Abstract
Cytokinins control key processes during plant growth and development, and cytokinin receptors CYTOKININ RESPONSE 1/WOODEN LEG/ARABIDOPSIS HISTIDINE KINASE 4 (CRE1/WOL/AHK4), AHK2, and AHK3 have been shown to play a crucial role in this control. The involvement of cytokinins in signaling the status of several nutrients, such as sugar, nitrogen, sulfur, and phosphate (Pi), has also been highlighted, although the full physiological relevance of this role remains unclear. To gain further insights into this aspect of cytokinin action, we characterized a mutant with reduced sensitivity to cytokinin repression of a Pi starvation-responsive reporter gene and show it corresponds to AHK3. As expected, ahk3 displayed reduced responsiveness to cytokinin in callus proliferation and plant growth assays. In addition, ahk3 showed reduced cytokinin repression of several Pi starvation-responsive genes and increased sucrose sensitivity. These effects of the ahk3 mutation were especially evident in combination with the cre1 mutation, indicating partial functional redundancy between these receptors. We examined the effect of these mutations on Pi-starvation responses and found that the double mutant is not significantly affected in long-distance systemic repression of these responses. Remarkably, we found that expression of many Pi-responsive genes is stimulated by sucrose in shoots and to a lesser extent in roots, and the sugar effect in shoots of Pi-starved plants was particularly enhanced in the cre1 ahk3 double mutant. Altogether, these results indicate the existence of multidirectional cross regulation between cytokinin, sugar, and Pi-starvation signaling, thus underlining the role of cytokinin signaling in nutrient sensing and the relative importance of Pi-starvation signaling in the control of plant metabolism and development.
Cytokinins are structurally diverse plant hormones with important roles in growth and development. Since their discovery as plant cell division promoting substances, an ever expanding number of roles have been attributed to these hormones. These include their well-established roles in the control of root branching and growth, shoot initiation, leaf differentiation, chloroplast biogenesis, and senescence (for review, see Mok and Mok, 1994, 2001; Haberer and Kieber, 2001). Several reports have also implicated cytokinins in responses related to the status of nutrients such as sugar, nitrogen, phosphorous, and sulfur (for review, see Sakakibara, 2003; Franco-Zorrilla et al., 2004; Maruyama-Nakashita et al., 2004a).
Significant progress has been recently made toward the elucidation of the molecular details of cytokinin signaling, leading to a model for signal transduction involving a His-Asp phosphorelay cascade that is similar to bacterial two-component systems (for review, see Hutchinson and Kieber, 2002; Hwang et al., 2002; Heyl and Schmulling, 2003). Molecular and genetic analyses have shown that the CYTOKININ RESPONSE 1/WOODEN LEG/ARABIDOPSIS HISTIDINE KINASE 4 (CRE1/WOL/AHK4) gene encodes a cytokinin receptor (Mähönen et al., 2000; Inoue et al., 2001; Suzuki et al., 2001; Franco-Zorrilla et al., 2002). CRE1 is a hybrid His kinase that, upon perception of the hormone, is activated through autophosphorylation at a His residue and triggers a cascade of phosphorelay reactions. In Arabidopsis (Arabidopsis thaliana), there are two additional genes, AHK2 and AHK3, coding for His kinase proteins with close homology to CRE1 (Inoue et al., 2001; Ueguchi et al., 2001; Yamada et al., 2001). The recent isolation and analysis of single, double, and triple mutants of members of the CRE1 family have proven the important overlapping, but distinct, role of these cytokinin receptors in root growth, leaf development, formation and maintenance of meristem activity, and reproductive growth (Higuchi et al., 2004; Nishimura et al., 2004). However, the full physiological roles of cytokinins and cytokinin receptors are not yet understood, particularly with regard to their participation in the modulation of responses to plant nutrient status.
The involvement of cytokinins in nutrient signaling responses was first suggested by studies in which cytokinin levels were found to decrease after phosphate (Pi) or nitrate starvation (Salama and Wareing, 1979; Horgan and Wareing, 1980). In the case of Pi, we have shown that exogenous application of cytokinins represses the induction of many Pi starvation-responsive genes (Martín et al., 2000), and this effect is impaired in cre1 mutants, implicating cytokinin two-component signaling in the negative regulation of Pi-starvation responses (Franco-Zorrilla et al., 2002). Similar results regarding cytokinins and CRE1 have been reported for high affinity sulfate transporter genes (Maruyama-Nakashita et al., 2004b). The observation that cytokinins affect only Pi-starvation responses that are dependent upon whole plant Pi status (controlled by systemic signals), and not local Pi-dependent responses (such as increased root hair number and length), led us to suggest that the cytokinin signaling pathway could be a candidate for the systemic repression signaling system (Martín et al., 2000).
Cytokinin signaling may also be involved in sugar sensing. Loss-of-function mutations in the HEXOKINASE 1 (HXK1) gene display decreased sensitivity to sugar as well as increased cytokinin sensitivity. Reciprocally, constitutive activation of cytokinin signaling confers decreased sugar sensitivity (Moore et al., 2003). In summary, there is good evidence for the interaction between cytokinins and the sensing of several nutrients, although its significance remains unclear. Moreover, links among the sensing of different nutrients are emerging, with sugar sensing occupying a central position. Indeed, interactions between sugar and nitrogen sensing have been documented (Coruzzi and Zhou, 2001; Price et al., 2004) and some reports indicate a relationship between sugars, sulfate, and Pi (Sadka et al., 1994; Nielsen et al., 1998; Ciereszko et al., 2001; Ciereszko and Kleczkowski, 2002; Hammond et al., 2003; Lejay et al., 2003; Wu et al., 2003; Maruyama-Nakashita et al., 2004a). Pi starvation, for instance, was shown to decrease the expression of several sugar-induced genes in different plants, and sugars have been shown to induce the expression of a Pi transporter in Arabidopsis (Sadka et al., 1994; Nielsen et al., 1998; Ciereszko et al., 2001; Ciereszko and Kleczkowski, 2002; Lejay et al., 2003). Additionally, microarray expression analysis of Pi-starved plants showed that the expression of genes involved in carbohydrate metabolism is altered under this stress (Hammond et al., 2003; Wu et al., 2003). All these data prompted us to investigate further the possible interactions between sugar, Pi, and cytokinin signaling.
