Abstract
Understanding the function of detoxifying enzymes in plants toward xenobiotics is of major importance for phytoremediation applications. In this work, Arabidopsis (Arabidopsis thaliana; ecotype Columbia) seedlings were exposed to 0.6 mm acetochlor (AOC), 2 mm metolachlor (MOC), 0.6 mm 2,4,6-trinitrotoluene (TNT), and 0.3 mm hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX). In vivo glutathione (GSH) conjugation reactions of AOC, MOC, RDX, and TNT were studied in root cells using a multiphoton microscope. In situ labeling with monochlorobimane, used as a competitive compound for conjugation reactions with GSH, confirmed that AOC and MOC are conjugated in Arabidopsis cells. Reverse transcription-PCR established the expression profile of glutathione S-transferases (GSTs) and nitroreductases enzymes. Genes selected for this study were AtGSTF2, AtGSTU1, AtGSTU24, and two isoforms of 12-oxophytodienoate reductase (OPR1 and OPR2). The five transcripts tested were induced by all treatments, but RDX resulted in low induction. The mRNA level of AtGSTU24 showed substantial increase for all chemicals (23-fold induction for AOC, 18-fold for MOC, 5-fold for RDX, and 40-fold for TNT). It appears that GSTs are also involved in the conjugation reactions with metabolites of TNT, and to a lesser extent with RDX. Results indicate that OPR2 is involved in plant metabolism of TNT (11-fold induction), and in oxidative stress when exposed to AOC (7-fold), MOC (9-fold), and RDX (2-fold). This study comprises gene expression analysis of Arabidopsis exposed to RDX and AOC, which are considered significant environmental contaminants, and demonstrates the importance of microscopy methods for phytoremediation investigations.
Uptake is a necessary prerequisite for close contact between the pollutant and the detoxifying enzymes of plants that are localized in the cytosol of living cells. The presence and activity of this complex array of enzymes is crucial for degradation of chemicals under consideration for phytoremediation (Coleman et al., 1997a). However, very little is known about the exact number of plant enzymes potentially involved in the metabolism of xenobiotic compounds and their capacity to bind and metabolize them (Schalk et al., 1997; Scwitzguebel and Vanek, 2003).
The sequential metabolic steps of xenobiotics in plant metabolism are grouped in three main phases known as phase I (conversion), phase II (conjugation), and phase III (compartmentation; Sandermann, 1994; Ohkawa et al., 1999). Initial transformations in phase I include a number of oxidation, reduction, and hydrolysis reactions. Oxidation is the most often observed, while certain organics, such as nitroaromatics, tend to favor reduction processes (Hannink et al., 2002). Conjugations of xenobiotics represent phase II transformation reactions, which are usually mediated by a sugar moiety or tripeptide, such as malonic acid, d-Glc, glutathione, Cys, and other amino acids (Sandermann, 1994). Conjugation reactions result in the formation of products that are generally much less toxic, more stable, soluble, polar, and higher in molecular weight than the original compounds (Edwards, 1998; Wolfe and Hoehamer, 2003). There are several enzyme classes known to catalyze conjugation reactions, and glutathione S-transferases (GSTs; EC 2.5.1.18) are abundant enzymes found in all plant organs that are involved in these reactions (Pflugmacher and Sandermann, 1998; Pflugmacher et al., 2000). After xenobiotics are conjugated, they are converted in phase III to secondary conjugates or to insoluble bound residues that are deposited in the vacuole or other compartments of plant cells (Ohkawa et al., 1999). In plants, vacuolar compartmentation is a critical step in the detoxification of organic metabolites because it removes conjugated products from vulnerable sites of the cytosol and further processing of the conjugates may take place in the vacuolar matrix (Coleman et al., 1997b, 2002).
In higher plants, reduced glutathione (GSH) and GSTs play crucial roles in the degradation of several pollutants. The major metabolic reaction involved with the degradation of chloroacetanilide herbicides in plants is direct conjugation with GSH or its homolog homoglutathione (Jablonkai and Hatzios, 1993). This conjugation process is mediated by soluble GSTs, which leads to an immediate decrease of the cellular GSH pool (Coleman et al., 1997b; Cobbett, 2000). The level of GSH in plants is important in defense reactions against biotic and abiotic stresses (May et al., 1998). The efficiency at which cells can raise the cytoplasmic GSH pool after depletion may influence their degree of stress tolerance or susceptibility to herbicides (May et al., 1998; DeRidder et al., 2002).
Conjugation processes with 2,4,6-trinitrotoluene (TNT) have been observed in studies with periwinkle (Catharanthus roseus) and parrot feather (Myriophyllum periwinkle; Bhadra et al., 1999; Bhadra et al., 2001). In these studies, the isolated conjugates to TNT were comprised of sugars, amino acids, and organic acids available for substitution at the amino group reactive sites of reduced TNT metabolites. Plants are able to attach Glc (by glucosyltransferases) or malonate (by malonyltransferases) to amino groups, thus forming the TNT conjugates (Coleman et al., 1997a). Recent studies with serial analysis of gene expression (SAGE) in Arabidopsis (Arabidopsis thaliana) roots exposed to TNT suggest that GSTs are the enzymes primarily responsible for conjugation reactions involving TNT metabolites (Ekman et al., 2003).
Prior to conjugation, phase I reduction reactions that target the aromatic nitro groups for further transformation are very frequent and have also been reported in plant tissues (Bhadra et al., 1999; Larson et al., 1999). Enzymes involved in the reduction of aromatic nitro groups, the so-called nitroreductases, are classified by activity in the presence of molecular oxygen into two classes, Type I or Type II, each with distinctive properties (Peterson et al., 1979). The reduction of the nitro group by Type I enzymes is insensitive to molecular oxygen, whereas Type II is oxygen sensitive (Peterson et al., 1979). The reducing enzymes in plants involved on TNT transformation are classified as type II nitroreductases, which include cytochrome P450 reductase, cytochrome c reductase, aldehyde oxidase, glutathione reductase (GR), xanthine dehydrogenase, succinic dehydrogenase, and quinone reductase (Bryant and Deluca, 1991). Additional studies suggest that such reduction reactions with TNT may also be catalyzed by NADPH-dependent flavoenzymes, such as 12-oxophytodienoate reductase (OPR) and nitrate ester reductase (French et al., 1999; Ekman et al., 2003).
Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) is grouped as a nitramine explosive that is more difficult to transform than TNT (Subramanian and Shanks, 2003). Few investigations indicate that RDX can be removed from aqueous media by various plant species, where it accumulates in plant tissues as bound residues (Larson et al., 1999; Burken et al., 2000; Bhadra et al., 2001; Mezzari et al., 2004). Recent studies with plant tissues exposed to radiolabeled RDX have suggested that conjugation reactions of nonidentified RDX metabolites may occur in the plant cell (Just and Schnoor, 2004; Mezzari et al., 2004). Therefore, further identification of these metabolites is necessary to elucidate the sequence of transformations and fate of RDX within plant cells.
