Abstract
Wild-type Arabidopsis (Arabidopsis thaliana L. Heynh.) roots growing on a tilted surface of impenetrable hard-agar media adopt a wave-like pattern and tend to skew to the right of the gravity vector (when viewed from the back of the plate through the medium). Reversible root-tip rotation often accompanies the clockwise and counterclockwise curves that form each wave. These rotations are manifested by epidermal cell file rotation (CFR) along the root. Loss-of-function alleles of ROOT HAIR DEFECTIVE3 (RHD3), a gene previously implicated in the control of vesicle trafficking between the endoplasmic reticulum and the Golgi compartments, resulted in an almost complete suppression of epidermal CFR, root skewing, and waving on hard-agar surfaces. Several other root hair defective mutants (rhd2-1, rhd4-1, and rhd6-1) did not exhibit dramatic alterations in these root growth behaviors, suggesting that a generalized defect in root hair formation is not responsible for the surface-dependent phenotypes of rhd3. However, similar alterations in root growth behavior were observed in a variety of mutants characterized by defects in cell expansion (cob-1, cob-2, eto1-1, eto2-1, erh2-1, and erh3-1). The erh2-1 and rhd3-1 mutants differed from other anisotropic cell expansion mutants, though, by an inability to respond to low doses of the microtubule-binding drug propyzamide, which normally causes enhanced left-handed CFR and right skewing. We hypothesize that RHD3 may control epidermal CFR, root skewing, and waving on hard-agar surfaces by regulating the traffic of wall- or plasma membrane-associated determinants of anisotropic cell expansion.
The primary roots of Arabidopsis (Arabidopsis thaliana L. Heynh.) seedlings display different growth behaviors depending on the conditions to which they are exposed. If grown within a homogenous environment (such as liquid or penetrable agar media), primary roots will grow downward in response to gravity (Blancaflor and Masson, 2003). When forced to grow on the surface of impenetrable hard-agar media, they will display more complex growth behaviors with characteristics dictated by the angle of the surface (Okada and Shimura, 1990). On surfaces tilted slightly forward, the roots of commonly used ecotypes will skew their growth to the right of the vertical when viewed through the agar medium, as per the established convention (Rutherford and Masson, 1996; Simmons et al., 1996). If the surface is tilted backward, roots will form waves in addition to skewing. This pattern results from the root tip pressing on the surface when gravitropism directs downward growth (Okada and Shimura, 1990, 1992; Rutherford and Masson, 1996; Simmons et al., 1996; Thompson and Holbrook, 2004). Root waving is usually accompanied by a reversible rotation of the tip about its axis, which manifests itself by a twisting of epidermal cell files (cell file rotation [CFR]) along the root (Okada and Shimura, 1990). While curving to the right, roots tend to display left-handed CFR. By contrast, roots curving to the left display right-handed CFR (Rutherford and Masson, 1996; Simmons et al., 1996). This trend is exacerbated when seedlings are grown on horizontal hard-agar surfaces, a condition that promotes counterclockwise root coiling and left-handed CFR (Migliaccio and Piconese, 2001).
While a strong correlation has been observed between CFR handedness and direction of root curving, the cause-effect relationship between them remains controversial. Even though CFR precedes root curvature (Mochizuki et al., 2005), suggesting the former might contribute to the latter, conditions have also been identified in which no CFR could be observed despite strong waving and skewing (Migliaccio and Piconese, 2001; Buer et al., 2003) or where CFR occurs in the absence of curving (Mullen et al., 1998; Sedbrook et al., 2002). It has been proposed that CFR illustrates a circumnutation-like process that contributes to waving and skewing on hard-agar surfaces (Rutherford and Masson, 1996; Simmons et al., 1996; Thompson and Holbrook, 2004). Alternatively, CFR and root bending may be mechanical consequences of impedance to root-tip movement by the agar surface (Thompson and Holbrook, 2004) or a secondary effect of the helical circumnutational movement of roots due to the necessity of discharging the energy accumulated by a flattening of the helix on the agar surface (Migliaccio and Piconese, 2001).
The cellular basis for root CFR remains poorly understood. One model proposes that CFR might result from differences in anisotropic cell expansion between epidermis and more internal tissues (Furutani et al., 2000). A radial gradient in anisotropic cell expansion might generate mechanical tensions between tissues, resulting in tissue breakage and plant death unless the epidermal cell files twist around the ground tissue. The handedness of CFR could originate in the helical alignment of cortical microtubules in epidermal cells at the basal side of the central elongation zone (CEZ). There, microtubules shift their alignment from transverse in most elongating cells of the CEZ to longitudinal in the mature zone (Baskin et al., 1994; Furutani et al., 2000). This helical alignment of cortical microtubules in CEZ epidermal cells would dictate a parallel deposition of cellulose microfibrils in the wall, as proposed by the cellulose synthase constraint model (Giddings and Staehelin, 1991), thereby restraining expansion to a direction perpendicular to these networks at an angle to the longitudinal axis of the root and resulting in CFR.
Several observations support a role for cortical microtubules at the basal side of the CEZ in controlling CFR. First, most CFR- and root-skewing mutants have defects in microtubule organization and/or turnover dynamics. For example, the spr1/sku6, spr2, and wvd2-1 mutants, which show right-handed CFR and skew to the left on hard-agar surfaces, display left-handed cortical microtubule arrays in CEZ epidermal cells (Furutani et al., 2000; Yuen et al., 2003). These mutations affect various microtubule-interacting proteins (Buschmann et al., 2004; Nakajima et al., 2004; Sedbrook et al., 2004). Conversely, the lefty1 and lefty2 mutants, which harbor destabilized cortical microtubule networks due to defects in the α-tubulin genes TUA6 and TUA4, respectively, display enhanced right-skewing phenotypes associated with right-handed cortical microtubule arrays in epidermal cells of the basal CEZ (Thitamadee et al., 2002). Similarly, altering the dynamics of microtubule turnover in wild-type Arabidopsis with drugs (oryzalin or propyzamide) or by transformation with constructs directing expression of green fluorescent protein-tubulin or tubulin-binding proteins results in right-handed helical arrangement of cortical microtubules in CEZ epidermal cells, exacerbated left-handed CFR, and enhanced root skewing to the right on agar surfaces (Furutani et al., 2000; Hashimoto, 2002).
However, recent observations suggest a more complex relationship between orientation of cortical microtubules, cellulose microfibrils, and CFR. First, analysis of the temperature-sensitive mor1-1 mutant, which affects a microtubule-binding protein (Sugimoto et al., 2003), showed that root CFR can occur under restrictive temperature despite microtubule disruption. Further analysis of mor1-1 rsw1-1 double mutants also suggested that microtubules can promote axial growth by a mechanism separate from cellulose microfibril alignment (Sugimoto et al., 2003). Second, roots of seedlings homozygous for sku6, an allele of spr1, displayed transverse microtubule alignment in basal CEZ cells despite maintaining a substantial left-handed CFR (Sedbrook et al., 2004). Third, the degree of pitch of cortical microtubules in wild-type root basal CEZ did not correlate with the direction and amount of CFR (Sedbrook et al., 2004). Fourth, genetically or pharmacologically induced left-handed CFR always appears to override right-handed CFR, constituting an intermediate step toward increased microtubule disruption and enhanced radial swelling (Wasteneys, 2004).
These contradictory results led Wasteneys (2004) to propose that CFR handedness might originate in an inherent torsional handedness in roots distinct from the alignment of cortical microtubules or cellulose microfibrils. In this model, the correlation observed in handedness between the helical alignment of cortical microtubules in CEZ cells and CFR could simply reflect signaling between cortical microtubule arrays and wall components. CFR might be induced either by an influence of cortical microtubules on anisotropic cell expansion or by constraints in the cell wall modulating the alignment of cortical microtubules near the plasma membrane (Fisher and Cyr, 1998).
If the latter model is correct, one predicts the existence of mutations that affect CFR without altering organization of the cortical microtubule network in expanding cells of the CEZ. Such mutations could affect regulators of anisotropic cell expansion unrelated to the cortical microtubules. Mutations affecting the trafficking of wall biogenesis precursors or wall-remodeling enzymes, the synthesis of wall biogenesis enzymes, or the activity of factors determining wall extensibility would fall into this category. Alternatively, the inherent torsional handedness proposed in this model may be related to the helical patterns resulting from periclinal divisions of lateral root cap/epidermis initials in the root meristem (Rutherford and Masson, 1996; Cnops et al., 2000; Wasteneys and Collings, 2004).
