Abstract
Mitochondria play a crucial role in regulating cellular energy homeostasis and cell death, making them essential organelles. Maintaining proper cellular functions relies on the removal of damaged mitochondria through a process called mitophagy. Mitophagy is associated with changes in the pH value and has implications for numerous diseases. To effectively monitor mitophagy, fluorescent probes that exhibit high selectivity and sensitivity based on pH detection have emerged as powerful tools. In this review, we present recent advancements in the monitoring of mitophagy using small-molecule fluorescence pH probes. We focus on various sensing mechanisms employed by these probes, including intramolecular charge transfer (ICT), fluorescence resonance energy transfer (FRET), through bond energy transfer (TBET), and photoelectron transfer (PET). Additionally, we discuss disease models used for studying mitophagy and summarize the design requirements for small-molecule fluorescent pH probes suitable for monitoring the mitophagy process. Lastly, we highlight the remaining challenges in this field and propose potential directions for the future development of mitophagy probes.
Keywords: mitochondria, lysosome, mitophagy, small molecules, fluorescent probes, pH, response mechanisms, disease models
1. Introduction
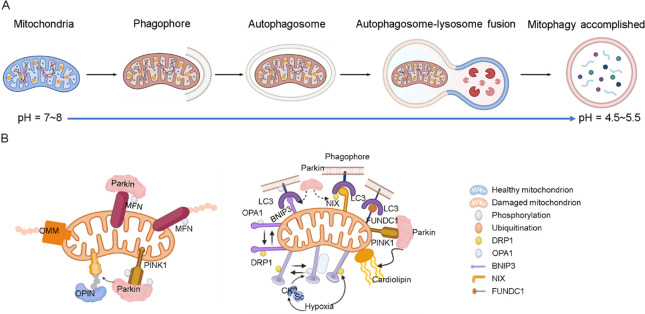
Mitochondria are vital organelles in eukaryotic cells, serving as energy factories for cells and playing a crucial role in cellular energy supply, signal transduction, reactive oxygen species (ROS) production, calcium ion storage, cell differentiation, and cell death.1−5 Mitochondria quantity and quality are maintained through the selective removal of damaged and excess mitochondria, a process known as mitophagy. This process involves the separation of damaged mitochondria into a double-membrane vesicle called an autophagosome, which subsequently binds to lysosomes, in which lysosomal enzymes break down the enclosed material into amino acids or small molecules that can be reused by the cell (Figure 1A).6−8 Mitophagy alters the microenvironment of the mitochondria, including viscosity, polarity, and pH. Due to the participation of two organelles, including mitochondria and lysosomes, the change in pH is particularly obvious. Under normal physiological conditions, mitochondria remain weakly alkaline environment (pH = ∼8), but during mitophagy, damaged mitochondria in lysosomes become weakly acidic (pH = ∼4.5).9−12 This shift in pH is a significant factor in the process of mitophagy.
Figure 1.
(A) Mitophagy process. (B) PINK1 and E3 Parkin-mediated pathway (left) and outer-membrane mitophagy receptor-mediated pathway (right).
There are two primary pathways that regulate mitophagy: the pathway involving protein kinase 1 (PINK1) and the ubiquitin ligase (E3) Parkin, and the pathway involving BCL2 interacting protein 3 (BNIP3), B-cell leukemia/lymphoma 2 (Bcl-2)/adenovirus 19 kDa interacting protein 3-like (Bnip3L), and FUN14 domain-containing 1 (FUNDC1) nuclear fission phagocytosis receptor proteins (Figure 1B).13,14 In the PINK-Parkin pathway, PINK is enriched in the outer mitochondrial membrane (OMM) and becomes activated through phosphorylation.15,16 Parkin then is recruited and localized to the OMM to bind with PINK1. Subsequently, microtubule-associated protein 1 light chain 3 (LC3) binds to form autophagosomes, which eventually fuse with lysosomes.17,18
Receptor-mediated mitophagy involves mitophagy receptor proteins such as BNIP3, Bnip3L, FUNDC1, located in the OMM, which bind to LC3 to facilitate the clearance of damaged mitochondria.3,19 The phosphorylation level of FUNDC1 is regulated by Casein kinase II (CK2) under hypoxic conditions, which affects mitochondrial dynamics and the onset of mitophagy. Mitophagy receptors also actively regulate the degradation and release of optic atrophy 1 (OPA1) from damaged mitochondrial membranes and complement dynamin-related protein 1 (DRP1) on mitochondria, accelerating the removal of damaged mitochondria.20,21 Abnormal mitophagy can lead to various diseases including neurodegeneration, metabolic disorders, muscular dystrophy, liver disease, cardiovascular disease, and cancer. For instance, in Parkinson’s disease (PD), damaged mitochondria accumulate in nerve cells, resulting in the accumulation of ROS and cell death due to inhibition of the mitophagy process. Therefore, it is crucial to monitor mitophagy effectively to prevent the onset of disease.22,23
Fluorescent imaging has become an important method for monitoring mitophagy in recent years due to its high sensitivity, exceptional spatiotemporal resolution, and strong applicability to live cells and tissues. Several fluorescent imaging strategies/tools are currently used for this purpose such as immunofluorescence and fluorescent probes.24,25 However, immunofluorescence can be complex, be time-consuming, and have high requirements for operational skills.26 Fluorescent probes, on the other hand, are more stable and sensitive, allowing for more accurate and rapid monitoring, and are easy to use. Small-molecule fluorescent probes, in particular, are simple in structure, easy to design/modify, and good in biocompatibility and have attracted increasing attention for monitoring the mitophagy process.27−32 These probes offer a more effective way for monitoring changes that occur during mitophagy compared with other probes.
Considering that pH variation is a key characteristic of mitophagy,33 pH can serve as an indicator for monitoring this process. Therefore, pH-sensitive small-molecule fluorescent probes have been demonstrated effective for this purpose.34,35 Furthermore, monitoring pH changes during mitophagy can also provide valuable insights into the role of mitochondria in physiology and pathology as abnormalities in mitophagy are linked to various diseases. As a small-molecule fluorescent probe based on pH detection to monitor mitophagy processes, it typically consists of two key components: a pH-sensitive fluorescent moiety and a mitochondrial targeting moiety. Most of the small-molecule fluorescent probes to date developed to monitor mitophagy have been based on simple and easily modifiable structures such as xanthenes, BODIPY, naphthalimide, benzoxazole, and trichothecenes.36
Various mitochondria-targeting units, such as triphenylphosphine cation (TPP+), pyridinium, dequalinium, guanidine salt, rhodamine, and transition metal complexes, are commonly used in research.37 Many of these compounds belong to a class known as dissociated domain lipophilic cations. Their lipid solubility allows them to effectively cross cellular and mitochondrial membranes, and their positive charge facilitates their entry into the mitochondrial membrane, as the mitochondria exhibit a negative electrochemical potential across their membranes. Consequently, these properties enhance the mitochondrial accumulation of probes.38,39
Small-molecule fluorescent probes that track mitophagy have proven to be useful in the diagnosis and prognosis of disease. However, there is a dearth of literature on designing small-molecule fluorescent probes that can monitor mitophagy in response to changes in pH. Given the critical role of pH detection in monitoring the process of mitophagy, a timely review article summarizing recent advances in the use of small-molecule fluorescent pH probes for monitoring mitophagy could provide valuable insights for readers in chemistry, biology, pharmaceutical sciences, and medicine. This review paper focuses on small-molecule fluorescence probes for monitoring mitophagy, with an emphasis on the significance of different charge transfer mechanisms in the design of these probes. Special attention will be given to their electron transfer modes and the methodologies employed in establishing cell models for mitophagy. The goal is to encourage the development of small-molecule fluorescent pH probes for monitoring mitophagy, which may be useful for detecting mitochondria-related diseases in the future due to their rapid response and high sensitivity.
2. Disease Models for Mitophagy Investigation
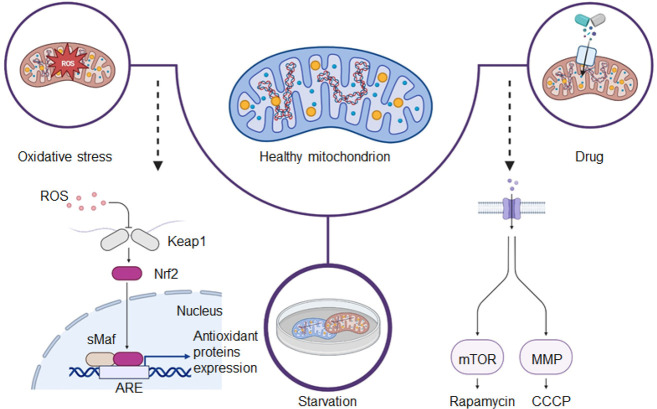
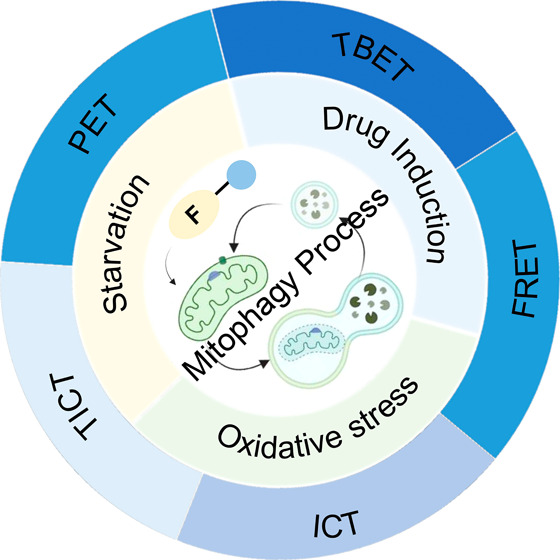
Understanding the pathogenesis of human diseases is a complex and time-consuming process, as the human body is a challenging subject to study. Constructing effective disease models allows us to deliberately modify factors that are difficult or impossible to exclude under natural conditions. Through such models, we can obtain experimental results that facilitate a deeper and more accurate comprehension of the mechanisms underlying human disease development. Furthermore, these models can help in the development of more effective monitoring and prevention tools, enabling us to tackle diseases more efficiently. Mitophagy, the process of the selective degradation of mitochondria, is generally characterized by low activity under normal physiological conditions, making its detection challenging. Nonetheless, it is possible to enhance mitophagy through human intervention. Several methods have been identified to increase mitophagy activity (Figure 2), including culturing cells in serum-free conditions, introducing compounds like cyanide 3-chlorophenylhydrazone (CCCP) or rapamycin, and subjecting cells to hydrogen peroxide (H2O2)-induced oxidative stress, mimicking starvation and drug-induced models, respectively. Validating the effectiveness of fluorescent probes in detecting mitophagy can be achieved by employing these different mitophagy models.
Figure 2.
Illustration of three pathways used to construct mitophagy models.
2.1. Starvation-Induced Mitophagy Models
Live cells can resist starvation environments by increasing their mitophagy levels (Figure 2).40 The starvation model is typically implemented by culturing cells in a serum-free medium or Earle’s Balanced Salt Solution (EBSS) medium for extended durations. Under starvation conditions, the cellular ROS levels significantly increase. Additionally, DNA damage receptor proteins from the nucleus are recruited to the mitochondria to maintain aerobic mitochondrial respiration. Energy receptors at the mitochondria participate in this process by phosphorylating DNA damage receptor proteins, thereby contributing to the maintenance of aerobic mitochondrial respiratory activity. Finally, phosphorylated DNA damage receptor proteins play a role in energy-deprivation-induced mitophagy by influencing the formation of the mitophagy complex.41 Serum-free medium refers to the culture of cells without the inclusion of serum, which may necessitate the addition of growth factors or cytokines to sustain the cells in a basal state of survival. This approach can promote mitophagy due to nutrient deficiency in the medium. Similarly, EBSS medium, which lacks serum and amino acids, induces mitophagy by simulating a state of starvation.