In this study, we report the isolation of a mutant displaying reduced sensitivity to cytokinin repression of a Pi starvation-responsive gene and show that it corresponds to a mutant allele of the cytokinin receptor AHK3. This gene displays partial redundancy with CRE1 in several cytokinin responses, including the repression of Pi-starvation responses, as well as sugar sensitivity. Studies on the expression of Pi starvation-responsive genes in wild type, and cre1 and ahk3 mutants, suggest that a prominent role for CRE1 and AHK3 in systemic repression of Pi starvation is unlikely. Remarkably, these studies provide evidence for a functional link between cytokinin, sugar, and Pi-starvation signaling, involving the CRE1 and AHK3 receptors.
RESULTS
Isolation and Characterization of a Cytokinin Hyposensitive ahk3 Mutant
With the aim of identifying genes responsible for the repression by cytokinins of the expression of Pi starvation-responsive genes, we previously screened for mutants defective in the cytokinin-mediated repression of the Pi starvation-responsive IPS1-β-glucuronidase (GUS) reporter gene. In that study, we recovered 10 mutant lines, with seven displaying high cytokinin insensitivity and showing mutations at CRE1 (Franco-Zorrilla et al., 2002). One of the three lines not corresponding to CRE1 was less sensitive to cytokinin repression than the wild type at lower cytokinin concentrations (Fig. 1). This mutant exhibited insensitivity to this hormone in additional cytokinin response assays (see below; Fig. 1).
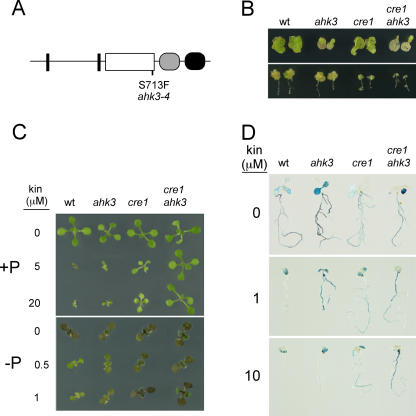
Figure 1.
Isolation and characterization of the ahk3 mutant. A, Scheme of the AHK3 predicted protein, where biologically relevant domains are represented as follows: transmembrane domains, black rectangles; His kinase, white box; pseudo-receiver domain, gray oval; and receiver domain, black oval. The point mutation in ahk3-4 causing a Ser to Phe substitution is indicated. B, Calli growth assays. Cotyledon (top) and root (bottom) explants from the same lines as in A were assayed for calli formation in the presence of 100 ng mL−1 2,4-dichlorophenoxyacetic acid and 100 (cotyledons) or 125 (roots) ng mL−1 kinetin. C, Plant growth and the effect of kinetin. Wild-type (wt), ahk3, cre1, and double cre1 ahk3 mutants were cultivated for 9 d in Pi-sufficient (+P; top) and in Pi-deficient (−P; bottom) media supplemented with kinetin at different concentrations for 9 d. D, IPS1-GUS expression in wild-type (wt), ahk3, cre1, and double cre1 ahk3 mutants. Plants were cultivated in Pi-lacking medium supplemented with kinetin for 9 d and histochemical analysis of GUS activity was performed.
The mutation segregated as a codominant trait in F2 segregating populations obtained from backcrosses with the wild type (not shown). Mapping of the mutation revealed that it was within a 1.1-Mb region on chromosome 1 flanked by simple sequence length polymorphism (SSLP) markers ZFPG and nga392. The AHK3 gene is within this interval (At1g27320; Ueguchi et al., 2001) and sequence analysis revealed a G to A transition in this gene in the mutant line. This mutation generates a Ser to Phe change at amino acid 713 of the predicted AHK3 protein. This is located at the C-terminal end of the His kinase catalytic domain and within the G2 motif, which is involved in ATP/ADP binding (Fig. 1A; Stock et al., 2000; Wolanin et al., 2002). Transformation of the mutant with a 7.8-kb genomic fragment containing the AHK3 gene rescued the wild-type phenotype in 11 out of 13 independent transgenic lines homozygous for the construct (data not shown). These results confirmed that the mutant was an allele of AHK3, which we have designated ahk3-4 (hereafter referred to as ahk3). For further analysis, the ahk3 mutant was backcrossed four times. To evaluate the effects of the ahk3 mutation, we performed several phenotypic tests related to cytokinin responses. Given the high similarity between AHK3 and CRE1 (81% amino acid similarity; being the highest similarity among cytokinin receptors), these analyses were also carried out initially (i.e. experiments in Fig. 1) for two strong cre1 alleles, cre1-3 and cre1-7 (Franco-Zorrilla et al., 2002), and the corresponding double mutants cre1-3 ahk3 and cre1-7 ahk3. Due to the similar phenotypes shown by the two cre1 alleles in these experiments, either alone or in combination with ahk3, experiments presented in subsequent sections were carried out only on the cre1-3 allele (hereafter referred to as cre1) and the corresponding double mutant (cre1 ahk3). Here, we show only the results from this cre1 allele. We first examined callus formation and growth from cotyledon and root explants. Qualitative differences were observed between the ahk3 and cre1 mutants (Fig. 1B). ahk3 cotyledons displayed higher insensitivity to cytokinins than cotyledons of cre1, whereas the opposite was found when root explants were used in these assays. For both types of explants, the response of the double mutant was lower than that of any of the single mutants. These results are very similar to those reported for putative loss of function T-DNA insertion alleles (Higuchi et al., 2004; Nishimura et al., 2004), suggesting that ahk3-4 is also a strong (if not a loss of function) allele. Next, we evaluated plant growth under two Pi regimens (Pi sufficient and Pi starvation) in the presence of different amounts of cytokinin (kinetin). No difference was observed between the mutants and the wild type when grown in the absence of cytokinin (Fig. 1C). At all kinetin concentrations tested, ahk3 plants showed only a slight insensitivity to the hormone, intermediate between that of cre1 and wild-type plants. This is seen in the growth of rosettes, as well as in anthocyanin accumulation in Pi-starved plants, which in the wild type is dramatically reduced by cytokinins (Fig. 1C). However, the double mutant revealed more than additive effects of cre1 and ahk3 on the development of the plants grown in the presence of kinetin, which were much more conspicuous under high concentrations of the hormone (Fig. 1C). Similar results were obtained for the repression of the Pi-responsive IPS1:GUS reporter gene (Fig. 1D). Altogether, these results indicate overlapping but distinct functions of AHK3 and CRE1 and their involvement in the negative control of Pi-starvation responses by cytokinins.