Screening of genome-coded sequences is a powerful tool in identifying the degradation pathways of specific enzymes involved in the metabolism of xenobiotics. In addition, changes in gene expression can be informative and useful in developing transgenic plants that respond to contaminants for phytoremediation enhancement and phytosensing. Phytosensors are engineered plants that have an inducible promoter fused to a reporter gene, such as that encoding green fluorescent protein (Patel et al., 2004). While phytoremediation is characterized by the use of plants for in situ treatment of contaminated areas, phytosensors are plant sentinels that produce a phenotypic response to specific environmental stimuli, including xenobiotic exposure (Patel et al., 2004).
Numerous enzyme sequences have been identified in Arabidopsis, but little progress has been made in matching specific enzymes with their function on xenobiotic substrates. There are only a few studies with Arabidopsis GST (AtGST) and gene expression in response to treatment with metolachlor (MOC), TNT, and other xenobiotics (Wagner et al., 2002; Ekman et al., 2003). These studies did not include a nitramine exposure analysis; therefore, the gene expression pattern of Arabidopsis in response to RDX exposure requires further study.
In situ labeling with the fluorescent dye monochlorobimane (MCB) was used to observe possible conjugation reactions of GSH with chloroacetanilide herbicides and explosive chemicals in Arabidopsis root cells. MCB binds to GSH and forms a glutathione S-bimane (GSB) conjugate, which changes in fluorescent intensity at one distinct wavelength and has an excitation peak at 395 nm (Meyer and Fricker, 2000). GSH can be depleted by conjugation to xenobiotics such as MCB or 1-chloro-2,4-dinitrobenzene (CDNB; Coleman et al., 1997b). Multiphoton microscopy was used to quantify GSH using MCB to study in vivo conjugation processes with GST-catalyzed conjugation reaction in Arabidopsis root cells and the competition between binding sites with acetochlor (AOC), MOC, TNT, and RDX.
Reverse transcription (RT)-PCR was also conducted to study the expression profile of AtGST and OPR genes in Arabidopsis plants exposed to AOC, MOC, TNT, and RDX. Genes selected for this study were AtGSTF2, AtGSTU1, AtGSTU24, OPR1, and OPR2.
This article reports on (1) in vivo analysis of GSH conjugation and storage reactions with AOC, MOC, RDX, and TNT; (2) monitoring changes in transcript levels of AtGST and OPR in Arabidopsis; (3) the involvement of specific GST and OPR isoforms associated with selected xenobiotic-induced activity; and (4) the importance of understanding antioxidative defense mechanisms in the Arabidopsis plant model for further application in phytoremediation processes.
RESULTS
Toxicity Tests
To examine the effect of the selected chemicals on Arabidopsis biomass growth, seedlings were exposed to different concentrations of AOC (0.313, 0.625, 1.25, 2.5, and 5 mm), MOC (0.625, 1.25, 2.5, 5, and 10 mm), RDX (0.02, 0.078, 0.313, 1.25, and 5 mm), and TNT (0.2, 0.4, 0.8, 1.6, and 3.2 mm). Young seedlings were examined for signs of stress (chlorosis and necrosis) and survival rates during the 5 d of exposure. Biomass growth inhibition was observed in all treatments.
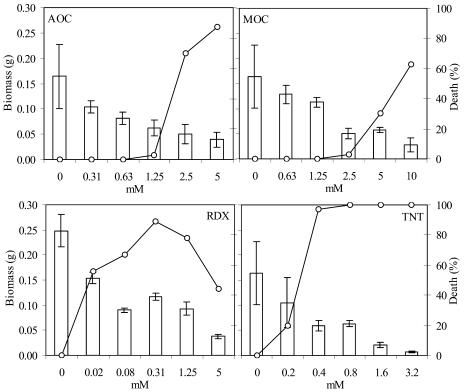
For the 5 d of exposure, the survival percentage of Arabidopsis was only 13% and 37% when exposed to AOC and MOC at the highest concentrations (Fig. 1). TNT was toxic at the four highest concentrations applied to Arabidopsis seedlings, where extensive chlorosis followed by necrosis was the main symptom observed. According to Pavlostathis et al. (1998), the phytotoxicity of TNT to all plant species appears to be marked by chlorosis and growth suppression. By contrast, RDX exposure was significantly less toxic than TNT and the other test chemicals, possibly because of the low solubility in the media.
Figure 1.
Toxicity test performed in Arabidopsis plants for selection of xenobiotic concentration to time course treatment study. Two-week-old Arabidopsis seedlings were exposed to a range of millimolar concentrations from AOC, MOC, RDX, and TNT. Biomass growth is represented as bars and death percentage is represented in the secondary y axis as dots.
Nitroreductase Gene Induction
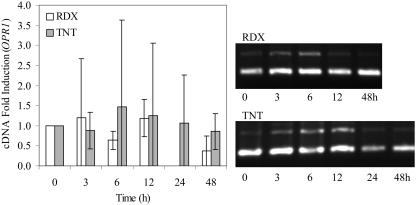
OPR is a FMN-dependent oxidoreductase in plants that can reduce aromatic nitro compounds and nitrate esters (Biesgen and Weiler, 1999; Matsui et al., 2004). There are three enzyme isoforms of OPR in plants, but only OPR1 and OPR2 are induced by environmental stresses (Biesgen and Weiler, 1999). To determine which chemicals can induce gene expression of both OPR isoforms in Arabidopsis, 2-week-old seedlings were exposed to the selected concentrations of 0.625 mm for AOC, 2 mm for MOC, 6 mm for TNT, and 0.313 mm RDX for 0, 3, 6, 12, 24, and 48 h. In this work, the maximum induction OPR1 and OPR2 was observed at 3 h for RDX and 6 h for TNT (Figs. 2 and 3). Thus, OPR1 showed a relatively low induction (less than 4-fold) toward all chemicals tested, especially for RDX, AOC, and MOC if compared with the control treatment (Fig. 2; Table I).
Figure 2.
Time course of OPR1 expression in Arabidopsis seedlings treated with 0.313 mm RDX and 0.6 mm TNT. The analysis of gene induction is represented in the bar chart using average values of semiquantitative RT-PCR triplicate analysis. The results are represented with respect to the 0 h (control) fold induction, which was analyzed from nonexposed plants. Total RNA was extracted from three independent experiments for each chemical exposure. β-ATP internal standard is represented as the top band of each treatment. The bottom band represents OPR1 expression under different time exposures. Bars represent the means and sd of triplicate determinations.
Figure 3.
Time course of OPR2 expression in Arabidopsis seedlings treated with 0.313 mm RDX and 0.6 mm TNT. The analysis of gene expression is represented in the bar chart using average values of semiquantitative RT-PCR triplicate analysis. The results are represented with respect to the 0 h (control) fold induction, which was analyzed from nonexposed plants. Total RNA was extracted from three independent experiments for each chemical exposure. β-ATP internal standard is represented as the bottom band of each treatment. The top band represents OPR2 expression under different time exposures. Bars represent the means and sd of triplicate determinations.
Table I.
