Lipid signaling. Curr Opin Plant Biol 7: 329–336 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang C, Sang Y, Qin C, Welti R (2002) Networking of phospholipases in plant signal transduction. Physiol Plant 115: 331–335 [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H-E, Rajashekar CB, Williams TD, Wang X (2002) Profiling membrane lipids in plant stress responses: role of phospholipase D-alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Westergren T, Dove SK, Sommarin M, Pical C (2001) AtPIP5K1, an Arabidopsis thaliana phosphatidylinositol phosphate kinase, synthesizes PtdIns(3,4)P2 and PtdIns(4,5)P2 in vitro and is inhibited by phosphorylation. Biochem J 359: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisstock JC, Wiradjaja F, Waters JE, Garung R (2002) The structure and function of catalytic domains within inositol polyphosphate 5-phosphatases. IUBMB Life 53: 15–23 [DOI] [PubMed] [Google Scholar]
- Wu Y, Cosgrove DJ (2000) Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J Exp Bot 51: 1543–1553 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee B-h, Ishitani M, Lee H, Zhang C, Zhu J-K (2001) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Janmey P, Frigeri LG, Han W, Fujita J, Kawakami Y, Apgar JR, Kawakami T (1999) Pleckstrin homology domains interact with filamentous actin. J Biol Chem 274: 19752–19761 [DOI] [PubMed] [Google Scholar]
- Yin HL, Janmey PA (2003) Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol 65: 761–789 [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang C, Qin C, Wood T, Olafsdottir G, Welti R, Wang X (2003) The oleate-stimulated phospholipase D, PLD{delta}, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15: 2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison WH III, Ye Z-H (2004) FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. Plant Cell 16: 3242–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye Z-H (2003) The SAC domain-containing protein gene family in Arabidopsis. Plant Physiol 132: 544–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye Z-H (2004) Molecular and biochemical characterization of three WD-repeat-domain-containing inositol polyphosphate 5-phosphatases in Arabidopsis thaliana. Plant Cell Physiol 45: 1720–1728 [DOI] [PubMed] [Google Scholar]