Molecular Characterization of ahk3
To evaluate the effects of the ahk3-4 mutation at the molecular level, we performed northern-blot analyses to monitor the expression of the primary cytokinin response genes A-type ARABIDOPSIS RESPONSE REGULATORs (ARRs; Fig. 2A) and of Pi starvation-induced genes (Fig. 2B). Two sets of A-type ARR genes were used as markers: ARR4 and ARR6, which are mainly induced in shoots (D'Agostino et al., 2000; To et al., 2004) and ARR15 and ARR16, whose expression in roots is particularly reduced in cre1-1 mutants (Kiba et al., 2002). Additionally, we examined the expression of the cyclin CYCD3, which has been shown to be induced by cytokinins after 24 h (Riou-Khamlichi et al., 1999). In the absence of hormone, basal expression of all the genes tested was quite similar in wild-type and mutant plants. However, after kinetin treatment, the induction of all these genes was compromised in mutant plants, albeit to different extents (Fig. 2A). In particular, ARR16 showed a similar dramatic reduction in expression in any of the single cre1 and ahk3 mutants and in the double mutant, suggesting that expression of this gene requires the cooperation between CRE1 and AHK3. Molecularly, such cooperation could be via the formation of a heterodimer, given the likely action of these receptors as dimers as suggested by interallelic complementation studies with cre1 (wol) alleles (García-Ponce de León et al., 2004).
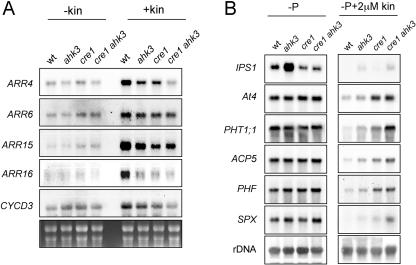
Figure 2.
Molecular characterization of ahk mutants. A, Northern analysis of expression of type cytokinin-induced genes in wild type (wt), ahk3, cre1, and the corresponding double mutant (cre1 ahk3). RNA was extracted from plants grown for 6 d and treated with or without kinetin (15 μm) for an additional day. Total RNA was isolated and RNA blots containing 15 μg of RNA per sample were subsequently hybridized to probes corresponding to the cytokinin-induced genes indicated. B, Influence of kinetin on the expression of Pi starvation-responsive genes in roots. Plants were grown in absence of Pi supplemented or not with 2 μm kinetin, and root material was collected after 9 d. Total RNA was isolated from roots, and RNA blots containing 15 μg of RNA per sample were subsequently hybridized to probes corresponding to the Pi starvation-responsive genes indicated.
The effect of the cre1 and ahk3 mutations on CYCD3 expression is noteworthy, since previously it was found that cytokinin responsiveness of CYCD3 was dependent on regulatory phosphorylation (Riou-Khamlichi et al., 1999). Our finding suggests that regulatory phosphorylation acts downstream of canonical cytokinin signaling.
To test the effect of these mutations on the expression of Pi starvation-responsive genes, RNAs were obtained from roots of plants grown for 7 d in the absence of Pi and in the presence or absence of kinetin. We analyzed the expression of several Pi starvation-responsive genes, the related nonprotein coding genes IPS1 and At4 (Burleigh and Harrison, 1999; Martín et al., 2000), the high affinity Pi transporter gene PHT1;1 (Muchhal et al., 1996), the purple acid phosphatase gene ACP5 (del Pozo et al., 1999), and PHF and SPX (At3g52190 and At2g45130, respectively; F. Scaglia, R. Bustos, and J. Paz-Ares, unpublished data). In this experiment (Fig. 2B), all genes except IPS1 responded similarly to Pi starvation in the absence of cytokinins in wild-type and mutant plants. However, in the presence of cytokinins, while Pi-starvation genes were dramatically down-regulated in the wild type, in ahk3 and especially cre1 mutants, and even more the cre1 ahk3 double mutant, they displayed reduced down-regulation. These data support mostly additive roles of both receptors in the cytokinin repression of the Pi-starvation response. Further investigation is required to explain the higher expression of IPS1 in ahk3 and its reduced expression in cre1.
Systemic Down-Regulation of Pi-Starvation Responses in ahk Mutants
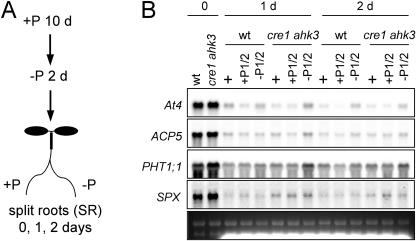
In roots, most nutrient deficiency responses, including Pi starvation, depend on the whole plant status of the nutrient in question rather than on the external concentration of the nutrient. As a result, if one part of the root system receives enough nutrient to satisfy the needs of shoot growth, the corresponding nutrient starvation response will be systemically down-regulated in the remaining part of the root system (see, for instance, Drew and Saker, 1984; Scheible et al., 1997; Liu et al., 1998; Burleigh and Harrison, 1999; Lappartient et al., 1999). We previously highlighted a parallel between this phenomenon and the repression of the Pi-starvation responses by cytokinins (Martín et al., 2000). To test the role of cytokinin signaling in systemic down-regulation of Pi-starvation responses, we conducted split root experiments with wild type and the double mutant cre1 ahk3, in which one part of the root system of Pi-starved plants was placed in Pi-rich media, whereas the other part was placed in media lacking this nutrient, as schematically represented in Figure 3A. We analyzed the expression of some Pi starvation-inducible genes in a time-course experiment by RNA-gel blot analysis (Fig. 3B). Repression of Pi starvation-induced gene expression in the roots of both genotypes was observed within the first day of the treatment, regardless of whether the split roots were grown in Pi-rich or in Pi-lacking media. In the double mutant, a slight decrease in repression could be observed, but relative to the wild type, it was not specific for the split roots growing in Pi-lacking media.