OPR1 and OPR2 fold induction of 2-week-old Arabidopsis seedlings exposed to 0.625 mm AOC and 2 mm MOC
| Time
|
OPR1 (±sd)
|
OPR2 (±sd)
|
||
|---|---|---|---|---|
| AOC (0.625 mm) | MOC (2 mm) | AOC (0.625 mm) | MOC (2 mm) | |
| h | ||||
| 0 | 1.00 (±0.00) | 1.00 (±0.00) | 1.00 (±0.00) | 1.00 (±0.00) |
| 3 | 1.66 (±2.25) | 0.77 (±0.54) | 6.61 (±2.91) | 8.07 (±2.30) |
| 6 | 0.76 (±1.13) | 0.62 (±0.78) | 5.23 (±2.55) | 8.98 (±2.29) |
| 12 | 1.54 (±2.50) | 0.74 (±1.04) | 6.01 (±2.59) | 8.14 (±2.77) |
| 24 | 1.95 (±3.21) | 0.37 (±0.40) | 4.68 (±1.72) | 7.32 (±2.39) |
| 48 | 1.22 (±1.65) | 2.17 (±2.42) | 7.43 (±4.73) | 7.19 (±1.28) |
The results presented are the means of triplicate determinations.
In contrast to OPR1, higher OPR2 induction was individually observed for all of the chemicals tested in the time course experiment (Figs. 2 and 3; Table I). Induction of OPR2 isoform was much more distinguished with a maximum of 11-fold increase for TNT-exposed Arabidopsis at 6 h (Fig. 3). These results suggest that this NADPH-dependent flavoenzyme may be involved in reduction reactions of TNT-exposed plants. In the case of RDX exposure, there was a relatively low induction (less than 4-fold) for both OPR genes at 3 h. Therefore, it is very likely that these reductive enzymes may not be involved on RDX transformation reactions in Arabidopsis.
Induction values of both OPR genes from AOC and MOC exposure were lower than in Arabidopsis exposed to TNT and, also, time course variable (Table I). The fluctuating data may reflect the absence of reductive reactions involved in the transformation of parent chloroacetanilide compounds, which are not commonly observed (Tal et al., 1995). Thus, the time course data showed an overall tendency for inducing OPR genes after 24 h of exposure. Therefore, phase I transformation enzymes from plants exposed to chloroacetanilide herbicides does not include OPR, and the induction of this enzyme activity is probably related to stress response (Biesgen and Weiler, 1999).
GST Gene Induction
To elucidate the induction of a few AtGST genes from Arabidopsis exposed to AOC, MOC, RDX, and TNT, transcript abundances were tested for AtGSTF2 (At4g02520), AtGSTU1 (At1g17170), and AtGSTU24 (At2g29490). The selected AtGST represent genes from the enzyme isoforms that are part of the large phi (GSTF) and tau (GSTU) classes, which are plant specific (Wagner et al., 2002).
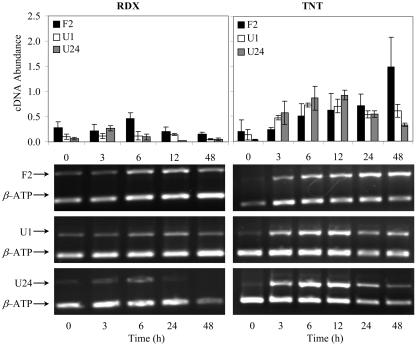
Results from this work showed that the selected GSTs from Arabidopsis were induced by all four chemical treatments. Interestingly, the highest induction observed to all chemical treatments was for AtGSTU24. Previous studies with SAGE from Arabidopsis exposed to 0.06 mm TNT suggest that GST enzymes are primarily involved on TNT metabolite conjugation reaction, where induction reached 28-fold increase for AtGSTU1 and 11-fold for AtGSTU24 (Ekman et al., 2003). In this work, induction of AtGSTU24 in Arabidopsis exposed to 0.6 mm TNT showed an average of 40-fold increase in gene expression at 6 h from triplicate experiments (Fig. 4). The other two AtGST genes also showed considerable induction of 9- and 15-fold for U1 and F2, respectively, from TNT exposure. AtGSTF2 expression profile average was higher than U1, which presumably should be the most activated gene according to Ekman et al. (2003). However, gene induction responses obtained in this investigation may be reflecting the higher concentration used in the study.
Figure 4.
PCR-based expression of AtGSTF2 (represented as F2), AtGSTU1 (U1), and AtGSTU24 (U24) in 2-week-old Arabidopsis plants exposed to 0.313 mm RDX and 0.6 mm TNT. Total RNA was extracted from Arabidopsis at the times indicated on the graphs (in hours). Five grams of this total RNA, obtained from different tissues, was converted to cDNA with random primers. An internal standard β-ATP was used as control, and samples were subjected to semiquantitative RT-PCR. Zero-hour exposure represents control (nonexposed plants). Bars represent the means and sd of triplicate determinations.
Similar induction response was obtained for 0.313 mm RDX treatment, where AtGSTU24 was induced to the highest level (5-fold) and AtGSTU1 was the lowest (2-fold; Fig. 4). These values occurred at 3 h of exposure, reflecting an early stage of defense against RDX. However, studies using cell suspensions of Jimsonweed (Datura innoxia) exposed to solubility limits of RDX and octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine (HMX) showed no effect on toxicity or viability, which implies that phytotoxicity to RDX may result primarily from the transformation products (Lucero et al., 1999). The low AtGST gene induction observed at a short time of exposure supports the idea of low toxicity effect. Even though lower AtGST induction was observed for seedlings exposed to RDX, there is a possibility that conjugation reactions may occur with RDX metabolites.
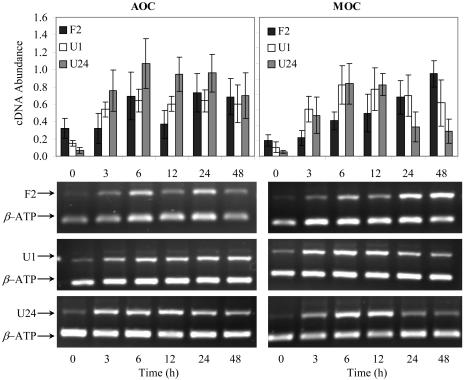
Arabidopsis exposed to chloroacetanilide herbicides showed a variation in cDNA abundance for F2, U1, and U24 genes (Fig. 5). AOC induced all of the three selected genes, and the highest values occurred for AtGSTU24, with a 23-fold increase at 6 h. Both AtGSTF2 and AtGSTU1 showed elevated induction of about 5-fold at 24 h. Literature research supports the involvement of the AtGSTF2 gene activation for conjugation with MOC, where a 0.5 mm concentration causes a response of 16-fold induction after 3 h of exposure (Wagner et al., 2002). For AOC and MOC exposure, AtGSTF2 has substantial induction; and yet this investigation showed that AtGSTU24 was the most expressed. Overall, induction values for F2 and U1 were higher for MOC-exposed plants than AOC. The highest induction values for F2 and U1 were 15 at 48 h and 12 at 6 h, respectively. AtGSTU24 showed the highest induction value at 18-fold.
Figure 5.
