Abstract
Two types of methionine (Met) sulfoxide reductases (Msr) catalyze the reduction of Met sulfoxide (MetSO) back to Met. MsrA, well characterized in plants, exhibits an activity restricted to the Met-S-SO-enantiomer. Recently, a new type of Msr enzyme, called MsrB, has been identified in various organisms and shown to catalytically reduce the R-enantiomer of MetSO. In plants, very little information is available about MsrB and we focused our attention on Arabidopsis (Arabidopsis thaliana) MsrB proteins. Searching Arabidopsis genome databases, we have identified nine open reading frames encoding proteins closely related to MsrB proteins from bacteria and animal cells. We then analyzed the activity and abundance of the two chloroplastic MsrB proteins, MsrB1 and MsrB2. Both enzymes exhibit an absolute R-stereospecificity for MetSO and a higher catalytic efficiency when using protein-bound MetSO as a substrate than when using free MetSO. Interestingly, we observed that MsrB2 is reduced by thioredoxin, whereas MsrB1 is not. This feature of MsrB1 could result from the lack of the catalytical cysteine (Cys) corresponding to Cys-63 in Escherichia coli MsrB that is involved in the regeneration of Cys-117 through the formation of an intramolecular disulfide bridge followed by thioredoxin reduction. We investigated the abundance of plastidial MsrA and B in response to abiotic (water stress, photooxidative treatment) and biotic (rust fungus) stresses and we observed that MsrA and B protein levels increase in response to the photooxidative treatment. The possible role of plastidic MsrB in the tolerance to oxidative damage is discussed.
Oxygen is essential to all aerobic organisms but can also lead to many harmful effects (Davies, 1995). Proteins are easily damaged by reactive oxygen species, with Met being one of the amino acid residues most susceptible to oxidation (Dann and Pell, 1989). The generation of Met sulfoxide (MetSO) is mediated by various biological oxidants such as hydrogen peroxide, hydroxyl radicals, ozone as well as by metals, and results in modifications of activity and conformation for many proteins (Gao et al., 1998; Davis et al., 2000). Oxidation of Met residues is readily reversed by the action of an enzyme initially referred as peptide MetSO reductase (PMSR), which catalyzes the thioredoxin-dependent reduction of MetSO back to Met (Brot et al., 1981). The enzyme is present in most living organisms and has been described as belonging to the minimal set of proteins sufficient for cell life (Mushegian and Koonin, 1996). PMSR has a protective role against oxidative damage (Moskovitz et al., 1997). Indeed, pmsr null mutants of Escherichia coli and Saccharomyces cerevisiae show a decreased resistance toward oxidative stress conditions (Moskovitz et al., 1995, 1997). Accordingly, PMSR overexpression in S. cerevisiae or in human T-cells results in an increased resistance to oxidative treatments (Moskovitz et al., 1998). The first PMSR has been renamed Met sulfoxide reductase A (MsrA) and has been shown to be specific to the Met-S-SO-enantiomer (Sharov et al., 1999). Note that another enzyme, biotin sulfoxide reductase BisC from E. coli, has been very recently reported to reduce free Met-S-SO (Ezraty et al., 2005). In the last years, a new type of Msr enzyme, termed MsrB, has been identified and shown to catalytically reduce the R-enantiomer of MetSO (Grimaud et al., 2001; Kryukov et al., 2002; Kumar et al., 2002; Olry et al., 2002). Both Msr activities are essential for organisms, since Met oxidation results in a racemic mixture of R- and S-enantiomers. It is noteworthy that MsrB proteins do not display any sequence or structural homology with MsrA. In Neisseria gonorrhoeae, the protein PILB is composed of three subdomains, a domain with thioredoxin-like activity in the N terminus, a domain displaying MsrA activity, and the last one strikingly similar to human SelX, a selenoprotein that exhibits MsrB activity (Olry et al., 2002; Wu et al., 2005). Selenoproteins contain the rare amino acid selenocysteine instead of Cys. This new type of Msr activity has also been reported for SelX homologs in E. coli (Grimaud et al., 2001), Staphylococcus aureus (Singh et al., 2001), and mouse (Moskovitz et al., 2002).
With regard to the plant kingdom, Sanchez et al. (1983) obtained evidence for Msr activity in a wide variety of plant species and showed that the activity was mainly localized in the stroma of leaf chloroplasts. These authors proposed that thioredoxin (Trx) could constitute the physiological electron donor to Msr enzymes and that these proteins may participate in the plastidic antioxidant network. Msr activity has been described for decades, but only recent studies, using the model plant Arabidopsis (Arabidopsis thaliana), revealed the complexity of the multigenic family coding for MsrA proteins. Sadanandom et al. (2000) reported the presence of at least five MsrA genes encoding different protein isoforms (three cytosolic, one chloroplastic, and one targeted to a secretory pathway). The cytosolic isoform is present in all tissues, whereas the chloroplastic enzyme is largely confined to green tissues and prominent in leaves. Arabidopsis mutants, lacking a functional cytosolic MsrA gene (PMSR2, At5g07460), exhibit reduced growth and slower development under short-day conditions, but not under long-day conditions (Bechtold et al., 2004). PMSR2 has been proposed to repair oxidized proteins under long-night periods to avoid a loss of energy and of carbon resources resulting from an increased rate of protein turnover. Msr needs only a source of reductant mediated through interaction with Trx. In connection with this feature, a plastidic MsrA protein has been isolated as a Trx target in Chlamydomonas reinhardtii (Lemaire et al., 2004). In Arabidopsis, the abundance of plastidic MsrA (pMsrA) is increased by high light and methyl viologen treatments but not by ozone (Romero et al., 2004). The involvement of pMsrA in the tolerance to oxidative stress has been shown using transgenic Arabidopsis plants under- or overexpressing pMsrA subjected to various oxidative treatments (Romero et al., 2004). The first potential pMsrA substrate identified is a small heat shock protein, Hsp21 (Gustavsson et al., 2002). Hsp21 has a conserved N-terminal region highly rich in Met residues, and keeping these residues in a reduced form is required to maintain the chaperone-like activity of the protein (Sundby et al., 2005).
Until now, very few studies concerning plant MsrB proteins have been reported. Three genes showing strong similarity with SelX selenoproteins have been described in Arabidopsis (Rodrigo et al., 2002). The expression of one of these genes is enhanced in response to water deficit and oxidative stress (Rodrigo et al., 2002). Very recently, a MsrB was isolated in potato (Solanum tuberosum) as a target of CDSP32 (chloroplastic drought-induced stress protein of 32 kD; Rey et al., 2005). CDSP32 is a peculiar plant Trx induced under severe environmental stress conditions (Rey et al., 1998; Broin et al., 2000) and composed of two typical Trx modules, with only one active redox disulfide center in the C-terminal domain (Rey et al., 1998). CDSP32 has been shown to play an essential role in the protection of the photosynthetic apparatus against oxidative damage (Broin et al., 2002; Broin and Rey, 2003).
In this work, we describe the characteristics of the MsrB family in Arabidopsis that appears even more complex than the MsrA family. We cloned the two cDNAs encoding the predicted plastidic MsrB proteins, named MsrB1 and MsrB2. The catalytic activity of recombinant proteins has been studied using free or protein-bound MetSO substrates and dithioerythritol (DTE) or Trx as electron donors. We then investigated the abundance of the two plastidic MsrB proteins, in comparison with that of pMsrA, in the different organs of Arabidopsis plants. We also checked protein level during abiotic stresses in Arabidopsis leaves subjected to water deficit and photooxidative treatments or during a biotic stress, in poplar (Populus × interamericana) leaves infected by the rust fungus Melampsora larici-populina.
RESULTS
The MsrB Gene Family in Arabidopsis
Searching the Arabidopsis genome sequence databases (The Arabidopsis Information Resource, Munich Information Center for Protein Sequences), we identified nine open reading frames encoding proteins closely related to the MsrB proteins from Drosophila melanogaster, Mus musculus, or S. cerevisiae. All genes are located in chromosome IV except At1g53670. Accession numbers and location in each chromosome are shown in Table I. Noteworthy, MsrB genes in chromosome IV are grouped in two clusters, the first comprising At4g04800, At4g04810, At4g04830, and At4g04840 and the second At4g21830, At4g21840, At4g21850, and At4g21860. To gain insight into functionality of the genes, the expressed sequence tag (EST) database at the National Center for Biotechnology Information (NCBI) site was searched and cDNA were detected for all of them. Only one EST was found for At4g04810 and At4g21840, whereas At4g21850 and At4g21860 seem to be highly expressed with 22 and 24 ESTs, respectively. These data indicate that the nine MsrB genes are expressed and that they display dramatic differences in expression. The genes were named MsrB1-MsrB9 according to their predicted subcellular location (chloroplast, secretory pathway, and cytosol) and also according to the order in which they are located in each chromosome.
Table I.
MsrB proteins in Arabidopsis
GenBank accession number for each protein.
Location indicates the position of the first and last nucleotides of each gene in the chromosome.
Predicted subcellular compartment of each protein by Predotar, TargetP, and ChloroP softwares.