Figure 3.
Split-root assays of systemic repression of Pi-starvation responses in ahk mutants. A, Schema of the split root experiment. First plants were grown for 10 d in complete medium and then transferred for an additional 2 d to a Pi-lacking medium. Subsequently, plants were transferred for 1 or 2 d to split plates with two independent compartments placing part of the root system in each of these compartments. B, Northern analysis of systemic repression of Pi-starvation response in the cre1 ahk3 double mutant. Plants were grown for 1 or 2 d with the roots split in two compartments having Pi-sufficient (P1/2) media and Pi-lacking media (−P1/2), respectively. As control, both parts of the split root system were placed in Pi-rich media (+). In this experiment, only wild-type (wt) and double mutant cre1 ahk3 RNA were prepared from each part of the split root harvested independently when they where placed in different media, and 10 μg of RNA sample were used in the experiment. The blot was hybridized with the probes corresponding to the Pi starvation-responsive genes indicated.
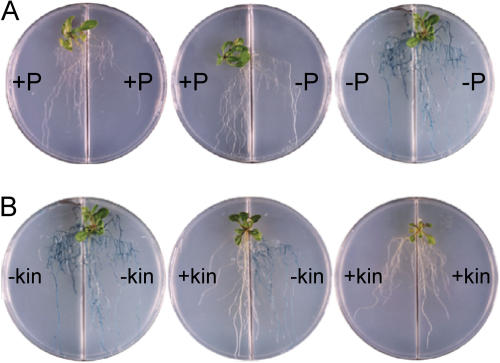
We also examined whether externally added cytokinins could trigger systemic repression using a similar split root assay with wild-type plants harboring the IPS1-GUS reporter gene (Fig. 4). When the roots of Pi-starved plants were split between Pi-containing and Pi-lacking media, the expression of the IPS1-GUS was negligible in both parts of the root, indicating systemic down-regulation (Fig. 4A). In contrast, when the roots were split and transferred to media lacking Pi with or without cytokinin, then repression of IPS1-GUS occurred only in the part of the root system in contact with the hormone (Fig. 4B). This indicates that exogenously added kinetin does not translocate efficiently throughout the whole plant and that local perception of cytokinin is necessary to block the Pi-starvation response.
Figure 4.
Effect of cytokinin on repression of Pi starvation-responsive IPS1-GUS in split root assays. Ten-day-old transgenic plants harboring the IPS1-GUS gene (Martín et al., 2000) were Pi starved for 2 d, then transferred to split plates with the compartments containing media with the compositions as indicated and, after 3 d, histochemical analysis of GUS activity was performed. A, The compartments had Pi-sufficient (+P) or Pi-deficient (−P) media. B, The compartments had media lacking Pi, but each was supplemented or not with 10 μm kinetin (kin).
Sugar Sensitivity of ahk Mutants
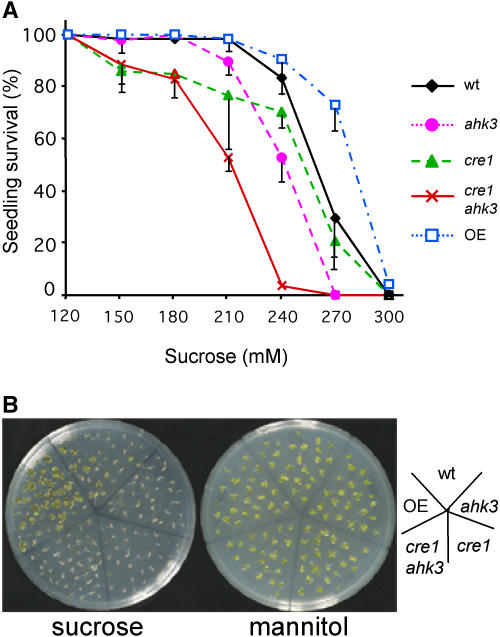
Recently, an antagonistic interaction between cytokinin and HXK1-dependent sugar signaling has been described (Moore et al., 2003). We therefore investigated whether CRE1 and AHK3 could be directly involved in such sugar-cytokinin cross talk. We evaluated sugar sensitivity of single mutants, the double mutant cre1 ahk3, and a transgenic line highly overexpressing AHK3 (driven from the cauliflower mosaic virus 35S promoter; see “Materials and Methods”). The overexpressing line used was chosen out of three showing the highest level of AHK3 RNA, and concomitantly resulting in enhanced response (albeit moderate) to cytokinin in root growth assays and repression of IPS1:GUS expression (data not shown). Plants were germinated and grown in complete medium with varying concentrations of Suc for 10 d. Plants ectopically expressing AHK3 were more resistant to high Suc concentrations than the wild type and, reciprocally, cytokinin-insensitive mutants displayed higher Suc sensitivity, in particular the cre1 ahk3 double mutant (Fig. 5). Control experiments in which mannitol was substituted for the Suc were carried out in parallel, and no decrease in plant survival was observed, excluding differences attributable to osmotic effects (Fig. 5B). Thus, these results confirm the antagonistic interaction between cytokinin and Suc and involve CRE1 and AHK3 in this effect.
Figure 5.
ahk mutants display Suc sensitivity. A, Seedlings were grown for 10 d in media with increasing concentrations of Suc in a range from 60 to 360 mm and scored after this time for seedling survival. Suc concentrations below 120 mm did not cause lethality in any of the genotypes tested and above 300 mm prevented seedlings survival. Genotypes used are the same as in previous figures and OE represents one 35S-AHK3 overexpressing line. Vertical bars represent sds from two replica experiments in which 40 seedlings per genotype and experiment were scored. B, Ten-day-old plants from the same genotypes grown at 275 mm Suc or mannitol.