Expression of AtGSTF2 (F2), AtGSTU1 (U1), and AtGSTU24 (U24) in 2-week-old Arabidopsis seedlings exposed to 0.625 mm AOC and 2 mm MOC. Total RNA was extracted from Arabidopsis at the times indicated on the graph (in hours). Five grams of total RNA was converted to cDNA with random primers. An internal standard β-ATP was used as control and samples were subjected to semiquantitative RT-PCR. Zero-hour exposure represents control (nonexposed plants). Bars represent the means and sd of triplicate determinations.
Confocal Microscopy Studies
MCB has been used as a model xenobiotic to study in vivo conjugation reactions with GSH and its sequestration to the plant cell vacuole in phase III transformation pathway (Coleman et al., 1997b; Fricker et al., 2000; Fricker and Meyer, 2001; Meyer and Fricker, 2002). It is known that MCB easily enters the cell through diffusion and it is nonfluorescent until conjugated to GSH, when it becomes fluorescent with an excitation peak at 395 nm (Coleman et al., 1997b). Because MCB forms the fluorescent adduct with GSH specifically and preferentially over other thiols (Coleman et al., 1997b), in vivo conjugation reactions of AOC, MOC, RDX, and TNT were observed based on competition to available GSH (Fig. 6).
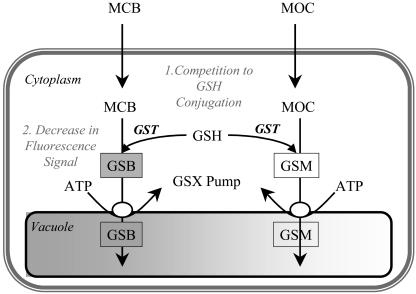
Figure 6.
Schematic diagram of the GHS-based detoxification pathway in plants. MCB is used as a model substrate for conjugation to GSH by a GST in the cytoplasm and subsequent sequestration in the vacuole by a glutathione S-conjugate (GSX) pump. MCB is not fluorescent until conjugation to GSH displaces the chloride-leaving group and gives rise to a fluorescent GSB. If another xenobiotic substrate for conjugation to GSH is present, such as MOC, competition to GSH will occur and consequently, lower the fluorescent signal from GSB (adapted from Fricker and Meyer, 2001).
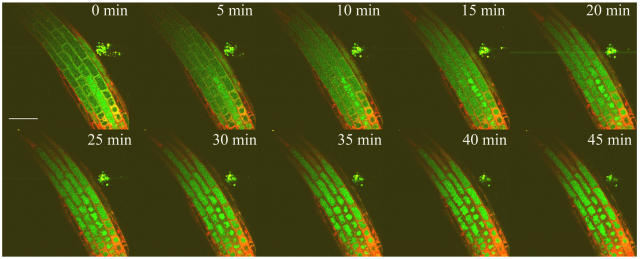
Fluorescence signal from GSB conjugate was imaged in optical sections of intact Arabidopsis roots through time. The GSB fluorescent intensity (green fluorescent color) was observed to increase initially in the cytoplasm and then transferred into the vacuole over time in Arabidopsis root cells (Fig. 7). To quantify the conjugate formed in the root cell, fluorescent intensities were calibrated against GSB standards imaged under the same conditions as previously described by Meyer and Fricker (2000). To prevent subsequent transfer of GSB to the vacuole, sodium azide (NaN3) was included as a negative control in the labeling protocol to inhibit the ATP-dependent vacuolar glutathione-xenobiotic conjugate pump (Fricker et al., 2000). The propidium iodide (PI; red) fluorescent probe was used to label cell walls and also used as an indicator for viability of cells, since it stains nuclei of dead cells.
Figure 7.
Confocal imaging of GSH conjugation to MCB in Arabidopsis root. GSH in root of 5-d-old Arabidopsis seedlings was labeled with 100 μm MCB to give a green fluorescent GSB conjugate and imaged as a time series of optical sections collected at 5-min intervals over 45 min. Fifty micromolar PI was added to solution prior to imaging, and labeled the cell walls and nuclei of dead cells in red. Bar = 50 μm.
The decrease in fluorescent level was analyzed in samples exposed to CDNB, AOC, MOC, RDX, and TNT, which are chemicals that may compete for the available GSH (Fig. 8). To address this point, the fluorescent intensity from GSB conjugates was measured in a time course experiment with exposed tissues to selected xenobiotics. The fluorescent intensity was compared to controls for positive (MCB/PI-exposed tissues) and negative response (MCB/PI/NaN3-exposed tissues).
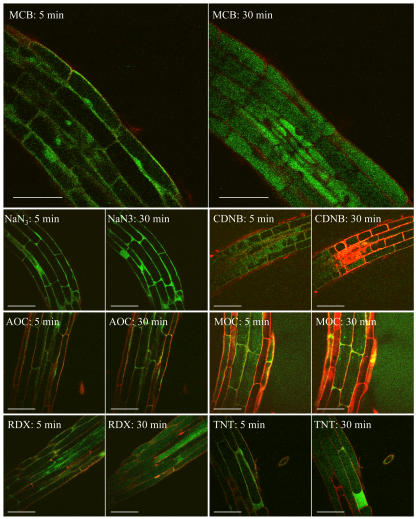
Figure 8.
Confocal imaging of GSB formation and sequestration (green fluorescent signal) at 5 and 30 min in Arabidopsis root cells exposed to selected xenobiotics. Comparison between MCB (positive control; formation and compartmentation of GSB in vacuole), NaN3 (negative control; formation of GSB without compartmentation), CDNB, AOC, MOC, RDX, and TNT (competitive xenobiotics for glutathione conjugation; formation of GSB at a lower extent). Bars represent 50 μm.
The GSB formation from Arabidopsis root tissues exposed to CDNB, AOC, and MOC showed a reduced fluorescent intensity, and thus competition to GSH conjugation reactions occurred (Fig. 8). CDNB is a classical substrate for the glutathione S-conjugate, and it can diffuse freely into cells where it is conjugated with GSH to form 2,4-dinitrophenyl-glutathione (Elferink et al., 1993). In the presence of 10 mm CDNB, Arabidopsis root cells showed similar decrease in the synthesis of GSB, and consequently low fluorescence signal was produced. In the case of chloroacetanilide herbicides, similar responses occurred, since conjugation reactions with GSH is a common degradation pathway for these chemicals (Jablonkai and Hatzios, 1993).
Results from the in vivo analysis of GSH conjugation reactions with RDX and TNT showed that GSB is formed, thus conjugation to these selected explosive compounds does not occur (Fig. 8). Interestingly, it was observed that some root cells showed the accumulation of GSB conjugate in the cytosol and no transfer to the cell vacuole. These results were very similar to what was observed for roots exposed to NaN3, where vacuole transport is inhibited because of blocked ATP synthesis. In addition, the concentration of GSB conjugates formed in root cells exposed to TNT and RDX was significantly higher than roots exposed to the positive control solution (Fig. 9).
Figure 9.
Time course analysis of GSB formation and accumulation in the vacuole of Arabidopsis root cells. NaN3 was used to avoid compartmentation of GSB in the vacuole (negative control); AOC, MOC, RDX, TNT, and CDNB were used as substrate competitors for conjugation with glutathione. sds are from triplicate determinations and each replicate represents data from one cell in a single root.