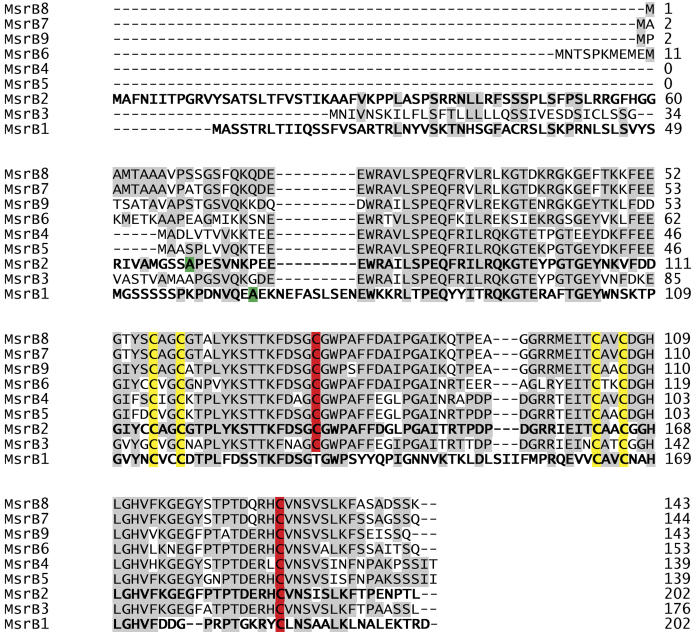
A detailed alignment of the proteins encoded by Arabidopsis MsrB genes is shown in Figure 1. The proteins display different sizes, ranging from 139 to 202 amino acids. Multiple alignment of MsrB sequences revealed that all members have four conserved Cys residues, which are organized in two CxxC motifs and could potentially be involved in zinc fixation as proposed for Drosophila MsrB (Kumar et al., 2002). MsrB protein sequences also contain one strictly conserved Cys residue (Cys-117 in E. coli). A second Cys (Cys-63 in E. coli) is present in eight of them, MsrB1 possessing a Thr residue instead of this Cys. Moreover, MsrB1 also exhibits a peculiar feature with another Cys located in position 47, between the two Cys potentially involved in zinc fixation. According to prediction software (TargetP and Predotar), MsrB3 is predicted to be addressed to a secretory pathway. MsrB1 and MsrB2, which have N-terminal extensions of 65 and 68 residues, respectively, are predicted to be addressed to the chloroplast. As shown in Figure 1, MsrB proteins from Arabidopsis are highly conserved. MsrB4, MsrB5, and MsrB6, assumed to be cytosolic enzymes, display 87% to 97% identity. These three MsrB share about 70% to 80% identity with MsrB2 without the predicted N-terminal extension, but only 40% with predicted mature MsrB1. Thus, the sequence of MsrB1 appears as the most divergent in comparison with the eight others. For example, it exhibits an extension of nine amino acids in the N-terminal region. Thereafter, we focused our studies on the two predicted plastidic MsrB with regard to their activity and expression characteristics.
Figure 1.
Multiple sequence alignment of Arabidopsis MsrB proteins. Sequences were obtained through the NCBI, and the alignment of amino acid sequences was performed using the ClustalW software. Amino acids are given using standard single-letter designation, and dashes indicate gaps. Numbers indicate protein length in amino acids. MsrB typical amino acids are indicated as follows: the catalytic Cys (Cys-117 and Cys-63) in red, and the putative zinc-coordinating Cys (Cys-45 and Cys-48, Cys-92 and Cys-95) in yellow. The numbering of amino acid residues is based on the E. coli MsrB sequence. The first amino acid of plastidial MsrB mature proteins, as predicted by the ChloroP software, is represented in green. Other conserved residues are highlighted in gray.
Production and Purification of MsrB1 and MsrB2 Recombinant Proteins
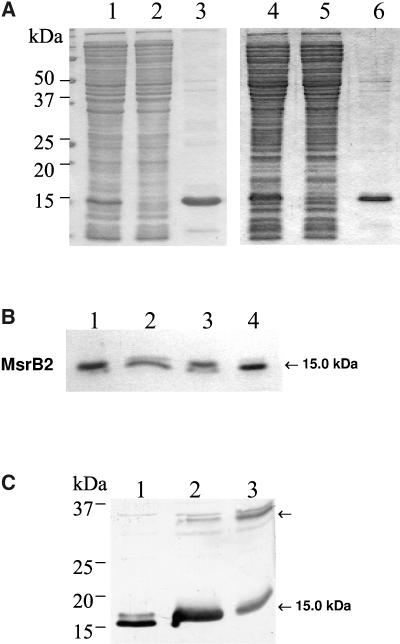
We investigated the biological activities of Arabidopsis MsrB1 and B2 proteins by producing recombinant proteins in E. coli. Both MsrB proteins were expressed as mature proteins without the transit peptide predicted by the ChloroP software and with an N-terminal poly-His tag to allow purification using affinity chromatography as shown in Figure 2A. Fractions collected during the purification steps were analyzed using SDS-PAGE. Recombinant MsrB1 and MsrB2 exhibited apparent molecular masses of approximate 15 to 16 kD. Note that MsrB2 was visualized as two distinct bands using Coomassie Blue and western analyses. In order to test whether the two forms correspond to different redox states or to degradation products, we carried out incubation experiments of the MsrB2 protein in the presence of reducing reagents (Fig. 2B), followed by SDS-PAGE and revelation by Coomassie Blue or western analysis. Without any reagent, MsrB2 was visualized as two bands, the most abundant form being the one with the higher apparent molecular mass (lane 1). When MsrB2 was incubated with reducing reagents (DTE or dithiothreitol [DTT]), the ratio between the two forms was modified with the most abundant form corresponding to the band with the lower apparent molecular mass (lanes 2 and 4). If urea, a denaturing reagent, was added to the MsrB2 protein, no change was noted when compared to the untreated protein (lane 3). We conclude, from these experiments, that the two bands correspond to two redox states of MsrB2, the one with the apparent lower molecular mass corresponding to a reduced form. During purification of recombinant MsrB2, we also noticed that the protein has the ability to form dimers in the absence of reducing reagent (Fig. 2C). This feature of MsrB2 was enhanced by adding diamide that favors the formation of disulfide bridges. These results indicate that MsrB2 forms homodimers in vitro through disulfide bridges.
Figure 2.
A, SDS-PAGE analysis of fractions collected during the purification of recombinant MsrB proteins. Lane 1, Crude protein extract of E. coli cells transformed with the pSD80 vector harboring the Arabidopsis MsrB1 gene and induced with 0.1 mm IPTG. Lane 2, Protein extract nonfixated on the Ni affinity-column. Lane 3, MsrB1 protein eluted from the Ni-resin using a buffer containing 500 mm imidazole. Lane 4, Crude protein extract of E. coli cells transformed with the pQE-30 vector harboring the Arabidopsis MsrB2 gene and induced with 0.1 mm IPTG. Lane 5, Similar to lane 2. Lane 6, MsrB2 protein eluted from the Ni-resin using a buffer containing 500 mm imidazole. B, Characterization of the redox state of recombinant Arabidopsis MsrB2. Recombinant Arabidopsis MsrB2 protein was incubated in vitro with reducing reagents (DTE or DTT) or with urea, a denaturing reagent, separated using SDS-PAGE, and revealed by Coomassie Blue. Lane 1, MsrB2; Lane 2, +DTE; Lane 3, +urea 6 m; Lane 4, +DTT. C, SDS-PAGE and western-blotting analysis of recombinant Arabidopsis MsrB2. Crude protein extracts of E. coli cells transformed with the pQE-30 harboring Arabidopsis MsrB2 gene were subjected to SDS-PAGE (13% acrylamide gel), electrotransferred on to a nitrocellulose membrane, and immunoblotted with anti-MsrB polyclonal antibodies. Lane 1, +DTT; Lane 2, −DTT; Lane 3, −DTT, +diamide.
Activity of Recombinant Arabidopsis MsrB1 and MsrB2
To compare the efficiencies of the two MsrB, a steady-state kinetic analysis was performed using an HPLC method, dabsyl-MetSO, a protein-bound-like MetSO substrate, and DTE as a reductant. In our assay conditions, MsrB1 and MsrB2 exhibited Michaelis-Menten kinetics (data not shown). Km values for the substrate were close since they ranged from 55 μm for MsrB2 to 80 μm for MsrB1 (Table II). Turnover numbers (kcat) were 0.07 s−1 and 0.02 s−1 for MsrB1 and MsrB2, respectively. These results indicate that MsrB1 has a kcat and a catalytic efficiency (kcat/Km) 2.5- to 3-fold greater than that of MsrB2, using protein-bound MetSO. Altogether, these data show that both MsrB exhibit Msr activities, with MsrB1 being more efficient than MsrB2.
Table II.