Sugar Control of Pi Starvation-Responsive Genes in ahk Mutants
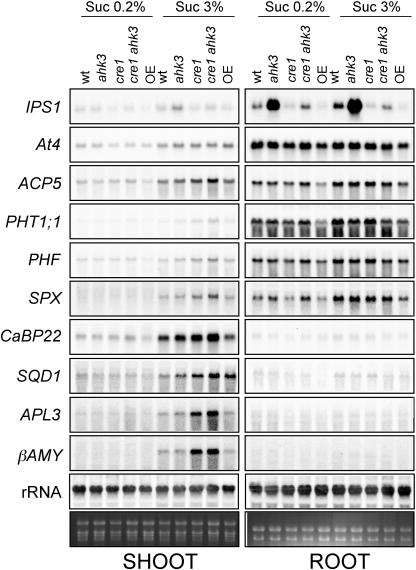
There is some evidence supporting the interaction between sugar and Pi sensing (Sadka et al., 1994; Nielsen et al., 1998; Ciereszko et al., 2001; Ciereszko and Kleczkowski, 2002; Hammond et al., 2003; Lejay et al., 2003; Wu et al., 2003). These indications, together with the antagonistic interaction between sugar and cytokinins, prompted us to evaluate the effects of sugars and Pi starvation on gene expression in the cytokinin-insensitive mutants, as well as in the transgenic line overexpressing AHK3. For this purpose, RNA was extracted from plants grown for 7 d in the presence of low Suc and then transferred for 3 d to media lacking Pi and containing high or low Suc before harvesting shoots and roots separately (Fig. 6). Pi starvation-responsive genes whose expression was analyzed were the same as in previous experiments (Fig. 2), as well as SQD1 (Essigmann et al., 1998) and CaBP22 (At2g41090; F. Scaglia, R. Bustos, and J. Paz-Ares, unpublished data). Additionally, we analyzed the expression of two sugar-inducible genes, APL3 and β-AMY (Mita et al., 1995; Sokolov et al., 1998).
Figure 6.
Effect of Suc on the expression of Pi starvation-responsive genes in ahk mutants. Plants of the genotypes indicated were grown for 7 d in Pi-rich media containing a low Suc concentration (0.2%) and transferred for additional 3 d to Pi-lacking media containing low (0.2%) or high (3%) Suc concentration. RNA was prepared from shoots and roots separately and 15 μg was loaded in each lane. Blots were hybridized to the probes corresponding to Pi starvation-responsive genes (IPS1, At4, ACP5, PHT1;1, PHF, SPX, CaBP22, and SQD1) and sugar-responsive genes (APL3 and β-AMY).
Despite some differences in the behavior of the various genes analyzed, the overall picture is that high Suc enhances the expression not only of sugar-induced genes but also of Pi starvation-responsive genes, particularly in shoots, but also in roots (when expression was detected), and the effect of sugars in shoots of Pi-starved plants was particularly enhanced in the cre1 ahk3 double mutant (Fig. 6). We note that shoots are sensitive to Pi starvation and in fact the expression of several of the genes examined in this experiment (At4, IPS1, ACP5, PHT1;1, and SQD1) has been demonstrated to be Pi starvation responsive in shoots (Muchhal et al., 1996; Essigmann et al., 1998; Burleigh and Harrison, 1999; del Pozo et al., 1999; Martín et al., 2000). The nonprotein coding genes, At4 and IPS1, deviated from the behavior displayed by all other Pi starvation-responsive genes examined, although they showed differences in expression levels in mutants relative to wild-type plants, at least in some of the conditions/tissue tested and displayed responsiveness to sugars (at least in some mutants). The reasons for the different behavior of these presumably regulatory, nonprotein coding genes, needs to be further investigated.
Some differences were observed between the expression of sugar-responsive and Pi starvation-responsive genes in relation to the cre1 and ahk3 mutants. Thus, the expression of sugar-responsive genes was more highly enhanced in the double mutant than Pi starvation-responsive genes, and ahk3 had a negligible contribution to the increase in expression of these genes, whereas in the case of Pi starvation-responsive genes, the effect of ahk3, although not so important as that of cre1, was evident (Fig. 6).
In the cases in which the effect of the cre1 or ahk3 mutations on overall gene expression was not evident (i.e. in shoots from low-Suc grown plants and in roots from high- or low-Suc grown plants), AHK3 overexpression resulted in a small but consistent decrease of the expression of all sugar and Pi starvation-inducible genes. Altogether, these results provide a clear indication of an interaction between cytokinin, sugar, and Pi-starvation signaling and suggests that the role of cytokinin signaling in the control of sugar and Pi starvation-responsive genes may be quite broad and complex and not exclusive for shoots of plants grown under a high-Suc, low-Pi regimen.
DISCUSSION
Many studies have shown that plants have nutrient-specific signaling mechanisms to adapt their growth and development to changing nutritional conditions. One example of these is the controlling system of Pi starvation, in which the transcription factor PHR1 plays a key regulatory role (Rubio et al., 2001). This study reveals an additional level of complexity in the control of Pi-starvation responses, as indicated by the multidirectional interactions between Pi starvation, sugar, and cytokinin signaling, involving the cytokinin receptors CRE1 and AHK3. This finding further underlines the relevance of cytokinins in relation to nutrient responses and the prominent role of Pi-starvation signaling in the control of plant metabolism and development.
Cytokinin Interactions with Sugar and Pi-Starvation Signaling
Our studies have shown that plants with impaired cytokinin receptors CRE1 and AHK3 display increased sugar sensitivity in seedling survival tests and enhanced expression of both Pi starvation- and sugar-responsive genes in shoots of high sugar grown plants (see “Results” and Figs. 5 and 6). In the case of sugar signaling, it has recently been reported that there is a bidirectional antagonistic interaction between sugars and cytokinins. In this study, however, the effect of cytokinins on sugar sensing was based on studies with transgenic plants overexpressing genes constitutively activating cytokinin signaling (Moore et al., 2003). Our results showing that the cre1-3 and ahk3-3 mutants, particularly in combination, display increased Suc sensitivity both in seedling survival and gene expression assays provide a clear demonstration on the involvement of these two genes in the sugar cytokinin interaction.
It is noteworthy that our study revealed an effect of the cre1 mutation on the β-AMY expression, whose sugar responsiveness has been attributed to the HXK1-independent sugar-sensing pathway (Xiao et al., 2000). This extends the effect of cytokinins on sugar-responsive genes beyond the sugar sensing dependent of HXK1 (Smeekens, 2000; Rolland et al., 2002; Moore et al., 2003), at least during Pi starvation. A similar conclusion on a bidirectional interaction can be drawn for Pi starvation and cytokinins, since it was shown that Pi starvation reduces cytokinin signaling by decreasing both cytokinin content and CRE1 expression (Salama and Wareing, 1979; Horgan and Wareing, 1980; Franco-Zorrilla et al., 2002).