DISCUSSION
Plant toxicity studies published to date have established that nitroaromatic compounds are notoriously difficult to metabolize since a wide variety of plant species from diverse plant families appear to degrade TNT only partially (Best et al., 1999). In addition to explosives compound studies, previous investigations of RDX toxicity in plants have indicated that this chemical is much less toxic than TNT (Lucero et al., 1999; Thompson et al., 1999). It was observed in hybrid poplars (Populus deltoides × Populus nigra, DN34) over a 14-d hydroponic exposure that RDX concentrations of up to 94 μm did not affect the growth or transpiration rates of the cuttings (Thompson et al., 1999). Moreover, according to studies performed with cell cultures of Jimsonweed by Lucero et al. (1999), RDX levels of 173 to 270 μm did not decrease cell growth, indicating that this explosive did not induce toxicity (Lucero et al., 1999). During these studies, cell suspensions of Jimsonweed showed no effect on viability for HMX and RDX exposures in excess of the reported solubility limits (5 mg of HMX per liter and up to 60 mg of RDX per liter). This study with Arabidopsis is in agreement to the low toxicity of RDX.
The main phytotoxic action of chloroacetanilide herbicides is protein synthesis inhibition (WSSA, 1994). The toxicity response between the two chloroacetanilide herbicides correlates to literature information, where AOC is more phytotoxic than MOC (WSSA, 1994).
Numerous studies have confirmed that a large number of xenobiotic-metabolizing enzymes are induced by a diversity of chemicals in order to achieve tolerance and survival of the plant cell (Schroder and Collins, 2002). Reduction, though probably not the primary transformation process of nitroaromatic compounds, appears to be significant and ubiquitous (Subramanian and Shanks, 2003). Nitroaromatic reductions are commonly observed, such as TNT reduction to hydroxylaminodinitrotoluenes, aminodinitrotoluenes (ADNT), and the rarely observed in diaminonitrotoluene derivatives (Burken et al., 2000). The specific enzymes involved in TNT reduction reactions have not been revealed, but recent investigations with Arabidopsis exposed to TNT have reported a 10-fold induction of OPR1 (Ekman et al., 2003). Based on our results, OPR2 seems to be also a good candidate for reduction reactions involving TNT-exposed plants. However, the level of OPR1 and OPR2 induction obtained could be involved in stress responses rather than reduction reactions of TNT.
Although a growing number of studies on transformation reactions with TNT have been reported, just a few have been focused on the other widely used explosive, RDX. This nitramine is of significant cause for health and environmental concern, and, to our knowledge, this study constitutes the first investigation on transcriptome-level responses to RDX exposure in plants. In this work, it was observed that OPR enzymes from Arabidopsis exposed to RDX was not significantly induced (below 4-fold). Because reductive reactions with AOC and MOC are not very common (Tal et al., 1995), OPR induction responses toward these herbicides may also be involved on cell stress response.
GSTs have a wide and overlapping ability to bind compounds of diverse structures and physical properties, though the specificity for xenobiotics is very low (Clarke et al., 1998). GST transcriptome-level responses of AtGSTF2, AtGSTU1, and AtGSTU24 in Arabidopsis exposed to TNT and RDX were significantly induced. Between these, the AtGSTU24 was the most induced in Arabidopsis at a very short time of exposure. Similar results were obtained for Arabidopsis exposed to AOC and MOC. Overall, the AtGSTU24 enzyme isoform seems to have low binding specificity to the selected compounds, warranting further investigations on this enzyme's function.
It is known that GSTs are the main enzymes involved on the conjugation reactions of chloroacetanilide herbicides. However, in the case of explosive compounds, just a few reports have suggested the involvement of GST in conjugation reactions with TNT and RDX metabolites. Recent investigations have shown that safeners can also enhance levels of GSH and GST (Deng and Hatzios, 2002; DeRidder et al., 2002). Safeners are generally applied to crops in order to increase herbicide tolerance. Many safeners are aromatic compounds with nitrogen-containing functional groups, which are structurally very similar to TNT and RDX (DeRidder et al., 2002). Therefore, based on this statement, it is possible that GST reactions in response to TNT and RDX exposure may occur.
Most studies propose that conjugation of nitroaromatic metabolites involve the addition of a molecule of Glc or malonate to the newly formed amino groups via N-glycosylation (Hatzios and Penner, 1982). This reaction was demonstrated when Bhadra et al. (1999) showed that in periwinkle hairy root cultures, TNT is transformed into 4-amino-2,6-dinitrotoluene (4-ADNT), followed by the N-linked addition of Glc to the newly formed amino group of 4-ADNT. However, according to Ekman et al. (2003), GST-mediated conjugation reactions with TNT is suggested, and the results obtained in this work also confirm the induction of the three GST isoforms in Arabidopsis. In the case of RDX, it has been suggested that conjugation processes occur for metabolites, such as formaldehyde, via S-formylglutathione, a glutathione-dependent formaldehyde dehydrogenase enzyme (Just and Schnoor, 2004), and the induction of GST genes imply the involvement of this enzyme on conjugation processes of RDX. It now appears feasible that GSTs could be involved in the conjugation reactions of RDX (not previously reported) and TNT metabolites.
In vivo analysis of GSH conjugation reactions with RDX and TNT is a notable approach for the elucidation of probable degradation pathways of these compounds. In vivo analyses indicate that GSHs are not the main enzymes involved on conjugation processes of RDX and TNT metabolites. In fact, GSB conjugate concentration was significantly higher for plants exposed to RDX and TNT than the positive control MCB (Fig. 9). This response may be related to a possible induction of GR enzymes, since they may also be involved in the NADPH-dependent elimination of nitro group compounds, which are known to have type II nitroreductase activity (Shah and Spain, 1996). GR enzymes are also responsible for maintaining the GSH levels for cell defense against oxidative damage (Foyer et al., 2001). In addition to that, studies with SAGE from Arabidopsis exposed to TNT have showed that GRs are prominent among the enzymes likely to protect against oxidative stress (Ekman et al., 2003). Therefore, this elevated concentration of free GSH in Arabidopsis root cells may have resulted in more conjugation reactions with MCB, thus increasing fluorescent intensity.
The observed inhibition of GSB vacuolar transportation is directly related to low ATP production. The significance of this response is unclear, and this was not observed on experiments with hybrid poplar (P. deltoides × P. nigra, DN34) roots (data not shown). A possible insight into the responsible mechanisms may be partially related to GSH biosynthesis, since it requires ATP for production, and the level of GSH in Arabidopsis is much lower than poplars (May et al., 1998). Arabidopsis is a common weed, and these plants are very susceptible to herbicides because of the low cytoplasmic GSH pool (May et al., 1998; DeRidder et al., 2002). Thus, further analysis is required to provide important information on this response in Arabidopsis.
In conclusion, the identification of the genes involved in the metabolism of AOC, MOC, RDX, and TNT will provide valuable targets for future rounds of genetic engineering in order to enhance the natural properties of environmentally relevant plant enzymes to remediate contaminated ecosystems. Using fluorescent probes for in vivo studies also emphasize the important new insights that imaging techniques can bring to the study of metabolism at the cellular level in an intact system. In addition, an interesting approach to connect both genetics and microscopy technique is also available for the development of phytosensors. Whole promoters or defined cis-regulatory elements from genes specifically induced by xenobiotics can be fused to a reporter gene, such as green fluorescent protein (Kooshki et al., 2003). The phytosensors could be valuable to monitor and detect environmental contamination.