In this article, we report the isolation and characterization of the wave dampened6 (wvd6) mutation, which inhibits root waving, skewing, and CFR on hard-agar surfaces without altering the organization of the cortical microtubule network in cells of the CEZ. We show that mutations affecting several aspects of cell expansion regulation also inhibit CFR, root skewing, and waving on hard-agar surfaces. Tested mutants include erh3-1, which is defective in the organization of cortical microtubules (Burk et al., 2001); cob-1 and cob-2, deficient in the deposition of crystalline cellulose (Schindelman et al., 2001); rhd1-1/reb1, affected in the galactosylation of wall xyloglucan and type-II arabinogalactan in root epidermal cells (Seifert et al., 2002); and eto1-1 and eto2-1, which overproduce ethylene (Shibaoka, 1994; Chae et al., 2003), a hormone that alters the alignment of cortical microtubules (Shibaoka, 1994) and regulates root waving (Buer et al., 2003). However, the wvd6 and erh2 mutants differ from other cell expansion mutants in that they lack the left-handed CFR and enhanced root skewing normally seen in response to low doses of propyzamide. wvd6 affects ROOT HAIR DEFECTIVE3 (RHD3), a gene that regulates root hair development and anisotropic cell expansion in all organs of the plant (Wang et al., 1997). The amino terminus of the 89-kD RHD3 protein contains GTP-binding motifs (Wang et al., 1997), and this protein has been implicated in controlling the transport of vesicles between the endoplasmic reticulum and the Golgi apparatus of most cell types, thereby contributing to cell wall biogenesis and anisotropic cell expansion (Hu et al., 2003; Zheng et al., 2004). We propose that RHD3 might regulate root growth behavior on hard-agar surfaces by mediating the transport of wall- or plasma membrane-associated determinants of anisotropic cell expansion.
RESULTS
Identification and Phenotypic Characterization of wvd6
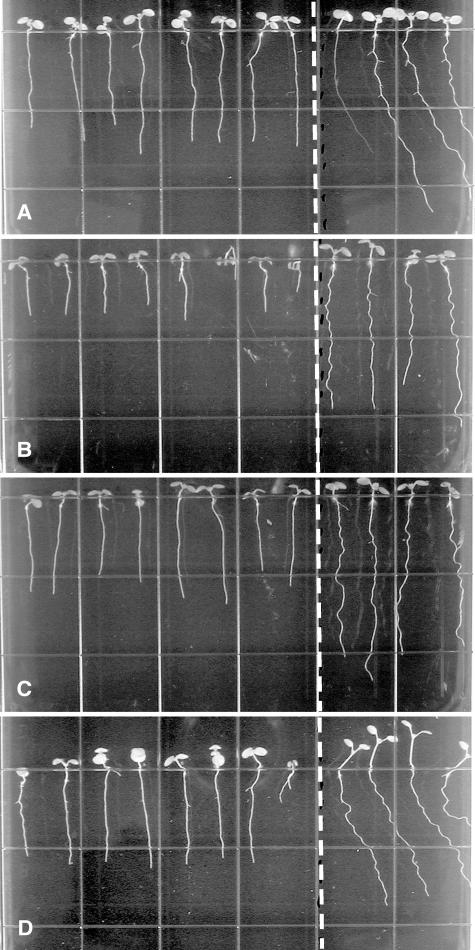
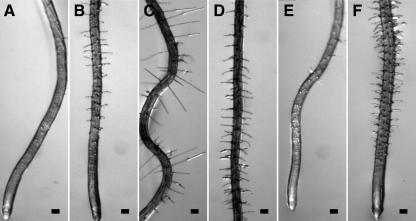
The wvd6 mutant was identified by screening a collection of T-DNA-mutagenized seedlings for altered root growth phenotypes on inclined agar surfaces (Sedbrook et al., 2002). Under these conditions, the roots of wild-type Wassilewskija (Ws) seedlings adopt a wavy pattern of growth and skew moderately to the right, and these movements are accompanied by reversible CFR at the tip (Simmons et al., 1995; Rutherford and Masson, 1996). wvd6 roots, on the other hand, displayed a dramatic reduction in skewing and waving in this assay, and did not show much evidence of CFR (Figs. 1A and 2B). Also, wvd6 roots were noticeably shorter than their wild-type counterparts (Fig. 1A). Comparison of wvd6 and wild-type root growth over a 48-h period indicated that wvd6 roots reached a length that was only 66% of that reached by wild-type Ws roots (Table I). A slight but statistically significant increase in root diameter was also observed (Table I). Additionally, the length of mature wvd6 root hairs (Fig. 2D) was dramatically shorter than wild type (Fig. 2C).
Figure 1.
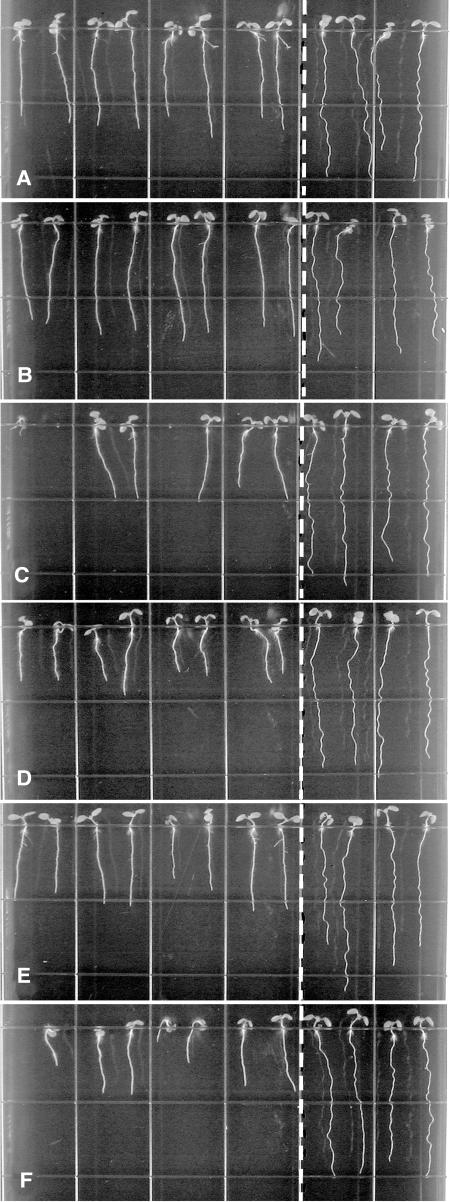
Root-waving phenotypes of wvd6 and rhd3. A, wvd6 (Ws ecotype); B, rhd3-1 (Col ecotype); C, rhd3-2 (Col ecotype); and D, rhd3-3 (No-0 ecotype). Mutant seedlings were plated left of the dotted line, while wild-type seedlings of the same ecotype were plated on the right. Seedlings were grown for 8 to 9 d on 1.5% (w/v) agar-solidified medium, tilted backward 30° from the vertical. Images were taken from the bottom of the plates through the medium.
Figure 2.
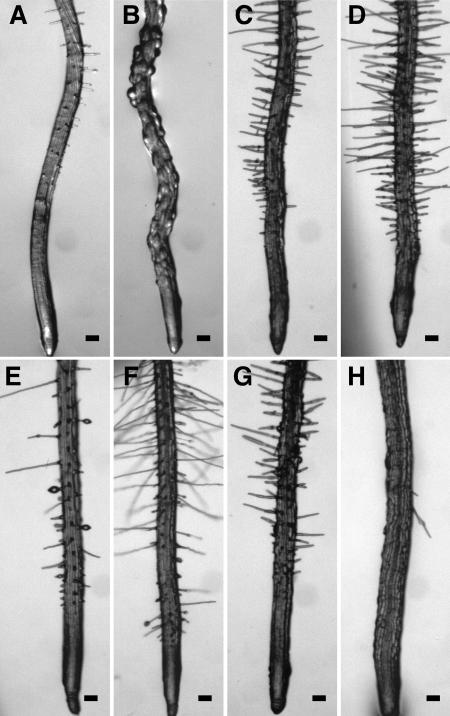
Epidermal CFR phenotypes of wvd6 and rhd3 roots. Seedlings were grown on a tilted agar surface and photographed 9 DAG. Root images were taken either at the tip (A, B, E, and F) or at a position within the mature zone where root hair length was maximal (C and D). A and C, Wild-type Ws; B and D, wvd6; E, wild-type No-0; and F, rhd3-3. Scale bars = 100 μm.
Table I.