It is intriguing that the effect of cre1 and ahk3 mutations on the expression of sugar and Pi starvation-responsive genes is only evident under conditions of high sugar and in the shoot, but not in the root (Fig. 5). Such a restricted role of cytokinins in Pi-starvation signaling is in conflict with reports associating the increase of the root-to-shoot growth ratio of plants during Pi starvation with the observed reduction of cytokinin signaling under these conditions (Kuiper, 1988; Kuiper et al., 1988). In addition, it is plausible that senescence, a Pi-mobilizing condition that can be stimulated by Pi starvation, involves reduced cytokinin signaling. A simple explanation of these apparent differences is that there is additional cytokinin receptor function in the cre1 ahk3 double mutant probably mediated by the third cytokinin receptor, AHK2, whose partial functional redundancy with CRE1 and AHK3 is differentially manifested in different parts of the plant and under different sugar regimens. This possibility would be in line with the recently reported existence of both redundant and specific functions for CRE1, AHK2, and AHK3 in relation to plant growth and development (Higuchi et al., 2004; Nishimura et al., 2004). Actually, the fact that Pi starvation-responsive genes are repressed by cytokinins in roots (Martín et al., 2000; see also Figs. 1, 2, and 3) and the existence of some cytokinin sensitivity in the double cre1 ahk3 mutant, at least concerning cytokinin repression of Pi starvation-responsive genes (Figs. 1 and 2), suggests that AHK2 also plays a role in the control of nutrient related responses. Moreover, hyperexpression of AHK3 does result in consistent, albeit moderate, reduction of nutrient-responsive gene expression both in roots, independent of the sugar concentration in the growth media, and to some extent also in shoots from low sugar grown plants (see Fig. 6). The analysis of the triple mutant cre1 ahk2 ahk3 could confirm this hypothesis on the role of AHK2 regarding nutrient related responses. However, the severe growth and developmental defects of the triple mutant (Higuchi et al., 2004; Nishimura et al., 2004) complicates the analysis of Pi-starvation and sugar responses. The isolation of weak mutants of these AHK genes might help to overcome these limitations.
We have also investigated the potential role of cytokinins in long-distance repression signaling of Pi-starvation responses. However, the analysis of systemic repression remaining in the cre1 ahk3 mutant did not support any significant role for these receptors per se in long-distance signaling of whole Pi status (Fig. 3). Additionally, exogenous cytokinins themselves are unable to systemically repress Pi-starvation responses (Fig. 4). Altogether, these results make a prominent role of cytokinins in systemic repression unlikely.
Interaction between Pi Starvation and Sugar Signaling
Phosphorus is an essential macronutrient, and plants have evolved an adaptive system to cope with growth under P limiting conditions, involving both developmental and metabolic adaptations (for review, see Raghothama, 1999; Abel et al., 2002; Rausch and Bucher, 2002; Franco-Zorrilla et al., 2004). A primary determinant of metabolic adaptation is the close linkage between Pi, the form in which phosphorus is assimilated, and sugar metabolism. Pi plays a key role in the coupling of light and dark reactions in photosynthesis and in the export of trioses from the chloroplasts. Pi is also a substrate or a product in many reactions of sugar metabolism and an effector of key enzymes of starch and Suc synthesis. Paralleling this close metabolic link, it was previously shown that Pi starvation induced the expression of several Suc-responsive genes (Nielsen et al., 1998; Sadka et al., 1994; Ciereszko et al., 2001; Ciereszko and Kleczkowski, 2002). Our results showing that expression of most if not all the Pi starvation-induced genes is enhanced by Suc (Fig. 6) indicate that the Pi-starvation/sugar regulatory interaction is bidirectional and not exclusive of the high affinity Pi transporter previously reported (Lejay et al., 2003).
One possible explanation for the highest expression of sugar and Pi starvation-responsive genes when Pi-starvation and high sugar conditions are combined could be that these genes are actually Pi starvation responsive and that high sugar further reduces cellular Pi levels by increasing the levels of sugar Pi (Sadka et al., 1994). However, this metabolic interpretation involving a Pi-starvation signal is insufficient alone. For instance, several mutants have been isolated displaying increased sugar sensitivity as assayed not only by the expression of sugar-responsive genes whose expression is enhanced by Pi starvation, such as β-AMY and APL3, but also in other sugar sensitivity assays, and these mutants do not display increased hexose content (Baier et al., 2004). Moreover, the cytokinin to sugar and cytokinin to Pi-starvation regulatory interactions mentioned above provide evidence for a formal link between sugar and Pi-starvation signaling, at least through cytokinin signaling.
Interplay between Cytokinin, Sugar, and Pi Signaling
In summary, our results demonstrate the bidirectional antagonistic interactions between cytokinin and both sugar and Pi-starvation signaling involving CRE1 and AHK3 and probably AHK2, as well as a positive bidirectional interaction between sugar and Pi-starvation signaling. These intricate interconnections between cytokinin, sugar, and Pi-starvation signaling place Pi-starvation signaling high in the regulatory hierarchy controlling plant metabolism and development in accord with the physiological importance of Pi. Such regulatory cross talk allows not only Pi-starvation responses and Pi acquisition to be fine-tuned according to the status of the key signaling metabolites, sugars (whose rate synthesis is primarily determined by factors affecting photosynthesis such as light, CO2, nitrate, cytokinins), but also metabolism and development to be adjusted to Pi status. For instance, low Pi will enhance sugar responses, such as those leading to starch production and releasing Pi from sugar Pi, and reduce cytokinin signaling, thereby increasing the root-to-shoot growth ratio and concomitantly the soil Pi scavenging potential, as well accelerating senescence, a Pi-mobilizing process. The engineering of plants for better Pi use efficiency will be dependent on appreciating this regulatory cross talk and the molecular mechanisms that underpin it.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) L. Heynh ecotypes used in this study were Columbia and Landsberg erecta. Growth conditions and media were as previously described (Franco-Zorrilla et al., 2002) except where indicated for Suc or mannitol concentrations.