MATERIALS AND METHODS
Chemicals
AOC, MOC, and TNT were supplied by Chem Service (West Chester, PA). RDX was synthesized in house using formaldehyde, ammonium hydroxide, and fuming nitric acid. All chemicals and solutions were obtained from Fisher Scientific (Pittsburgh), Sigma-Aldrich (St. Louis), or as indicated.
Plant Material
Seeds of Arabidopsis (Arabidopsis thaliana L. Heyn; ecotype Columbia) were surface sterilized and placed onto petri plates containing half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) with 0.8% (w/v) agar. Seeds germinated and grew for 2 weeks at 21°C under standard conditions (approximately 200 μmol photons m2 s−1, 16-h-day/8-h-night cycle) with plates incubated in vertical position. After 2 weeks, the young seedlings were transferred to the sterile Murashige and Skoog liquid medium containing various concentrations of AOC, MOC, TNT, and RDX, and grown for another 5 d under standard conditions.
Toxicity Tests and Time Course Exposure Tests
Stock solutions of 0.5 m AOC, 1 m MOC, 0.5 m RDX, and 0.32 m TNT were prepared in dimethyl sulfoxide (DMSO). Toxicity tests were assessed by adding the DMSO stock solution in autoclaved half-strength Murashige and Skoog medium to yield five dosages with 10% DMSO in 2-fold geometric progressions (4-fold to RDX). The highest concentrations tested were 5 mm for AOC, 10 mm for MOC, 5 mm for RDX, and 3.2 mm for TNT. All toxicity tests were carried on agar plate solutions, except for RDX. Because RDX is very hydrophobic, the 2-week-old Arabidopsis were transferred into autoclaved 50-mL glass baby food jars with plastic lids containing half-strength Murashige and Skoog liquid medium. These were incubated with continuous agitation at 80 rpm to provide a homogeneous concentration media for plant exposure. The biomass growth and wilting response was analyzed after 5 d of exposure.
Selected concentrations from the toxicity tests, based on biomass and wilting stress threshold response, were applied to a time course experimental approach for each chemical. The time course exposure test was performed for 0, 3, 6, 12, 24, and 48 h. The concentrations selected for AOC, MOC, RDX, and TNT were 0.6 mm, 2 mm, 0.6 mm, and 0.3 mm, respectively. Concentrations were established based on preliminary experiments, where plants showed very small wilting percentage on their leaves (data not shown). Due to the low solubility in water (log octanol-water coefficient of 0.9), a concentration of 0.313 mm RDX was used on liquid media to obtain a more homogeneous distribution and to avoid turbidity problems. All procedures for the time course experiment were carried out in triplicate with each replicate containing 10 plants.
In Vivo Studies Using Multiphoton and Confocal Microscopy
Experiments were performed with 5-d-old Arabidopsis seedlings. Plant samples were transferred to a drop of fluorescent dye solution on a microscope slide and covered with a coverslip using adhesive tape as spacers to avoid crushing the roots. The coverslip was sealed to the microscope slide with melted valap (vaseline, lanolin, paraffin, at a ratio of 1:1:1) to avoid evaporation of dye solution and consequent movement of root parts during time course experiments.
The in situ labeling of GSH was accomplished using MCB (Molecular Probes, Eugene, OR) as a fluorescent marker and PI (Molecular Probes) as a fluorescent dye for cell walls and indicator for cell viability. Stock solutions of 100 mm MCB were prepared in DMSO and stored at −20°C. PI was prepared as a 5-mm aqueous-stock solution.
The positive control solution was prepared with aliquots from MCB stock, which were thawed immediately prior to use and diluted in deionized water to a final concentration of 100 μm. PI was used at a final concentration of 50 μm. The negative control solution was freshly prepared with NaN3, and added to the positive control solution at a final concentration of 5 mm. NaN3 depletes ATP levels and thereby inhibits vacuolar sequestration of GSB.
Working solutions containing CDNB, AOC, MOC, RDX, and TNT were freshly prepared and added to the positive control solution at a final concentration of 10 mm. All of the chemicals were tested to compete for the labeling of GSH with MCB and consequently identify conjugation reactions between these selected compounds.
After addition of specific solution treatment, Arabidopsis root tissues were immediately imaged using a Radiance 2100 MP confocal and multiphoton microscope (Bio-Rad Laboratories, Hercules, CA). The multiphoton microscope was attached to a Nikon Eclipse E-800 upright microscope and images were obtained using a 60× water-immersion lens. Excitation was achieved with a green 543-nm HeNe laser (Liconix, Santa Clara, CA) coupled into the scan head by a fiber optic. Fluorescence and excitation of GSB was achieved with a Mai Tai laser set to record at λem = 770 nm and PI fluorescence at λem > 585 nm with a long pass filter.
To follow the labeling kinetics over time, slides with root samples from Arabidopsis were placed on the microscope stage immediately after exposure to the dye solution. Optical (x, y) sections of root cells were collected with a sampling rate at 5 min over 30 min.
The GSB conjugate fluorescence intensity was measured from a 3-fold geometric calibration curve containing five different concentrations of GSB ranging from 0.012 to 1 mm. The calibration standards were prepared using a 10 mm fresh stock solution of GSB, which was made from 10 mm MCB and 100 mm GSH (G 1404; Sigma-Aldrich) in the presence of GST (Equine Liver GST, G 6511; Sigma-Aldrich). Fluorescence of GSB in intact cells was calibrated against GSB standard imaged using identical instrument settings.
Image processing and analysis was performed with the public domain NIH Image program and Bio-Rad z-series function of Image J version 1.32 (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image).Graphical analysis was performed with Excel (Microsoft, Seattle).
RNA Isolation and RT-PCR Reaction
Samples were ground in liquid nitrogen and total RNA was isolated according to manufacturer's protocol using TRIzol reagent (Invitrogen, Carlsbad, CA). About 50 to 200 mg of plant tissue was used for RNA extraction. The thawed slurry was transferred to a 2-mL microcentrifuge tube containing TRIzol, vortexed for 30 s, and incubated at room temperature for 5 min. Chloroform was added into each tube, mixed gently for 15 s, and cell debris was pelleted via centrifugation at 12,000g for 15 min (4°C). The supernatant containing the RNA fraction was extracted and further precipitated at 12,000g for 10 min (4°C) with isopropanol and 1.2 sodium citrate/0.8 m NaCl solution. The pellet was washed and centrifuged at 9,500g for 7 min (4°C) with 75% (v/v) ethanol.