2.2. Drug-Induced Models
Mitophagy can be induced by various agents (Figure 2), including rapamycin, CCCP, and carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP). Rapamycin, a macrolide antibiotic, is effective in promoting mitophagy across different cell types by interrupting the mTOR signaling pathway.42 CCCP, a potent mitochondrial oxidative phosphorylation uncoupling agent, enhances mitophagy by increasing the permeability of the inner mitochondrial membrane protons (H+). This causes a loss of membrane potential on both sides of the inner mitochondrial membrane, thereby facilitating efficient mitophagy.43 Similarly, FCCP serves as another uncoupling agent for mitochondrial oxidative phosphorylation. It disrupts adenosine triphosphate (ATP) synthesis by transporting H+ ions across the inner mitochondrial membrane, ultimately leading to the depolarization of the mitochondrial membrane potential and induction of mitophagy.44
In the drug-induced models, initial colocalization experiments are conducted to validate the probe’s specific targeting of mitochondria, a prerequisite for monitoring mitophagy within these organelles. Subsequently, cells are exposed to both the probe and a mitophagy inducer, and the resulting samples are analyzed by using fluorescence imaging techniques. To show even more proof that the probe is capable of monitoring the process of mitophagy, control experiments can be conducted by adding drugs to inhibit the mitophagy activity. These experiments can be carried out in reverse to demonstrate that the probe specifically detects mitophagy. Several drugs, such as chloroquine (CQ), liensinine, and antimalarial drugs, are known to inhibit mitophagy. CQ functions by increasing the pH in acidic lysosomes, leading to the inactivation of acidic lysosomal hydrolases. This inhibition prevents the fusion and degradation of intracellular autophagic lysosomes, thereby inhibiting the onset of mitophagy.5 Major isoquinoline alkaloid liensinine effectively prevents autophagosomes from fusing with lysosomes, which causes buildup of autophagosomes and mitophagosomes. It achieves this effect by potentially blocking the recruitment of RAB7A, a member of the RAS oncogene family, to lysosomes, but not to autophagosomes.45
2.3. Oxidative Stress/Hypoxia Models
An imbalance between oxidative and antioxidant activity in the body that favors oxidation is what causes oxidative stress. It causes neutrophil infiltration into the inflammation, a rise in the release of proteases, the generation of numerous oxidative intermediates, and excessive ROS production will induce the onset of mitophagy (Figure 2), as well as cells and tissues exhibiting a variety of physiological and pathological responses.46 ROS encompasses species such as the superoxide anion (O2–), hydroxyl radical (·OH), and H2O2,47 with H2O2 being the most significant mitophagy-triggering ROS. Additionally, conditions such as hypoxia or exposure to the specific drug phorpol-12-myrice-13-acetate (PMA) can lead to oxidative stress, making them valuable conditions for constructing experimental models.48
3. Fluorescent Probes Based on Different Energy Transfer Mechanisms
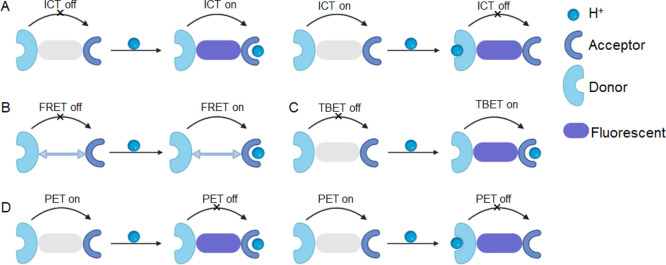
Designing small-molecule fluorescent probes to monitor mitophagy by detecting changes in mitochondrial pH has become a predominant strategy, with numerous probes reported in the literature. These probes can be classified based on their design into four categories (Figure 3): intramolecular charge transfer (ICT, including twisted intramolecular charge transfer, TICT), fluorescence resonance energy transfer (FRET), through bond energy transfer (TBET), and photoinduced electron transfer (PET). The design rationale for each mechanism is discussed in detail to provide a comprehensive understanding of how pH changes during the mitophagy process.
Figure 3.
Schematic representation of the fluorescent mechanisms discussed in this review. (A) Intramolecular charge transfer (ICT). (B) Fluorescence resonance energy transfer (FRET). (C) Through-bond energy transfer (TBET). (D) Photoinduced electron transfer (PET).
3.1. ICT-Based Small-Molecule Fluorescent pH Probes
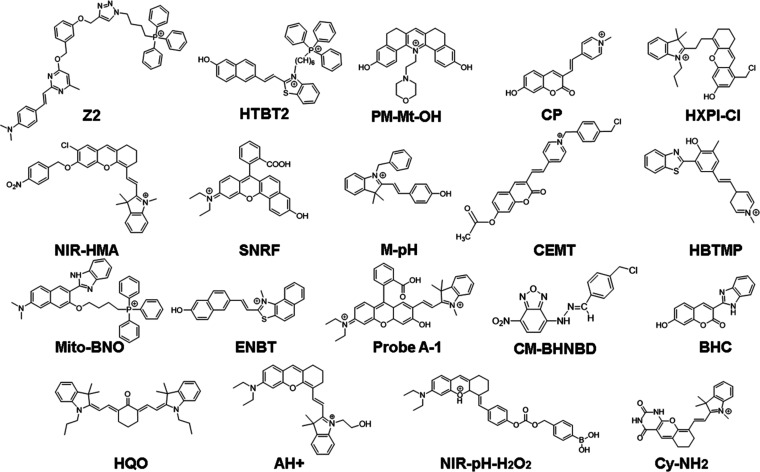
Small-molecule fluorescent probes designed to monitor mitophagy by detecting changes in mitochondrial pH often rely on the ICT mechanism. These probes typically have a conjugative bridge connecting the donor and acceptor groups, forming a D–π–A conjugate system (Figure 3A). The fluorescence intensity and emission wavelength of the probe are strongly influenced by the electron-donating and -withdrawing characteristics of each group in the probe molecules. A strong ICT effect typically results in long wavelengths and strong fluorescence intensity, while a weak ICT results in short wavelengths and weak fluorescence intensity.49,50 Protonation of the electron donor group in an acidic environment or deprotonation of the electron acceptor group in a basic environment can weaken the ICT process, leading to reduced fluorescence intensity and/or blue-shifted the emission wavelength (Figure 3A left), whereas deprotonation of electron-donating groups in an alkaline environment or protonation of electron-withdrawing groups in an acidic environment can enhance the ICT process, leading to an increase in the fluorescence intensity and/or a redshift in the emission wavelength (Figure 3A right).51−53 Numerous ICT-based small-molecule fluorescent pH probes have been developed for effectively monitoring mitophagy effectively. Their molecules are shown in Figure 4, and their unique characteristics are summarized in Table 1.
Figure 4.
Small-molecule fluorescent pH probes based on the ICT mechanism.
Table 1. Parameters of Interest for ICT-Based Small-Molecule Fluorescent pH Probes.
| probes | MW | λex (nm)(pH) | λem (nm)(pH) | pKa | solution | model | refs |
|---|---|---|---|---|---|---|---|
| Z2 | 761 | 450 | 550 | 4.62 | PBS buffer | CCCP | (54) |
| HBTB2 | 644 | 436/566 | 612 | 8.04 | PBS buffer | H2O2 | (55) |
| PM-Mt-OH | 429 | 410/490 | 580/657 | PBS buffer with 0.1% DMSO | rapamycin | (56) | |
| CP | 279 | 400(4)/467(9) | 528(4)/606(9) | 5.88 | Britton-Robinson buffer | CCCP | (5) |
| HXPI-Cl | 460 | 694(8.5)/610(4.5) | 714(8.5)/678(4.5) | 5.77 | PBS buffer | rapamycin | (8) |
| NIR-HMA | 554 | 570 | 675(3)/710(8) | 6.50 | hypoxia | (57) | |
| SNRF | 439 | 510/560 | 586/628 | 8.3 | PBS buffer | rapamycin | (58) |
| M-pH | 354 | 435(3)/535(8) | 552(3)/584(8) | 6.87 | PBS buffer | sodium selenite/BSO | (59) |
| CEMT | 482 | 480 | 558(4.98)/620(9) | Britton-Robinson buffer | CE | (60) | |
| HBTMP | 488 | 375 | 470(8)/580(5) | 6.82 | Britton-Robinson buffer | CCCP | (61) |
| Mito-BNO | 620 | 405 | 468(8)/520(4) | 5.23 | PBS buffer | CQ/CCCP | (62) |
| ENBT | 368 | 428(4.42)/503(11.40) | 595(4.42)/700(11.40) | 7.98 | PBS buffer | CCCP | (63) |
| probe A-1 | 570 | 480 | 558(8)/698(3) | 8.26 | 30% ethanol | serum-free medium | (64) |
| BHC | 278 | 390/420 | 480 | 4.2/7.0 | PBS buffer | serum-free medium | (65) |
| HQO | 520 | 530(8.23)/710(6.59) | 650(8.23)/750(6.59) | 7.15 | 10 mM SDS in HEPES | liensinine/rapamycin | (1) |
| CM-BHNBD | 346 | 495 | 516 | 3.91 | Britton-Robinson buffer | oxidative stress | (66) |
| AH+ | 469 | 670 | 727 | 7.60 | citrate–phosphate buffer | FCCP | (67) |
| NIR-pH-H2O2 | 540 | 560 | 680 | 6.70 | PBS buffer | PMA | (68) |
| Cy-NH2 | 399 | 500 | 550(8)/670(3) | 3.85 | PBS buffer | serum-free media | (69) |
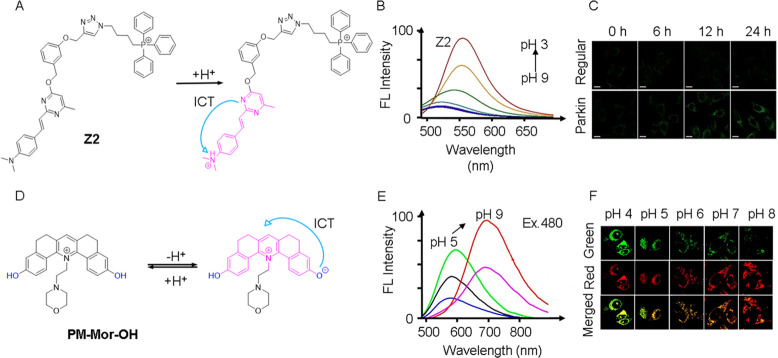
For example, in 2022, our group developed a pyrimidine-based fluorescent probe (Z2) for monitoring mitophagy.54 The probe utilized a D–π–A photoelectron molecular structure with dimethylaniline serving as the electron donor group, an alkene bond as the conjugative bridge, and a pyrimidine as the strong electron-withdrawing group. By targeting mitochondria via TPP+ (Figure 5A), Z2 exhibited a unique characteristic achieved by introducing a benzyl alcohol component to the molecular structure, which possesses a strong electron-withdrawing effect to inhibit the ICT process and quench fluorescence. During mitophagy, the decrease in pH led to the protonation of the dimethylaniline moiety. It transformed the dimethylaniline fraction from an electron-donating state to a strong electron-withdrawing organic salt. Consequently, charge transfer from the pyrimidine to the dimethylaniline caused fluorescence to be activated, as evidenced by the increased intensity at 450 nm with decreasing pH (Figure 5B). The fluorescence quantum yield also exhibited a corresponding increase from 0.07 to 0.62 when the pH was dropped from 9 to 3. The probe demonstrated a clear “turn-on” fluorescence response under acidic conditions. To validate the ability of Z2 to monitor mitophagy, both HeLa cells (as a control) and Parkin-overexpressed HeLa cells (Parkin-HeLa cells) were pretreated with the mitophagy-inducing agent (CCCP) and imaged at different time points (Figure 5C). The fluorescence intensity of Z2 progressively increased with longer treatment times, indicating an elevated degree of mitophagy and a decrease in pH.
Figure 5.
(A) Molecular structure and pH sensing mechanism of probe Z2. (B) Fluorescence spectra of Z2 (λex = 450 nm). (C) HeLa cells and Parkin-HeLa cells pretreated with 10 μM CCCP, and then stained with Z2. λex/em = 458/480–520 nm. Scale bar = 20 μm. (A–C) Reproduced with permission from ref (54). Copyright 2022 Wiley. (D) Molecular structure and pH sensing mechanism of probe PM-Mor-OH. (E) Ratiometric emission spectra of PM-Mor-OH (λex = 458 nm) in different pH solutions. (F) Imaging of HeLa cells under different pH conditions treated with PM-Mor-OH. Scale bar = 10 μm. (D–F) Reproduced from ref (56). Copyright 2022 American Chemical Society.
SNRF (Figure 4) as a probe has been successfully used to observe the mitophagy process induced by rapamycin.58 The probe is a rhodamine derivative synthesized from 1,6-dihydroxynaphthalene and 4-diethylaminoketoic acid. The researchers discovered that, in comparison to the probe-only treated cells, the rapamycin and probe coincubated cells had higher yellow fluorescence when they were fluorescently imaged at various times. The changes became more and more obvious over time, which indicated that the probe could monitor the mitophagy process induced by rapamycin.
In the study on the Mito-BNO probe (Figure 4),62 the probe itself is composed of a benzimidazole, a naphthalene derivative, and a TPP+. When the pH decreased, the benzimidazole group became protonated, generating ICT. The researchers initially demonstrated the probe’s ability to selectively target mitochondria and its potential for monitoring rapamycin-induced mitophagy through colocalization experiments. To further validate the probe’s effectiveness, two control groups were established in the study. Control A received only the probe, while control B had the probe added and was cultured in a serum-free medium. CQ and rapamycin were subsequently added to the cells cultured with the probe, and fluorescence images were captured. In control A, only the blue channel showed fluorescence. This result was consistent with the findings observed when cells were treated with CQ. This similarity suggests that CQ inhibited the lysosome-dependent mitophagy process. In control B, the fluorescence intensity of the green channel was significantly increased, a phenomenon that suggests that the fluorescence of the probe may be associated with rapamycin-induced mitophagy. By combining the results of these two control experiments, it can be confirmed that the change in fluorescence ratio is indicative of mitophagy, thereby establishing the probe’s effectiveness in monitoring the mitophagy process.