Genetic Analysis and Positional Cloning of AHK3
ahk3-4 mutant plants were backcrossed four times to wild-type plants (Columbia). To map the mutation, we obtained an F2 segregating population derived from a cross between the mutant and the Landsberg erecta ecotype. DNA was prepared from 48 plants showing the mutant phenotype and used to analyze linkage of the ahk3 mutation to previously described SSLP (Bell and Ecker, 1994) and cleaved amplified polymorphic sequence (CAPS; Konieczny and Ausubel, 1993). AHK3 was mapped to chromosome 1, between SSLP markers ZFPG and nga392, which defined a 1.1-Mb region.
Binary Constructs and Plant Transformation
A 7.8-kb genomic DNA fragment containing the AHK3 gene and 2,628 upstream the ATG start codon and 915 bp and downstream the stop codon was obtained by PCR using bacterial artificial chromosome F17A16 DNA as template and Expand High Fidelity Polymerase (Roche Applied Science, Mannheim, Germany). The oligonucleotides employed were GTTTCCgtcgACTACATTCACGAAGTGCAAGG and AAGGGgTCGacTACTGCAACTCACCGTGAACG, where some nucleotides were substituted (small letters) to generate SalI recognition sites (underlined). The PCR product was digested with SalI and cloned in the SalI site of the pCAMBIA1300 vector, generating gAHK3. The vector gAHK3 was introduced into the C58 strain of Agrobacterium tumefaciens and Arabidopsis plants were transformed as described (Bechtold et al., 1993). Transgenic plants were selected on Murashige and Skoog medium supplemented with hygromicin (40 μg/mL) and carbenicillin (50 μg/mL). To obtain a cDNA fragment corresponding to AHK3, a two-step procedure was followed. First, two overlapping cDNA fragments corresponding to the 5′ and 3′ halves of AHK3 were obtained through reverse transcription-PCR, using AHK3-specific oligonucleotides CGGAATTCCGAGAATATGGGCTGG and CTATAAACAAGTTCACATAAGG in the same reaction from 10 mg of RNA from seedlings as template and using Superscript Reverse Transcriptase (Ambion, Austin, TX). First strand cDNAs were subjected to PCR with oligonucleotide pairs GGGGGTTGATCGTGTATTCAAGTGGTGGATG-CCAACCTTGGGAGTACTCGAGAATCC (5′-AHK3) and ATATCCAGGACGCATGGAGGCACAGG-ACTACTTCAAGATGCATAGG (3′-AHK3) using Expand High Fidelity Polymerase. PCR products were cloned into pGEMT vector (Promega, Madison, WI). In a second step, the 3′-AHK3 fragment was excised with XhoI-NotI and cloned into the XhoI-NotI sites of 5′-AHK3, generating an in-frame fusion for the complete AHK3 cDNA. Finally, AHK3 cDNA was excised with SmaI and NotI, blunt ended with Klenow (Roche), and cloned into the SmaI site of the pBIB vector downstream the 35S-cauliflower mosaic virus promoter (Becker, 1990) and introduced in A. tumefaciens (Bechtold et al., 1993).
Cytokinin Response Assays
In the calli induction experiments, plants were grown in complete medium for 7 d and cotyledons or 1-cm root pieces were excised and placed onto calli induction media as indicated in the text. For RNA-blot analysis of kinetin sensitivity, plants were grown in Murashige and Skoog for 6 d and transferred to Whatman paper soaked in liquid Murashige and Skoog medium supplemented or not with 15 μm kinetin for an extra day and RNA prepared from whole seedlings.
Northern Analysis and Probes
RNA extraction was carried out with the RNAwiz reagent (Ambion) following manufacturer's instructions. RNA electrophoresis, transfer to nylon membrane, and hybridization were performed following standard procedures (Sambrook et al., 1989). Genes analyzed included AHK3 and CRE1 (Franco-Zorrilla et al., 2002); the cytokinin-induced ARR4, ARR6, ARR15, ARR16, and CYCD3 (Riou-Khamlichi et al., 1999; D'Agostino et al., 2000; Kiba et al., 2002; To et al., 2004); the Pi starvation-responsive At4, IPS1, ACP5, AtPT1, SQD1 (Muchhal et al., 1996; Essigmann et al., 1998; Burleigh and Harrison, 1999; del Pozo et al., 1999; Martín et al., 2000), and PHF, SPX, and CaBP22 (At3g52190, At2g45130 and At2g41090, respectively; F. Scaglia, R. Bustos, and J. Paz-Ares, unpublished data); and the sugar-induced genes APL3 and β-AMY (Mita et al., 1995; Sokolov et al., 1998). Probes corresponding to ARR6 and ARR15, IPS1, At4, PHT1;1, ACP5, CRE1, APL3, and β-AMY were obtained as previously described (Martín et al., 2000; Franco-Zorrilla et al., 2002; Baier et al., 2004; García-Ponce de León et al., 2004). Probes for AHK3 and to PHF were amplified from cDNA with the following oligonucleotide pairs: ATATCCAGGACGCATGGAGGCACAGG-CTTAAGCAATGAGATTGCC (AHK3) and ATGGAGATTGAAGAAGCGAG-TTACATAATCTTTCTATAGG (PHF). Probes for ARR4, ARR16, CYCD3, SPX, CaBP22, and SQD were PCR amplified from genomic DNA with oligonucleotide pairs: GCTCGTCTATGGCCAGAGAC-CCAGAATAGTTCCACTAATC (ARR4); ATGGCTCTCAGAGATTTATC-TTGAGCTCAATCATTTAACC (ARR16); TTTAGTCCCCCACAATGGCG-TCGAGCTTTCGATTATGGAG (CYCD3); GATGAAGTTTGGAAAGAGG-GACAACAACATCATGGAATAGG (SPX); TCAAGGCCGAGAGTTCGTAG- TCTAACATACCAGCCAGAGG (CaBP22); ATGGCGCATCTACTTTCAGC- ACACAGAACCGGTTAAGTGC (SQD1).