The amount of total RNA was determined by UV spectrophotometry. Reverse transcriptase (Moloney Murine Leukemia Virus, Invitrogen) was used to synthesize first-strand cDNA from 5 μg of total RNA according to the manufacturer's instructions. The cDNAs produced by RT were amplified with a pair of gene-specific primers (10 pmol for each primer) for each gene. The nuclear gene β-ATP that encodes the β-subunit of mitochondrial ATP synthase was used as an internal standard, since the expression is not affected by toxicity treatment. PCR was performed using standard reaction conditions provided by Promega (Madison, WI). PCR reactions for all genes were subjected to 5 min at 94°C, followed by 26 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 54°C, and extension for 1 min 15 s at 72°C with a 7-min terminal extension step at 72°C. PCR primers were designed based on the published enzyme sequences in the GenBank database (Table II). Primers specificity was confirmed using the BLAST program (BLAST 2.0; http://www.ncbi.nlm.nih.gov/BLAST/).
Table II.
Listing of the primer sets used for RT-PCR analysis
Specific enzyme genes were selected based on observed positive induction toward similar chemicals from previous studies (Wagner et al., 2002; Ekman et al., 2003). Primers were designed using the Integrated DNA Technologies homepage (http://biotools.idtdna.com/Primerquest/).
| Name | Locus | Primer Sequence | Product Size |
|---|---|---|---|
| bp | |||
| AtGSTF2 | At4g02520 | S: 5′-CGGACACCCAGCTTCCATTGCCA-3′ | 617 |
| R: 5′-TCACTGAACCTTCTCGGAAGCTG-3′ | |||
| AtGSTU1 | At1g17170 | S: 5′-TGAGGACAAGAATTGCTCTGGC-3′ | 563 |
| R: 5′-TGACCTTCTCTGACTCAGGCAG-3′ | |||
| AtGSTU24 | At2g29490 | S: 5′-GGCGTGCCATACGAATACTTGG-3′ | 571 |
| R: 5′-GTCTCTGCAATTTTGGTCATGC-3′ | |||
| OPR1 | At1g76680 | S: 5′-AGCGGGTTTCAGCCAAATGGAA-3′ | 516 |
| R: 5′-GTGTGAGGACAAGCATGTAC-3′ | |||
| OPR2 | At1g76690 | S: 5′-CGCGGTTTTCAGCCAAGGAG-3′ | 517 |
| R: 5′-GTGTGAGAACACGCTGCTAT-3′ |
About 5 μL of the PCR products were resolved in 1% agarose gel by electrophoresis and stained with ethidium bromide. Each tier on the cDNA agarose gel was run with one control lane (0 h exposure). The resolved PCR products were imaged by UV illuminator and digitally photographed (DC120 digital camera; Eastman Kodak, Rochester, NY). The intensities of the cDNA bands were quantified by computerized image analysis and the public domain NIH Image program with the gel-analyzer function of Image J version 1.32 (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image). Relative mRNA levels were then determined by taking the ratio of the band intensity specific for the gene probe of interest minus the band intensity of the β-ATP internal control.
Sequence data from this article were obtained from the EMBL/GenBank data libraries under accession numbers NM 116486, NM 101578, NM 128503, NM 202428, and NM 106319.
Acknowledgments
We are grateful to Tom Moninger and Professor Kenneth Moore from the Central Microscopy Research Facility for providing support and technical assistance on the confocal and multiphoton microscope. The technical support of Timothy Lin, Bo Cheng, Sonna Bristle, and Hsiao Ping Peng from the Department of Biological Sciences for RT-PCR analysis is greatly appreciated.
This work was supported by the Strategic Environmental Research and Development Program, the Center for Health Effects of Environmental Contamination, and Conselho Nacional de Desenvolvimento Científico e Tecnológico of the Ministry for Science and Technology of Brazil (scholarship to M.P.M.). The study was conducted as part of a W.M. Keck Foundation project.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.056168.
References
- Best EPH, Sprecher SL, Larson SL, Fredrickson HL, Bader DF (1999) Environmental behavior of explosives in groundwater from the Milan Army Ammunition Plant in aquatic and wetland plant treatments: uptake and fate of TNT and RDX in plants. Chemosphere 39: 2057–2072 [DOI] [PubMed] [Google Scholar]
- Bhadra R, Wayment DG, Hughes JB, Shanks JV (1999) Confirmation of conjugation processes during TNT metabolism by axenic plant roots. Environ Sci Technol 33: 446–452 [Google Scholar]
- Bhadra R, Wayment DG, Williams RK, Barman SN, Stone MB, Hughes JB, Shanks JV (2001) Studies on plant-mediated fate of the explosives RDX and HMX. Chemosphere 44: 1259–1264 [DOI] [PubMed] [Google Scholar]
- Biesgen C, Weiler EW (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208: 155–165 [DOI] [PubMed] [Google Scholar]
- Bryant C, Deluca M (1991) Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae. J Biol Chem 266: 4119–4125 [PubMed] [Google Scholar]
- Burken JG, Shanks JV, Thompson PL (2000) Phytoremediation and plant metabolism of explosives and nitroaromatic compounds. In JC Spain, JB Hughes, H Knackmuss, eds, Biodegradation of Nitroaromatic Compounds and Explosives, Ed 1. Lewis Publishers, Boca Raton, FL, pp 239–276
- Clarke ED, Greenhow DT, Adams D (1998) Metabolism-related assays and their application to agrochemical research: reactivity of pesticides with glutathione and glutathione transferases. Pestic Sci 54: 385–393 [Google Scholar]
- Cobbett CS (2000) Phytochelatin biosynthesis and function in heavy-metal detoxification. Curr Opin Plant Biol 3: 211–216 [PubMed] [Google Scholar]
- Coleman JOD, Blake-Kalff MMA, Davies TGE (1997. a) Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci 2: 144–151 [Google Scholar]
- Coleman JOD, Frova C, Schroder P, Tissut M (2002) Exploiting plant metabolism for the phytoremediation of persistent herbicides. Environ Sci Pollut Res Int 9: 18–28 [DOI] [PubMed] [Google Scholar]
- Coleman JOD, Randall R, Blake-Kalff MMA (1997. b) Detoxification of xenobiotics in plant cells by glutathione conjugation and vacuolar compartmentalization: a fluorescent assay using monochlorobimane. Plant Cell Environ 20: 449–460 [Google Scholar]
- Deng F, Hatzios KK (2002) Characterization and safener induction of multiple glutathione S-transferases in three genetic lines of rice. Pestic Biochem Physiol 72: 24–39 [Google Scholar]
- DeRidder B, Dixon DP, Beussman DJ, Edwards R, Goldsbrough PB (2002) Induction of glutathione S-transferases in Arabidopsis by herbicide safeners. Plant Physiol 130: 1497–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RJ (1998) Targeting antipeptide antibodies towards cytochrome P450 enzymes. In IR Phillips, EA Shepard, eds, Methods in Molecular Biology, Vol 107. Humana Press, Totowa, NJ, pp 239–249 [DOI] [PubMed]