In 2020, Song et al. designed and synthesized a mitochondrial biocompatible fluorescent probe HTBT2 (Figure 4) by introducing TPP+ as a mitochondria-targeting unit into the molecule.55 HTBT2 showed a significant Stokes shift and a long emission band, primarily due to the strong ICT process from the naphthol group to benzothiazole. It demonstrated orange-red fluorescence in neutral and acidic environments. However, in weakly basic environments, the hydroxyl group underwent deprotonation, forming naphthoxide (TBT), which led to fluorescence quenching due to unstable charge separation in the ICT state. The fluorescence intensities of HTBT2 at 612 nm indicated a linear correlation with pH values between 8.70 and 7.20. The determined pKa value of HTBT2 was 8.04 ± 0.02, which closely corresponds to the pH of mitochondria (pH ∼ 8.0). Remarkably, HTBT2 accurately distinguished between cancerous and healthy cells by detecting pH changes in living cells. These findings strongly suggest that HTBT2 holds great potential for investigating physiological processes associated with pH alterations.
In addition to targeting mitochondria using the TPP+ fraction, another approach involves using the morpholine fraction. Samanta et al. have developed a fluorescent ratiometric probe named PM-Mor-OH (Figure 5D),56 which is a conjugated pyridine derivative of the morpholine ligand. Normally, probes with the morpholine fraction localize to lysosomes, but interestingly, PM-Mor-OH demonstrated a specific affinity for mitochondria. This probe existed in two distinct forms within mitochondria and mitolysosomes, characterized by protonation and deprotonation, respectively. Under excitation at 480 nm, a shift in pH from 8.5 to 4.0 (Figure 5E) led to a noticeable alteration in ratiometric emission intensity, transitioning from 657 nm (red) to 580 nm (green). Thus, the probe exhibited both an “on” and “ratiometric” response, indicating its capability to respond to changes in pH. The fluorescence image in Figure 5F vividly illustrated the variation in emission intensity in the green and red channels as a result of 488 nm laser irrdiation. Notably, during mitophagy, PM-Mor-OH underwent protonation in an acidic environment, resulting in an enhanced green fluorescence. These results strongly support the conclusion that PM-Mor-OH is a ratiometric fluorescent probe, effectively enabling the monitoring of mitophagy.
In 2015, Yu et al. developed a mitochondria-targeting ratiometric fluorescent probe (CP, Figure 4) based on pyridinium-functional 7-hydroxycoumarin linked by a double bond.5 The design of CP aimed to leverage the excellent electron absorption and mitochondrial targeting properties of the pyridine cation, along with its ability to form a conjugated system with coumarins, to achieve a strong ICT effect. Regarding its optical properties, CP exhibited high sensitivity to changes in pH. As the pH decreased, there was a blue shift in the maximum absorption band, while an enhancement at 606 nm was followed by a decrease at 528 nm as the pH increased. This characteristic enabled ratiometric detection. The pKa value of the CP was determined to be 5.88, and the quantum yields under pH 4.0 and pH 9.0 conditions were determined to be 0.0584 and 0.0159, respectively. The researchers conducted various experiments to test the pH sensitivity of CP, including manipulating mitochondrial acidification, lysosomal alkalinization, increasing H2O2-induced ATP hydrolysis acidification, and augmenting the glutathione (GSH) content with N-acetyl-l-cysteine (NAC) to promote cellular redox stability. This study represented the first exploration of ratiometric fluorescent probes for detecting mitochondrial pH in living cells, providing valuable insights for the design of future probes for studying mitophagy.
Using the starvation model, the researchers are able to verify the capability of the probes to monitor the mitophagy process. For example, researchers employed this model to investigate the labeling of the mitophagy process using the Cy-NH2 probe (Figure 4).69 The Cy-NH2 probe comprises a xanthenesemi-cyanine and a terminal indole with a primary amine. At physiological pH, the probe exhibited a solely green fluorescence. However, under acidic conditions, the primary amine was protonated. As a result, the green fluorescence declined but the near-infrared (NIR) fluorescence was restored. To induce mitophagy, cells were cultured in a serum-free medium, and real-time fluorescence images were captured. During mitophagy, lysosomes and mitochondria merged to form autolysosomes, causing an increase in the fluorescence intensity of the red channel and a decrease in the intensity of the green channel. This observation confirms that the probe underwent protonation and highlights its potential for monitoring mitophagy.
Apart from a serum-free culture, EBSS can also induce mitophagy by creating a starvation environment. An observation made regarding the NIR-HMA probe (Figure 4),57 it was observed that NIR-HMA accumulated specifically in mitochondria, where its 4-nitrobenzene group was converted to an amino group by nitroreductase and then eliminated through a 1,6-rearrangement mechanism. To investigate mitophagy induced by the EBSS medium, researchers incubated cells containing NIR-MAOH as a contrast probe with EBSS. Fluorescence imaging revealed a strong fluorescent signal from the probe, indicating its potential utility for monitoring mitophagy induced by EBSS.
TICT is a fluorescence quenching mechanism that is deemed an “extreme form” of ICT. It is a photophysical phenomenon that occurs in molecules with a donor–acceptor structure, usually connected by a single bond between the two donor and acceptor planes. When excited, these molecules undergo conformational twisting in a polar environment and relax to their ground state without radiation emission. The formation of TICT depends on two factors: a spatial resistance and an electron push–pull effect.70 In 2020, Tan et al. conducted an investigation and developed a probe CM-BHNBD (Figure 4),66 which was designed to accurately detect pH levels across a wide range, operating through an proton-driven TICT mechanism. The design of CM-BHNBD involved utilizing the fluorophore NBD (4-nitrobenzo-2-oxa-1,3-diazole), with the inclusion of a benzyl chloride moiety for mitochondrial fixation purposes. This modification was aimed at forming a covalent bond between the probe and mitochondrial proteins,71 preventing its leakage from mitochondria and facilitating its preferential accumulation within these organelles. Under acidic conditions, the protonated nitrogen atoms of the probe adopted a planar conjugated p−π structure, triggering the occurrence of a TICT mechanism upon sensing H+. The researchers introduced CM-BHNBD into cultured cells in the presence of MTR (MitoTracker Red) and exposed them to various pH buffers. The CM-BHNBD’s fluorescence intensity demonstrated an increase as the pH decreased. The colocalization of CM-BHNBD and MTR in the mitochondria was verified by merged images. By analyzing these colocalization images, the researchers determined that CM-BHNBD could offer pH-dependent linear signals within the pH range of 7.00 to 2.00. Furthermore, the study validated the capability of the probe to monitor mitophagy through ratio fluorescence measurements.
3.2. FRET-Based Small-Molecule Fluorescent pH Probes
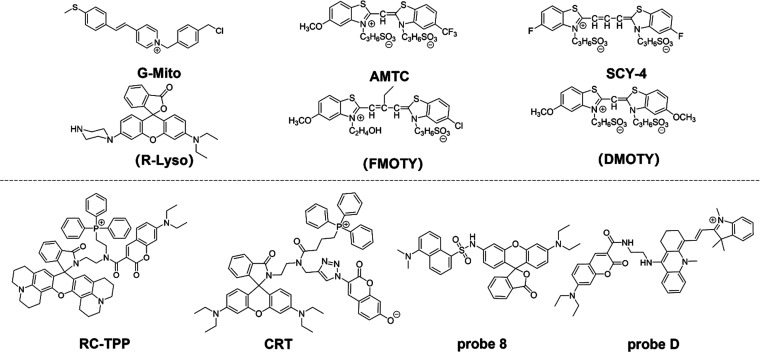
Through intermolecular interactions, the nonradiative process of FRET allows energy to be transferred from the excited state of the donor to the excited state of the acceptor. When a donor absorbs a specific frequency of photons, it becomes excited to a higher electronic energy state. Before the electron returns to its ground state, it transfers energy to the adjacent acceptor via dipole interactions (Figure 3B).72−75 For this process to occur successfully, The FRET donor and acceptor must be physically close to one another, and the donor fluorescent molecule’s emission spectrum must successfully overlap the acceptor fluorescent molecule’s absorption spectrum. Additionally, their excitation light must be sufficiently separated, and the dipole moment orientations of the donor and acceptor must align in a specific way.76,77 As shown in Figure 6, it illustrates representative molecules utilized as mitophagy probes based on the FRET mechanism. The top panel shows FRET molecular pairs, while the bottom panel features one-molecule probes. Table 2 summarizes the unique characteristics of reported FRET-based small-molecule fluorescent pH probes.
Figure 6.
Small-molecule fluorescent pH probes based on the FRET mechanism.
Table 2. Parameters of Interest for FRET-Based Small-Molecule Fluorescent pH Probes.
| probe | MW | λex (nm)(pH) | λem (nm)(pH) | pKa | solution | model | refs |
|---|---|---|---|---|---|---|---|
| G-Mito | 402 | 405 | 550 | PBS buffer | H2O2/CQ | (27) | |
| R-Lyso | 456 | 550 | 590 | PBS buffer | H2O2/CQ | (27) | |
| FMOTY | 567 | 430 | 480 | Tris-HCl buffer | serum-free medium | (78) | |
| AMTC | 624 | 550 | 590 | Tris-HCl buffer | serum-free medium | (78) | |
| SCY-4 | 588 | 559 | 581 | PBS buffer | rapamycin | (79) | |
| DMOTY | 586 | 405 | 510 | PBS buffer | rapamycin | (79) | |
| RC-TPP | 1064 | 425/585 | 475/615 | Britton-Robinson buffer | CCCP | (80) | |
| CRT | 1056 | 400(9)/560(3) | 475(9)/590(3) | DMSO | PMA | (81) | |
| probe 8 | 620 | 405 | 472/576 | 6.96 | PBS buffer | H2O2 | (82) |
| probe D | 682 | 436(3)/540(7) | 470(3)/660(7) | 6.01 | PBS buffer | (83) |
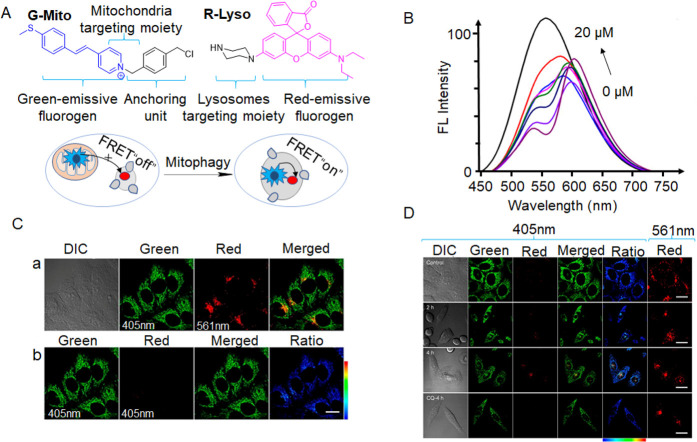
Many probes designed based on the FRET mechanism take advantage of the fact that lysosomes and mitochondria are both connected to the process of mitophagy. The specificity of the light signal generated only when mitochondria and lysosomes fuse makes it an excellent indicator of mitophagy. For example, in 2022, Tian et al. designed two fluorescent probes (G-Mito and R-Lyso) for directly visualizing mitophagy in a two-color manner (Figure 7A).27 The FRET process was examined by mixing G-Mito (10 μM) with various R-Lyso (0–20 μM) concentrations and recording their fluorescence spectra under 405 nm excitation (Figure 7B). As the R-Lyso concentration increased, the emission at 550 nm of G-Mito decreased, while the emission at 590 nm increased, indicating that the FRET process occurred once the two molecules were combined. It was expected that before mitophagy G-Mito resided primarily in mitochondria and emitted green fluorescence while R-Lyso localized in lysosomes and emitted faint red fluorescence under excitaion. At high levels of mitophagy, the red fluorescence of R-Lyso increased significantly as G-Mito was sent into the lysosome in the proximity of R-Lyso, triggering the FRET process (Figure 7A). The feasibility of the G-Mito and R-Lyso pair for mitophagy monitoring was examined in live cells by costaining experiments (Figure 7C). Bright green fluorescence of G-Mito under 405 nm excitation in the microfilament form indicated localization to mitochondria, while speckled red luminescent spots of R-Lyso under 561 nm excitation suggested lysosomal staining (panel a in Figure 7C). Live cells costained with the two probes under 405 nm excitation showed intense minimal red emission and green fluorescence (panel b in Figure 7C), indicating that the FRET process was inhibited due to the two probes’ differing subcellular localizations. The cells treated with H2O2 exhibited a remarkable increase in fluorescence in the red channel, indicating significant enhancement in the mitophagy process and the recovery of the FRET process (Figure 7D). These findings demonstrate that both probes effectively visualized mitophagy in a two-color approach.
