Abstract
G-to-A hypermutation has been sporadically observed in human immunodeficiency virus type 1 (HIV-1) proviral sequences from patient peripheral blood mononuclear cells (PBMC) and virus cultures but has not been systematically evaluated. PCR primers matched to normal and hypermutated sequences were used in conjunction with an agarose gel electrophoresis system incorporating an AT-binding dye to visualize, separate, clone, and sequence hypermutated and normal sequences in the 297-bp HIV-1 protease gene amplified from patient PBMC. Among 53 patients, including individuals infected with subtypes A through D and at different clinical stages, at least 43% of patients harbored abundant hypermutated, along with normal, protease genes. In 70 hypermutated sequences, saturation of G residues in the GA or GG dinucleotide context ranged from 20 to 94%. Levels of other mutants were not elevated, and G-to-A replacement was entirely restricted to GA or GG, and not GC or GT, dinucleotides. Sixty-nine of 70 hypermutated and 3 of 149 normal sequences had in-frame stop codons. To investigate the conditions under which hypermutation occurs in cell cultures, purified CD4+ T cells from normal donors were infected with cloned NL4-3 virus stocks at various times before and after phytohemagglutinin (PHA) activation. Hypermutation was pronounced when HIV-1 infection occurred simultaneously with, or a few hours after, PHA activation, but after 12 h or more after PHA activation, most HIV-1 sequences were normal. Hypermutated sequences generated in culture corresponded exactly in all parameters to those obtained from patient PBMC. Near-simultaneous activation and infection of CD4+ T cells may represent a window of susceptibility where the informational content of HIV-1 sequences is lost due to hypermutation.
The survival of species depends on a favorable balance between the beneficial and harmful aspects of mutation. There is loss of the information content of nucleotide sequences once mutation rates are increased beyond a tolerable error threshold (15). Some RNA viruses seem to tolerate mutation rates near this threshold, existing, not as a specific sequence, but as a quasispecies (12, 15, 26). When viral mutation rates approach the maximum value compatible with viability (14), what results is an intolerance to even small increases in mutation rates (27). How surprising, then, that for some of the same viruses with the highest mutation rates, another mutation process has been described in which remarkable levels of one specific type of nucleotide substitution are observed. These sequences are referred to as hypermutants and are a product of one specific mutation at rates far beyond viability. Two main types of hypermutation, which differ with respect to the type of substitution observed, have been described for viral sequences. A-to-G hypermutation occurs mostly in measles virus and vesicular stomatitis virus (8, 9, 49), and three cases in nonlentiviral retroviruses (avian leukosis virus and spleen necrosis virus) have been reported (16, 23, 33). G-to-A hypermutation is found primarily in the lentivirus family of retroviruses, along with two other examples in satellite tobacco mosaic virus (37) and hepatitis B virus (22). G-to-A hypermutation in the lentivirus human immunodeficiency virus type 1 (HIV-1) is the subject of this report.
G-to-A hypermutation is defined as a mutational process in which G-to-A transitions far exceed all other mutations in viral sequences (60). In published reports, up to 60% of G's may be replaced by A's along a hypermutated sequence (64). G-to-A hypermutation occurs specifically within the GpA or GpG dinucleotide context (18, 60). Although G-to-A hypermutation was first described for spleen necrosis virus (SNV) by Pathak and Temin (50), G-to-A hypermutation has been mostly found among the lentiviral group of retroviruses, including HIV-1 (4, 11, 18, 21, 39, 40, 47, 60), HIV-2 (19), simian immunodeficiency virus (SIV) (30), equine infectious anemia virus (EIAV) (52), and caprine arthritis-encephalitis virus (CAEV) (64).
The susceptibility of lentiviruses to hypermutation is thought to be a property of their reverse transcriptase (RT), which, compared to nonlentiviral RTs, has a great capacity to elongate beyond nucleotide mismatches (51) and an increased ability to hypermutate in the presence of unbalanced nucleotide pools in vitro (43). This susceptibility is further evidenced by the elevated A content of lentivirus genomes (7). Retroviral genomes form a bimodal distribution with respect to base composition that follows taxonomic groups (71).