Enzymatic kinetic parameters of MsrB1 or MsrB2 by following the reduction of dabsyl-MetSO in the presence of DTE
KSulfoxide and kcat/KSulfoxide should be respectively divided and multiplied by two, considering that only the R isomer is a substrate of MsrB.
| KSulfoxide | kcat | kcat/KSulfoxide | |
|---|---|---|---|
| μm | s−1 | m−1s−1 | |
| B1 | 78.53 ± 2.17 | 0.07 ± 0.004 | 891 ± 78 |
| B2 | 54.06 ± 1.79 | 0.02 ± 0.002 | 370 ± 48 |
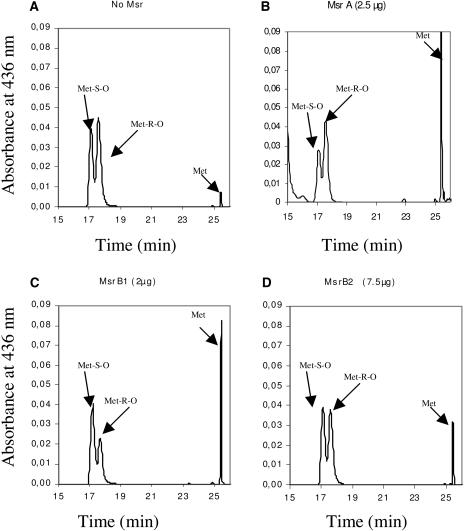
Then, we investigated the stereospecificity of the two MsrB proteins, using HPLC and the dabsyl-MetSO substrate, as shown in Figure 3. In the absence of Msr, MetSO was separated into two pools corresponding to the R and S forms (Fig. 3A). The Met-R-SO form was always found to be slightly more abundant than the S form. The proportion of reduced Met, before the addition of Msr, was about 2%. As a control, we assayed the activity of the recombinant poplar pMsrA. The chromatogram shows a noticeable decrease of the Met-S-SO form together with a strong increase of the peak corresponding to Met, whereas no change in the Met-R-SO amount was noticed (Fig. 3B). These data confirm the specific pMsrA activity toward Met-S-SO. When similar experiments were performed with MsrB1 (Fig. 3C) and MsrB2 (Fig. 3D), we observed that the peaks corresponding to Met-R-SO decreased, whereas the Met-S-SO amount did not vary. Using equimolar amounts of the two catalysts, the decrease in peak height was substantially more pronounced when assaying MsrB1 activity. These data are fully consistent with MsrB1 having a 2.5-fold higher catalytic efficiency than MsrB2 (Table II) using dabsyl-MetSO as a substrate. From these results, we conclude that both MsrB proteins exhibit an absolute R-stereospecificity for MetSO.
Figure 3.
Substrate specificity of poplar pMsrA and Arabidopsis MsrB1 and MsrB2 proteins. Reduction of dabsyl-Met-S-sulfoxide and dabsyl-Met-R-sulfoxide was followed using HPLC assays and monitored by measuring the A436. Location of dabsyl-MetSO substrates (Met-S-O and Met-R-O) and dabsyl Met product (Met) are indicated by arrows. A, Separation of the two diastereomers of MetSO. B, DTE-dependent reduction of dabsyl-Met S-sulfoxide by poplar pMsrA. C, DTE-dependent reduction of dabsyl-Met R-sulfoxide by Arabidopsis MsrB1. D, DTE-dependent reduction of dabsyl-Met R-sulfoxide by Arabidopsis MsrB2.
Finally, the nature of the physiological reductant of MsrB was tested using Trx as an electron donor instead of DTE. The Msr activity was assayed by following spectrophotometrically the oxidation of NADPH at 340 nm in the presence of free MetSO or N-acetyl-MetSO as substrates, and a Trx system constituted of various poplar cytosolic Trx (either Trx h1, or h3, or h5; Gelhaye et al., 2004) in combination with the Arabidopsis NADPH-linked Trx reductase. MsrB1 was not able to reduce free MetSO or N-acetyl-MetSO in the presence of the Trx reducing system whatever the Trx isoforms and the concentrations used (data not shown). On the contrary, we observed that the MsrB2 protein can be reduced by all Trx h tested, with a Km value around 8 μm for Trx h1 (Table III; data not shown). MsrB2 displayed dramatic differences depending on the substrate used, since the Km values for sulfoxides ranged from 2 mm with N-acetyl-MetSO to 30 mm with free MetSO, reflecting the much lower ability of MsrB2 to reduce free MetSO. The kcat values found with MsrB2 were 1.37 and 2.94 s−1 with free MetSO and N-acetyl-MetSO, respectively. Based on these kinetic parameters, we conclude that MsrB2 preferentially reduces protein-bound substrates like N-acetyl-MetSO with a catalytic efficiency (kcat/Km) 30-fold higher compared to that observed with free MetSO. All these results indicate that MsrB1 and MsrB2 exhibit Msr activities with different kinetic parameters according to the type of substrate and use specific electron donors.
Table III.
Enzymatic kinetic parameters of MsrB2 using N-acetyl-MetSO and free MetSO as substrates by following NADPH oxidation in the presence of the Trx reducing system
Apparent Km for Trx h1, MetSO, or N-acetyl-MetSO were determined at saturating concentration of the other substrate. KSulfoxide and kcat/KSulfoxide should be respectively divided and multiplied by two, considering that only the R isomer is a substrate of MsrB.
| KSulfoxide | kcat | kcat/KSulfoxide | KTrx h1 | |
|---|---|---|---|---|
| mm | s−1 | m−1s−1 | μm | |
| Free MetSO | 28.980 ± 4.98 | 1.37 ± 0.07 | 47 ± 2 | 8.32 ± 1.44 |
| N-acetyl-MetSO | 2.16 ± 0.28 | 2.94 ± 0.12 | 1,361 ± 55 |
Western Analysis of pMsrA, MsrB1, and MsrB2 in Leaf Proteins and Localization in Chloroplast
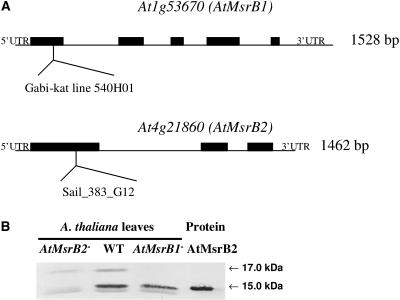
Using a serum raised against a synthetic peptide corresponding to a part of MsrB1 (from residue 89 to residue 103), two main bands, with apparent molecular masses of around 15 and 17 kD (Fig. 4B, lane 2), were revealed in leaf soluble proteins. Both proteins were not revealed when using the preimmune serum (data not shown). We hypothesized that the two proteins could correspond to MsrB1 and MsrB2 proteins since the MsrB1 sequence part selected for raising the serum displays 73% amino acid identity with the corresponding MsrB2 sequence. Both recombinant proteins were recognized by the serum, which nevertheless appeared more specific for MsrB1 (data not shown). To determine precisely the identity of the proteins recognized by the serum in plants, we characterized Arabidopsis knockout mutants for MsrB1 and MsrB2 genes. The MsrB1 mutant, corresponding to a transgenic T-DNA insertion plant, was obtained from Max Planck Institute (Germany) and termed GABI-Kat line 540H01 (Rosso et al., 2003). The MsrB2 mutant was obtained from the Syngenta Arabidopsis Insertion Library (SAIL) collection and provided through the Arabidopsis Biological Resource Center (ABRC) as the SAIL_383_G12 line (Sessions et al., 2002). For both mutants, T-DNA insertions were precisely mapped by PCR (Fig. 4A) and homozygous lines were obtained (data not shown). Using reverse transcription-PCR, MsrB1 and MsrB2 transcripts were undetected in GABI and SAIL homozygous mutant lines, respectively (data not shown). Western analyses were carried out using leaf soluble proteins prepared from mutants (Fig. 4B). The 17-kD protein was only present in the MsrB2 mutant, whereas the 15-kD band was only detected in the MsrB1 mutant. From these data, we conclude that MsrB1 and MsrB2 proteins correspond to the 17- and 15-kD bands, respectively.
Figure 4.
Western analysis of MsrB1 and MsrB2 abundances in AtMsrB1 and AtMsrB2 T-DNA insertion mutant lines of Arabidopsis. A, Genetic mapping of AtMsrB1 and AtMsrB2 T-DNA mutants. T-DNA insertions within the AtMsrB1 (Gabi-kat line 540H01) and the AtMsrB1 (Sail_383_G12) mutant lines were precisely mapped by PCR. Exons are shown by black boxes. B, Western analysis of MsrB1 and MsrB2 abundances in leaf soluble proteins from wild-type Arabidopsis plants and from AtMsrB1 and AtMsrB2 homozygous mutant plants. Lane AtMsrB2−, AtMsrB2 insertion line; WT, wild type; Lane AtMsrB1−, AtMsrB1 insertion line; Protein AtMsrB2, purified recombinant AtMsrB2 fused with a 6×His-tag as shown in Figure 2A. Leaf soluble proteins (30 μg per lane) and recombinant AtMsrB2 (5 ng per lane) were separated by SDS-PAGE. The serum raised against the MsrB1 synthetic peptide was used diluted 1:1,000.
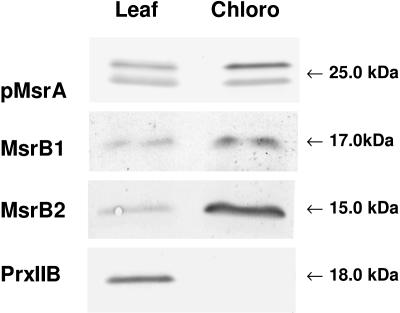
We then investigated the subcellular localization of the MsrB1 and MsrB2 proteins. Arabidopsis leaf chloroplasts were purified on Percoll gradients and immunoblotting experiments were carried out on leaf and chloroplastic soluble proteins using the serum recognizing both MsrB1 and MsrB2 proteins (Fig. 5). We first checked the purity of the chloroplast preparation using a serum raised against the cytosolic type-IIB peroxiredoxin (Bréhelin et al., 2003). The protein was revealed at a high level in leaves, but was not detected in chloroplastic proteins, showing the absence of cytosolic contamination in purified chloroplasts. With regard to MsrB1 and MsrB2, western data exhibited a higher abundance for both proteins in chloroplast samples. This result is consistent with the predictions obtained using TargetP and Predator software. Note also that the potato MsrB1 protein was isolated from purified potato chloroplasts, as a potential target of the CDSP32 Trx (Rey et al., 2005). We conclude from these data that MsrB1 and MsrB2 are located in the chloroplast.