Figure 7.
(A) G-Mito and R-Lyso’s chemical compositions and proposed response mechanism to detect mitophagy. (B) Combined, normalized fluorescence spectra of G-Mito (10 μM) and R-Lyso (0–20 μM) with a 405 nm excitation. (C) G-Mito and HepG2 cell coculture images (2 μM, λex = 405 nm) and R-Lyso (4 μM, λex = 561 nm) for 30 min. (D) Images of H2O2-treated HepG2 cells using G-Mito (2 μM, λex = 405 nm) and R-Lyso (4 μM, λex = 561 nm). Scale bar = 20 μm. Reproduced from ref (27). Copyright 2021 American Chemical Society.
In a similar fashion, in 2021, Sun and colleagues developed two FRET-based probes (FMOTY and AMTC, Figure 6) targeting mitochondria and lysosomes, respectively.78 The probe pair exhibited an FRET effect specifically when mitochondria fused with lysosomes during mitophagy. FMOTY and AMTC were not inherently associated with mitochondria or lysosomes and did not generate background signals that could interfere with FRET. Fluorescent signals were exclusively generated in the presence of mitophagy, establishing these molecules as highly specific FRET pairs for monitoring mitophagy. However, the cyan dye used in the FRET probes has a relatively low fluorescence quantum yield, resulting in only partial overlap between the acceptor and donor dyes. Consequently, higher probe concentration was required for effective cellular imaging.
Apart from the dual-probe method that monitors mitophagy by targeting mitochondria and lysosomes with probes emitting different fluorescence, the FRET mechanism can also be observed by using a one-molecule probe. In 2017, Han et al. developed a probe named RC-TPP, which exhibited distinct response signals in mitochondria and lysosomes (Figure 8A).80 This probe consists of rhodamine-lactam, coumarin, and TPP+. Coumarin fluorescence remained constant, while rhodamine-lactam under acidic conditions underwent ring opening to generate strong fluorescence. The probe specifically targeted mitochondria through TPP+. Rhodamine fluorescence was minimal at pH 7.0–9.0, but as the pH decreased from 6.5 to 4.5, (Figure 8B), rhodamine fluorescence increased due to the proton-mediated opening of intramolecular spirolactams. To confirm the ability of RC-TPP to monitor the mitophagy process, the researchers introduced both CCCP, a mitophagy-inducing drug, and the probe into cultured cells (Figure 8C). As expected, cells in control groups exhibited predominantly blue coumarin fluorescence signal localized in the mitochondria, whereas CCCP-treated cells displayed reduced blue fluorescence mainly from coumarin and a significantly enhanced red fluorescent signal mainly from rhodamine. These observations validated that RC-TPP can fluorescently visualize the mitophagy process by responding differently to the temporal sequence of mitophagy induced by CCCP. This innovative approach offered a novel paradigm for investigating pH variations during mitophagy.
Figure 8.
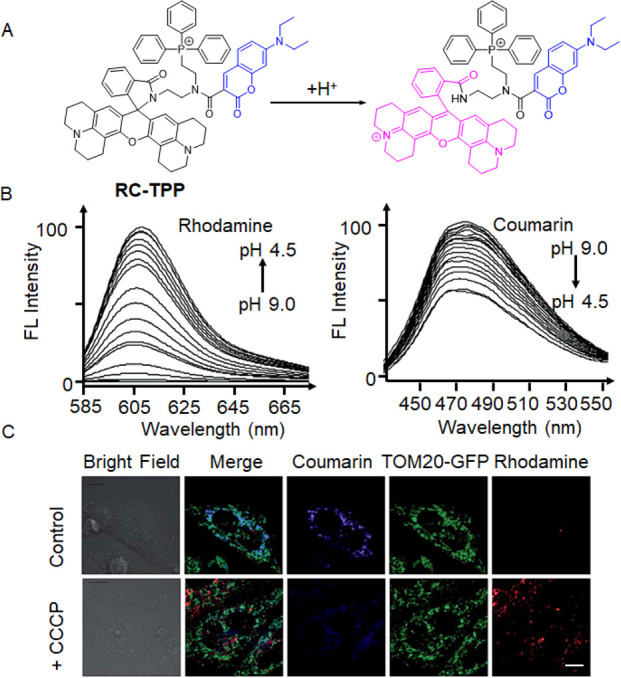
(A) Intramolecular spirolactam of RC-TPP is fluorogenically opened by a proton to release the amide form. (B) RC-TPP (20 μM) fluorescence emission in a buffer with a pH range of 4.5 to 9.0 (λex = 425 nm for coumarin and λex = 585 nm for rhodamine). (C) RC-TPP-loaded Tom20-GFP+ HeLa cells were treated without or with CCCP (20 μM). Colocalization of Tom20-GFP (green) and mitochondrial RC-TPP (blue) is shown in cyan. Scale bar = 10 mm. Reproduced with permission from ref (80). Copyright 2017 Royal Society of Chemistry.
Mitochondria in their normal state produce ROS for normal cellular physiological responses, whereas in a state of hyperoxia or hypoxia, an irregular production of ROS can cause mitochondrial damage and thus induce mitophagy to occur.84 Therefore, in experiments, it is possible to evaluate whether a probe can monitor mitophagy by creating a model of oxidative stress. For instance, in experiments involving the CRT probe (Figure 6),81 synthesized from 7-hydroxycoumarin, rhodamine B, and TPP+, researchers initially confirmed the probe’s ability to target mitochondria through colocalization experiments in cells. The probe was then introduced into two groups of cells cultured in a serum-free medium and treated with PMA, respectively. Fluorescence imaging of the two groups of cells showed nearly identical fluorescence, indicating that PMA may induce mitophagy, and the probe effectively monitored this process. This observation suggests that reduced oxidative stress greatly suppressed mitophagy, thus confirming that oxidative stress induced by PMA treatment can induce mitophagy.
Similar to elevated ROS levels, long-term hypoxia also increases the level of cellular oxidative stress, which damages the mitochondria and triggers mitophagy. For example, in the experiment on the NIR-HMA probe (Figure 4),57 the researchers established an anoxic model (0.1% O2) as well as a control group in a normal oxygen environment (20% O2). Cells were cultured under both conditions and exposed to the probe. Fluorescence imaging showed weak fluorescence in the normal oxygen environment but significantly enhanced fluorescence in the low oxygen environment, confirming the occurence of hypoxia-induced mitophagy.
3.3. TBET-Based Small-Molecule Fluorescent pH Probes
TBET is an additional energy transfer mechanism used in the construction of small-molecule fluorescent pH probes. This mechanism involves the covalent bonding of the donor and acceptor components within the molecular structure of the probe (Figure 3C).85 The acceptor group receives energy from the donor group via electronically conjugated rigid structures, often facilitated by a π-system linker such as a phenyl or triple bond.86 Both FRET and TBET involve the transfer of energy between pairs of fluorophores, while the other serves as the energy acceptor and the former serves as the energy donor. However, there are significant differences in their mechanisms and requirements. The intensity of the donor’s fluorescence is reduced while the acceptor’s fluorescence is increased in FRET because the donor’s excitation energy is directly transferred to the acceptor fluorophore through a nonradiative mechanism. Spatial proximity is crucial for FRET to occur effectively. In contrast, TBET operates through a different mechanism and is not subject to the same spatial proximity requirements as FRET. In addition, unlike FRET, TBET does not rely on the donor and acceptor’s spectrum overlap. Through electronically conjugated rigid π-system connections, the excitation energy of the donor fluorophore is transmitted to the acceptor, twisting the donor and acceptor groups out of coplanarity. This unique mechanism, coupled with the energy transfer occurring primarily through chemical bonding, leads to higher energy transfer efficiency and the potential for an easier ratiometric fluorescence response in TBET. Distinct mechanism and composition principles of TBET make it advantageous for efficient energy transfer and potential applications in ratiometric fluorescence response.87 However, TBET often encounters interference from ICT and FRET, making it difficult to definitively characterize this process in many instances. Compared to FRET-based probes, TBET-based probes are less commonly reported due to the inherent challenges associated with this mechanism. Table 3 provides a summary of two well-defined TBET probes along with their significant sensing properties.
Table 3. Parameters of Interest Are Based on TBET Small-Molecule Fluorescent pH Probes.
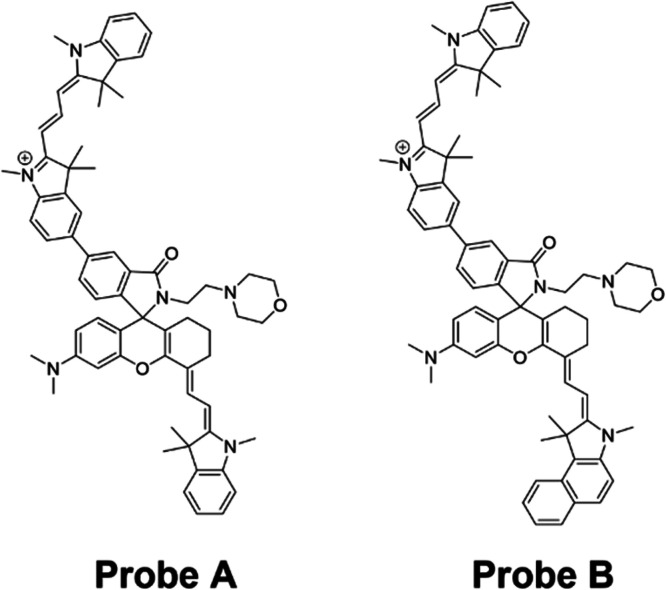
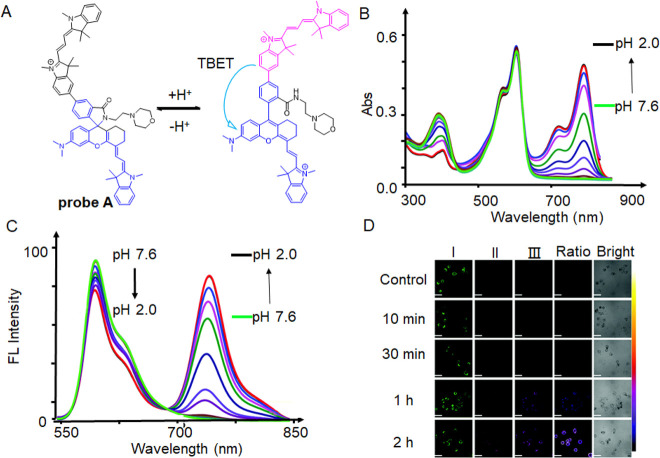
Probes A and B (Figure 9) share a similar structural composition, both consisting of a cyanine dye functioning as the donor and utilizing hemicyanine as the acceptor. These probes employ TBET as a mechanism for ratiometrically detecting mitochondrial pH.88 Probe A, for instance, has a positively charged cyan dye donor that is coupled to a negatively charged NIR semicyan acceptor via a biphenyl bridge that is mediated by a spironolactam switch that prevents the π-conjugation of the donor and acceptor (Figure 10A). The addition of morpholine functional groups to the molecule enhances the hydrophilicity of the probe. Additionally, it lowers the pKa value of the receptor, resulting in the suppression of its fluorescence, specifically in the weakly alkaline environment of mitochondria. Under weakly acidic conditions, the spironolactam group within the hemicyanine acceptor molecule underwent ring opening. This ring-opening process enabled efficient TBET from the hemicyanine acceptor molecule to the cyanine donor molecule. In the context of mitophagy, the hemicyanine acceptor molecule exhibited fluorescence due to the formation of a helical lactam ring resulting from the ring opening under acidic conditions. Simultaneously, the fluorescence intensity of the cyanine donor decreased as the mitophagy progressed. At basic or neutral pH, the probe displayed two primary absorption peaks at 380 and 558 nm (Figure 10B). However, when the pH gradually decreased, an additional absorption peak appeared at 715 nm due to acidic conditions resulting in the hemicyanine receptor’s spirolactam ring opening, which increased π-conjugation. The probe displayed only one emission peak at 588 nm (λex = 520 nm) under weakly basic or neutral pH conditions. However, in an acidic environment, a new emission peak at 740 nm was seen (λex = 520 nm; Figure 10C). Defective mitochondria eventually joined with lysosomes during mitophagy to create acidic autolysosomes, which caused the pH to drop. To verify its capability to monitor mitophagy in live cells, HeLa cells were subjected to incubation with probe A in a serum-free culture medium, while varying the duration of starvation from 10 min to 2 h. During this time, a notable decrease in the visible donor fluorescence was observed in channel I, whereas the near-infrared acceptor fluorescence in channel II exhibited a gradual increase (Figure 10D). Following 1 h of incubation, there was a noticeable color shift in the ratio image between the visible fluorescence in channel I and the near-infrared fluorescence in channel II, appearing as a dark blue color. Subsequently, after 2 h of starvation, the color transitioned to a shade of purple-brown (Figure 10D).