Hypermutation in HIV-1 was first detected during propagation of HIV-1 virus populations in vitro (11, 21, 60). Since then, several groups have recovered hypermutated HIV-1 sequences from clinical samples, confirming that they occur in vivo (4, 18, 40, 47). Typically, mixtures of hypermutated and normal sequences are found, with hypermutated sequences in the minority (4, 18, 19, 30, 39, 52, 67). With respect to genome region, hypermutation was initially found in small subregions of gag, env, nef, and U3/R elements of the long terminal repeat (LTR) (11, 21, 60), but others have found hypermutation throughout the genome (4).
Some progress has been made in elucidating the biochemical basis for hypermutation. First thought to be the work of a mutant RT (18, 50), hypermutation has now been shown to be a property of wild-type RT (42) operating under suboptimal conditions. G-to-A hypermutation is thought to occur when HIV minus-strand DNA synthesis takes place simultaneously with an increased intracellular concentration of dTTP relative to dCTP (60). Attempts have been made to generate hypermutants in a cell-free system in vitro with RNA, purified RT, and strongly biased deoxynucleoside triphosphate (dNTP) pools (43, 44, 59, 61). While G-to-A hypermutants were produced, they lacked the marked preference for the GpA and GpG dinucleotide contexts that typify hypermutants produced in vivo or during virus cultivation in vitro. An intermediate approach where RT in virus particles was allowed to complete first-strand DNA synthesis in the presence of exogenously provided, highly biased dNTP pools also resulted in loss of the GpA and GpG dinucleotide preference (62). Hypermutants were also recovered at low frequency (0.5%) from cell cultures after the addition of deoxythymidine to supernatants, which resulted in an elevated intracellular [dTTP]/[dCTP] ratio (62), but because all possible dinucleotide contexts were not represented in the sequence that was studied, a complete analysis of context preference was not possible. The cell-free experiments described above were conducted using relative concentrations of dTTP and dCTP that differed by 3 to 4 orders of magnitude. More subtle perturbations in the intracellular environment may trigger dNTP pool biases sufficient to generate hypermutation while maintaining the proper dinucleotide context (67). Indeed, a 40- fold [dTTP]/[dCTP] ratio produced hypermutants in which the GpA context preference was maintained (62), and hypermutants were recovered from 1 to 2% of unstimulated and phytohemagglutinin (PHA)-stimulated peripheral blood mononuclear cells (PBMC) without manipulation of dNTP pools (62).
Imbalanced and fluctuating nucleotide pools are a key element of many types of mutation, including hypermutation. Normal dNTP pools are highly asymmetric in mammalian cells (reviewed in reference 45). dNTP pool biases are mutagenic during DNA replication (34–36, 46) and are able to affect RT error rates in vitro and retroviral mutation rates in vivo (2, 29, 32, 53). Since the composition of dNTP pools changes as cells progress through the cell cycle (5, 20, 45), the timing of HIV-1 infection with respect to T-cell activation and entry into the cell cycle could be an important factor determining the generation of hypermutants. [dTTP]/[dCTP] ratios at different stages of cell activation varied from 1.3:1 to 6:1 (3, 5, 10, 20, 58). In a retrovirus-based shuttle vector, G-to-A transitions predominated (31, 32) and were presumably a result of even these modest fluctuations in pools during the cell cycle.
At this point understanding of hypermutation is still limited, in part because of the lack of a successful systematic screening method. For example, a method based on the blue/white β-galactosidase complementation assay yielded a low recovery of hypermutants (0.5 to 2%) (62), not higher than that obtained when clones were randomly picked and sequenced. Indeed, many hypermutated sequences have been submitted to databases without being perceived as such (63). Given the lack of a specific genome location for hypermutation, the fact that hypermutants are almost always buried in a large excess of normal sequences, and the difficulty of precisely reproducing hypermutation in vitro, it is not surprising that this mutational process has been regarded as erratic, rare, and of minor importance in the HIV-1 life cycle. Here we describe the design and application of powerful new methods for systematic detection and recovery of hypermutants and their application to clinical samples and to the products of HIV-1 infection in cell culture. The results call for a reassessment of the frequency of hypermutation in vivo, clarify the conditions that generate hypermutants in cell culture, and importantly, highlight a vulnerability of HIV-1 that could be exploited for clinical benefit.
MATERIALS AND METHODS
Clinical samples.