- Ekman DR, Lorenz WW, Przybyla AE, Wolfe NL, Dean JFD (2003) SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene. Plant Physiol 133: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink RPJO, Bakker CTM, Jansen PLM (1993) Glutathione conjugate transport by human colon adenocarcinoma cells (CACO2 cells). Biochem J 290: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Theodoulou FL, Delrot S (2001) The functions of inter- and intracellular glutathione transport systems in plants. Trends Plant Sci 6: 486–492 [DOI] [PubMed] [Google Scholar]
- French CE, Rosser SJ, Davies GJ, Nicklin S, Bruce NC (1999) Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase. Nat Biotechnol 17: 491–494 [DOI] [PubMed] [Google Scholar]
- Fricker MD, May M, Meyer AJ, Sheard N, White NS (2000) Measurement of glutathione levels in intact roots of Arabidopsis. J Microsc 198: 162–173 [DOI] [PubMed] [Google Scholar]
- Fricker MD, Meyer AJ (2001) Confocal imaging of metabolism in vivo: pitfalls and possibilities. J Exp Bot 52: 631–640 [PubMed] [Google Scholar]
- Hannink NK, Rosser SJ, Bruce NC (2002) Phytoremediation of explosives. Crit Rev Plant Sci 21: 511–538 [Google Scholar]
- Hatzios KK, Penner D (1982) Metabolism of C14-labeled buthidazole in corn (Zea mays L.) and redroot pigweed (Amaranthus retroflexus L.). Weed Res 22: 337–343 [Google Scholar]
- Jablonkai I, Hatzios KK (1993) In vitro conjugation of chloroacetanilide herbicides and atrazine with thiols and contribution of nonenzymatic conjugation to their glutathione-mediated metabolism in corn. J Agric Food Chem 41: 1736–1742 [Google Scholar]
- Just CL, Schnoor JL (2004) Phytophotolysis of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) in leaves of reed canary grass. Environ Sci Technol 38: 290–295 [DOI] [PubMed] [Google Scholar]
- Kooshki M, Mentewab A, Stewart CN (2003) Pathogen inducible reporting in transgenic tobacco using a GFP construct. Plant Sci 165: 213–219 [Google Scholar]
- Larson SL, Jones RP, Escalon L, Parker D (1999) Classification of explosives transformation products in plant tissue. Environ Toxicol Chem 18: 1270–1276 [Google Scholar]
- Lucero ME, Mueller W, Hubstenberger J, Phillips GC, O'Connell MA (1999) Tolerance to nitrogenous explosives and metabolism of TNT by cell suspensions of Datura innoxia. In Vitro Cell Dev Biol Plant 35: 480–486 [Google Scholar]
- Matsui H, Nakamura G, Ishiga Y, Toshima H, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y (2004) Structure and expression of 12-oxophytodienoate reductase (subgroup I) genes in pea, and characterization of the oxidoreductase activities of their recombinant products. Mol Genet Genomics 271: 1–10 [DOI] [PubMed] [Google Scholar]
- May MJ, Vernoux T, Leaver C, Van Montagu M, Inze D (1998) Glutathione homeostasis in plants: implications for environmental sensing and plant development. J Exp Bot 49: 649–667 [Google Scholar]
- Meyer AJ, Fricker MD (2000) Direct measurement of glutathione in epidermal cells of intact Arabidopsis roots by two-photon laser scanning microscopy. J Microsc 198: 174–181 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Fricker MD (2002) Control of demand-driven biosynthesis of glutathione in green Arabidopsis suspension culture cells. Plant Physiol 130: 1927–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzari MP, Van Aken B, Yoon JM, Just CL, Schnoor JL (2004) Mathematical modeling of RDX and HMX metabolism in poplar (Populus deltoides × Populus nigra, DN34) tissue culture. Int J Phytoremediation 6: 323–345 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ohkawa H, Imaishi H, Shiota N, Yamada T, Inui H (1999) Cytochrome P450s and Other Xenobiotic Metabolizing Enzymes in Plants, Vol 233. The Royal Society of Chemistry, Cambridge, UK
- Patel N, Cardoza V, Christensen E, Rekapalli L, Ayalew M, Stewart CN (2004) Differential gene expression of Chlamydomonas reinhardtii in response to 2,4,6-trinitrotoluene (TNT) using microarray analysis. Plant Sci 167: 1109–1122 [Google Scholar]
- Pavlostathis SG, Comstock KK, Jacobson ME, Saunders FM (1998) Transformaton of 2,4,6-trinitrotoluene by the aquatic plant Myriophyllum spicatum. Environ Toxicol Chem 17: 2266–2273 [Google Scholar]
- Peterson FJ, Mason RP, Hovsepian J, Holtzman JL (1979) Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J Biol Chem 254: 4009–4014 [PubMed] [Google Scholar]
- Pflugmacher S, Sandermann H (1998) Cytochrome P450 monooxygenases for fatty acids and xenobiotics in marine macroalgae. Plant Physiol 117: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugmacher S, Schroder P, Sandermann H (2000) Taxonomic distribution of plant glutathione S-transferases acting on xenobiotics. Phytochemistry 54: 267–273 [DOI] [PubMed] [Google Scholar]
- Sandermann H (1994) Higher-plant metabolism of xenobiotics: the green liver concept. Pharmacogenetics 4: 225–241 [DOI] [PubMed] [Google Scholar]
- Schalk M, Pierrel MA, Zimmerlin A, Batard Y, Durst F, Werck-Reichhart D (1997) Xenobiotics: substrates and inhibitors of the plant cytochrome P450. Environ Sci Pollut Res Int 4: 229–234 [DOI] [PubMed] [Google Scholar]
- Schroder P, Collins C (2002) Conjugating enzymes involved in xenobiotic metabolism of organic xenobiotics in plants. Int J Phytoremediation 4: 247–265 [Google Scholar]
- Scwitzguebel JP, Vanek T (2003) Some fundamental advances for xenobiotic chemicals. In SC McCutcheon, JL Schnoor, eds, Phytoremediation: Transformation and Control of Contaminants. John Wiley & Sons, Hoboken, NJ, pp 123–157
- Shah MM, Spain JC (1996) Elimination of nitrite from the explosive 2,4,6-trinitrophenylmethylnitramine (tetryl) catalyzed by ferredoxin NADP oxidoreductase from spinach. Biochem Biophys Res Commun 220: 563–568 [DOI] [PubMed] [Google Scholar]
- Subramanian M, Shanks JV (2003) Role of plants in the transformation of explosives. In SC McCutcheon, JL Schnoor, eds, Phytoremediation: Transformation and Control of Contaminants. John Wiley & Sons, Hoboken, NJ, pp 389–408
- Tal JA, Hall JC, Stephenson GR (1995) Nonenzymatic conjugation of fenoxaprop-ethyl with glutathione and cysteine in several grass species. Weed Res 35: 133–139 [Google Scholar]
- Thompson PL, Ramer LA, Schnoor JL (1999) Hexahydro-1,3,5-trinitro-1,3,5-triazine translocation in poplar trees. Environ Toxicol Chem 18: 279–284 [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49: 515–532 [DOI] [PubMed] [Google Scholar]
- Wolfe NL, Hoehamer CF (2003) Enzymes used by plants and microorganisms to detoxify organic compounds. In SC McCutcheon, JL Schnoor, eds, Phytoremediation: Transformation and Control of Contaminants. John Wiley & Sons, Hoboken, NJ, pp 159–187
- WSSA (1994) Herbicide Handbook, Ed 7. Weed Science Society of America, Champaign, IL