Figure 9.
Small-molecule fluorescent pH probes based on the TBET mechanism.
Figure 10.
(A) Ratiometric fluorescent probe A and its structural responses to pH changes. (B) Absorption spectra of probe A at different pH values. (C) Fluorescence spectra of probe A (10 μM, λex = 520 nm) with pH changing from 7.6 to 2.0. (D) HeLa cells were imaged using ratiometric fluorescence with probe A in a serum-free solution at different periods of donor and acceptor excitation with scale bars of 50 μm. By dividing the visible fluorescence in the first channel by the near-infrared fluorescence in the second channel, ratio images were created. Reproduced with permission from ref (88). Copyright 2020 Royal Society of Chemistry.
3.4. PET-Based Small-Molecule Fluorescent pH Probes
PET is an important mechanism for energy transfer. In a typical PET system, the analyte-receptor component consists of an electron donor linked to a fluorophore through a spacer group. The fluorophore absorbs and emits fluorescence, while the receptor binds to the target of interest. The spacer group plays a crucial role in connecting the electron donor and fluorophore, forming a covalent connection system. This system serves the dual function of recognizing the target and providing the optical signal in the form of light.89 In PET fluorescent probes, fluorescence quenching occurs due to light-induced electron transfer between the fluorophore and the acceptor unit, where electrons are transferred from the electron donor to the electron acceptor. As a result, the probe does not show any fluorescence until it binds to the target (Figure 3D). In PET-based pH probes, the fluorophore remains quenched under neutral or basic conditions due to the PET, which can be inhibited by protonation, thus turning on the fluorescence.90Figure 11 shows representative examples of fluorescent probes based on the PET mechanism. Additionally, Table 4 provides a summary of the crucial sensing parameters associated with PET-based small-molecule fluorescent pH probes.
Figure 11.
Small-molecule fluorescent pH probes based on the PET mechanism.
Table 4. Based on PET and Other Relevant Parameters of Small-Molecule Fluorescent pH Probes.
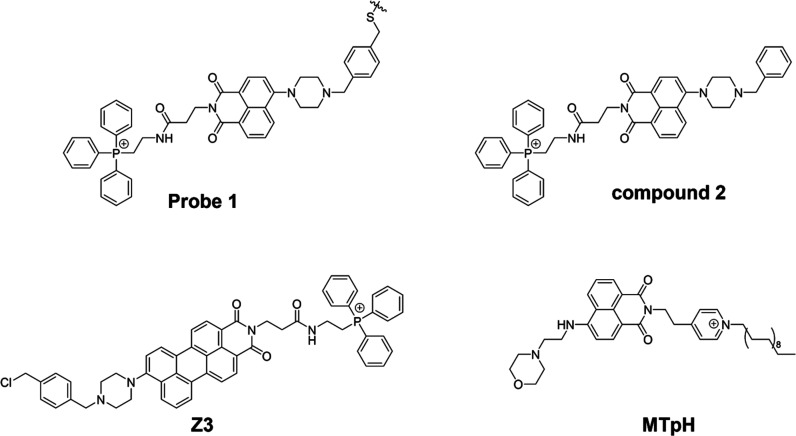
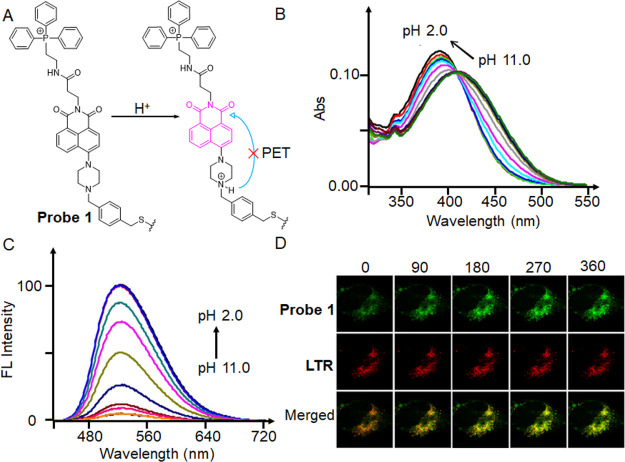
In 2014, Kim et al. developed a fluorescent probe made of piperazine, TPP+, and benzyl chloride that is pH-sensitive (Figure 12A).91 The benzyl chloride functional group can anchor within the mitochondria, while the TPP+ group is responsible for mitochondrial targeting. The piperazine-based naphthalimide group is a fluorophore that provides an “off-on” fluorescent signal via protonation-induced suppression of the neutral form of PET at acidic pH levels. However, the researchers acknowledged that the fluorescence response of probe 1 could potentially be affected by the benzyl chloride portion because it might be hydrolyzed or interact with other biologically relevant substances. To address this concern and minimize interference, they utilized compound 2 (Figure 11), which is an analogue of probe 1 but lacks the benzyl chloride functional groups. By using compound 2, the researchers aimed to evaluate the effect of pH without potential complications arising from the benzyl chloride portion. The effect of pH was then investigated by absorption spectra (Figure 12B) and fluorescence spectra (Figure 12C) of compound 2. The absorption peak gradually changed from 411 to 391 nm, with an evident isosbestic point at 409 nm, as the pH value dropped from 11 to 2. With a drop in pH, the fluorescence intensity at 525 nm grew monotonously. To monitor mitophagy, a starvation model was established in HeLa cells. During mitophagy, damaged mitochondria might be phagocytosed by autolysosomes, and starved cells were then labeled with probe 1 and LTR (LysoTracker Red) (Figure 12D). As the starvation time increased, the green fluorescence of probe 1 gradually increased and overlapped with the fluorescence of LTR, a sign that the mitochondria had become more acidic. Therefore, probe 1 was capable of measuring changes in mitochondrial pH as well as monitoring mitophagy.
Figure 12.
(A) Ratiometric fluorescent probe 1 and its structural response to pH changes. (B) Absorption and (C) fluorescence spectra of compound 2, an analogue of probe 1 lacking the benzyl chloride functionality, recorded at different pH values. (D) Time course (0–360 s) images of nutrient-deprived cells. With a 488 nm excitation wavelength and 510–550 and 570–660 nm band-path emission filters, all pictures were captured. Reproduced from ref (91). Copyright 2014 American Chemical Society.
While most probes targeting mitochondria rely on negative mitochondrial membrane potentials, they often leak from the mitochondria and may not be suitable for tracking the evolution of mitochondrial pH over time. Therefore, to solve these problems, current research concentrates on creating immobilized fluorescent probes.92 In recent years, numerous pH probes have been created by researchers to investigate mitochondrial immobilization. Traditionally, a typical immobilization method utilizes the reaction between benzyl chloride with the thiol proteins in mitochondria. However, this immobilization method could cause irreversible damage to the mitochondrial structure function. In 2021, Yu’s team devised a solution for this problem by utilizing the fluorescent group naphthalimide to create a pH-sensitive fluorescent probe named MTpH (Figure 11).93 This probe consists of a pyridinium with a long alkyl chain and a nonconjugated morpholine component. The design of MTpH allows for immobilization within the mitochondria through interaction of the long chain with the mitochondrial membrane via physical hydrophobic interactions. This approach effectively addresses the problem at hand. MTpH responded sensitively to a wide range of pH changes and restored fluorescence by preventing the protonation of morpholine, which prevented it from being converted into naphthalimide during the PET process.94 It was validated by staining cells with MTpH and LTDR (LysoTracker Deep Red) in PBS, followed by CCCP and gastrin A-induced mitotic processes.
4. Conclusions and Perspective
As an energy-supplying organelle, a vital role for appropriate mitophagy in the cell and the entire living body and abnormal autophagy is closely associated with the development of many diseases. Therefore, real-time monitoring of cellular mitophagy is of great importance for its application in life sciences and diseases diagnosis in the clinical setting. In recent years, there has been great development of small-molecule fluorescent probes that can dynamically monitor mitophagy have been developed. Most of the probes are designed according to the differences in the microenvironment of different stages of cellular mitophagy, such as pH, viscosity, and polarity. Mitophagy is often monitored by assessing and observing changes in pH, as alterations in the pH are particularly notable in this process. Among the probes, a part only targets mitochondria containing autophagosomes without labeling lysosomes, showing high detection accuracy. Some probes target mitochondria and lysosomes separately and fluoresce when mitophagy occurs, which improves the accuracy of monitoring the pH changes during mitophagy. Moreover, some ratiometric probes can show different fluorescence changes depending on the microenvironment in which they are placed, allowing a molecular probe to visualize the various stages of mitophagy.
In the current review, we outline four mechanisms, namely, ICT (including TICT), FRET, TBET, and PET, that are utilized in the development of mitophagy probes. Over the past decade, it has been observed that the majority of probes are constructed based on ICT and FRET mechanisms, with the design of ICT-based probes being more predominant. In the case of FRET probes, employing a single molecular FRET response approach offers a novel perspective for studying alterations in mitophagy processes through pH changes as opposed to using separate molecules that target mitochondria and lysosomes individually for monitoring mitophagy. TBET is an effective means of an effective Stoke shift and high sensitivity sensing. The majority of TBET probes are based on the rhodamine system, which has a high energy conversion efficiency, and its structural systems are very straightforward for fluorescence probes using TBET technology. However, they also have certain limitations. TBET-based probes pose challenges in introducing response sites for analyte detection due to the limited number of active sites on the molecule. Further, it is difficult for TBET systems to completely avoid interference from the ICT and FRET processes. Therefore, future probe studies of TBET still need further development.95,96 PET-based fluorescent probes have a very strong quenching effect on fluorescence, thus providing advantages such as an exceptional signal-to-noise ratio, remarkable sensitivity, and high temporal and spatial resolution. However, these probes suffer from proton interference, which can have a significant impact on the PET response, and lowering the pKa value is expected to overcome these effects.75
In the design of probes for monitoring mitophagy, lipophilic cations are usually introduced as targeting groups so that the probes can collect in the mitochondrial matrix under the influence of transmembrane potential after passing through the phospholipid bilayer. The probe molecules’ cation content also increases their water solubility, allowing them to pass through the cell membrane and into the interior of the cell. However, targeting groups still need to be optimized. For example, TPP+ is the most commonly used mitochondrial targeting group, and with the increase of probe concentration, the toxicity of TPP+ will become non-negligible. Compared to TPP+, studies have demonstrated the feasibility of mitochondrial targeting using low toxicity pyridines or biomolecular targeting employing amino acid peptides.37,39,97 Hence, for the future development of probes targeting mitochondria, these motifs can be considered as alternatives to TPP+ for achieving efficient mitochondrial targeting.
Next, the sensitivity of the probes to the mitochondrial pH changes is still insufficient. Most current pH probes follow the Henderson–Hasselbalch equation because the pKa value (acidity coefficient) is an inherent property of the substance, and most pH probes have high sensitivity at pKa = 6–8 to match the pH of the mitochondria.98−101 Interestingly, the pH within the mitochondria undergoes only a slight change prior to the onset of mitophagy, yet this small pH variation significantly impacts its biological function. However, the pH response range of the mitophagy process is quite wide, spanning approximately two pH units. As a result, the Henderson–Hasselbalch equation is not capable of accurately predicting these subtle pH changes. On the other hand, Hill-type pH probes demonstrate greater accuracy and sensitivity in detecting minor pH fluctuations, particularly in the case of acid and weak base transitions. However, the development of guidelines for designing small-molecule Hill-type pH probes that are highly responsive is still a challenge. In order to create responsive sensing systems, two parameters must be carefully managed: the conversion range (identifying the conversion midpoint within the desired narrow response range) and the conversion width. However, due to limited precision and overall complexity, adjusting the midpoint may hinder the practical implementation of these probes.102−104 Thus, when designing small-molecule pH probes, it is crucial to create probes that accurately respond to the desired pH fluctuation range while also considering that the pKa values of the probes fall within the appropriate conversion range. One of the primary objectives to enhance the sensitivity of the pH probe is to investigate its capability to monitor early and late stages of mitophagy. This is based on understanding that different stages of mitophagy exhibit different pH values. The ability of the pH probe to differentiate between these various stages holds significant value as it has the potential to provide valuable insights into the progression and dynamics of mitophagy.