Purified PBMC from 53 HIV-1-positive individuals were the source of DNA for analysis. Twenty-six samples were from patients hospitalized in Tanzania in 1996 with symptoms compatible with AIDS (25). Fourteen others were from individuals who seroconverted to HIV-1 while in the U.S. Military between 1997 and 1998 (6). Thirteen people were participants in the San Francisco Men's Health Study between 1985 and 1988 (38, 66). The latter two groups were in the early, asymptomatic stage of HIV-1 infection, and the sample was typically drawn within 6 months of HIV-1 seroconversion.
PCR.
DNA extracted from patient PBMC or virus cultures was the template for PCR amplification of HIV-1 protease sequences. Nested PCR primers (28) were used either without modification, for amplification of normal sequences, or with modifications designed to increase their homology to hypermutated sequences. The primers for amplification of hypermutants contained either mixed bases (hyp primers) or G-to-A replacements (hypa primers) at some of the sites (GpA or GpG) susceptible to hypermutation. Each PCR amplification was run in duplicate, once with normal primers and a second time with an equal mixture of hyp and hypa primers. The first round of PCR was conducted with 2 mM dNTPs, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 0.4 μM each primer or primer mixture, and 1.25 U of Amplitaq.
A touchdown approach, with incremental decreases in annealing temperature, was used in the initial 14 cycles of the first-round PCR (13, 24, 55). The first cycle consisted of denaturation at 95°C for 20 s, annealing at 53°C for 30 s, and extension at 72°C for 60 s. With each cycle the annealing temperature was decreased by 1°C, until the final temperature of 40°C, the remaining 21 cycles used this annealing temperature. The second-round PCR was carried out, using 1 μl of the first-round product and 0.4 μM primers DP16 and DP17, for 35 cycles like the first-round PCR, but with a constant annealing temperature of 55°C.
The HA yellow gel system.
PCR products were analyzed in parallel 1% agarose gels, with or without HA yellow, a compound which consists of the DNA ligand bisbenzamide covalently linked to polyethylene glycol (PEG) (Hanse Analytik, Bremen, Germany). Bisbenzamide binds preferentially to AT-rich regions in DNA (in order from highest to lowest preference, AATT and AAAA, TAAT, ATAT, TATA, and TTAA) (1) and, when coupled to PEG, retards DNA mobility during gel electrophoresis according to AT content (41, 48, 65). HA yellow was incorporated at 1 U/ml after the gel solution was cooled to 65°C. Electrophoresis was at 80 V in 1× Tris-borate-EDTA (TBE) for 90 min. Products were visualized by poststaining with ethidium bromide.
Isolation and molecular cloning of normal and hypermutated sequences.
We attempted to recover both normal and hypermutated sequences from every sample, but different approaches were used depending on the HA yellow gel profile of the bulk PCR product. The PCR product was cloned directly if it was separated into normal and hypermutated bands of nearly equal intensity, or if there was only one band. If the hypermutated and normal bands were of unequal intensity, the two populations were excised from the gel, extracted using the Qiaquick Spin Purification Gel Extraction Kit from Qiagen (Valencia, Calif.), and cloned separately. When hypermutated bands were faint, the two populations were excised and extracted as above, but the hypermutated sequences were reamplified with second-round primers DP16 and DP17 prior to cloning.
In experiments with virus cultivation in cell cultures, enrichment for hypermutated sequences by predigestion of the cellular DNA with restriction endonucleases ScrF1(CCAG∗G∗A) and AvaII (G∗G∗ACC), which preferentially cleave normal, as opposed to hypermutated, HIV-1 protease sequences, was used to recover rare hypermutated sequences from some samples. The G residues susceptible to hypermutation within the recognition sequences (underlined) are indicated by asterisks. DNA was codigested with ScrF1 and AvaII at 37°C overnight prior to PCR amplification of HIV-1 protease.
Ligation into a plasmid vector, transformation of Escherichia coli, and plasmid purification were done using the TOPO TA cloning kit and One Shot Chemically Competent Cells from Invitrogen (San Diego, Calif.) and the Qiawell 8 ultra plasmid kit (Qiagen), respectively, as directed by the vendors.
DNA sequencing.
Eight hundred nanograms of purified plasmids was digested using 20 U of EcoR1 (New England Biolabs, Beverly, Mass.) for 1 h at 37°C to release the cloned protease gene, which was then rescreened on agarose gels with HA yellow. At least one clone, and often several clones, representing all of the different mobilities found within each sample was selected for sequencing.