Quantification of root elongation and radial expansion phenotypes
Elongation, Mean root growth assessed over a 48-h interval starting at 5 DAG. Diameter, Mean diameters of roots were obtained within their mature zones 8 DAG. Data are presented as means ± ses. The percentages of mutant root elongation or root diameter, relative to the mean wild-type values, are provided in the columns to the right, with the P values for two-tailed t-test analyses between mutant and wild-type root data given below in parentheses. The ecotypes of wvd6 and rhd3-3 are Ws and No-0, respectively. The ecotype of the remaining mutants is Col. n, sample size.
| Elongation | % Wild-Type Elongation | Diameter | % Wild Type Diameter | |
|---|---|---|---|---|
| mm | μm | |||
| Ws | 9.2 ± 0.2 (n = 31) | 100 | 104.1 ± 1.1 (n = 12) | 100 |
| wvd6 | 6.1 ± 0.1 (n = 34) | 66.4% (P = 8.5E-17) | 117.1 ± 2.2 (n = 12) | 112.5% (P = 8.5E-05) |
| No-0 | 8.7 ± 0.3 (n = 30) | 100 | 101.2 ± 1.9 (n = 12) | 100 |
| rhd3-3 | 5.0 ± 0.1 (n = 33) | 56.8% (P = 3.5E-13) | 136.2 ± 1.6 (n = 12) | 134.5% (P = 2.3E-12) |
| Col | 10.3 ± 0.1 (n = 84) | 100 | 115.2 ± 1.8 (n = 12) | 100 |
| rhd1-1 | 3.6 ± 0.1 (n = 24) | 34.7% (P = 6.2E-66) | 113.8 ± 3.3 (n = 12) | 98.8% (P = 0.71) |
| cob-1 | 7.1 ± 0.1 (n = 28) | 68.7% (P = 6.9E-29) | 121.1 ± 2.2 (n = 12) | 105.1% (P = 0.048) |
| cob-2 | 7.3 ± 0.2 (n = 29) | 70.6% (P = 1.0E-16) | 125.4 ± 2.0 (n = 12) | 108.9% (P = 9.8E-04) |
| erh2-1 | 4.7 ± 0.1 (n = 28) | 45.5% (P = 9.5E-51) | 135.0 ± 2.5 (n = 12) | 117.2% (P = 3.2E-06) |
| erh3-1 | 3.8 ± 0.1 (n = 18) | 36.9% (P = 3.3E-50) | 203.0 ± 3.8 (n = 12) | 176.2% (P = 8.4E-13) |
| eto1-1 | 6.1 ± 0.3 (n = 25) | 59.8% (P = 3.8E-13) | 124.3 ± 1.8 (n = 12) | 107.9% (P = 1.6E-03) |
| eto2-1 | 4.0 ± 0.2 (n = 31) | 39.1% (P = 3.2E-35) | 132.2 ± 3.3 (n = 12) | 114.7% (P = 3.2E-04) |
The wvd6 Mutation Is Caused by a Reciprocal Translocation That Disrupts RHD3
The roots of F1 progeny resulting from crosses between wvd6 and wild-type (Ws) plants were phenotypically indistinguishable from wild type on inclined agar surfaces. Analysis of the F2 generation indicated that wvd6 is recessive and segregates as a single nuclear mutation (data not shown). Analysis of the root-waving and kanamycin-resistance (kanr) phenotypes of 89 F4 families (derived from a plant heterozygous for wvd6 in the F2 generation) showed that wvd6 cosegregated with the neomycin phosphotransferaseII (kanr) marker found on the pGKB5 T-DNA (Bouchez et al., 1993). Southern-blot analysis of EcoRI-, EcoRV-, HincII-, or ScaI-digested wvd6 genomic DNA using a fragment corresponding to the BASTA gene as a probe indicated the presence of two concatameric T-DNAs arranged in a head-to-tail configuration (data not shown).
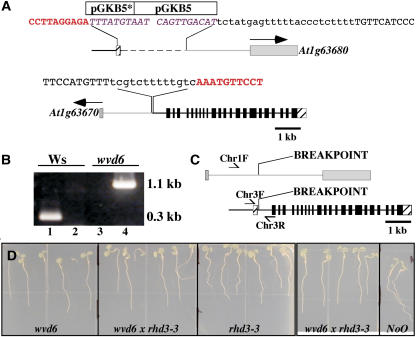
Molecular cloning and characterization of sequences flanking the T-DNA inserts in wvd6 detected a reciprocal translocation between chromosomes 1 and 3, with one translocated chromosome harboring the tandem T-DNA insertion and the other lacking any apparent T-DNA sequences (Fig. 3A). The translocation breakpoint in the tandem T-DNA-carrying chromosome is located within the first intron of RHD3 adjacent to the T-DNA insert (Fig. 3, A and C).
Figure 3.
wvd6 is caused by a translocation that disrupts RHD3. A, Structure of the wvd6 translocation. The top image represents the arrangement of DNA on the translocated chromosome carrying the chromosome 3 centromere, while the bottom image represents the DNA arrangement on the chromosome carrying the chromosome 1 centromere. The 5′ and 3′ untranslated regions of RHD3 are represented by striped boxes and the coding region of RHD3 by black boxes. Gray boxes are used to represent the predicted first exons of At1g63670 and At1g63680, with arrows indicating the direction of transcription. The gray lines and boxes correspond to DNA originating from chromosome 1, and the black lines and boxes correspond to DNA originating from chromosome 3. Note that the pGKB5 T-DNA inserts are not drawn to scale. The asterisk (*) denotes the truncated T-DNA. The sequences shown represent the various junctions between chromosome 1, chromosome 3, and pGKB5. Sequences in red bold correspond to RHD3 (chromosome 3); sequences in purple italics are from pGKB5; uppercase black sequences are from chromosome 1; and lowercase black sequences are of unknown origin. B, PCR-based confirmation of the wvd6 translocation. Wild-type Ws genomic DNA served as the template for the PCR reactions loaded in lanes 1 and 2, and wvd6 genomic DNA was used for lanes 3 and 4. The primer pair used for lanes 1 and 3 was Chr3F and Chr3R, while Chr1F and Chr3R were used for lanes 2 and 4 (see “Materials and Methods”). Their positions on wild-type chromosomes 1 (top) and 3 (bottom) are depicted by arrows in C, which also shows the translocation breakpoints in the wvd6 mutant. D, From left to right, root growth behavior of 7-d-old wvd6, wvd6 × rhd3-3 (F1), rhd3-3, wvd6 × rhd3-3 (F1), and wild-type No-0 seedlings grown on tilted hard-agar surfaces showing that wvd6 and rhd3-3 do not complement each other.
To verify that other loss-of-function mutations at RHD3 also affect root growth behavior on hard-agar surfaces, we analyzed root waving, skewing, and CFR of mutant seedlings carrying the previously identified mutant alleles rhd3-1, rhd3-2, and rhd3-3 (Schiefelbein et al., 1997). rhd3-1 and rhd3-2 are missense mutations within exons 18 (A → V) and 8 (D → N) of the gene, respectively, whereas rhd3-3 is a frameshift mutation within exon 5 and is likely a null (Wang et al., 1997). We found that rhd3-1 and rhd3-2 completely suppress CFR, root waving, and skewing on tilted hard-agar surfaces, whereas rhd3-3 seedlings display only minimal root skewing, waving, and CFR under the same conditions (Figs. 1, B–D, and 2, E and F; Table II). Moreover, like wvd6 roots, rhd3-1, rhd3-2, and rhd3-3 roots grew to be shorter than wild-type roots on hard-agar media (Fig. 1, A–D; Table I).
Table II.
Mean root skewing in the presence or absence of propyzamide
Angles of root skewing were measured for seedlings grown on 1.5% agar plates tilted backward 30°, 9 DAG. Plates contained 0.05% DMSO and either 0 or 3 μm propyzamide, as indicated. Negative values represent slanting to the left of the vertical and positive values to the right of the vertical, as viewed from the bottom of the plate. All results are mean values ± ses, and n = sample size. Statistical significance of differences in mean root-skewing angles between mutant and wild-type seedlings are indicated as two-tailed t-test P values in the third and fifth columns for the 0 and 3 μm propyzamide treatments, respectively. Δ values were obtained by subtracting the mean root-skewing angle on 0 μm propyzamide from the mean angle on 3 μm propyzamide. The P values in the sixth column indicate the statistical significance of the observed differences (two-tailed t-test analyses). PPM, Propyzamide. –, Not determined.
| Skew Angle (°) | P Values: Mutant versus Wild Type | Skew Angle (°) | P Values: Mutant versus Wild Type | Δ (°) | |
|---|---|---|---|---|---|
| 0 μm PPM | 0 μm PPM | 3 μm PPM | 3 μm PPM | ||
| Ws | 21.8 ± 1.1 (n = 31) | 30.1 ± 0.9 (n = 30) | 8.4 (P = 1.8E-07) | ||
| wvd6 | −1.6 ± 0.8 (n = 44) | P = 4.7E-29 | 0.5 ± 0.8 (n = 46) | P = 2.98E-36 | 2.1 (P = 0.065) |
| No-0 | 23.0 ± 1.1 (n = 35) | 33.6 ± 1.5 (n = 34) | 10.6 (P = 4.0E-07) | ||
| rhd3-3 | 3.4 ± 0.7 (n = 43) | P = 1.4E-22 | 2.8 ± 0.9 (n = 39) | P = 1.33E-23 | −0.6 (P = 0.58) |
| Col | 2.9 ± 0.3 (n = 153) | 17.2 ± 0.8 (n = 132) | 14.3 (P = 2.2E-36) | ||
| rhd1-1 | −1.2 ± 1.2 (n = 43) | P = 0.0015 | 9.5 ± 2.2 (n = 35) | P = 0.0023 | 10.7 (P = 8.7E-05) |
| cob-1 | 4.3 ± 0.7 (n = 45) | P = 0.047 | 23.3 ± 1.1 (n = 39) | P = 2.54E-05 | 19.0 (P = 4.1E-22) |
| cob-2 | 7.5 ± 0.7 (n = 45) | P = 6.5E-10 | 21.5 ± 0.9 (n = 42) | P = 0.0005 | 14.0 (P = 2.1E-20) |
| erh2-1 | 2.2 ± 0.8 (n = 41) | P = 0.43 | −0.1 ± 0.8 (n = 39) | P = 5.82E-29 | −2.3 (P = 0.057) |
| erh3-1 | 0.8 ± 1.5 (n = 18) | P = 0.17 | – | – | – |
| eto1-1 | 6.6 ± 1.3 (n = 33) | P = 0.008 | 9.9 ± 0.9 (n = 34) | P = 1.09E-07 | 3.3 (P = 0.041) |
| eto2-1 | 8.4 ± 1.4 (n = 42) | P = 0.0004 | 13.2 ± 0.9 (n = 29) | P = 0.0017 | 4.9 (P = 4.6E-03) |
To determine whether wvd6 and rhd3 are allelic, we crossed homozygous wvd6 and rhd3-3 plants. The F1 progeny of these crosses showed root growth patterns similar to those of wvd6 and rhd3-3 single mutants (Fig. 3D). Since both wvd6 and rhd3-3 are recessive mutations, their inability to complement strongly suggests that the RHD3 disruption is the cause of the wvd6 phenotype.