Overall, we highlight the significant progress in developing fluorescent probes based on energy transfer mechanisms for biological applications that will be challenged and advance with the development of improved measuring techniques, which call for probes to detect pH changes in vitro and in vivo for comprehending the function of mitophagy in some disorders. However, using fluorescent probes in clinical settings still presents significant challenges. The depth of certain human tissues presents a significant challenge that needs to be addressed in the design of fluorescent molecular probes for clinical applications. it is crucial to develop probes with improved tissue penetration and high resolution. Currently, most fluorescent probes currently absorb and emit in the visible range, which limits their potential for disease diagnosis. Consequently, the design of small molecule fluorescent probes with extended emission wavelengths to the NIR region represents an important future direction. In addition, the majority of fluorescent probes lack a targeting ligand and rely on nonspecific blood transport to reach the affected area, which can potentially affect imaging sensitivity. Therefore, incorporating a specific targeting ligand in fluorescent probes is essential for enhancing imaging accuracy and sensitivity. Another critical consideration is reducing the phototoxicity associated with fluorescent probes. High levels of phototoxicity can cause significant alterations to cellular structures and may result in detected states that differ from the physiological response. Thus, minimizing the phototoxicity of fluorescent probes is crucial to ensure accurate representation of cellular and physiological conditions.105 Fluorescent probes have a long way to go before they can be used. To create more useful fluorescence probes, it is still vital to research novel designs and response mechanisms. In the future, the design of small molecules with simpler structures, longer excitation as well as emission wavelengths, less phototoxicity, better transmembrane properties, and dual-band emission in the near-infrared region should become an increasingly popular research direction.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (22077101, L.L.), Fundamental Research Funds for the Central Universities (L.L.), President Funding of XMU (20720230047, D.A.X.), and Startup Program of XMU (D.A.X. and L.L.).
Glossary
Vocabulary Terms
- Fluorescent probes
A fluorescent probe is a molecular sensing tool designed to convert specific biological and chemical events into measurable fluorescent signals. Typically a fluorescent probe consists of three main components: the fluorophore, the connecting component, and the recognition unit.
- Energy transfer mechanisms
Energy transfer involves the change in fluorescence signal when the probes interact with an object. Based on the energy transfer mode, the response mechanism can be classified into intramolecular charge transfer (ICT), twisted intramolecular charge transfer (TICT), fluorescence resonance energy transfer (FRET), through bond energy transfer (TBET), and photoinduced electron transfer (PET).
- Disease models
When disease-causing factors act upon animals under specific conditions, they result in pathological damage to animal tissues, organs, or the whole body. This leads to a range of functional, metabolic, and structural changes. These basic pathological processes serve as ideal methods for studying disease mechanisms and conducting drug screening.
- Triphenylphosphine cation (TPP+)
TPP+ is an organic cation that are frequently utilized as targeting groups in fluorescent probes for mitochondrial localization.
- Reactive oxygen species (ROS)
ROS are highly reactive chemicals that contain oxygen. They include various peroxides, superoxides, and hydroxyl radicals.
Author Contributions
# Y.L. and D.Z.: Equal contribution to this work.
The authors declare no competing financial interest.
References
- Liu Y.; Zhou J.; Wang L.; Hu X.; Liu X.; Liu M.; Cao Z.; Shangguan D.; Tan W. A Cyanine Dye to Probe Mitophagy: Simultaneous Detection of Mitochondria and Autolysosomes in Live Cells. J. Am. Chem. Soc. 2016, 138, 12368–12374. 10.1021/jacs.6b04048. [DOI] [PubMed] [Google Scholar]
- She M.-T.; Yang J.-W.; Zheng B.-X.; Long W.; Huang X.-H.; Luo J.-R.; Chen Z.-X.; Liu A.-L.; Cai D.-P.; Wong W.-L.; et al. Design Mitochondria-Specific Fluorescent Turn-on Probes Targeting G-Quadruplexes for Live Cell Imaging and Mitophagy Monitoring Study. Chem. Eng. J. 2022, 446, 136947–136961. 10.1016/j.cej.2022.136947. [DOI] [Google Scholar]
- Ding W. X.; Yin X. M. Mitophagy: Mechanisms, Pathophysiological Roles, and Analysis. Biol. Chem. 2012, 393, 547–564. 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.; Xiao H.; Cheng Y.; Zhang W.; Huang F.; Zhang W.; Wang H.; Tang B. A Near-Infrared-Emitting Fluorescent Probe for Monitoring Mitochondrial pH. Chem. Commun. 2014, 50, 7184–7187. 10.1039/c4cc01390e. [DOI] [PubMed] [Google Scholar]
- Wu M. Y.; Li K.; Liu Y. H.; Yu K. K.; Xie Y. M.; Zhou X. D.; Yu X. Q. Mitochondria-Targeted Ratiometric Fluorescent Probe for Real Time Monitoring of pH in Living Cells. Biomaterials 2015, 53, 669–678. 10.1016/j.biomaterials.2015.02.113. [DOI] [PubMed] [Google Scholar]
- Gui L.; Yuan Z.; Kassaye H.; Zheng J.; Yao Y.; Wang F.; He Q.; Shen Y.; Liang L.; Chen H. A Tumor-Targeting Probe Based on a Mitophagy Process for Live Imaging. Chem. Commun. 2018, 54, 9675–9678. 10.1039/C8CC04246B. [DOI] [PubMed] [Google Scholar]
- Iwashita H.; Torii S.; Nagahora N.; Ishiyama M.; Shioji K.; Sasamoto K.; Shimizu S.; Okuma K. Live Cell Imaging of Mitochondrial Autophagy with a Novel Fluorescent Small Molecule. ACS Chem. Biol. 2017, 12, 2546–2551. 10.1021/acschembio.7b00647. [DOI] [PubMed] [Google Scholar]
- Li X.; Hu Y.; Li X.; Ma H. Mitochondria-Immobilized Near-Infrared Ratiometric Fluorescent pH Probe to Evaluate Cellular Mitophagy. Anal. Chem. 2019, 91, 11409–11416. 10.1021/acs.analchem.9b02782. [DOI] [PubMed] [Google Scholar]
- Li M.; Huang Y.; Song S.; Shuang S.; Dong C. Piperazine-Based Mitochondria-Immobilized pH Fluorescent Probe for Imaging Endogenous ONOO– and Real-Time Tracking of Mitophagy. ACS Appl. Bio Mater. 2022, 5, 2777–2785. 10.1021/acsabm.2c00213. [DOI] [PubMed] [Google Scholar]
- Chen W.; Han J.; She J.; Wang F.; Zhu L.; Yu R. Q.; Jiang J. H. Simultaneous Imaging of Lysosomal and Mitochondrial Viscosity During Mitophagy Using Molecular Rotors with Dual-Color Emission. Chem. Commun. 2020, 56, 7797–7800. 10.1039/D0CC00868K. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Zhang Y.; Zhao L.; Xu L.; Han H.; Huang Y.; Fei Q.; Sun Y.; Ma P.; Song D. A Novel Water-Soluble Near-Infrared Fluorescent Probe for Monitoring Mitochondrial Viscosity. Talanta 2021, 233, 122592–122598. 10.1016/j.talanta.2021.122592. [DOI] [PubMed] [Google Scholar]
- Wang T. R.; Chen Q.; Tang M. Y.; Zhang Y.; Shen S. L.; Cao X. Q. Visual Monitoring of the Mitochondrial pH Changes During Mitophagy with a NIR Fluorescent Probe and Its Application in Tumor Imaging. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 280, 121496–121504. 10.1016/j.saa.2022.121496. [DOI] [PubMed] [Google Scholar]
- Lu X.; Altshuler-Keylin S.; Wang Q.; Chen Y.; Henrique Sponton C.; Ikeda K.; Maretich P.; Yoneshiro T.; Kajimura S. Mitophagy Controls Beige Adipocyte Maintenance through a Parkin-Dependent and Ucp1-Independent Mechanism. Sci. Signal. 2018, 11, 8526–8534. 10.1126/scisignal.aap8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Wang M.; Ruan Y.; Tan J.; Wang H.; Yang T.; Li J.; Zhou Q. Ginkgolic Acids Impair Mitochondrial Function by Decreasing Mitochondrial Biogenesis and Promoting Fundc1-Dependent Mitophagy. J. Agric. Food Chem. 2019, 67, 10097–10106. 10.1021/acs.jafc.9b04178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C.; Przedborski S. Mitophagy: The Latest Problem for Parkinson’s Disease. Trends Mol. Med. 2011, 17, 158–165. 10.1016/j.molmed.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Jin S. M.; Youle R. J. Pink1- and Parkin-Mediated Mitophagy at a Glance. J. Cell Sci. 2012, 125, 795–799. 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zungu M.; Schisler J.; Willis M. S. All the Little Pieces Regulation of Mitochondrial Fusion and Fission by Ubiquitin and Small Ubiquitin-Like Modifer and Their Potential Relevance in the Heart. Circ. J. 2011, 75, 2513–2521. 10.1253/circj.CJ-11-0967. [DOI] [PubMed] [Google Scholar]
- Youle R. J.; Narendra D. P. Mechanisms of Mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14. 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B.; Zhang B.; Zhai R.; Wang L.; Ding Y.; Li H.; Bai H.; Wang Z.; Peng B.; Li L.; et al. Two-Photon Fluorescence Imaging of Mitochondrial Viscosity with Water-Soluble Pyridinium Inner Salts. New J. Chem. 2022, 46, 2487–2494. 10.1039/D1NJ05020F. [DOI] [Google Scholar]
- Zhang D.; He Y.; Wang J.; Wu L.; Liu B.; Cai S.; Li Y.; Yan W.; Yang Z.; Qu J. Mitochondrial Structural Variations in the Process of Mitophagy. J. Biophotonics 2022, 15, e202200006 10.1002/jbio.202200006. [DOI] [PubMed] [Google Scholar]
- Um J.-H.; Yun J. Emerging Role of Mitophagy in Human Diseases and Physiology. BMB Rep. 2017, 50, 299–307. 10.5483/BMBRep.2017.50.6.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V. V.; Suzuki H.; Marinkovic M.; Lang V.; Kato R.; Kawasaki M.; Buljubasic M.; Sprung M.; Rogova N.; Wakatsuki S.; et al. Phosphorylation of the Mitochondrial Autophagy Receptor Nix Enhances Its Interaction with Lc3 Proteins. Sci. Rep. 2017, 7, 1131–1142. 10.1038/s41598-017-01258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina-Varela R.; Castillo N.; Videla L. A.; Del Campo A. Impact of Mitophagy and Mitochondrial Unfolded Protein Response as New Adaptive Mechanisms Underlying Old Pathologies: Sarcopenia and Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2020, 21, 7704–7730. 10.3390/ijms21207704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Methods for Monitoring Autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2491–2502. 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Jeong S.; Widengren J.; Lee J. C. Fluorescent Probes for Sted Optical Nanoscopy. Nanomaterials 2022, 12, 21–36. 10.3390/nano12010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu G.; Zhang R.; Shi X.; Park H.; Xie S.; Kwok R. T. K.; Lam J. W. Y.; Tang B. Z. Aie Luminogens as Fluorescent Bioprobes. Trends Anal. Chem. 2020, 123, 115769–115796. 10.1016/j.trac.2019.115769. [DOI] [Google Scholar]