Both strands of the DNA were sequenced using fluorescent dye terminators present in the PRISM Ready Reaction Dyedeoxy Terminator kit with FS Taq (Applied Biosystems, Foster City, Calif.), and an Applied Biosystems Automated Sequencer as directed by the manufacturer. Sequences were assembled with Sequencher software (Applied Biosystems).
Determination of HIV-1 subtype.
The genetic subtypes of normal protease sequences from the 53 patients were established by phylogenetic analysis (see Table 2). Sequences were manually aligned with reference sequences of HIV-1 subtypes A through J. Components of the PHYLIP package (17), including SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE, were used to assign each sequence to a known subtype, if possible. All of the asymptomatic patients from the United States harbored subtype B. Among the AIDS patients more than half harbored subtype C; the remainder harbored mostly subtype A or D. One patient was dually infected with subtypes A and D, and one other harbored an unclassified strain, possibly an AD recombinant (data not shown).
TABLE 2.
Evaluation of patient PBMC for the presence of hypermutated HIV-1 sequences
| Country | Study | Date | Clinical Stage | Subtypea | No. of patients | No. (%) of patients with hypermutationb |
|---|---|---|---|---|---|---|
| United States | San Francisco Men's Health Study | 1985–1988 | Asymptomatic | B | 13 | 5 (38) |
| U.S. Military Seroconverters | 1997–1998 | Asymptomatic | B | 14 | 7 (50) | |
| Tanzania | Sentinel Surveillance | 1996 | AIDS | A | 7 | 4 (57) |
| C | 14 | 3 (21) | ||||
| D | 3 | 2 (67) | ||||
| Other | 2 | 2 (100) | ||||
| Total | 53 | 23 (43) |
The genetic subtype of the normal HIV-1 protease sequence was determined by phylogenetic analysis (data not shown). Other, mixed infection or unclassified.
Hypermutated HIV-1 sequences could be recovered from primary PBMC as described in Materials and Methods.
Preparation and characterization of virus stocks.
Virus stocks generated from an infectious molecular clone of HIV-1 subtype B isolate NL4-3 (56, 57) were used to infect PBMC from a seronegative donor. Cells were activated with 1 μg of PHA/ml for 72 h, infected for 24 h, washed twice, and maintained in RPMI 1640 with 15% fetal calf serum (FCS), 1% PenStrep (Quality Biological, Gaithersburg, Md.), 1% l-glutamine, and 20 U of interleukin-2 (IL-2)/ml. Culture supernatants were assayed for p24 antigen (HIV-1 p24 Antigen Assay Kit; Coulter Corporation, Miami, Fla.) at 3-day intervals. Culture supernatants collected near the peak of p24 antigen production were treated with 50 U of DNAse I/ml (Boehringer Mannheim) for 30 min at room temperature and filtered sterilized (pore size, 0.22 μm). Viral stocks of at least 20,000 50% tissue culture infective doses (TCID50)/ml were stored in liquid nitrogen until use.
To verify that the virus stocks were free of hypermutated sequences, RNA was extracted from 200 μl of virus stocks using the Nuclisens Extraction Kit (Organon Teknika), and RT-PCR was performed using the Reverse Transcription System (Promega, Madison, Wis.) with primer DP11. Amplification of the resulting cDNA was done by nested DNA PCR as described above. The PCR product was evaluated for the presence of hypermutated sequences on HA yellow gels and by sequencing. All virus stocks were free of hypermutants by these assays.
Cell culture.
Fresh leucopacks were obtained from HIV-1-seronegative donors (RH Laboratories, Baltimore, Md.). PBMC were isolated by density gradient centrifugation over Ficoll-Hypaque (Pharmacia, Stockholm, Sweden). CD4+ T cells were purified by negative selection on magnetic beads with the MACS CD4+ T Cell Isolation Kit (Miltenyi Biotec, Auburn, Calif.). Prior to infection, an aliquot of the purified CD4+ T cells was fixed with 2% formaldehyde and stained with fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-conjugated monoclonal antibodies specific for cell surface molecules CD3, CD4, CD8, CD14, CD20, CD56, and HLA-DR (Becton Dickinson, San Jose, Calif.). Cell populations were analyzed by flow cytometry on a BD FACScan (Becton Dickinson). The majority of the cells in each experiment were CD4+ T cells (mean, 81%; range, 69 to 99%) representing most of the CD3+ T-cell subset (mean, 83%; range, 76 to 100%). The other major cell type was B cells (CD20+; mean, 10.8%; range, 4.5 to 14.9%). Monocytes (CD14+), CD8+ T cells (CD8+), and NK cells (CD56+) were, on average, 2 to 4% of the population. Between 14.6 and 17.7% of the cells were HLA-DR positive.