Differential Root Growth Behaviors in Root Hair and Root Expansion Mutants
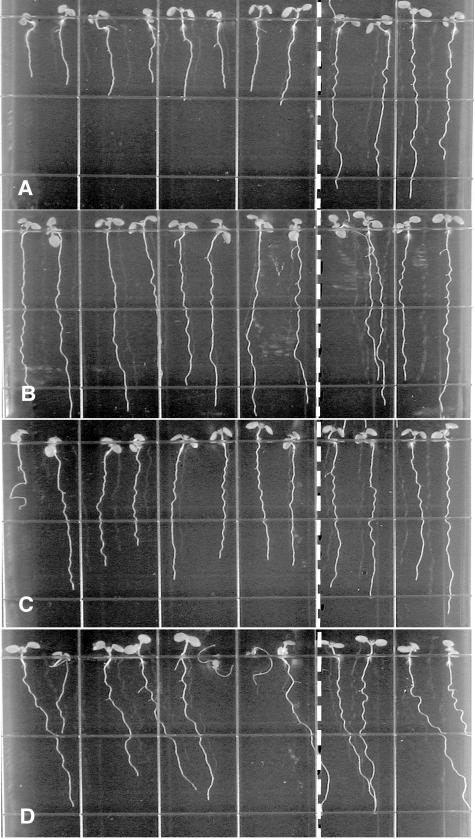
Root hairs may help anchor the root to its substratum, thereby restricting root movement to its subapical region. Therefore, we tested the possibility that improper root hair development might influence root waving by analyzing the root growth patterns of several root hair defective mutants (rhd1-1, rhd2-1, rhd4-1, and rhd6) on tilted hard-agar surfaces. The roots of rhd2-1 and rhd4-1, which develop bulges at the base of their hairs (Schiefelbein and Somerville, 1990), exhibit a wavy growth phenotype similar to that of wild-type Columbia (Col) roots (Fig. 4, B and C). Similarly, rhd6 roots, which are defective in root hair initiation (Masucci and Schiefelbein, 1994), also retain the ability to wave (Fig. 4D). Only rhd1-1 roots, which form bulging trichoblastic cells (Schiefelbein and Somerville, 1990), consistently fail to develop a normal wavy growth pattern under these conditions (Fig. 4A). This analysis indicates that the ability to form root hairs does not significantly affect root waving.
Figure 4.
Root-waving phenotypes of root hair defective mutants. A, rhd1-1 (Col ecotype); B, rhd2-1 (Col ecotype); C, rhd4-1 (Col ecotype); and D, rhd6 (Ws ecotype). Mutant seedlings were plated left of the dotted line, while wild-type seedlings of the same ecotype were plated on the right. Seedlings were grown for 8 to 9 d on 1.5% (w/v) agar-solidified medium, tilted backward 30° from the vertical. Images were taken from the bottom of the plates through the medium.
Unlike rhd2-1, rhd4-1, and rhd6, the rhd1-1 mutant exhibits a severe root elongation defect but no alteration in root radial expansion (Fig. 4A; Table I). rhd1-1 roots are not straight, like rhd3 roots, but instead exhibit rhythmic curves that resemble root waving, albeit with greatly reduced amplitude (Fig. 6B). Unfortunately, the cell-bulging phenotype of the mutant prevents determination of CFR. Thus, the extent of similarity between rhd1-1 and wild-type root waving remains uncertain.
Figure 6.
Epidermal CFR phenotypes of root expansion mutants. Seedlings were grown on an agar surface tilted backward by 30° and photographed 9 DAG. All root images were taken at the tip. A, Wild-type Col; B, rhd1-1; C, cob-1; D, cob-2; E, eto1-1; F, eto2-1; G, erh2-1; and H, erh3-1. Scale bars =100 μm.
We also analyzed the root-waving phenotypes of several mutants defective in root anisotropic cell expansion, but not in root hair development. Under our growth conditions, root elongation is significantly reduced for cob-1, cob-2 (Hauser et al., 1995), eto1-1, eto2-1 (Guzman and Ecker, 1990), erh2-1, and erh3-1 (Schneider et al., 1997; Fig. 5, A–E; Table I). The roots of erh3-1 also exhibit a strong radial expansion phenotype (Fig. 6H), whereas more subtle (but statistically significant) radial expansion defects are observed in the case of cob-1, cob-2, eto1-1, eto2-1, and erh2-1 roots (Fig. 6, C–G; Table I). Note that our growth conditions (1.5% Suc) do not permit for the dramatic radial root-tip swelling phenotype of cob-1 and cob-2 mutants reported on media containing higher levels (4.5%) of Suc (Hauser et al., 1995).
Figure 5.
Root-waving phenotypes of root expansion mutants. A, cob-1; B, cob-2; C, eto1-1; D, eto2-1; E, erh2-1; and F, erh3-1. The ecotype background of all of these mutants is Col. Mutant seedlings were plated left of the dotted line, while wild-type Col seedlings were plated on the right. Seedlings were grown for 8 to 9 d on 1.5% (w/v) agar-solidified medium, tilted backward 30° from the vertical. Images were taken from the bottom of the plates through the medium.
With the exception of wvd6 (Ws ecotype) and rhd3-3 (Nossen [No-0] ecotype), all root-expansion mutants examined here were isolated in the Col background. The roots of wild-type Col seedlings typically exhibit little rightward root skewing on hard-agar surfaces under our growth conditions (Table II; Rutherford and Masson, 1996). While we find that erh2-1 and erh3-1 roots also display low or insignificant levels of root skewing, cob-1, cob-2, eto1-1, and eto2-1 roots skew slightly more to the right than wild type (Table II). Root epidermal CFR (Fig. 6, C–H) and root waving (Fig. 5, A–F) are strongly suppressed in all mutants.
Organization of the Microtubule Network in Radial Expansion Mutants
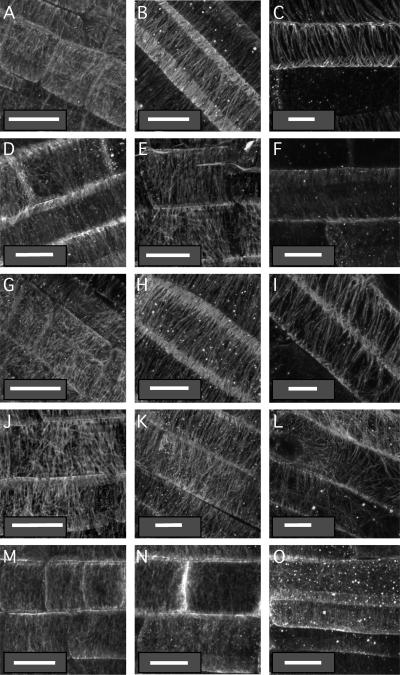
The orientation of cortical microtubules within the root CEZ may govern CFR and the direction of cell elongation (Furutani et al., 2000). Thus, we analyzed epidermal microtubule orientation in the root maturation zone (region where root hairs initiate and develop), the CEZ (defined here as a subapical zone of the root located distally to the site of root hair initiation and including cells that take a clearly oblong shape), and distal elongation zone (DEZ; located between the root CEZ and its promeristem) of Col, rhd3-1, cob-1, erh2-1, and eto1-1 roots. The epidermal cortical microtubule orientation of wild-type Col roots shifts from disorganized in the DEZ (Fig. 7A), and often transverse in the CEZ (Fig. 7B), to right-handed helical in the maturation zone where root hairs initiate (Fig. 7C). In general, root epidermal microtubule organization in cob-1 and rhd3-1 is similar to wild type (Fig. 7, D–I). However, under our growth conditions, we do observe a substantially shorter CEZ in all mutants compared to wild type. The CEZ of cob-1 is longest among all mutants, and most of its epidermal cells display transverse microtubules (Fig. 7, G–I; data not shown). On the other hand, the CEZ of eto1-1 roots is much smaller than wild type due to rapid transition from DEZ through the mature zone (Fig. 7, M–O; data not shown). Reduced CEZ is also observed in rhd3-1 and erh2-1 roots, although the reduction is not as strong (Fig. 7, D–F and J–L). In summary, although we observed significant differences in the size of the CEZ within these mutants compared to wild type, cells within specific zones of the root tip retain wild-type cortical microtubule orientations.