- Lu Q.; Li W.; Chen K.; Tian M. Monitoring Mitophagy Via the Fret Mechanism: Visualizing Mitochondria, Lysosomes, and Autolysosomes in Three Different Sets of Fluorescence Signals. Anal. Chem. 2021, 93, 9471–9479. 10.1021/acs.analchem.1c01237. [DOI] [PubMed] [Google Scholar]
- Sairi A. S.; Kuwahara K.; Sasaki S.; Suzuki S.; Igawa K.; Tokita M.; Ando S.; Morokuma K.; Suenobu T.; Konishi G.-i. Synthesis of Fluorescent Polycarbonates with Highly Twisted N,N-bis(dialkylamino)anthracene AIE Luminogens in the Main Chain. RSC Adv. 2019, 9, 21733–21740. 10.1039/C9RA03701B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong K.; Deng L.; Zhao J.; Yan X.; Sun T.; Li J.; Tang L. A Novel Near-Infrared Fluorescent Probe for Highly Selective Recognition of Hydrogen Sulfide and Imaging in Living Cells. RSC Adv. 2018, 8, 23924–23929. 10.1039/C8RA03457E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munan S.; Kottarathil S.; Joseph M. M.; Jana A.; Ali M.; Mapa K.; Maiti K. K.; Samanta A. IndiFluors: A New Full-Visible Color-Tunable Donor-Acceptor-Donor (D1-A-D2) Fluorophore Family for Ratiometric pH Imaging During Mitophagy. ACS Sens. 2022, 10.1021/acssensors.1c02381. [DOI] [PubMed] [Google Scholar]
- Bai Y.; Wang Y.; Cao L.; Jiang Y.; Li Y.; Zou H.; Zhan L.; Huang C. Self-Targeting Carbon Quantum Dots for Peroxynitrite Detection and Imaging in Live Cells. Anal. Chem. 2021, 93, 16466–16473. 10.1021/acs.analchem.1c03515. [DOI] [PubMed] [Google Scholar]
- Wang J.; Liu W.; Luo G.; Li Z.; Zhao C.; Zhang H.; Zhu M.; Xu Q.; Wang X.; Zhao C.; et al. Synergistic Effect of Well-Defined Dual Sites Boosting the Oxygen Reduction Reaction. Energy Environ. Sci. 2018, 11, 3375–3379. 10.1039/C8EE02656D. [DOI] [Google Scholar]
- Cicchini M.; Karantza V.; Xia B. Molecular Pathways: Autophagy in Cancer-A Matter of Timing and Context. Clin. Cancer Res. 2015, 21, 498–504. 10.1158/1078-0432.CCR-13-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A.; Komatsu M.; Mizushima N. Autophagy-Monitoring and Autophagy-Deficient Mice. Autophagy 2017, 13, 1619–1628. 10.1080/15548627.2017.1343770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.; Bu Y.; Wang J.; Qu M.; Zhang J.; Zhu X.; Liu G.; Wu Z.; Chen G.; Zhou H. A Convenient Fluorescent Probe for Monitoring Lysosomal pH Change and Imaging Mitophagy in Living Cells. Sens. Actuators B Chem. 2021, 330, 129363–129370. 10.1016/j.snb.2020.129363. [DOI] [Google Scholar]
- Niu L. Q.; Huang J.; Yan Z. J.; Men Y. H.; Luo Y.; Zhou X. M.; Wang J. M.; Wang J. H. Fluorescence Detection of Intracellular pHChanges in the Mitochondria-Associated Process of Mitophagy Using a Hemicyanine-Based Fluorescent Probe. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 207, 123–131. 10.1016/j.saa.2018.09.015. [DOI] [PubMed] [Google Scholar]
- Wang H.; Fang B.; Peng B.; Wang L.; Xue Y.; Bai H.; Lu S.; Voelcker N. H.; Li L.; Fu L.; et al. Recent Advances in Chemical Biology of Mitochondria Targeting. Front. Chem. 2021, 9, 683220–683236. 10.3389/fchem.2021.683220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka J.; Joseph J.; Sikora A.; Hardy M.; Ouari O.; Vasquez-Vivar J.; Cheng G.; Lopez M.; Kalyanaraman B. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem. Rev. 2017, 117, 10043–10120. 10.1021/acs.chemrev.7b00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N.; Liu Y. Y.; Li Y.; Zhuang J. B.; Cui R. R.; Gong Q.; Zhao N.; Tang B. Z. Fine Tuning of Emission Behavior, Self-Assembly, Anion Sensing, and Mitochondria Targeting of Pyridinium-Functionalized Tetraphenylethene by Alkyl Chain Engineering. ACS Appl. Mater. Interfaces 2018, 10, 24249–24257. 10.1021/acsami.8b04113. [DOI] [PubMed] [Google Scholar]
- Kuma A.; Hatano M.; Matsui M.; Yamamoto A.; Nakaya H.; Yoshimori T.; Ohsumi Y.; Tokuhisa T.; Mizushima N. The Role of Autophagy During the Early Neonatal Starvation Period. Nature 2004, 432, 1032–1036. 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Yao W.; Li Y.; Wu L.; Wu C.; Zhang Y.; Liu J.; He Z.; Wu X.; Lu C.; Wang L.; et al. Atg11 Is Required for Initiation of Glucose Starvation-Induced Autophagy. Autophagy 2020, 16, 2206–2218. 10.1080/15548627.2020.1719724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y.; Zheng K.; Clark J.; Swerdlow R. H.; Pulst S. M.; Sutton J. P.; Shinobu L. A.; Simon D. K. Rapamycin Drives Selection against a Pathogenic Heteroplasmic Mitochondrial DNA Mutation. Hum. Mol. Genet. 2014, 23, 637–647. 10.1093/hmg/ddt450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. S.; Choi S. E.; Koh H. C. PGAM5 Regulates Pink1/Parkin-Mediated Mitophagy via Drp1 in CCCP-Induced Mitochondrial Dysfunction. Toxicol. Lett. 2018, 284, 120–128. 10.1016/j.toxlet.2017.12.004. [DOI] [PubMed] [Google Scholar]
- Mlejnek P.; Dolezel P. Loss of Mitochondrial Transmembrane Potential and Glutathione Depletion Are Not Sufficient to Account for Induction of Apoptosis by Carbonyl Cyanide 4-(Trifluoromethoxy) Phenylhydrazone in Human Leukemia K562 Cells. Chem. Biol. Interact. 2015, 239, 100–110. 10.1016/j.cbi.2015.06.033. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Li G.; Zheng Y.; Shen H. M.; Hu X.; Ming Q. L.; Huang C.; Li P.; Gao N. A Novel Autophagy/Mitophagy Inhibitor Liensinine Sensitizes Breast Cancer Cells to Chemotherapy through DNM1L-Mediated Mitochondrial Fission. Autophagy 2015, 11, 1259–1279. 10.1080/15548627.2015.1056970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefa U.; J eong N. Y.; Song I. O.; Chung H. J.; Kim D.; J ung J.; Huh Y. Mitophagy Links Oxidative Stress Conditions and Neurodegenerative Diseases. Neural Regen. Res. 2019, 14, 749–756. 10.4103/1673-5374.249218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak V. I. Free Radicals, Reactive Oxygen Species, Oxidative Stress and Its Classification. Chem. Biol. Interact. 2014, 224, 164–175. 10.1016/j.cbi.2014.10.016. [DOI] [PubMed] [Google Scholar]
- Damascena H. L.; Silveira W. A. A.; Castro M. S.; Fontes W. Neutrophil Activated by the Famous and Potent Pma (Phorbol Myristate Acetate). Cells 2022, 11, 2889–2907. 10.3390/cells11182889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liese D.; Haberhauer G. Rotations in Excited Ict States - Fluorescence and Its Microenvironmental Sensitivity. Isr. J. Chem. 2018, 58, 813–826. 10.1002/ijch.201800032. [DOI] [Google Scholar]
- Yang G.; Li S.; Wang S.; Hu R.; Feng J.; Li Y.; Qian Y. Novel Fluorescent Probes Based on Intramolecular Charge- and Proton-Transfer Compounds. Pure Appl. Chem. 2013, 85, 1465–1478. 10.1351/PAC-CON-13-02-06. [DOI] [Google Scholar]
- Misra R.; Bhattacharyya S. P.. Intramolecular Charge Transfer Theory and Applications; Wiley, 2018. [Google Scholar]
- Pal A.; Karmakar M.; Bhatta S. R.; Thakur A. A Detailed Insight into Anion Sensing Based on Intramolecular Charge Transfer (Ict) Mechanism: A Comprehensive Review of the Years 2016 to 2021. Coord. Chem. Rev. 2021, 448, 214167–214221. 10.1016/j.ccr.2021.214167. [DOI] [Google Scholar]
- Patrizi B.; Cozza C.; Pietropaolo A.; Foggi P.; Siciliani de Cumis M. Synergistic Approach of Ultrafast Spectroscopy and Molecular Simulations in the Characterization of Intramolecular Charge Transfer in Push-Pull Molecules. Molecules 2020, 25, 430–449. 10.3390/molecules25020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Du W.; Shao T.; Li Z.; Zhang D.; Wang L.; Fang Z.; Li J.; Wu Q.; Zhang C.; et al. et al., Pyrimidine-Based Fluorescent Probe for Monitoring Mitophagy via Detection of Mitochondrial pH Variation. ChemBioChem. 2022, 23, e202200217 10.1002/cbic.202200217. [DOI] [PubMed] [Google Scholar]
- Lin B.; Fan L.; Zhou Y.; Ge J.; Wang X.; Dong C.; Shuang S.; Wong M. S. A Benzothiazolium-Based Fluorescent Probe with Ideal pKa for Mitochondrial pH Imaging and Cancer Cell Differentiation. J. Mater. Chem. B 2020, 8, 10586–10592. 10.1039/D0TB01253J. [DOI] [PubMed] [Google Scholar]
- Munan S.; Ali M.; Yadav R.; Mapa K.; Samanta A. Pet- and Ict-Based Ratiometric Probe: An Unusual Phenomenon of Morpholine-Conjugated Fluorophore for Mitochondrial Ph Mapping During Mitophagy. Anal. Chem. 2022, 94, 11633–11642. 10.1021/acs.analchem.2c02177. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Teng L.; Chen L.; Ma H.; Liu H. W.; Zhang X. B. Engineering of a Near-Infrared Fluorescent Probe for Real-Time Simultaneous Visualization of Intracellular Hypoxia and Induced Mitophagy. Chem. Sci. 2018, 9, 5347–5353. 10.1039/C8SC01684D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W.; Yu J.; Li Z.; Sun X.; Wang Y.; Wu Y.; Qin J.; Zhang Y.; Xiao H.; Zhang X. A Mitochondrial Suitable Ratiometric Fluorescent Probe for Tracking pH Change During Mitophagy. Sens. Actuators B Chem. 2022, 369, 132323–132329. 10.1016/j.snb.2022.132323. [DOI] [Google Scholar]
- Xiao H.; Dong Y.; Zhou J.; Zhou Z.; Wu X.; Wang R.; Miao Z.; Liu Y.; Zhuo S. Monitoring Mitochondrial pH with a Hemicyanine-Based Ratiometric Fluorescent Probe. Analyst 2019, 144, 3422–3427. 10.1039/C9AN00422J. [DOI] [PubMed] [Google Scholar]
- Jiang A.; Chen G.; Xu J.; Liu Y.; Zhao G.; Liu Z.; Chen T.; Li Y.; James T. D. Ratiometric Two-Photon Fluorescent Probe for in Situ Imaging of Carboxylesterase (Ce)-Mediated Mitochondrial Acidification During Medication. Chem. Commun. 2019, 55, 11358–11361. 10.1039/C9CC05759E. [DOI] [PubMed] [Google Scholar]
- Fu G.; Yin G.; Niu T.; Wu W.; Han H.; Chen H.; Yin P. A Novel Ratiometric Fluorescent Probe for the Detection of Mitochondrial pH Dynamics During Cell Damage. Analyst 2021, 146, 620–627. 10.1039/D0AN01240H. [DOI] [PubMed] [Google Scholar]
- Cheng F.; Qiang T.; Ren L.; Liang T.; Gao X.; Wang B.; Hu W. Observation of Inflammation-Induced Mitophagy During Stroke by a Mitochondria-Targeting Two-Photon Ratiometric Probe. Analyst 2021, 146, 2632–2637. 10.1039/D1AN00208B. [DOI] [PubMed] [Google Scholar]
- Lin B.; Wei Y.; Hao Y.; E S.; Shu Y.; Wang J. β-Naphthothiazolium-Based Ratiometric Fluorescent Probe with Ideal pKa for pH Imaging in Mitochondria of Living Cells. Talanta 2021, 232, 122475–122484. 10.1016/j.talanta.2021.122475. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Xia S.; Mikesell L.; Whisman N.; Fang M.; Steenwinkel T. E.; Chen K.; Luck R. L.; Werner T.; Liu H. Near-Infrared Hybrid Rhodol Dyes with Spiropyran Switches for Sensitive Ratiometric Sensing of pH Changes in Mitochondria and Drosophila Melanogaster First-Instar Larvae. ACS Appl. Bio Mater. 2019, 2, 4986–4997. 10.1021/acsabm.9b00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X.; Liu Z.; Yang Y.; Li H.; Qi X.; Ren W. X.; Deng M.; Lu M.; Wu J.; Liang S. A Mitochondria-Targeted Two-Photon Fluorescent Probe for Sensing and Imaging pH Changes in Living Cells. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 224, 117435–117442. 10.1016/j.saa.2019.117435. [DOI] [PubMed] [Google Scholar]