Cells were either left unstimulated or stimulated with 1 μg of PHA (Pharmacia)/ml for various intervals before, during, or after virus infection at a multiplicity of infection of 0.02 for 2 h at 37°C. Cells were washed twice and maintained in RPMI 1640 containing 10% autologous plasma, and IL-2 at 20 U/ml (Amersham). Autologous plasma, necessary to prevent activation of the unstimulated CD4+ T cells by foreign antigens, was used throughout. Supernatants from cell cultures were evaluated for p24 antigen production using the Coulter HIV-1 p24 Antigen Assay Kit.
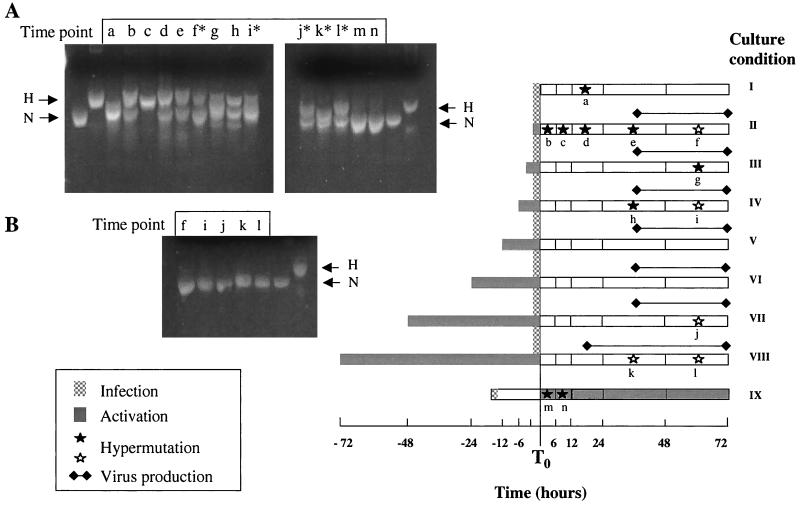
Nine different culture conditions were established (see Fig. 6) as follows: Culture I, virus infection without PHA stimulation; culture II, PHA and virus added simultaneously for 2 h; cultures III through VIII, PHA stimulation preceding virus infection by 3, 6, 12, 24, 48, or 72 h, respectively; culture IX, addition of virus for 2 h, washing away of virus, and PHA addition 24 h later. Cells and culture supernatants were collected at 6, 12, 24, 48, and 72 h after infection, with the exception of culture condition IX, where collection took place at 6, 12, 24, 48, and 72 h after addition of PHA.
FIG. 6.
Relationship of hypermutation to T-cell activation in virus cultures. CD4+ T cells isolated from the PBMC of normal donors were infected with an NL4-3 virus stock for 2 h at a multiplicity of infection of 0.02 (stippled gray bar) under varying conditions of PHA activation (gray bars). Condition I, infection without addition of PHA; condition II, simultaneous infection and PHA addition; conditions III to VIII, infection with prior PHA activation for 3 to 72 h respectively; condition IX, infection and addition of PHA 24 h later. Cultures were sampled at intervals and assayed for the presence of p24 antigen in the culture supernatant (solid lines) and for the presence of hypermutated HIV-1 DNA sequences. Filled stars, samples where hypermutated sequences were abundant; open stars, samples with rare hypermutated sequences. The appearance of PCR products on HA yellow gels is shown on the right. Time points are labeled “a” through “n” at the tops of the gels and in the diagram. Samples a, b, c, d, e, g, and h are bulk PCR products. Samples from time points at which hypermutation was rare (f, i, j, k, and l) are shown both as bulk PCR products (B) and after pre-enrichment by ScrF1 and AvaII digestion (A). Time points not marked by stars or asterisks yielded only normal sequences both before and after enrichment (not shown).
Sequence analysis.