Figure 5.
Western analysis of pMsrA, MsrB1, MsrB2, and PrxIIB abundances in leaf and plastidic soluble proteins from Arabidopsis. Leaf and Chloro, leaf and chloroplastic soluble proteins, respectively, from Arabidopsis control wild-type plants. Soluble proteins (10 μg per lane for MsrA and PrxIIB blots and 25 μg per lane for MsrB1 and MsrB2 blots) were separated by SDS-PAGE. The sera raised against poplar pMsrA and Arabidopsis MsrB were used diluted 1:1,000. The serum raised against Arabidopsis PrxIIB (type-IIB peroxiredoxin) was provided by Dr. Y. Meyer (CNRS-Université de Perpignan) and used diluted 1:20,000.
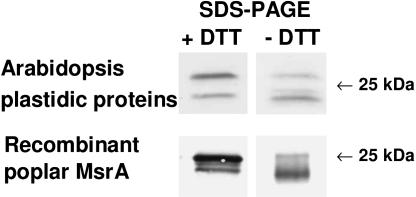
Using a serum raised against the recombinant poplar plastidic MsrA (pMsrA) protein, two bands at around 25 kD were revealed in leaves and in chloroplasts from Arabidopsis, but only the highest one was found to be more abundant in the plastidic fraction. We then investigated whether the redox state could modify the pMsrA migration properties by running SDS-PAGE gel without DTT and western analysis (Fig. 6). We observed that under nonreducing conditions, the serum mainly reveals the lower form of pMsrA in plastidic Arabidopsis proteins, whereas the higher form was more abundant following a DTT treatment. Interestingly, a similar migration shift was noticed for the recombinant poplar pMsrA protein, when migration was carried out with or without DTT. Consequently, we propose that the two pMsrA forms revealed in plastidic samples correspond to two redox forms, the reduced form migrating with a higher apparent molecular mass.
Figure 6.
Western analysis of Arabidopsis plastidic MsrA and of poplar recombinant pMsrA separated in the presence or absence of DTT during SDS-PAGE. Western analysis was carried out using the serum raised against poplar recombinant pMsrA protein diluted 1:1,000. Extraction of soluble plastidic proteins was performed in the absence of β-mercaptoethanol. 25 and 0.1 μg proteins per lane for Arabidopsis leaf proteins and poplar recombinant pMsrA, respectively.
Abundance of Plastidic Msr Proteins in Arabidopsis Organs
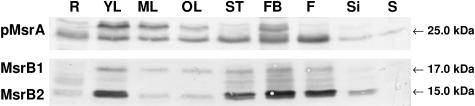
The abundance of MsrB1 and MsrB2 proteins was investigated in various plant tissues from Arabidopsis by western analysis and compared to that of pMsrA (Fig. 7). In most organs, except mature and old leaves, MsrB2 was found to be more abundant than MsrB1. The highest MsrB2 protein levels were detected in developing organs such as young leaves and floral buds, and noticeable amounts were also observed in stems and in flowers. MsrB1 exhibits a pattern of expression close to that of MsrB2 with higher protein amounts in young leaves and in floral buds. It is noteworthy that the abundance of both MsrB proteins decreases with leaf age. MsrB proteins were much less abundant in siliques and in roots and were not detected in seeds. pMsrA showed two different patterns depending on which form was observed. The reduced form with the higher apparent molecular mass was found, as for MsrB proteins, to be the most abundant in young developing leaves and in floral buds. This form was revealed at a very low level in roots and in stems and was undetected in flowers, siliques, and seeds. The lower pMsrA form was present at a substantial level in most organs, but was revealed to a much lesser extent in siliques and in seeds.
Figure 7.
Western analysis of pMsrA, MsrB1, and MsrB2 abundances in the different organs of Arabidopsis plants. Soluble proteins (25 μg per lane) were separated by SDS-PAGE. R, Root; YL, young leaf; ML, mature leaf; OL, old leaf; ST, stem; FB, flower bud; F, flower; Si, silique; S, seed. The sera raised against poplar pMsrA and Arabidopsis MsrB were used diluted 1:1,000. The bands corresponding to pMsrA, MsrB1, and MsrB2 were revealed at 25, 17, and 15 kD, respectively.
Abundance of Plastidic Msr Proteins under Stress Conditions
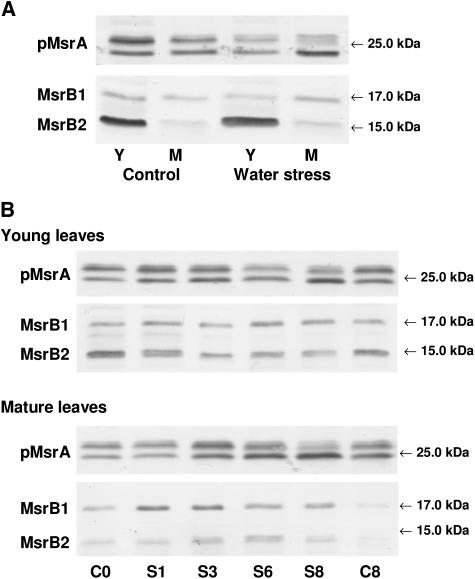
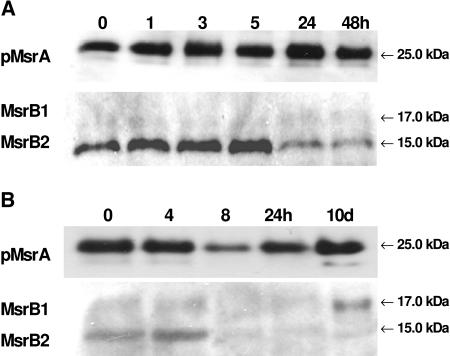
We then analyzed the abundance of the plastidic MsrA and B proteins under various stress conditions. First, we subjected Arabidopsis plants to water deficit. After 6 d, plants displayed severe wilting symptoms, which mainly concerned mature leaves (data not shown). No noticeable change was noticed in MsrB1 and MsrB2 amounts in young and mature leaves under water stress (Fig. 8A). With regard to the plastidic MsrA protein, the reduced form appeared to be less abundant in young and in mature leaves of water-stressed plants compared to controls. The abundance of the oxidized form decreased in young leaves, but substantially increased in mature leaves upon water deficit.
Figure 8.
Western analyses of pMsrA, MsrB1, and MsrB2 abundances in young and mature leaves of Arabidopsis plants subjected to abiotic stress conditions. A, Water deficit. Y, Young developing green leaves; M, mature leaves; Control, well-watered plants; Water stress, plants subjected to removal of water for 6 d (relative water content around 70%). B, Photooxidative treatment (high light 1,400 μE m−2 s−1 and low temperature, 8°C, for 8 d. C, Control conditions for 0 or 8 d; S, photooxidative treatment for 1, 3, 6, or 8 d. Immunoblotting was performed on 25 μg protein per lane using the pMsrA and MsrB antisera diluted 1:1,250 and 1:1,000, respectively. The bands corresponding to pMsrA, MsrB1, and MsrB2 were revealed at 25, 17, and 15 kD, respectively.
Arabidopsis plants were then subjected to photooxidative stress by exposure to high light (1,400 μE m−2 s−1) and low temperature (8°C) for 8 d. After 3 d, mature and old leaves exhibited symptoms as bleaching and necrosis and were more damaged than young leaves (data not shown). The treatment also resulted in an anthocyanin accumulation occurring earlier in young leaves than in mature leaves. In young leaves, a slight decrease in MsrB2 level was observed during the treatment, whereas the MsrB1 amount was not noticeably modified (Fig. 8B). In mature leaves, a substantial increase in MsrB1 abundance occurred from the first day of treatment and the protein amount was much higher during the whole stress period than in control conditions. Compared to MsrB1, the MsrB2 level was found to increase later and to a lesser extent in response to the photooxidative treatment. An increase in the level of the pMsrA reduced form was observed during the first 3 d of treatment in young leaves. Then, the reduced form became much less abundant compared to the oxidized form. In mature leaves, an increase in the amount of the reduced form was observed from the third day of the stress period and a decrease was noticed thereafter. The pMsrA level is close in young and mature leaves after 8 d, with a much higher abundance of the oxidized form in both leaves. Note that this form was found to accumulate with some delay in mature leaves compared to young ones.