Figure 7.
Organization of the cortical microtubule network in wild-type and mutant root tips. Seedlings grown on vertical agar surfaces were subjected to the immunolocalization protocol described in “Materials and Methods” and stained with mouse antitubulin antibodies. Micrographs were taken in the middle of the distal elongation zone (meristem; left column, A, D, G, J, and M), in the middle of the central elongation zone (central column, B, E, H, K, and N), or at the maturing zone where root hairs initiate (right column, C, F, I, L, and O) of wild-type Col (A–C), rhd3-1 (D–F), cob-1 (G–I), erh2-1 (J–L), and eto1-1 (M–O) 5-d-old seedlings. White bars = 10 μm.
Effects of Propyzamide on Root Skewing and Cortical Microtubule Orientation
Enhanced rightward root skewing is observed when wild-type Landsberg erecta seedlings are grown on media containing 3 μm propyzamide, a β-tubulin-binding pronamide that affects microtubule dynamics and reorients microtubules in elongating epidermal cells to right-handed helices (Furutani et al., 2000; Nakamura et al., 2004). The latter observation was used to support a role for the alignment of cortical microtubules in CEZ epidermal cells in the control of root skewing on hard-agar surfaces (Furutani et al., 2000).
Under our conditions, 3 μm propyzamide has a similar effect on the skewing of wild-type No-0, Ws, and Col roots (Table II). This treatment also promotes dramatic increases in rightward skewing for cob-1, cob-2, and rhd1-1 roots (Table II), and minor, although statistically significant, increases in rightward skewing for eto1-1 and eto2-1 roots (Table II). On the other hand, propyzamide does not induce rightward skewing of erh2-1, rhd3-3, and wvd6 roots (Table II). We were unable to determine the skewing phenotype of erh3 under 3 μm propyzamide because mutant roots are too severely affected by this treatment (showing drastically increased radial expansion and decreased elongation) to permit accurate measurements of root skewing, even after 9 d of growth (data not shown). Severe disorganization of both cortical microtubule and cellulose microfibril alignment has been reported in the root cells of fra2 (Burk and Ye, 2002), which is allelic to erh3-1 (Webb et al., 2002) and is defective in a gene encoding a katanin-like microtubule-severing protein (Burk et al., 2001).
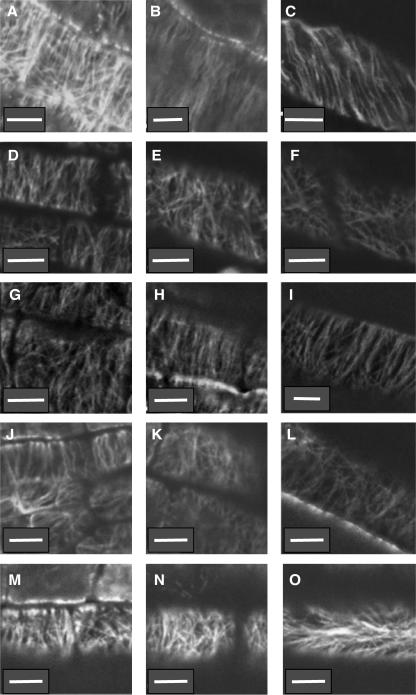
We analyzed the effects of 3 μm propyzamide on root epidermal cell cortical microtubule orientation of 5-d-old wild-type Col, rhd3-1, cob-1, erh2-1, and eto1-1 seedlings by whole-mount immunoconfocal microscopy (Fig. 8). Microtubule orientations in the DEZ, CEZ, and mature zone of cob-1 roots grown on 3 μm propyzamide are similar to that observed in wild-type Col roots and transits from disorganized in the DEZ and transverse in the CEZ to right-handed helical in the mature zone (Fig. 8, A–C and G–I, respectively). By contrast, the cortical microtubule network appears relatively disorganized in the CEZ and mature zones of propyzamide-treated rhd3-1, erh2-1, and eto1-1 seedlings (Fig. 8, E and F, K and L, and N and O, respectively).
Figure 8.
Organization of the cortical microtubule network in root tips of wild-type and mutant seedlings treated with 3 μm propyzamide. Seedlings grown on vertically oriented plates containing growth media with 3 μm propyzamide were used to analyze organization of root epidermal cell cortical microtubules by whole-mount immunoconfocal microscopy. Micrographs were taken in the middle of the distal elongation zone (left column, A, D, G, J, and M), in the middle of the central elongation zone (central column, B, E, H, K, and N), or at the maturing zone where root hairs initiate (right column, C, F, I, L, and O) of 5-d-old wild-type Col (A–C), rhd3-1 (D–F), cob-1 (G–I), erh2-1 (J–L), and eto1-1 (M–O) seedlings. White bars = 10 μm.
DISCUSSION
The wvd6 mutation suppresses root epidermal CFR, skewing, and waving on hard-agar surfaces and alters anisotropic cell expansion in all organs of the plant (Figs. 1 and 2; Tables I and II; data not shown). In this regard, wvd6 differs from most other root-skewing mutations characterized to date, which either enhance root skewing in the wild-type direction (sku1, sku2, sku3, sku5, lefty1, lefty2, mor1; Rutherford and Masson, 1996; Sedbrook et al., 2002; Thitamadee et al., 2002; Sugimoto et al., 2003) or promote right-handed CFR and leftward root skewing (spr1/sku6, spr2, wvd2-1; Furutani et al., 2000; Yuen et al., 2003; Sedbrook et al., 2004; Shoji et al., 2004). The complete elimination of root CFR, waving, and skewing in wvd6 suggests that WVD6/RHD3 is essential for development of these complex root growth behaviors on tilted hard-agar surfaces and has a distinct cellular function from proteins encoded by other root-skewing genes. wvd6 roots exhibit slightly reduced reorientation kinetics upon gravistimulation (data not shown). However, this defect is not likely to be responsible for the absence of rhd3 root skewing since reduced gravitropism would be expected to increase, rather than decrease, rightward root skewing (Simmons et al., 1995; Rutherford and Masson, 1996).
wvd6 is caused by a reciprocal translocation between chromosomes 1 and 3, with one of the recombined chromosomes carrying a tandemly duplicated T-DNA near the translocation breakpoint, which is located within the first intron of RHD3 (Fig. 3). It is common for complex chromosomal rearrangements of this nature to occur following T-DNA mutagenesis (Tax and Vernon, 2001; Guan et al., 2003). Several lines of evidence suggest that the wvd6 mutant phenotype is due to the disruption of RHD3. First, both wvd6 and previously characterized loss-of-function alleles of rhd3 possess reduced rates of root elongation (Wang et al., 1997). Second, wvd6 root hairs are significantly shorter than wild-type root hairs, a phenotype also observed in various alleles of rhd3 (Fig. 2). Third, the roots of rhd3-1 and rhd3-2 mutants neither wave nor skew on tilted hard-agar surfaces, whereas those of rhd3-3 display only minor waving and skewing under these conditions, which may reflect either the different ecotype background for this allele (No-0) compared to the other rhd3 alleles (Col), or allelic hypomorphy (Figs. 1, B–D, and 3D; Table II). Also, all rhd3 alleles suppress epidermal CFR (Fig. 2F; data not shown). Finally, F1 seedlings derived from a cross between homozygous wvd6 and rhd3-3 mutants display the mutant root growth phenotype on hard-agar surfaces despite the recessive nature of both alleles (Fig. 3D; data not shown).
It is unlikely that the suppressed CFR, skewing, and waving phenotypes of rhd3 roots result from the mutant's inability to form proper root hairs, since other root hair defective mutants (rhd2-1, rhd4-1, and rhd6-1) still retain the ability to wave. rhd1-1 is an exception, as its roots display waves with reduced amplitude. One possible explanation for the low-amplitude waving of rhd1-1 roots is that the mutant may retain the ability to modulate CFR, but is impaired in one or more of the growth processes responsible for the outward buckling of the subterminal region of waving roots or in the differential flank elongation that contributes to root waving (Simmons et al., 1995; Rutherford and Masson, 1996; Buer et al., 2003; Thompson and Holbrook, 2004). This phenotype could be related to the root elongation defect observed in this mutant (Fig. 4A; Table I). rhd1 is allelic to reb1, a mutation originally identified for its root epidermal cell-bulging phenotype (Baskin et al., 1992). RHD1 encodes an isoform of UDP-d-Glc 4-epimerase that is required for the production of UDP-Gal used to galactosylate xyloglucan and type-II arabinogalactan in roots (Seifert et al., 2002). rhd1 mutant root epidermal cell walls lack arabinosylated (1→6)-β-d-galactan and galactosylated xyloglucan, and mutant trichoblasts have a thinner outer wall, consistent with their propensity to swell (Andeme-Onzighi et al., 2002). Together, these results support a role for arabinogalactans and xyloglucans in the regulation of cell expansion, and the cell expansion defect associated with rhd1 may be responsible for the reduction in rhd1 root wave amplitude relative to wild type.