Sequences were examined for base composition, open reading frames, and the composition of the translated protein sequences with EDITSEQ (DNAStar, Madison, Wis.). Parameters of hypermutation were evaluated with the Hypermut Program Package (54) (http://www.hiv.lanl.gov/HYPERMUT/hypermut.html) as implemented at the Los Alamos HIV Sequence Database, Los Alamos, N. Mex. This program identifies mutations and their dinucleotide context with respect to a reference sequence that is provided with each alignment. For the patient samples, we used a sequence with the lowest percent A+T, a normal base composition, and an open reading frame as the normal reference for all sequences from that patient. For virus cultures, the sequence of NL4-3, obtained by RT-PCR and sequencing of the virus stock, was used as the reference.
Nucleotide sequence accession numbers.
All the HIV-1 sequences related to this work have been submitted to GenBank and were given accession numbers A Y036228 through A Y036577.
RESULTS
Amplification, detection, and recovery of hypermutated DNA sequences.
The replacement of G by A in hypermutated sequences increases their A+T content and decreases their homology with PCR primers based on normal sequences. Unless primer mismatching is addressed, the recovery of hypermutants will be severely limited. Once amplified, however, hypermutants should be easily detectable by their increased A+T content. Thus we addressed the issue of primer mismatching first. Table 1 shows the design of a PCR strategy specifically designed to permit more efficient recovery of hypermutated sequences. Nested primers which amplify a 297-bp segment encoding HIV-1 protease were examined for GA and GG dinucleotides, which are susceptible to hypermutation. New primers were designed with either incorporated GA or CT mixtures at some of these sites (hyp primers) or replaced G with A or C with T (hypa primers). The hyp and hypa primers were also shortened to bring potential mispairs closer to the 3′ end of the primer, where they could exert the maximal effect in destabilizing interaction with normal sequences.
TABLE 1.
Modified PCR primers for amplification of hypermutated sequences
| Primer | Function | Modification | Sequence type amplified | Location in HXB2 | Sequence (5′–3′)a |
|---|---|---|---|---|---|
| DP10 | 1st round, forward | None | Normal | 2198–2223 | GAA CTC CCT CTC AGA AGC AGG AGC CG |
| DP10hyp | 1st round, forward | Shortened, G+A mixed bases introduced | Normal and hypermutant | 2198–2223 | AA CTC CCT CTC ARA AGC ARR A |
| DP10hypa | 1st round, forward | Shortened, G-to-A replacements made | Hypermutant | 2198–2223 | AA CTC CCT CTC AAA AGC AAA A |
| DP11 | 1st round, reverse | None | Normal | 2598–2572 | CCA TTCCTG GCT TTA ATT TTA CTG GTA |
| DP11hyp | 1st round, reverse | Shortened, C+T mixed bases introduced | Normal and hypermutant | 2598–2572 | CCA TTY YTG GCT TTA ATT TTA CTG |
| DP11hypa | 1st round, reverse | Shortened, C to T replacements made | Hypermutant | 2598–2572 | CCA TTT TTG GCT TTA ATT TTA CTG |
| DP16 | 2nd round, forward | None | Normal and hypermutant | 2253–2274 | CCT CAA ATC ACT CTT TGG CAA C |
| DP17 | 2nd round, reverse | None | Normal and hypermutant | 2549–2529 | AAA ATT TAA AGT GCA GCC AAT |
Primer sequences DP10, DP11, DP16, and DP17 have been published previously (28, 52a). They amplify a 297-bp segment of the HIV-1 genome encoding the protease gene, as indicated by their location in the reference sequence HXB2 (GenBank accession number AF033819). Sequences susceptible to hypermutation (GA or GG in forward primers and TC or CC in reverse primers, respectively) are underlined. Some of these sequences were modified (indicated by boldface), either by incorporation of G+A (R) or C+T mixtures (Y), or by G-to-A or C-to-T mutation. Modified primers were also shortened to bring potential mismatches with normal sequences nearer to their 3′ ends and further destabilize their interaction with normal sequences.