We also followed Msr protein amount in poplar leaves infected by the rust fungus M. larici-populina, in the case of two different reactions, either compatible, i.e. the fungus is able to invade the plant cells, or incompatible, i.e. the fungus is killed after the plant generates reactive oxygen species, a process known as oxidative burst. As observed previously, the pMsrA was present under two forms, the reduced form being predominant (Fig. 9). The level of pMsrA increased slightly with time (after 1 h of infection) in the case of the incompatible reaction, whereas it is decreased during the compatible reaction with a minimum after 8 h of infection (Fig. 9, A and B). As in Arabidopsis, the anti-MsrB1 antibody was found to react with two isoforms of MsrB in poplar leaf proteins, corresponding in size to AtMsrB1 and AtMsrB2. Accordingly, two ESTs very similar to AtMsrB1 and AtMsrB2 have been found from Populus trichocarpa sequence genome databases. MsrB1 was hardly detected (Fig. 9). This may be due to a weak expression of the corresponding gene or to low cross reaction with the serum raised against the Arabidopsis protein. The MsrB1 amount seems to be unchanged except in the compatible reaction, where the level increased 10 d after infection (Fig. 9B). Conversely, the poplar MsrB2 was well recognized by the serum and the abundance of the protein decreased strongly after several hours of infection in both reactions (Fig. 9, A and B).
Figure 9.
Western analyses of plastidial MsrA and MsrB abundances in poplar leaves in response to a pathogenic attack by M. larici-populina during an incompatible (A) or a compatible (B) reaction. Immunoblotting was performed using 20 or 40 μg proteins per lane using the pMsrA and MsrB antisera diluted 1:1,000 and 1:500, respectively. The bands corresponding to pMsrA, MsrB1, and MsrB2 were revealed at 25, 17, and 15 kD, respectively. h, Hour after inoculation; d, day after inoculation.
DISCUSSION
This paper provides the first report on the activity and expression characteristics of two plant MsrB that reduce the R enantiomer of MetSO. These proteins belong to a complex multigenic family composed of nine members in Arabidopsis. Plants exhibit the largest number of Msr genes compared to bacteria and animal cells (Kryukov et al., 2002). Searching nucleotide sequence databases revealed that several eukaryotes, such as D. melanogaster, Caenorhabditis elegans, and S. cerevisiae, possess only one MsrA and one MsrB gene (Kryukov et al., 2002). In Arabidopsis, the MsrB family is more complex than the MsrA one, which is composed of five members. This complexity suggests that plant MsrB proteins exhibit specific expression or activity characteristics that could be required for a range of distinct physiological functions. This is illustrated by the fact that PMSR2, an Arabidopsis MsrA, was found to protect plants from oxidative damage especially during long dark periods (Bechtold et al., 2004).
In this work, we focused our attention on the two MsrB members that were predicted to be addressed to the chloroplast and whose localization has been confirmed by western analysis. Experiments carried out on Arabidopsis wild-type and knockout mutant plants indicate that MsrB2 and MsrB1 proteins exhibit 15- and 17-kD apparent molecular masses, respectively. These migration characteristics likely result from amino acid sequence differences, since an alignment of the two MsrB sequences shows a nine-residue extension in MsrB1. Moreover, we cannot exclude that the MsrB1 target signal could have a shorter length than the one predicted by the ChloroP software. It is noteworthy that MsrB1 does not present distinct redox states compared to MsrB2, which is detected under two forms. This major difference could be related to the lack in MsrB1 of the Cys-132, corresponding to Cys-63 in the E. coli protein, which could result in an incapacity to form an intramolecular disulfide bridge between Cys-132 and Cys-185. Interestingly, we observed that the pMsrA protein also exhibits two redox states. Previously, Sadanandom et al. (2000) reported that polyclonal antibodies, raised against pMsrA, recognized two bands in leaf proteins that were attributed to cytosolic and plastidial MsrA at 27 and 28 kD, respectively. Our experiments carried out, under reducing conditions and using leaf and chloroplast fractions without any cytosolic contamination, show identical band patterns, very close to those described by Sadanandom et al. (2000). These bands very likely correspond to two redox states of pMsrA and not to two different isoforms, since there is only one gene predicted to code for a plastidic MsrA in Arabidopsis. This hypothesis is further supported by the fact that under nonreducing conditions, the serum raised against pMsrA mainly reveals the lower oxidized form in plastidic Arabidopsis proteins, but also when using the recombinant pMsrA from poplar.
The finding that AtMsrB1 and AtMsrB2 proteins have Met-R-sulfoxide reductase activity was consistent with functional predictions based on sequence homologies. MsrB2 exhibits a higher catalytic efficiency with protein-bound MetSO than with free MetSO, as reported for PILB-MsrB from Neisseria meningitidis (Olry et al., 2002). No activity was detected with MsrB1 using free MetSO (data not shown). Interestingly, MsrB1 has a catalytic efficiency higher than that of MsrB2 when using dabsyl-MetSO as a substrate. The catalytic efficiency also depends on the nature of the protein-bound MetSO substrate, since MsrB2 is more efficient using N-acetyl-MetSO as a substrate. Experiments carried out to determine the nature of the electron donor to MsrB proteins have shown that MsrB1 cannot be reduced by Trx. Thus, recombinant plastidic MsrB proteins, which exhibit exactly the same size, display clear differences in terms of activity, substrate specificity, and reduction by Trx. Although we cannot totally exclude that these differences in enzymatic activity might originate from a wrong prediction in the cleavage site of the addressing peptides, they very likely result from the sequence features of the two proteins. MsrB1 lacks the catalytic Cys residue corresponding to Cys-63 in E. coli MsrB, which is involved in the regeneration of Cys-117 through the formation of an intramolecular disulfide bridge followed by Trx reduction (Olry et al., 2002). Other MsrB proteins from various organisms also display the substitution of the Cys-63 amino acid by a Thr (Kumar et al., 2002). Recently, Neiers et al. (2004) reported that Xanthomonas campestris MsrB, which does not possess Cys-63, is efficiently reduced by a Trx system and that Cys-31 located on another loop substitutes for Cys-63. AtMsrB1 has no Cys corresponding to Cys-31 in E. coli, but exhibits a Cys-53 localized between two Cys involved in zinc fixation, like a human MsrB (Neiers et al., 2004). The fact that MsrB1 is not reduced by a Trx system raises the question of the identity of the recycling process involved in its regeneration. As all experiments described here were carried out with cytosolic Trx, we cannot exclude that MsrB1 could be reduced by chloroplastic Trx. In potato chloroplasts, MsrB1 was recently found to be a potential target of CDSP32 by affinity chromatography (Rey et al., 2005). The CDSP32 protein is composed of two Trx modules, which could confer peculiar features and allow it to reduce MsrB1. Further studies are needed to determine the nature of the electron donor to MsrB1, particularly by assaying various types of chloroplastic Trx. This MsrB1 characteristic is not an exception, since 25% of the putative MsrB from various organisms have no Cys that can substitute for Cys-63 or Cys-31 (Neiers et al., 2004). MsrA enzymes are also divided into two classes with regard to the number of catalytic Cys. Most contain three Cys involved in MetSO reduction, but some such as Mycobacterium tuberculosis MsrA are characterized by only two functional Cys (Taylor et al., 2003).
Although MsrB1 and MsrB2 exhibit different catalytic properties, they display relatively similar expression patterns, since their abundance is much higher in developing tissues. In leaves, MsrB1 and MsrB2 show a decrease in abundance with ageing, as reported during senescence in human fibroblasts WI-38 (Petropoulos and Friguet, 2005). In agreement with data collected from EST databases, the MsrB2 protein level is higher compared to that of MsrB1, except in mature and old leaves. This pattern of protein abundance in leaves is consistent with those reported for the plastidic CDSP32 Trx (Broin et al., 2003). According to its redox state, pMsrA displays two distinct patterns. The reduced form exhibit a pattern similar to that of MsrB proteins, whereas the oxidized form is abundant in most organs except in siliques and seeds. Altogether, these data indicate that thiol antioxidative enzymes such as CDSP32 and Msr are particularly abundant in developing organs of plants.
When Arabidopsis plants were exposed to photooxidative stress, an increase in plastidial MsrB abundance was observed in mature leaves. Previously, the Arabidopsis AtSXL3 transcript, corresponding to MsrB9 in our study, has been reported to accumulate in seedlings subjected to dehydration and to oxidative stress generated in vitro by H2O2 treatment (Rodrigo et al., 2002). In our work, the pMsrA protein also exhibited an increased level after a few days of photooxidative stress. Accordingly, the pMsrA amount is also higher in Arabidopsis plants in response to high-light treatment for 48 h (Romero et al., 2004).
In poplar plants infected by two races of the rust fungus M. larici-populina, we observed that pMsrA and MsrB display different patterns of expression during compatible or incompatible reactions. In the case of the incompatible interaction, pMsrA and MsrB1 levels do not significantly change, whereas the MsrB2 amount decreases markedly after 24 h. In the compatible reaction, pMsrA and MsrB2 abundances substantially decrease after 8 h. Note that the poplar peroxiredoxin Q, which protects cells from oxidative damage by detoxifying peroxides, exhibits an expression pattern almost similar to that of pMsrA during infection by the rust fungus (Rouhier et al., 2004). These results indicate that the abundance of plastidic antioxidative enzymes is differentially regulated depending on the type of reaction and thus on the fungal races attacking the plants.