- Chen G.; Fu Q.; Yu F.; Ren R.; Liu Y.; Cao Z.; Li G.; Zhao X.; Chen L.; Wang H.; et al. Wide-Acidity-Range pH Fluorescence Probes for Evaluation of Acidification in Mitochondria and Digestive Tract Mucosa. Anal. Chem. 2017, 89, 8509–8516. 10.1021/acs.analchem.7b02164. [DOI] [PubMed] [Google Scholar]
- Mazi W.; Yan Y.; Zhang Y.; Xia S.; Wan S.; Tajiri M.; Luck R. L.; Liu H. A Near-Infrared Fluorescent Probe Based on a Hemicyanine Dye with an Oxazolidine Switch for Mitochondrial pH Detection. J. Mater. Chem. B 2021, 9, 857–863. 10.1039/D0TB02181D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X.; Wang Y.; Wang D.; Liu L.; Zhu W.; Zhang J.; Zha X. A Mitochondrial-Targetable Dual Functional near-Infrared Fluorescent Probe to Monitor pHand H2O2 in Living Cells and Mice. RSC Adv. 2020, 10, 26874–26879. 10.1039/D0RA03905E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.; Dai Y.; Gu B.; Liu M.; Yi Z.; Li Z.; Zhang Z.; He H.; Zeng R. A near Infrared Fluorescent Probe Based on Ict for Monitoring Mitophagy in Living Cells. Analyst 2020, 145, 1427–1432. 10.1039/C9AN02053E. [DOI] [PubMed] [Google Scholar]
- Sasaki S.; Drummen G. P. C.; Konishi G.-i. Recent Advances in Twisted Intramolecular Charge Transfer (TICT) Fluorescence and Related Phenomena in Materials Chemistry. J. Mater. Chem. C 2016, 4, 2731–2743. 10.1039/C5TC03933A. [DOI] [Google Scholar]
- Wang Y.; Krull I. S.; Liu C.; Orr J. D. Derivatization of Phospholipids. J. Chromatogr. B 2003, 793, 3–14. 10.1016/S1570-0232(03)00359-3. [DOI] [PubMed] [Google Scholar]
- Quan K.; Yi C.; Yang X.; He X.; Huang J.; Wang K. FRET-Based Nucleic Acid Probes: Basic Designs and Applications in Bioimaging. Trends. Analyt. Chem. 2020, 124, 115784–115795. 10.1016/j.trac.2019.115784. [DOI] [Google Scholar]
- Rowland C. E.; Brown C. W.; Medintz I. L.; Delehanty J. B. Intracellular FRET-Based Probes: a Review. Methods Appl. Fluoresc. 2015, 3, 042006–042033. 10.1088/2050-6120/3/4/042006. [DOI] [PubMed] [Google Scholar]
- Arai Y.; Nagai T. Extensive Use of Fret in Biological Imaging. Microscopy 2013, 62, 419–428. 10.1093/jmicro/dft037. [DOI] [PubMed] [Google Scholar]
- Tian X.; Murfin L. C.; Wu L.; Lewis S. E.; James T. D. Fluorescent Small Organic Probes for Biosensing. Chem. Sci. 2021, 12, 3406–3426. 10.1039/D0SC06928K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K.; Komatsu N.; Hirata E.; Kamioka Y.; Matsuda M. Stable Expression of Fret Biosensors: A New Light in Cancer Research. Cancer Sci. 2012, 103, 614–619. 10.1111/j.1349-7006.2011.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K.; Takakusa H.; Nagano T. Recent Advances in the Design of Small Molecule-Based FRET Sensors for Cell Biology. Trends Anal. Chem. 2004, 23, 407–415. 10.1016/S0165-9936(04)00608-9. [DOI] [Google Scholar]
- Guo X.; Li Q.; Xiang J.; Liu M.; Guan A.; Tang Y.; Sun H. A Hybrid Aggregate FRET Probe from the Mixed Assembly of Cyanine Dyes for Highly Specific Monitoring of Mitochondria Autophagy. Anal. Chim. Acta 2021, 1165, 338561–338569. 10.1016/j.aca.2021.338561. [DOI] [PubMed] [Google Scholar]
- Guo X.; Yang D.; Sun R.; Li Q.; Du H.; Tang Y.; Sun H. A Cyanine Dye Supramolecular FRET Switch Driven by G-Quadruplex to Monitor Mitophagy. Dyes Pigm. 2021, 192, 109429. 10.1016/j.dyepig.2021.109429. [DOI] [Google Scholar]
- Xue Z.; Zhao H.; Liu J.; Han J.; Han S. Responsive Hetero-Organelle Partition Conferred Fluorogenic Sensing of Mitochondrial Depolarization. Chem. Sci. 2017, 8, 1915–1921. 10.1039/C6SC04158B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Hu J.; Yang G.; Zhang X.; Zhang R.; Uvdal K.; Zhang Z.; Wu X.; Hu Z. Real-Time Tracking of Mitochondrial Dynamics by a Dual-Sensitive Probe. Sens. Actuators B Chem. 2020, 320, 128418. 10.1016/j.snb.2020.128418. [DOI] [Google Scholar]
- Qiu J.; Zhong C.; Liu M.; Xiong X.; Gao Y.; Zhu H. A Tunable pH Probe Scaffold Based on Sulfonamide Rhodamine and Its Application in Mitochondrial pH Research. Sens. Actuators B Chem. 2022, 371, 132606. 10.1016/j.snb.2022.132606. [DOI] [Google Scholar]
- Wan S.; Xia S.; Medford J.; Durocher E.; Steenwinkel T. E.; Rule L.; Zhang Y.; Luck R. L.; Werner T.; Liu H. A Ratiometric Near-Infrared Fluorescent Probe Based on a Novel Reactive Cyanine Platform for Mitochondrial pH Detection. J. Mater. Chem. B 2021, 9, 5150–5161. 10.1039/D1TB00643F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.; Jiang Y.; Guan Z.; Cao Y.; Sun S. C.; Liu H. FSH Protects Mouse Granulosa Cells from Oxidative Damage by Repressing Mitophagy. Sci. Rep. 2016, 6, 38090–38102. 10.1038/srep38090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D.; Zhu L.; Liu Z.; Lin W. Through Bond Energy Transfer (TBET)-Based Fluorescent Chemosensors. J. Photochem. Photobiol. C 2020, 44, 100371–100402. 10.1016/j.jphotochemrev.2020.100371. [DOI] [Google Scholar]
- Fan J.; Hu M.; Zhan P.; Peng X. Energy Transfer Cassettes Based on Organic Fluorophores: Construction and Applications in Ratiometric Sensing. Chem. Soc. Rev. 2013, 42, 29–43. 10.1039/C2CS35273G. [DOI] [PubMed] [Google Scholar]
- Lin W.; Yuan L.; Cao Z.; Feng Y.; Song J. Through-Bond Energy Transfer Cassettes with Minimal Spectral Overlap between the Donor Emission and Acceptor Absorption: Coumarin-Rhodamine Dyads with Large Pseudo-Stokes Shifts and Emission Shifts. Angew. Chem., Int. Ed. 2010, 49, 375–379. 10.1002/anie.200904515. [DOI] [PubMed] [Google Scholar]
- Xia S.; Wang J.; Zhang Y.; Whisman N.; Bi J.; Steenwinkel T. E.; Wan S.; Medford J.; Tajiri M.; Luck R. L.; et al. Ratiometric Fluorescent Probes Based on through-Bond Energy Transfer of Cyanine Donors to near-Infrared Hemicyanine Acceptors for Mitochondrial Ph Detection and Monitoring of Mitophagy. J. Mater. Chem. B 2020, 8, 1603–1615. 10.1039/C9TB02302J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.; Li M.; Fan J.; Peng X. Activity-Based Sensing and Theranostic Probes Based on Photoinduced Electron Transfer. Acc. Chem. Res. 2019, 52, 2818–2831. 10.1021/acs.accounts.9b00340. [DOI] [PubMed] [Google Scholar]
- Daly B.; Ling J.; de Silva A. P. Current Developments in Fluorescent PET (Photoinduced Electron Transfer) Sensors and Switches. Chem. Soc. Rev. 2015, 44, 4203–4211. 10.1039/C4CS00334A. [DOI] [PubMed] [Google Scholar]
- Lee M. H.; Park N.; Yi C.; Han J. H.; Hong J. H.; Kim K. P.; Kang D. H.; Sessler J. L.; Kang C.; Kim J. S. Mitochondria-Immobilized pH-Sensitive Off-Qn Fluorescent Probe. J. Am. Chem. Soc. 2014, 136, 14136–14142. 10.1021/ja506301n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Sun Q.; Huang Z.; Huang L.; Xiao Y. Immobilizable Fluorescent Probes for Monitoring the Mitochondria Microenvironment: a next Step from the Classic. J. Mater. Chem. B 2019, 7, 2749–2758. 10.1039/C9TB00043G. [DOI] [PubMed] [Google Scholar]
- Guo L.; Liu H.; Jin X.; Zhang Z.; Su J.; Yu X. Development of Reaction-Free and Mitochondrion-Immobilized Fluorescent Probe for Monitoring pH Change. Sens. Actuators B Chem. 2021, 341, 129962–129970. 10.1016/j.snb.2021.129962. [DOI] [Google Scholar]
- Koga N.; Tanioka M.; Kamino S.; Sawada D. Morpholine-Substituted Rhodamine Analogue with Multi-Configurational Switches for Optical Sensing of pH Gradient under Extreme Acidic Environments. Chem.—Eur. J. 2021, 27, 3761–3765. 10.1002/chem.202004254. [DOI] [PubMed] [Google Scholar]
- Gong Y. J.; Zhang X. B.; Zhang C. C.; Luo A. L.; Fu T.; Tan W.; Shen G. L.; Yu R. Q. Through Bond Energy Transfer: A Convenient and Universal Strategy toward Efficient Ratiometric Fluorescent Probe for Bioimaging Applications. Anal. Chem. 2012, 84, 10777–10784. 10.1021/ac302762d. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Tsao K.; Keillor J. W. Fluorogenic Protein Labelling: A Review of Photophysical Quench Mechanisms and Principles of Fluorogen Design. Can. J. Chem. 2015, 93, 389–398. 10.1139/cjc-2014-0405. [DOI] [Google Scholar]
- Zhu Z.; Wang Z.; Zhang C.; Wang Y.; Zhang H.; Gan Z.; Guo Z.; Wang X. Mitochondrion-Targeted Platinum Complexes Suppressing Lung Cancer through Multiple Pathways Involving Energy Metabolism. Chem. Sci. 2019, 10, 3089–3095. 10.1039/C8SC04871A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysandrou Y.; Newsome T.; Duty K.; Mohamed O.; Markiewicz J. T. Synthesis, Optical and Electronic Studies of a “Clickable” Quinoxaline-Based pH Sensor. J. Photochem. Photobiol., A 2022, 433, 114183–114191. 10.1016/j.jphotochem.2022.114183. [DOI] [Google Scholar]
- Xiao Y.; Huang Y.; Zeng Z.; Luo X.; Qian X.; Yang Y. Harnessing Thorpe-Ingold Dialkylation to Access High-Hill-Percentage pH Probes. J. Org. Chem. 2022, 87, 85–93. 10.1021/acs.joc.1c01887. [DOI] [PubMed] [Google Scholar]
- Webb B. A.; Chimenti M.; Jacobson M. P.; Barber D. L. Dysregulated pH: A Perfect Storm for Cancer Progression. Nat. Rev. Cancer 2011, 11, 671–677. 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- Zurawik T. M.; Pomorski A.; Belczyk-Ciesielska A.; Goch G.; Niedzwiedzka K.; Kucharczyk R.; Krezel A.; Bal W. Revisiting Mitochondrial pH with an Improved Algorithm for Calibration of the Ratiometric 5(6)-Carboxy-Snarf-1 Probe Reveals Anticooperative Reaction with H+ Ions and Warrants Further Studies of Organellar pH. PLoS One 2016, 11, e0161353 10.1371/journal.pone.0161353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Hu F.; Luo X.; Zhao M.; Sun Z.; Qian X.; Yang Y. Modulating the pKa Values of Hill-Type pH Probes for Biorelevant Acidic pH Range. ACS Appl. Bio. Mater. 2021, 4, 2097–2103. 10.1021/acsabm.0c01262. [DOI] [PubMed] [Google Scholar]
- Luo X.; Yang H.; Wang H.; Ye Z.; Zhou Z.; Gu L.; Chen J.; Xiao Y.; Liang X.; Qian X.; et al. Highly Sensitive Hill-Type Small-Molecule pH Probe That Recognizes the Reversed pH Gradient of Cancer Cells. Anal. Chem. 2018, 90, 5803–5809. 10.1021/acs.analchem.8b00218. [DOI] [PubMed] [Google Scholar]
- Nesterova I. V.; Nesterov E. E. Rational Design of Highly Responsive pH Sensors Based on DNA i-Motif. J. Am. Chem. Soc. 2014, 136, 8843–8846. 10.1021/ja501859w. [DOI] [PubMed] [Google Scholar]
- Han H. H.; Tian H.; Zang Y.; Sedgwick A. C.; Li J.; Sessler J. L.; He X. P.; James T. D. Small-Molecule Fluorescence-Based Probes for Interrogating Major Organ Diseases. Chem. Soc. Rev. 2021, 50, 9391–9429. 10.1039/D0CS01183E. [DOI] [PubMed] [Google Scholar]