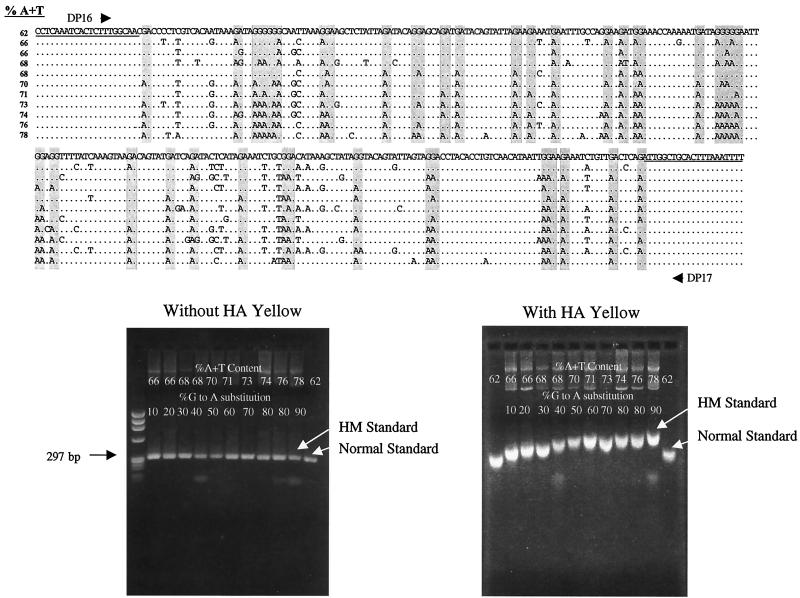
At the same time, a gel electrophoresis system was designed to permit detection of hypermutants. The dye HA yellow, in which consists of the DNA-binding ligand bisbenzamide coupled to PEG, was incorporated into 1% agarose gels. A series of cloned protease sequences representing different A+T contents, amplified and sequenced previously in the pilot phase of this project, were used for evaluation of the gel system (Fig. 1). These sequences represented a range of G-to-A substitution from 0 to 90% and had A+T content ranging from 62% (normal) to 78% (fully hypermutated). The cloned genes were all of equal length and mobility, as shown in the gel without HA yellow. In the presence of HA yellow, the clones were retarded in mobility according to the percent A+T. Sequences with as little as 1% increase in percent A+T, corresponding to 10% G-to-A substitution, were detectable by this approach.
FIG. 1.
Calibration of the HA yellow gel system. HIV-1 protease sequences amplified from patient PBMC and representing a range of G-to-A hypermutations were used to explore the performance of 1% agarose gels containing HA yellow, a dye that preferentially binds to AT-rich regions in DNA. The alignment of the 297-bp sequences, amplified with primers DP16 and DP17 and ranging from normal (62% A+T) to maximally hypermutated (78% A+T) is shown at the top. The GA and GG dinucleotides that are susceptible to hypermutation are shaded. The gel on the left shows the migration of these PCR products as a single band at 297 bp without HA yellow. On the right, a gel incorporating HA yellow is shown, illustrating the direct relationship between AT content and mobility. Sequences with as little as 10% G-to-A substitution (2nd lane) migrated differently than normal sequences. PCR products representing the normal and maximally hypermutated sequences were used as migration standards in subsequent HA yellow gels.
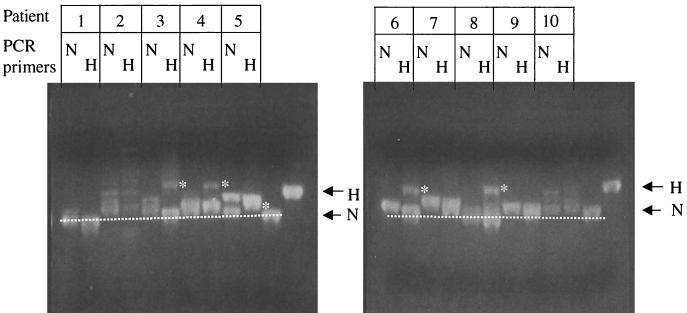
The performance of hypermutated primers (Table 1) was then assessed using DNA extracted from the PBMC of 10 early-stage, asymptomatic patients. Figure 2 shows a direct comparison of the PCR products obtained with normal or hypermutated primers, respectively. Among the 10 samples, 5 showed a substantial increase in hypermutated sequences when primers hyp and hypa were used (patients 3, 4, 5, 6, and 8; Fig. 2). Patients 2 and 10 yielded a variety of hypermutated bands with both normal and hypermutated primers. Patients 1, 7, and 9 showed essentially a single band at the normal position regardless of the primers used. Thus the use of primers hyp and hypa increased the number of samples with detectable hypermutation in the bulk PCR product on HA yellow gels from 2 of 10 to 7 of 10.