Our results, showing that both plastidic MsrB levels are higher in response to photooxidative stress, lead us to propose that the two proteins participate in the protection of chloroplasts against oxidative damage. These organelles need an efficient antioxidant machinery since they are the site of the photosynthetic process that can generate reactive oxygen species. In various cultivated plant species (cotton, pea, wheat, and potato) subjected to high temperature and water-deficit stress effects, protein MetSO contents have been investigated and no change was observed in leaves (Fergusson and Burke, 1994). All plants exhibited a decreased or unchanged Msr activity except wheat plants that showed an enhanced activity. These results suggest that plants have developed efficient systems that convert MetSO back to Met with no change in the overall Msr activity or that remove oxidized proteins by degrading them via the ubiquitin proteolytic pathway. But, the stress conditions used in this work could be considered relatively mild compared to a photooxidative stress (high light, low temperature). Accordingly, we did not observe substantial changes in Msr abundance during water deficit (Fig. 8A). Moreover, the study by Fergusson and Burke (1994) has not estimated the relative participation of each Msr type in the total Msr activity and did not take into account the different levels of regulation for Msr genes. Preliminary experiments carried out on the Arabidopsis MsrB1 knockout mutant subjected to a photooxidative treatment (high light combined to low temperature) did not reveal any difference of phenotype compared with wild-type plants (data not shown). The lack of phenotype could originate from the fact that MsrB2 expression could compensate for the absence of MsrB1. Similar data were reported previously for Arabidopsis atsxl3 knockout mutants that do not show an evident phenotype in response to oxidative stress, but a 2- to 3-fold increased protein-bound MetSO content (Rodrigo et al., 2002). Conversely, Arabidopsis plants overexpressing pMsrA have been reported to be more resistant to oxidative damage occurring in the chloroplast (Romero et al., 2004). Altogether, these data reveal the complexity of the regulation between MsrA and MsrB family members. In the future, it will be interesting to investigate the participation of each plastidic Msr during stress treatments by using knockout and overexpression mutants. Experiments currently carried out using Arabidopsis MsrB2 and double MsrB1-MsrB2 knockout mutants will allow us to gain insight into the respective roles of the two plastidic MsrB proteins.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) ecotype Columbia plants were grown in a growth chamber with an 8-h photoperiod, a photon flux density of 250 μE m−2 s−1, a temperature regime of 22°C/18°C (day/night), and a relative humidity of 55%. Water deficit was applied on 6-week-old plants by withholding watering for about 6 d. Photooxidative treatment was carried out by exposing 6-week-old plants to a high light intensity (1,400 μE m−2 s−1) at 8°C for 8 d. The infection of the beaupré cultivar of poplar (Populus × interamericana) by two races of Melampsora larici-populina and the protocol of protein extraction were described in Rouhier et al. (2004).
Isolation of Arabidopsis Homozygous Mutants
Arabidopsis knockout mutants for MsrB1 and MsrB2 genes were characterized as follows. The MsrB1 knockout mutant, corresponding to a transgenic T-DNA insertion plant, was obtained from Max Planck Institute (Germany) and termed GABI-Kat line 540H01 (Rosso et al., 2003). The MsrB2 knockout mutant was obtained from the SAIL collection and provided through the ABRC as SAIL_383_G12 line (Sessions et al., 2002). To determine if the insertions were in the positions predicted in the mutants and to isolate plant lines homozygous for T-DNA insertion mutations, Arabidopsis genomic DNA was extracted from leaves using DNeasy Plant Mini kit (Qiagen, Valencia, CA) and two PCR procedures were used. We used the forward and reverse primers to amplify a product from chromosomes without the insertion. The MsrB1 gene-specific primers were 5′-CGGGATCCGATTGTGTGATGGC-3′ and 5′-CCGCTCGAGCTCTCAATCTCTTG-3′. The MsrB2 gene-specific primers were 5′-CGGGATCCGCTGT TGTCAAGAG-3′ and 5′-CCGCTCGAGCACACCTATATTATC-3′. The other PCR used the reverse primer and the T-DNA left border primer to amplify the product from a chromosome carrying the insertion. The sequence of the left border primer used is 5′-TTCATAACCAATCTCGATACAC-3′.
Cloning of AtMsrB1 and AtMsrB2 cDNAs
A fragment containing AtMsrB1 coding region was synthesized by PCR using the clone RAFL16-88-B20 (AK117314), provided by RIKEN Genomic Sciences Center (Seki et al., 1998, 2002) as a template, and the specific primers (5′-GGAATTCATGCACCACCACCACCACCACGCTGAGAAGAATGAG-3′ and 5′-AACTGCAGTCAATCTCTTGTTTTCTCCAATGCGTT-3′).
AtMsrB2 was cloned using a reverse transcription-PCR strategy. Leaves of Arabidopsis plants were excised and frozen immediately in liquid nitrogen and stored at −80°C. RNA was extracted using TRIzol reagent (Invitrogen, Cergy-Pontoise, France) according to the manufacturer's instructions. A total of 1 μg of RNA was used for cDNA synthesis using a poly(A+) specific primer. AtMsrB2 specific primers (5′-CGCGGATCCGCTCCTGAATCG-3′ and 5′-CCCAAGCTTCAGGG TCGGATT-3′) were used to amplify by PCR the coding sequence corresponding to the MsrB2 mature protein.
Expression and Purification of Arabidopsis Plastidial Recombinant MsrB Proteins
Recombinant MsrB1 and MsrB2 proteins, containing an N-terminal 6×His-tag, were produced in Escherichia coli. The MsrB1 coding region without its N-terminal targeting sequence was ligated into the EcoRI and PstI restriction sites of the expression vector pSD80 (Patel and Dunn, 1995; Smith et al., 1996). The MsrB2 coding sequence was cloned into BamHI and HindIII restriction sites of the prokaryotic expression vector pQE-30 (Qiagen). E. coli cells (DH10β) were transformed with aliquots of the pSD80-MsrB1 ligation mixtures and grown in Luria-Bertani medium containing carbenicillin (50 μg mL−1) at 37°C. M15Rep4 strain of E. coli was transformed with the recombinant plasmid pQE-30 and grown in Luria-Bertani medium containing 50 μg mL−1 carbenicillin and 30 μg mL−1 kanamycin at 37°C. When A600 reached 0.5, isopropyl β-d-thiogalactoside was added to a final concentration of 0.1 mm and E. coli cells were further grown for 3 h at 37°C and for 5 h at room temperature, for producing MsrB2 and MsrB1, respectively. Cells were centrifuged at 9,850g for 10 min at 4°C and the pellet was immediately frozen at −80°C. The pellet was resuspended in buffer A (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 25 mm imidazole, 1 mm phenylmethylsulfonylfluoride) and the suspension was passed through the French press twice at 12,000 psi, and centrifuged at 150,000g for 30 min. The supernatant was applied onto a 4-mL Ni resin column (Amersham Biosciences, Uppsala) previously equilibrated with buffer A. After washing the column with buffer A, MsrB1 and MsrB2 were eluted with the same buffer containing 0.5 m imidazole. MsrB purity was verified using SDS-PAGE gels stained with Coomassie Brilliant Blue. Collected fractions of 4 mL were pooled and the protein was concentrated using a 10-kD molecular mass cutoff Centricon concentrator (Millipore, Bedford, MA). The fraction was added to a His-desalting column (Amersham Biosciences) and eluted with 50 mm phosphate buffer, pH 7.0, 100 mm KCl, 5 mm DTT. Protein concentration was determined spectrophotometrically at 280 nm using MsrB extinction coefficients.
Expression and Purification of Recombinant Poplar pMsrA
The nucleotidic sequence encoding the mature pMsrA form (GenBank accession no. AAS46232) was cloned by PCR into the expression plasmid pET-3d using a leaf cDNA library of Populus tremula × tremuloides as a template and the two following primers: 5′-CCCCCCATGGCTAACATCCTTAGCAAACTAGGC-3′ and 5′-CCCCGGATCCTTAGCCATAGCATCGGATTGGATC-3′ (cloning sites underlined). In addition, a codon for Ala (in bold) was inserted downstream the Met closest to the putative cleavage site and the corresponding N-terminal amino acid sequence was thus MANIL. The recombinant plasmid was used to transform the BL21(DE3) E. coli strain, which also contains the helper plasmid pSBET (Schenk et al., 1995). A culture of 5 L of a kanamycin (50 μg mL−1) and ampicillin (50 μg mL−1) resistant colony was grown at 37°C and induced by 0.1 mm isopropylthio-β-galactoside (IPTG) in the exponential phase. Bacteria were harvested by centrifugation, resuspended in a buffer 1 (30 mm Tris-HCl, pH 8, 1 mm EDTA, 200 mm NaCl) containing 20 mm DTT and lysed by sonication. After centrifugation (16,000g, 30 min), the soluble part was first precipitated between 0% and 50% of ammonium sulfate saturation. This fraction was then purified by exclusion size chromatography onto an ACA 44 column (Biosepra, Cergy Saint-Christophe, France) equilibrated in buffer 1. The fractions of interest were pooled, dialyzed to remove salts, and separated by DEAE Sepharose chromatography (Amersham Biosciences, Uppsala). The recombinant pMsrA was eluted around 100 mm NaCl using a linear gradient from 0 to 400 mm NaCl. The purified fractions were dialyzed to remove salts, adjusted to 1.6 m NaCl and applied to a Phenyl Sepharose column (Amersham Biosciences), equilibrated with buffer 2 (30 mm Tris-HCl, pH 8, 1 mm EDTA, 1.6 m NaCl). Recombinant proteins were eluted by a gradient from 1.6 to 0 m NaCl, dialyzed, and concentrated. Their purity was assessed using 15% SDS-PAGE. Protein concentration was estimated spectrophotometrically using a molar extinction coefficient of 25,700 m−1 cm−1.