Suppressed root waving on tilted hard-agar surfaces was also observed for other mutants possessing defects in cell expansion (cob-1, cob-2, eto1-1, eto2-1, erh2-1, and erh3-1), suggesting a role for controlled anisotropic cell expansion in the regulation of waving. Alternatively, the thicker roots carried by these mutants may be more resistant to the passive buckling imposed by surface-mediated impedance to root-tip mobility, a phenomenon that may contribute to waving (Migliaccio and Piconese, 2001; Thompson and Holbrook, 2004).
Among the mutants subjected to our study, erh3-1 seedlings have roots that do not skew significantly under our conditions (Table II). erh2-1 roots display a low level of skewing (2.2 ± 0.8; Table II), which is completely suppressed by propyzamide treatment (Table II). Interestingly, the difference in average root skewing for propyzamide-treated and control untreated erh2-1 roots is statistically insignificant (Table II), casting doubt on the significance of the minor skewing displayed by untreated erh2-1 seedlings. On the other hand, the cob and eto mutants exhibit a mild enhancement of rightward root skewing relative to wild-type Col (Table II). Neither of these mutants displays root CFR in the absence of propyzamide (data not shown). This observation is consistent with previous reports indicating a lack of correlation between CFR intensity and degree of skewing (Rutherford and Masson, 1996; Sedbrook et al., 2002; Buer et al., 2003).
Three-micromolar propyzamide enhances rightward root skewing and promotes left-handed CFR in wild-type Col and in eto1-1, eto2-1, cob-1, and cob-2 mutant seedlings, but has no effect on erh2-1 or erh3-1 root growth behavior (Table II). This result suggests a correlation between the ability of untreated seedlings to display root skewing on hard-agar surfaces and their propensity to respond to propyzamide by an enhancement of root skewing and a promotion of left-handed CFR.
The increased rightward skewing in the presence of 3 μm propyzamide was reported as being accompanied by a premature shift of cortical microtubules to a right-handed oblique alignment, a reduction in microtubule dynamic turnover, and increased microtubule catastrophe (Furutani et al., 2000; Nakamura et al., 2004). Although we have not characterized cortical microtubule dynamics, our experiments suggest the alignment of cortical microtubules in epidermal cells at different root-tip zones of propyzamide-treated cob-1 seedlings (Fig. 8, G–I) is similar to that found in propyzamide-treated wild-type Col seedlings (Fig. 8, A–C). Yet, the 3 μm propyzamide treatment resulted in enhanced root skewing and CFR in cob-1 relative to wild type (Table II; data not shown). We conclude that the alignment of cortical microtubules in CEZ epidermal cells is probably not an important determinant of root skewing and CFR levels, although we have not been able to eliminate the possibility of differential propyzamide effects on the cortical microtubule network between wild-type and mutant cells of the cortex or more internal root tissues (Furutani et al., 2000).
Both cob-1 and cob-2 are caused by the same missense mutation within the COBRA gene, which encodes a glycosylphosphatidylinositol-anchored protein necessary for cellulose biosynthesis. cob mutants are deficient in crystalline cellulose within cell walls at the root elongation zone (Schindelman et al., 2001). Taken together, these observations suggest that cellulose microfibrils are not important determinants of root skewing and CFR, in agreement with previous reports (Sugimoto et al., 2003; Wasteneys, 2004). Instead, our data suggest that cellulose microfibrils in the wall of CEZ cells might interfere with the development of CFR and root skewing. The combined effect of cob and propyzamide-induced alteration of cortical microtubule dynamics, which may both affect the integrity of cellulose microfibrils within the wall of CEZ cells (Schindelman et al., 2001; Wasteneys, 2004), might enhance root skewing and CFR in propyzamide-treated cob mutants (Table II).
Propyzamide had only a minor effect on the skewing of eto1-1 and eto2-1 roots (<5°). Both eto1-1 and eto2-1 are ethylene overproducers. Whereas eto2-1 affects the structural gene for isozyme 5 of 1-aminocyclopropane-1-carboxylic acid synthase (ACS5), an enzyme involved in ethylene biosynthesis (Vogel et al., 1998), eto1-1 is mutated in a gene that encodes a repressor of ACS activity and targets it for degradation (Wang et al., 2004). Ethylene treatments alter the organization of cortical microtubules in plant tissues (Shibaoka, 1994). Thus, the abnormally small CEZ observed in eto1-1 roots, with few cells exhibiting transverse cortical microtubules (Fig. 7, M and N), may be a consequence of abnormal ethylene levels in these tissues, and the reduced influence of propyzamide on eto1-1 and eto2-1 root skewing may derive from ethylene-related changes in cortical microtubule arrangement or turnover, along with a much smaller zone of responsive cells (CEZ). Our observations are in agreement with recent results showing that ethylene inhibits root skewing on hard-agar surfaces (Buer et al., 2003).
The most important observation in our study is that, unlike other cell expansion mutants, rhd3 and erh2 do not exhibit increased root skewing or CFR in the presence of propyzamide despite a dramatic propyzamide-induced disorganization of the cortical microtubules in the CEZ and mature zone. ERH2 has not been cloned. However, RHD3 encodes a putative GTP-binding protein (Wang et al., 1997) that functions in anterograde trafficking between the endoplasmic reticulum and the Golgi compartments (Zheng et al., 2004) and is required for cell wall biosynthesis and actin organization in developing fibers and xylem cells (Hu et al., 2003). Considering that (1) the alignment of cellulose microfibrils is necessary, but not sufficient, to restrict the radial expansion of cells; (2) the integrity of cellulose microfibrils may also be important (Wasteneys, 2004); and (3) the other plasma membrane and/or cell wall components may also control anisotropic cell expansion (Wiedemeier et al., 2002), we hypothesize that plasma membrane or cell wall determinants other than cellulose microfibrils might also be needed for the regulation of root skewing and CFR, as suggested by Wasteneys (2004). Such determinants would be delivered to the plasma membrane or cell wall by RHD3-controlled anterograde vesicular trafficking along actin microfilaments. Consequently, rhd3 mutant roots cannot skew and develop CFR on hard-agar surfaces, independently of whether they have been treated with microtubule-interacting drugs.
Under this model, the differential effect exerted by propyzamide on the alignment of CEZ cortical microtubules in different backgrounds (little or no effect in wild-type Col and mutant cob seedlings, and disorganization in rhd3, erh2, and eto1-1 roots; Fig. 8) would reflect different constraints exerted by mutant walls on the affected microtubular network (Fisher and Cyr, 1998), rather than a direct effect of propyzamide on microtubule alignment. Furthermore, the CFR- and skew-promoting effects of propyzamide in wild-type seedlings would be due to relaxation of the negative influence exerted by CEZ cellulose microfibrils on expressivity of the hypothesized wall or plasma membrane determinants, as discussed above. Such relaxation could derive from propyzamide-induced shortening of cellulose microfibrils (Wasteneys, 2004).
Because erh2-1 and rhd3 display similar root growth behaviors that are equally unresponsive to propyzamide treatments, it is reasonable to hypothesize that the ERH2 gene might encode a component of the RHD3 pathway. In the future, characterization of abundance and/or arrangement of plasma membrane and cell wall components in rhd3 and erh2 mutant roots relative to wild type, and molecular cloning of ERH2, may lead to a better understanding of the mechanisms regulating root CFR, waving, and skewing on hard-agar surfaces.
MATERIALS AND METHODS
Plant Stocks and Manipulation
The wvd6 mutant was isolated from the Versailles collection of T-DNA insertion mutants (Bouchez et al., 1993). rhd3-1, rhd3-2, rhd3-3, and rhd6 seeds were provided by John Schiefelbein (University of Michigan, Ann Arbor, MI); erh3 seeds were provided by Keith Roberts (John Innes Centre, Norwich, UK). Arabidopsis (Arabidopsis thaliana) wild-type seeds of the Ws ecotype were provided by Tim Caspar (DuPont, Wilmington, DE). All other plant stocks (cob1, cob2, eto1-1, eto2-1, rhd1-1, rhd2-1, rhd4-1, and ecotypes Col and No-0) were obtained from the Arabidopsis Biological Resource Center (ABRC; Ohio State University, Columbus, OH).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requester.
All techniques and conditions used to sterilize and germinate Arabidopsis seeds and to grow seedlings and plants were as described previously (Rutherford and Masson, 1996).