Acknowledgments
We thank Drs. Cathie Martin, Joseph Kieber, Jennifer Umphress, Carmen Castresana, Pilar Cubas, and Salomé Prat for critical reading of the manuscript. We also thank the contribution and enthusiasm of Dr. Roberto Solano in the early stages of this project. The excellent technical assistance of María Jesús Benito is greatly acknowledged.
This work was supported by the Spanish Ministry of Science and Education (ref. BIO2002–03568), by the Government of the Comunidad de Madrid (ref. 07B/0035/2002), and by the Government of the Comunidad de Madrid (postdoctoral fellowship to J.M.F.-Z.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060517.
References
- Abel S, Ticconi CA, Delatorre CA (2002) Phosphate sensing in higher plants. Physiol Plant 115: 1–8 [DOI] [PubMed] [Google Scholar]
- Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW (2004) Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth and developmental responses. Plant Physiol 134: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In Planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Paris Life Sci 316: 15–18 [Google Scholar]
- Becker D (1990) Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res 18: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ (1999) The down regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol 119: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszko I, Jonson H, Hurry V, Kleczkowski LA (2001) Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta 212: 598–605 [DOI] [PubMed] [Google Scholar]
- Ciereszko I, Kleczkowski LA (2002) Effects of phosphate deficiency and sugars on expression of rab18 in Arabidopsis: hexokinase-dependent and okadaic acid-sensitive transduction of the sugar signal. Biochim Biophys Acta 13: 43–49 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L (2001) Carbon and nitrogen sensing and signaling in plants: emerging matrix effects. Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Drew MC, Saker LR (1984) Uptake and long-distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley: evidence of non-allosteric regulation. Planta 160: 500–507 [DOI] [PubMed] [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, González E, Bustos R, Linhares F, Leyva A, Paz-Ares J (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 55: 285–293 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Martín AC, Solano R, Rubio V, Leyva A, Paz-Ares J (2002) Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J 32: 353–360 [DOI] [PubMed] [Google Scholar]
- García-Ponce de León BG, Franco-Zorrilla JM, Rubio V, Dahiya P, Paz-Ares J, Leyva A (2004) Interallelic complementation at the Arabidopsis CRE1 locus uncovers independent pathways for the proliferation of vascular initials and canonical cytokinin signalling. Plant J 38: 70–79 [DOI] [PubMed] [Google Scholar]
- Haberer G, Kieber JJ (2001) Cytokinins: new insights into a classic phytohormone. Plant Physiol 128: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Schmulling T (2003) Cytokinin signal perception and transduction. Curr Opin Plant Biol 6: 480–488 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mahönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan JM, Wareing PF (1980) Cytokinins and the growth responses of seedlings of Betula pendula Roth. and Acer pseudoplatanus L. to nitrogen and phosphorus deficiency. J Exp Bot 31: 525–532 [Google Scholar]
- Hutchinson CE, Kieber JJ (2002) Cytokinin signaling in Arabidopis. Plant Cell (Suppl) 14: S47–S59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J (2002) Two-component signal transduction pathways in Arabidopsis. Plant Physiol 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Mizuno T (2002) Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol 43: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kuiper D (1988) Growth responses of Plantago major L. ssp. pleiosperma (Pilger) to changes in mineral supply. Plant Physiol 87: 555–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper D, Schuit J, Kuiper PJC (1988) Effect of internal and external cytokinin concentrations on root growth and shoot to root ratio of Plantago major ssp. pleiosperma at different nutrient concentrations. Plant Soil 111: 231–236 [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass AD, Touraine B (1999) Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant J 18: 89–95 [DOI] [PubMed] [Google Scholar]
- Lejay L, Gansel X, Cerezo M, Tillard P, Muller C, Krapp A, von Wiren N, Daniel-Vedele F, Gojon A (2003) Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. Plant Cell 15: 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Muchhal US, Mukatira U, Kononowicz AK, Raghothama KG (1998) Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol 116: 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de la Peña A, Leyva A, Paz-Ares J (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24: 559–567 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H (2004. a) Regulation of high-affinity sulphate transporters in plants: towards systematic analysis of sulphur signalling and regulation. J Exp Bot 55: 1843–1849 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H (2004. b) A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J 38: 779–789 [DOI] [PubMed] [Google Scholar]
- Mita S, Suzuki-Fujii K, Nakamura K (1995) Sugar-inducible expression of a gene for β-amylase in Arabidopsis thaliana. Plant Physiol 107: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DWS, Mok MC (1994) Cytokinins: Chemistry, Activity and Function. CRC Press, Boca Raton, FL
- Mok DWS, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu Y-H, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor in nutrient, light and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG (1996) Phosphate transporter from higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TH, Krapp A, Röper-Schwarz U, Stitt M (1998) The sugar-mediated regulation of genes encoding the small subunit of Rubisco and the regulatory subunit of ADP glucose pyrophosphorylase is modified by phosphate and nitrogen. Plant Cell Environ 21: 443–454 [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martín SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadka A, DeWald DB, May GD, Park WD, Mullet JE (1994) Phosphate modulates transcription of soybean VspB and other sugar-inducible genes. Plant Cell 6: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H (2003) Nitrate-specific and cytokinin-mediated nitrogen signaling pathways in plants. J Plant Res 116: 253–257 [DOI] [PubMed] [Google Scholar]
- Salama AMSE-DA, Wareing PF (1979) Effects of mineral nutrition on endogenous cytokinins in plants of sunflower (Helianthus annus L.). J Exp Bot 30: 971–981 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Scheible WR, Lauerer M, Schulze ED, Caboche M, Stitt M (1997) Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco. Plant J 11: 671–691 [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Sokolov LN, Déjardin A, Kleczkowski LA (1998) Sugar and light/dark exposure trigger differential regulation of ADP-glucose pyrophosphorylase genes in Arabidopsis thaliana (thale cress). Biochem J 336: 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN (2000) Two-component signal transduction. Annu Rev Biochem 69: 183–215 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T (2001) The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- To JPC, Haberer G, Ferreira G, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C, Koizumi H, Suzuki T, Mizuno T (2001) Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol 42: 231–235 [DOI] [PubMed] [Google Scholar]
- Wolanin PM, Thomason PA, Stock JB (2002) Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol 3: 3013.1–3013.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC (2000) The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44: 451–461 [DOI] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]