Preparation of Dabsyl-MetSO and N-Acetyl-MetSO
Dabsyl-MetSO was prepared by incubating 50 mm Dabsyl-Met (Interchim, Montluçon, France) with 500 mm H2O2 overnight at room temperature. In these conditions, around 95% dabsyl-Met was oxidized to dabsyl-MetSO. After centrifugation at 3,250g for 10 min at room temperature, the supernatant was loaded on a C18 column (Sep-Pak C18 cartridges, Millipore) and dabsyl-MetSO was eluted with 5 mL acetonitrile. After evaporation of the solution under vacuum, the powder was dissolved in dimethyl sulfoxide. The actual concentration of dabsyl-MetSO was assayed by HPLC.
N-acetyl-MetSO was obtained by acetylation of free MetSO (Sigma, St. Louis) by anhydride acetic (Sanchez et al., 1983). Free MetSO (1 mmol) was mixed with 4 mL acetic glacial acid and 4 mL acetic anhydride for 2 h at room temperature under continual agitation. Then, 4 mL water were added, and the reaction mixture was frozen for 30 min at −80°C and then washed with 2 mL water before lyophilization overnight.
Assay for Msr Activity
MsrB activity in the presence of Trx was measured by following NADPH oxidation at 340 nm. A 500-μL cuvette was constituted of 30 mm Tris-HCl, pH 8.0, 1 mm EDTA, 200 μm NADPH, 1.3 μm Arabidopsis NADPH Trx reductase, saturating concentrations of poplar Trx h1 (42.5 μm) and free MetSO (100 mm) or N-acetyl-MetSO (20 mm) and 0.56 μm of MsrB2 or 1 to 10 μm MsrB1. The reaction was carried out at 30°C with a Cary 50 spectrophotometer (Varian, Palo Alto, CA). The catalytic parameters for Trx h1, free MetSO, or N-acetyl-MetSO were determined at saturating concentrations of the other enzyme substrate.
The activity of MsrB recombinant proteins was also determined by monitoring the reduction of the synthetic substrate, dabsyl-MetSO, in the presence of DTE. The reaction mixture, containing purified proteins MsrB1 and MsrB2 in 15 mm HEPES, pH 8, 10 mm MgCl2, 30 mm KCl, 20 mm DTE, 0.5 mm dabsyl-MetSO in a final volume of 100 μL was incubated for 1 h at 37°C. The reaction was stopped by adding 450 μL of ethanol:acetate buffer 29 mm, pH 4.16 (50:50, v/v) to 50 μL of the mixture. After centrifugation at 12,000g for 30 min at 4°C, 20 μL of supernatant were loaded on a C18 reverse phase column.
Reverse Phase HPLC of Dabsyl-Met
The method was modified from that of Minetti et al. (1994). The different solvents used were solvent A (acetate buffer 29 mm, pH 4.16) and solvent B (acetonitrile). The flow rate was 0.8 mL min−1 at room temperature. The detector wavelength was set at 436 nm for absorbance reading. The column utilized was a Symmetry C18 5 μm, 3 × 250 mm (Waters, Milford, MA).
The program used for routine analysis of enzymatic reaction mixtures, called program 1, starts with 20% solvent B, up to 66.7% solvent B in 10 min, up to 100% solvent B in 0.2 min, 100% B for 3.3 min, then back to 20% B in 0.2 min, equilibration at 20% solvent B for 8.3 min. The solvent A is replaced with solvent B to reached 100%. The whole program lasts 22 min. In these conditions, dabsyl-MetSO is eluted at 9.7 min and dabsyl-Met at 12.1 min.
The program 2 is used for discriminating dabsyl-MetSO diastereomers. It starts with 20% solvent B, up to 48% solvent B in 20 min, up to 100% solvent B in 1 min, at 100% B for 7 min, down to 20% B in 2.5 min, equilibration at 20% solvent B for 9.5 min. The whole program lasts 41 min. In these conditions, dabsyl-Met-S-SO, dabsyl-Met-R-SO, and dabsyl-Met elute at 17.1 min, 17.6 min, and 25.1 min, respectively.
Chloroplast Preparation
Crude chloroplasts were obtained from 5 to 7 g Arabidopsis leaves blended in 50 mL extraction buffer (50 mm Tris-HCl, pH 8.0, 20 mm EDTA, 0.33 m sorbitol, 28.6 mm β-mercaptoethanol). After filtration on Miracloth (Calbiochem, San Diego), the suspension was centrifuged at 2,500g for 3 min. After resuspension, chloroplasts were loaded on top of preformed Percoll gradients in 2-mL tubes and centrifuged at 4,000g for 7 min. Percoll gradients (50% Percoll in extraction buffer containing 0.1% bovine serum albumin) were preformed by centrifugation at 17,000g for 20 min. Intact chloroplasts, forming a ring in the gradient bottom part, were collected and washed with extraction buffer and centrifuged at 1,000g for 2 min. After an additional wash, chloroplasts were resuspended in storage buffer (10 mm Tris-HCl, pH 8.0, 2 mm EDTA, 15 mm NaCl, 1 mm phenylmethylsulfonylfluoride). Purified intact chloroplasts were lysed in a hypotonic medium containing protease inhibitors (50 mm Tris-HCl, pH 8.0, 5 mm β-amino caproic acid, 2 mm benzamidine, 1 mm phenylmethylsulfonylfluoride) for 60 min at 4°C with strong shaking and centrifuged at 10,000g for 10 min at 4°C. Stromal proteins were precipitated by adding 3 volumes of acetone to the supernatant.
Protein Extraction, SDS-PAGE, and Western-Blotting Experiments
For preparing leaf soluble proteins, leaf samples were ground in liquid nitrogen, and the powder was resuspended in 50 mm Tris-HCl, pH 8.0, 1 mm phenylmethylsulfonylfluoride, 50 mm β-mercaptoethanol, and centrifuged at 30,000g for 15 min at 4°C. Soluble proteins were precipitated at −20°C by adding 3 volumes of acetone to the supernatant. Soluble proteins from various organs (roots, leaves, stems, floral buds, flowers, siliques, and seeds) were prepared similarly. The protein content was determined using a method based on bicinchoninic acid (BC Assay Reagent, Interchim). Proteins were separated using SDS-PAGE (13%, w/v gel) and electrotransferred onto a nitrocellulose membrane (Pall Gelman Sciences, Ann Arbor, MI). Membranes were incubated with anti-pMsrA and anti-MsrB polyclonal antibodies diluted 1:1,250 and 1:1,000, respectively. The serum against MsrB1 was raised by preparing a synthetic peptide of 15 amino acids (ITRQKGTERAFTGEY) and coupling it to the keyhole limpet hemocyanin (Agro-Bio, Villeny, France). After four immunizations with the coupled peptide, the rabbit serum was collected. For purification of the antibody, a tablet of Complete EDTA free protein inhibitor (Roche, Meylan, France) was mixed with 15 mL of serum against MsrB1, diluted to a 40-mL volume with buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl), and sterilized by filtration. MsrB1 protein (4 mg) was fixed onto a HiTrap NHS-activated HP 1 mL column (Amersham Biosciences). The serum was bounded onto the column with a flow rate 0.2 mL min−1. Further steps of purification of the antibody were carried out following the manufacturer's recommendation. Bound antibodies were detected using an anti-rabbit IgG alkaline phosphatase conjugate (Sigma) diluted 1:10,000.
Antibodies against the recombinant poplar pMsrA were raised in rabbit and then purified from the serum onto an affinity column constituted of the recombinant pMsrA coupled to CnBr sepharose following a procedure described in Rouhier et al. (2004). For western blots using poplar proteins, 20 to 40 μg of proteins were separated using 15% SDS-PAGE gels and then blotted onto a polyvinylidene difluoride membrane (Millipore). The anti-pMsrA and anti MsrB antibodies were used diluted 1:1,000 and 1:500, respectively. The revelation was done using the Immun Star AP Goat anti-rabbit IgG Detection kit (BIO-RAD; dilution of secondary antibodies 1:3000).
Acknowledgments
We are very grateful to Stéphanie Verrier for helpful assistance in western experiments and to Dr. Gilles Peltier (CEA, DSV, DEVM-LEP) and Dr. Jean-Pierre Jacquot (Université de Nancy-INRA) for critical reading of the manuscript. We wish to thank the Groupe de Recherche Appliquée en Phytotechnologie team for technical assistance with controlled growth chambers. We also thank Dr. Yves Meyer (CNRS-Université de Perpignan) for providing us the antibody raised against Arabidopsis PrxIIB. We wish to thank the Max Planck Institute for providing the GABI-Kat line 540H01. The T-DNA mutants were generated in the context of the GABI-Kat program and provided by Bernd Weisshaar (MPI for Plant Breeding Research, Cologne, Germany). We are grateful to the ABRC that provided us the SAIL_383_G12 line obtained from the SAIL collection and to the RIKEN Genomic Sciences Center for providing the clone RAFL16-88-B20 (AK117314).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.062430.