To quantify root elongation and root diameter, we germinated seeds on square petri dishes containing one-half-strength Linsmaier and Skoog medium (Caisson Laboratories, Sugar City, ID) solidified with 1.5% (w/v) agar (type E; Sigma, St. Louis). The Suc content within the medium was 1.5% (w/v). The plates were sealed with paper surgical tape (Micropore; 3m, St. Paul) and kept in darkness at 4°C for 2 to 4 d. Afterward, the plates were transferred to a growth chamber at 22°C for a 16-h/8-h light/dark cycle (TC16; Conviron, Winnipeg, Manitoba, Canada) and positioned vertically. For quantification of root elongation, seedlings were photographed 5 d after germination (DAG) and again 48 h later, and total root growth was assessed over the 2-d interval. Eight-day-old seedlings were mounted on glass slides and imaged under a microscope to obtain pictures for the quantification of root diameters within their mature zones.
For assays of root waving and skewing, the plates were positioned vertically for the first 3 d of seedling growth, then inclined backward 30° and returned to the growth chamber. Seedlings were photographed 8 to 9 DAG. To quantify the mean angles of root skewing, we defined the vertical axis as 0°, and leftward or rightward deviation from the vertical as negative or positive values, respectively. The effect of propyzamide on root skewing was tested by germinating and growing wild-type and mutant seeds on media containing 3 μm propyzamide (Chem Service, West Chester, PA). To create propyzamide-containing growth medium, 6 mm propyzamide stock solutions were prepared in 100% dimethylsulfoxide (DMSO). The 0 μm propyzamide control plates also contained 0.05% DMSO. This amount of DMSO within the medium has a negligible effect on root waving and skewing (Yuen et al., 2003).
Kinetics of wvd6 root gravitropism was analyzed using previously published procedures (Sedbrook et al., 1999).
Molecular Cloning of wvd6
PCR amplification of the sequence immediately flanking the T-DNA left borders (LB) was performed via adaptor PCR (Siebert et al., 1995) utilizing a primer complementary to the LB of pGKB5 (5′-TGC CAG GTG CCC ACG GAA TAG TTT T-3′) with wvd6 genomic DNA, extracted as described previously (Klimyuk et al., 1993), and digested with EcoRV or ScaI prior to adaptor ligation. Using either restriction enzyme, only a single PCR band was obtained. The LB flanking sequence of the adaptor PCR product derived from ScaI-digested wvd6 DNA matched to an intergenic region on chromosome 1, 2.5 kb upstream of At1g63680 (a putative UDP-N-acetylmuramoylalanyl-d-Glu-2,6-diaminopimelate ligase) and 1.9 kb downstream of At1g63670 (a predicted gene of unknown function; Fig. 3A). On the other hand, the LB flanking sequence of the adaptor PCR product derived from EcoRV-digested wvd6 DNA corresponded to the T-DNA right border of pGKB5, thus confirming that the two T-DNA fragments were inserted in a head-to-tail arrangement (Fig. 3A).
To better clarify the genetic lesion in the wvd6 mutant, a wvd6 genomic DNA library was constructed by digesting wvd6 genomic DNA with EcoRI and ligating size-selected fragments (4–6 kb in length) into the EcoRI site of pZErO-2.1 vector DNA (Invitrogen, Carlsbad, CA). This library was screened for positive clones (Ausubel et al., 1994), utilizing as a probe a 4.5-kb EcoRI subfragment of a cosmid (cos47C) overlapping the T-DNA integration site within the At1g63670/At1g63680 intergenic region. A single positive clone (3E1A), containing an approximately 5-kb EcoRI insert, was identified. Sequencing of this insert revealed that one end corresponded to chromosome 1 intergenic sequence, while the other end matched a portion of the RHD3 gene on chromosome 3. A 13-bp fragment of unknown origin separated the chromosome 1 and 3 sequences (Fig. 3A). According to the sequence data obtained, the putative translocation breakpoint of RHD3 occurs within the first intron (Fig. 3A).
To confirm that the translocation suggested by the sequence of the 3E1A insert was truly indicative of the lesion in wvd6 genomic DNA, rather than being a cloning artifact, we designed three PCR primers. Two of the primers (Chr3F, 5′-CAT TCT CTT CGT CAT TCA TCA CTC TAT CT-3′, and Chr3R, 5′-GAA GAA AGA AGA ATC AAT CAC CTA CCA TC-3′) flank either end of the putative chromosome 3 breakpoint of wvd6 (Fig. 3C) and would be expected to yield a 0.3-kb PCR product when wild-type plant genomic DNA is utilized as the template for amplification, and none when wvd6 genomic DNA is used. Conversely, the primer Chr1F (5′-AGG AGG AGA GGG ACG TGA GAA TGA AAG AG-3′), which is complementary to a short stretch of chromosome 1 sequence located on one end of the putative translocation breakpoint, should not generate a PCR product when used along with Chr3R on wild-type genomic DNA (Fig. 3C). This same primer pair would be expected to yield a 1.1-kb PCR product if used on a wvd6 genomic DNA template. The results of our PCR analysis were exactly as predicted (Fig. 3B), indicating that the wvd6 genome did indeed contain a translocation between chromosomes 1 and 3.
To obtain a PCR fragment corresponding to the genomic DNA flanking the unresolved end of the T-DNA concatamer, reverse primers corresponding to the GUS (5′-TTG TTT GCC TCC CTG CTG CGG TTT TTC AC-3′) and NPT (5′-CAA GCG ACG CCC AAC CTG CCA TCA CGA GA-3′) cassettes of pGKB5 were paired with a forward primer situated 5′ of the RHD3 open reading frame (5′-TTA GTA AAG GAG AAG AGG ACG GCG AGG TG-3′). Only the 5′ RHD3/NPT pair yielded a PCR product (data not shown). Sequencing of this fragment revealed that a large truncation within the T-DNA had occurred, resulting in the complete loss of both the right border and GUS cassettes, and a partial loss of the OCS terminator sequence. This end of the T-DNA immediately flanks a region within the first exon of RHD3, indicating that T-DNA insertion was accompanied by a short deletion of RHD3 (Fig. 3A).
Image Analysis and Microscopy
Images of seedlings on plates were obtained with a Nikon Coolpix800 digital camera, with pictures taken from the bottom of the plate (through the agar medium). Images of root CFR were acquired with a Nikon 8008S camera attached to a dissecting microscope (Wild M3Z; Leica, Wetzlar, Germany), while images for the quantification of root diameter were acquired with a SPOT RT slider digital camera (National Diagnostics, Atlanta) attached to a Nikon Optiphot-2 microscope. Quantitative analyses of images to assess angle of root skewing, root elongation rate, or root diameter were performed using the public domain National Institutes of Health (NIH) image program (v1.62; http://rsb.info.nih.gov/nih-image), with obtained data exported to Microsoft Excel spreadsheets for statistical calculations.
To analyze the organization of the cortical microtubule network in elongating and maturing cells at the tips of wild-type and mutant roots, Col, rhd3-1, cob-1, and eto-1 seeds were germinated and grown for 5 d on vertical 0.8% agar-based GM media in the presence or absence of 3 μm propyzamide (from a 3 mm propyzamide stock solution in DMSO), as described in Rutherford and Masson (1996). The concentration of DMSO used here has a negligible effect on microtubule organization (data not shown). Seedlings were then fixed and subjected to the whole-mount immunofluorescence protocol described in Sugimoto et al. (2000), using the mouse anti-α-tubulin monoclonal antibody B-5-1-2 (Sigma T5168; 1:1,000 dilution) and the rabbit anti-mouse IgG-fluorescein isothiocyanate secondary antibody (Sigma F9137; 1:200 dilution). Paraformaldehyde (4%) was used in the fixation buffer for propyzamide-treated seedlings. Confocal images were acquired with a Zeiss LSM 510 confocal attached to a Zeiss Axiovert 100M inverted microscope, using either a 40× or 63× oil immersion objective (Department of Cell and Structural Biology, University of Illinois, Urbana, IL), and with a Bio-Rad MRC-1024 laser-scanning inverted confocal microscope (Bio-Rad Laboratories, Hercules, CA) using a 60× oil immersion objective (W.M. Keck Laboratory for Biological Imaging, University of Wisconsin, Madison, WI). Images were then processed using Image SXM software specially adapted by Dr. Steve Barrett to handle LSM 510 images (http://www.ImageSXM.org.uk).
Acknowledgments
We thank Herman Höfte for providing lines of the Versailles collection of T-DNA insertion mutants (Institut National de Recherche Agronomique, France), John Schiefelbein, Keith Roberts, Tim Caspar, and the ABRC (Ohio State University) for providing several of the mutants discussed in this article, and other members of the Masson laboratory for helpful discussions. We also thank the University of Wisconsin Biotechnology Sequencing Center for resolving our sequencing reactions, and the Keck Biological Imaging Laboratory at the University of Wisconsin, Madison, and the Department of Cell and Structural Biology at the University of Illinois for the use of their confocal microscopes.
This work was supported by the Fundamental Space Biology Program of the National Aeronautics and Space Administration (grant nos. NAG2–1189 and NAG2–1492 to P.H.M.), the U.S. Department of Agriculture/Hatch Funds (grant no. WIS04310 to P.H.M.), the National Institutes of Health (NIH; grant no. 1 R15 GM068489 to J.C.S.), the NIH Genetics Training Grant (no. 5T32GM07133), and the NIH National Research Service Award postdoctoral fellowship (5 F32 GM069184–02 to R.M.P.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.059774.