References
- Bechtold U, Murphy DJ, Mullineaux PM (2004) Arabidopsis peptide methionine sulfoxide reductase2 prevents cellular oxidative damage in long nights. Plant Cell 16: 908–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréhelin C, Meyer EH, de Souris JP, Bonnard G, Meyer Y (2003) Resemblance and dissemblance of Arabidopsis type II peroxiredoxins: similar sequences for divergent gene expression, protein localization, and activity. Plant Physiol 132: 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broin M, Besse I, Rey P (2003) Evidence for post-translational control in the expression of a gene encoding a plastidic thioredoxin during leaf development in Solanum tuberosum plants. Plant Physiol Biochem 41: 303–308 [Google Scholar]
- Broin M, Cuiné S, Eymery F, Rey P (2002) The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative damage. Plant Cell 14: 1417–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broin M, Cuiné S, Peltier G, Rey P (2000) Involvement of CDSP32, a drought-induced thioredoxin, in the response to oxidative stress in potato plants. FEBS Lett 467: 245–248 [DOI] [PubMed] [Google Scholar]
- Broin M, Rey P (2003) Potato plants lacking the CDSP32 plastidic thioredoxin exhibit over-oxidation of the BAS1 2-cys peroxiredoxin and increased lipid peroxidation in thylakoids under photooxidative stress. Plant Physiol 132: 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brot N, Weissbach J, Werth J, Weissbach H (1981) Enzymatic reduction of protein-bound methionine sulfoxide. Proc Natl Acad Sci USA 78: 2155–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann M, Pell E (1989) Decline of activity and quantity of ribulose-1,5-bisphosphate carboxylase/oxygenase by chloroplastic proteases requires ATP-hydrolysis. Planta 205: 459–466 [Google Scholar]
- Davies KJ (1995) Oxidative stress: the paradox of aerobic life. Biochem Soc Symp 61: 1–31 [DOI] [PubMed] [Google Scholar]
- Davis DA, Newcomb FM, Moskovitz J, Wingfield PT, Stahl SJ, Kaufman J, Fales HM, Levine RL, Yarchoan R (2000) HIV-2 protease is inactivated after oxidation at the dimer interface and activity can be partly restored with methionine sulphoxide reductase. Biochem J 346: 305–311 [PMC free article] [PubMed] [Google Scholar]
- Ezraty B, Bos J, Barras F, Aussel L (2005) Methionine sulfoxide reduction and assimilation in Escherichia coli: new role for the biotin sulfoxide reductase BisC. J Bacteriol 187: 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergusson DL, Burke JJ (1994) Methionyl sulfoxide content and protein-methionine-S-oxide reductase activity in response to water deficits or high temperature. Physiol Plant 90: 253–258 [Google Scholar]
- Gao J, Yin DH, Yao Y, Sun H, Qin Z, Schoneich C, Williams TD, Squier TC (1998) Loss of conformational stability in calmodulin upon methionine oxidation. Biophys J 74: 1115–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2004) The thioredoxin h system of higher plants. Plant Physiol Biochem 42: 265–271 [DOI] [PubMed] [Google Scholar]
- Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Bri C, Derrick PJ, Barras F (2001) Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J Biol Chem 276: 48915–48920 [DOI] [PubMed] [Google Scholar]
- Gustavsson N, Kokke B, Härndahl U, Silow M, Bechtold U, Poghosyan Z, Murphy D, Boelens W, Sundby C (2002) A peptide methionine sulfoxide reductase highly expressed in photosynthetic tissue in Arabidopsis thaliana can protect the chaperone-like activity of a chloroplast-localized small heat shock protein. Plant J 29: 545–553 [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN (2002) Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci USA 99: 4245–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, Koc A, Cerny RL, Gladyshev VN (2002) Reaction mechanism, evolutionary, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J Biol Chem 277: 37527–37535 [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Guillon B, Le Maréchal P, Keryer E, Miginiac-Maslow M, Decottignies P (2004) New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 101: 7475–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti G, Balduini C, Brovelli A (1994) Reduction of dabs-L-methionine-dl-sulfoxide by protein methionine sulfoxide reductase from polymorphonuclear leukocytes: stereospecificity towards the l-sulfoxide. Ital J Biochem 43: 273–283 [PubMed] [Google Scholar]
- Moskovitz J, Berlett BS, Poston JM, Stadtman ER (1997) The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA 94: 9585–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Flescher E, Berlett BS, Azare J, Poston JM, Stadtman ER (1998) Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc Natl Acad Sci USA 95: 14071–14075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Rahman MA, Strassman J, Yancey SO, Kushner SR, Brot N, Weissbach H (1995) Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol 177: 502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Singh VK, Requena J, Wilkinson BJ, Jayaswal RK, Stadtman ER (2002) Purification and characterization of methionine sulfoxide reductases from mouse and Staphylococcus aureus and their substrate stereospecificity. Biochem Biophys Res Commun 290: 62–65 [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV (1996) A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci USA 93: 10268–10273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiers F, Kriznik A, Boschi-Muller S, Branlant G (2004) Evidence for a new sub-class of methionine sulfoxide reductases B with an alternative thioredoxin recognition signature. J Biol Chem 279: 42462–42468 [DOI] [PubMed] [Google Scholar]
- Olry A, Boschi-Muller S, Marraud M, Sanglier-Cianferani S, Van Dorsselear A, Branlant G (2002) Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis. J Biol Chem 277: 12016–12022 [DOI] [PubMed] [Google Scholar]
- Patel AM, Dunn SD (1995) Degradation of Escherichia coli uncB mRNA by multiple endonucleolytic cleavages. J Bacteriol 177: 3917–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos I, Friguet B (2005) Protein maintenance in aging and replicative senescence: a role for the peptide methionine sulfoxide reductases. Biochim Biophys Acta 1703: 261–266 [DOI] [PubMed] [Google Scholar]
- Rey P, Cuiné S, Eymery F, Garin J, Court M, Jacquot J-P, Rouhier N, Broin M (2005) Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J 41: 31–42 [DOI] [PubMed] [Google Scholar]
- Rey P, Pruvot G, Becuwe N, Eymery F, Rumeau D, Peltier G (1998) A novel thioredoxin-like protein located in the chloroplast is induced by water deficit in Solanum tuberosum L. plants. Plant J 13: 97–107 [DOI] [PubMed] [Google Scholar]
- Rodrigo M-J, Moskovitz J, Salamini F, Bartels D (2002) Reverse genetic approaches in plants and yeast suggest a role for novel, evolutionarily conserved, selenoprotein-related genes in oxidative stress defense. Mol Genet Genomics 267: 613–621 [DOI] [PubMed] [Google Scholar]
- Romero HM, Berlett BS, Jensen PJ, Pell EJ, Tien M (2004) Investigations into the role of the plastidial peptide methionine sulfoxide reductase in response to oxidative stress in Arabidopsis. Plant Physiol 136: 3784–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Gualberto JM, Jordy MN, de Fay E, Hirasawa M, Duplessis S, Lemaire SD, Frey P, Martin F, et al (2004) Poplar peroxiredoxin Q. A thioredoxin-linked antioxidant functional in pathogen defense. Plant Physiol 134: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Poghosyan Z, Fairbairn DJ, Murphy DJ (2000) Differential regulation of plastidial and cytosolic isoforms of peptide methionine sulfoxide reductase in Arabidopsis. Plant Physiol 123: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J, Nikolau BJ, Stumpf PK (1983) Reduction of N-acetyl methionine sulfoxide in plants. Plant Physiol 73: 619–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Baumann S, Mattes R, Steinbiss HH (1995) Improved high level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare Arg tRNA's. Biotechniques 19: 196–200 [PubMed] [Google Scholar]
- Seki M, Carninci P, Nishiyama Y, Hayashizaki Y, Shinozaki K (1998) High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J 15: 707–720 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov VS, Ferrington DA, Squier TC, Schoneich C (1999) Diastereoselective reduction of protein-bound methionine sulfoxide by methionine sulfoxide reductase. FEBS Lett 455: 247–250 [DOI] [PubMed] [Google Scholar]
- Singh VK, Moskovitz J, Wilkinson BJ, Jayaswal RK (2001) Molecular characterization of a chromosomal locus in Staphylococcus aureus that contributes to oxidative defence and is highly induced by cell-wall-active antibiotic oxacillin. Microbiology 147: 3037–3045 [DOI] [PubMed] [Google Scholar]
- Smith SP, Barber KR, Dunn SD, Shaw GS (1996) Structural influence of cation binding to recombinant human brain S100b: evidence for calcium-induced exposure of a hydrophobic surface. Biochemistry 35: 8805–8814 [DOI] [PubMed] [Google Scholar]
- Sundby C, Härndahl U, Gustavsson N, Ahrman E, Murphy DJ (2005) Conserved methionines in chloroplasts. Biochim Biophys Acta 1703: 191–202 [DOI] [PubMed] [Google Scholar]
- Taylor AB, Benglis DM, Jr., Dhandayuthapani S, Hart PJ (2003) Structure of Mycobacterium tuberculosis methionine sulfoxide reductase A in complex with protein-bound methionine. J Bacteriol 185: 4119–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Neiers F, Boschi-Muller S, Branlant G (2005) The N-terminal domain of PILB from Neisseria meningitidis is a disulfide reductase that can recycle methionine sulfoxide reductases. J Biol Chem 280: 12344–12350 [DOI] [PubMed] [Google Scholar]