References
- Andeme-Onzighi C, Sivaguru M, Judy-March J, Baskin T, Driouich A (2002) The reb1-1 mutation of Arabidopsis alters the morphology of trichoblasts, the expression of arabinogalactan-proteins and the organization of cortical microtubules. Planta 215: 949–958 [DOI] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K (1994) Current Protcols in Molecular Biology—Updates 1994-2002. John Wiley and Sons, New York
- Baskin T, Betzner A, Hoggart R, Cork A, Williamson R (1992) Root morphology mutants in Arabidopsis thaliana. Aust J Plant Physiol 19: 427–437 [Google Scholar]
- Baskin T, Wilson J, Cork A, Williamson R (1994) Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol 35: 935–942 [PubMed] [Google Scholar]
- Blancaflor E, Masson P (2003) Plant gravitropism. Unraveling the ups and downs of a complex process. Plant Physiol 133: 1677–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez D, Camilleri C, Caboche M (1993) A binary vector based on BASTA resistance for in planta transformation of Arabidopsis thaliana. C R Acad Sci Ser III 316: 1188–1193 [Google Scholar]
- Buer C, Wasteneys G, Masle J (2003) Ethylene modulates root-wave responses in Arabidopsis. Plant Physiol 132: 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk D, Liu B, Zhong R, Morrison W, Ye Z-H (2001) A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Burk D, Ye Z (2002) Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 14: 2145–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H, Fabri C, Hauptmann M, Hutzler P, Laux T, Lloyd C, Schaffner A (2004) Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Curr Biol 14: 1515–1521 [DOI] [PubMed] [Google Scholar]
- Chae H, Faure F, Kieber J (2003) The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnops G, Wang X, Linstead P, Van Montagu M, Van Lijsebettens M, Dolan L (2000) Tornado1 and tornado2 are required for the specification of radial and circumferential pattern in the Arabidopsis root. Development 127: 3385–3394 [DOI] [PubMed] [Google Scholar]
- Fisher D, Cyr R (1998) Extending the microtubule/microfibril paradigm. Cellulose synthesis is required for normal cortical microtubule alignment in elongating cells. Plant Physiol 116: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Watanabe Y, Prieto R, Masakawa M, Suzuki K, Naoi K, Thitamadee S, Shikanai T, Hashimoto T (2000) The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127: 4443–4453 [DOI] [PubMed] [Google Scholar]
- Giddings T, Staehelin L (1991) Microtubule-mediated control of microfibril deposition: a re-examination of the hypothesis. In C Lloyd, ed, The Cytoskeletal Basis of Plant Growth and Form. Academic Press, San Diego, pp 85–100
- Guan C, Rosen E, Boonsirichai K, Poff K, Masson P (2003) The ARG1-LIKE2 (ARL2) gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway. Plant Physiol 133: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman P, Ecker J (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T (2002) Molecular genetic analysis of left-right handedness in plants. Philos Trans R Soc Lond B Biol Sci 357: 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M, Morikami A, Benfey P (1995) Conditional root expansion mutants of Arabidopsis. Development 121: 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhong R, Morrison W, Ye Z (2003) The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217: 912–921 [DOI] [PubMed] [Google Scholar]
- Klimyuk V, Carroll B, Thomas C, Jones J (1993) Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J 3: 493–494 [DOI] [PubMed] [Google Scholar]
- Masucci J, Schiefelbein J (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio F, Piconese S (2001) Spiralizations and tropisms in Arabidopsis roots. Trends Plant Sci 6: 561–565 [DOI] [PubMed] [Google Scholar]
- Mochizuki S, Harada A, Inada S, Sugimoto-Shirazu K, Stacey N, Wada T, Ishiguro S, Okada K, Sakai T (2005) The Arabidopsis WAVY GROWTH 2 protein modulates root bending in response to environmental stimuli. Plant Cell 17: 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J, Turk E, Johnson K, Wolverton C, Ishikawa H, Simmons C, Söll D, Evans M (1998) Root-growth behavior of the Arabidopsis mutant rgr1: roles of gravitropism and circumnutation in the waving/coiling phenomenon. Plant Physiol 118: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Furutani I, Tachimoto H, Matsubara H, Hashimoto T (2004) SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Naoi K, Shoji T, Hashimoto T (2004) Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol 45: 1330–1334 [DOI] [PubMed] [Google Scholar]
- Okada K, Shimura Y (1990) Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science 250: 274–276 [DOI] [PubMed] [Google Scholar]
- Okada K, Shimura Y (1992) Mutational analysis of root gravitropism and phototropism of Arabidopsis thaliana seedlings. Aust J Plant Physiol 19: 439–448 [Google Scholar]
- Rutherford R, Masson P (1996) Arabidopsis thaliana sku mutant seedlings show exaggerated surface-dependent alteration in root growth vector. Plant Physiol 111: 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J, Masucci J, Wang H (1997) Building a root: the control of patterning and morphogenesis during root development. Plant Cell 9: 1089–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J, Somerville C (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin T, Carpita N, Derbyshire P, McCann M, Benfey P (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Wells B, Dolan L, Roberts K (1997) Structural and genetic analysis of epidermal cell differentiation in Arabidopsis primary roots. Development 124: 1789–1798 [DOI] [PubMed] [Google Scholar]
- Sedbrook J, Carroll K, Hung K, Masson P, Somerville C (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J, Chen R, Masson P (1999) ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc Natl Acad Sci USA 96: 1140–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J, Ehrhardt D, Fisher S, Scheible W, Somerville C (2004) The Arabidopsis sku6/spiral1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G, Barber C, Wells B, Dolan L, Roberts K (2002) Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr Biol 12: 1840–1845 [DOI] [PubMed] [Google Scholar]
- Shibaoka H (1994) Plant hormone-induced changes in the orientation of cortical microtubules: alterations in the cross-linking between microtubules and the plasma membrane. Annu Rev Plant Physiol Plant Mol Biol 45: 527–544 [Google Scholar]
- Shoji T, Narita NN, Hayashi K, Asada J, Hamada T, Sonobe S, Nakajima K, Hashimoto T (2004) Plant-specific microtubule-associated protein SPIRAL2 is required for anisotropic growth in Arabidopsis. Plant Physiol 136: 3933–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23: 1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons C, Migliaccio F, Masson P, Caspar T, Söll D (1995) A novel root gravitropism mutant of Arabidopsis thaliana exhibiting altered auxin physiology. Physiol Plant 93: 790–798 [PubMed] [Google Scholar]
- Simmons C, Soll D, Migliaccio F (1996) Circumnutation and gravitropism cause root waving in Arabidopsis thaliana. J Exp Bot 46: 143–150 [Google Scholar]
- Sugimoto K, Himmelspach R, Williamson R, Wasteneys G (2003) Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15: 1414–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO (2000) New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax F, Vernon D (2001) T-DNA-associated duplication/translocations in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiol 126: 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S, Tuchihara K, Hashimoto T (2002) Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- Thompson M, Holbrook N (2004) Root-gel interactions and the root waving behavior of Arabidopsis. Plant Physiol 135: 1822–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Woeste K, Theologis A, Kieber J (1998) Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc Natl Acad Sci USA 95: 4766–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lockwood S, Hoetzel M, Schiefelbein J (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionary conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev 11: 799–811 [DOI] [PubMed] [Google Scholar]
- Wang K, Yoshida H, Lurin C, Ecker J (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Wasteneys G (2004) Progress in understanding the role of microtubules in plant cells. Curr Opin Plant Biol 7: 651–660 [DOI] [PubMed] [Google Scholar]
- Wasteneys G, Collings D (2004) Expanding beyond the great divide: the cytoskeleton and axial growth. In P Hussey, ed, The Plant Cytoskeleton in Cell Differentiation and Development. Blackwell, Oxford, pp 83–115
- Webb M, Jouannic S, Foreman J, Linstead P, Dolan L (2002) Cell specification in the Arabidopsis root epidermis requires the activity of ECTOPIC ROOT HAIR 3—a katanin-p60 protein. Development 129: 123–131 [DOI] [PubMed] [Google Scholar]
- Wiedemeier A, Judy-March J, Hocart C, Wasteneys G, Williamson R, Baskin T (2002) Mutant alleles of Arabidopsis RADIALLY SWOLLEN 4 and 7 reduce growth anisotropy without altering the transverse orientation of cortical microtubules or cellulose microfibrils. Development 129: 4821–4830 [DOI] [PubMed] [Google Scholar]
- Yuen C, Pearlman R, Silo-Suh L, Hilson P, Carroll K, Masson P (2003) WVD2 and WDL1 modulate helical organ growth and anisotropic cell expansion in Arabidopsis. Plant Physiol 131: 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I (2004) A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J 37: 398–414 [DOI] [PubMed] [Google Scholar]