FIG. 2.
Relative recovery of hypermutated sequences with normal and hypermutated PCR primers. Primers were designed to incorporate G-to-A substitutions in GA or GG dinucleotides (Materials and Methods) and compared to primers matched to normal sequences for their ability to amplify hypermutated sequences from the PBMC of 10 patients. A side-by side comparison of the PCR products using HA yellow gels is shown. N, normal primers; H, primers hyp and hypa. Hypermutated bands detected with primers hyp and hypa but not with normal primers are indicated for patients 3, 4, 5, 6, and 8 (asterisks). The positions of normal and fully hypermutated standards are indicated by arrows.
Hypermutation in patient PBMC.
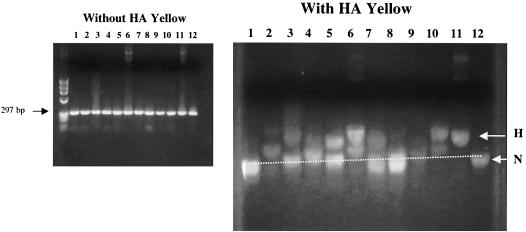
HIV-1 proviral DNA sequences from 53 HIV-1-positive patients were investigated (Table 2). The appearance of protease genes PCR amplified from patient PBMC is illustrated in Fig. 3. While all of the protease genes were of uniform length without HA yellow, they often separated into multiple bands of different A+T content on HA yellow gels. Sometimes all of the PCR product migrated at the normal position (Fig. 3, right, lane 1), but other PCR products split into multiple bands (lanes 2 to 7, 9, and 10). Notably, some samples had most of the PCR product retarded in the gel (lanes 2, 6, and 10). Other samples migrated mostly at the normal position but were smeared upward without distinct bands (Fig. 3, right, lane 8).
FIG. 3.
Abundance and variety of hypermutated PCR products amplified from patient PBMC. DNA extracted from patient PBMC was amplified in a touchdown, nested PCR with primers hyp and hypa as described in Materials and Methods. PCR products were compared to normal and hypermutated standards on HA yellow gels. While all products migrated as a single band at 297 bp without HA yellow (left), they were often split into several bands in the presence of HA yellow (right). Products were visualized by staining with ethidium bromide.
A combination of approaches was used to recover and sequence a range of normal and hypermutated sequences from each of 53 patient samples. These included use of both normal and hypermutated PCR primers, extraction of PCR products after separation in HA yellow gels, and reamplification of rare sequences extracted from HA yellow gels (see Materials and Methods). Multiple sequences were obtained from each patient, ranging from 1 to 28; the total number of sequences was 287.
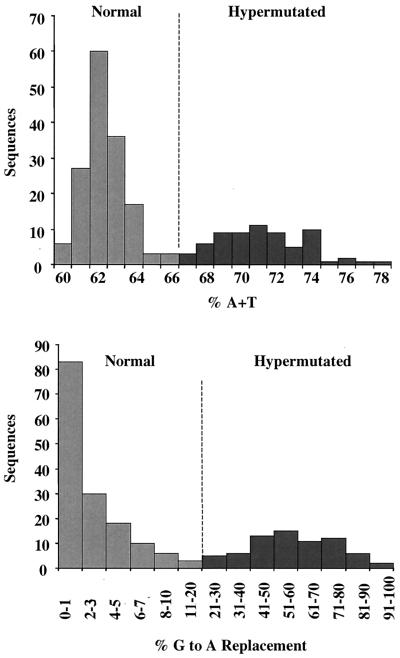
The distribution of A+T content among all patient sequences is shown in Fig. 4 (top). For this analysis, we eliminated identical sequences, which were sometimes found in within-patient comparisons, from the data set. Among 219 different sequences, the observed range of A+T content was 60 to 78%. The sequences formed a bimodal distribution, with 149 sequences in the range of 60 to 65% A+T and 70 sequences with higher A+T content.
FIG. 4.
Classification of sequences from patient PBMC. HIV-1 protease genes were amplified using DNA extracted from the PBMC of 53 patients ranging from the early, asymptomatic stage of HIV-1 infection to late-stage AIDS, and including infections with HIV-1 subtypes A through D (Table 2). Products PCR amplified with primers hyp and hypa were molecularly cloned and sequenced. In total, 287 sequences were obtained, of which 219 were unique. Among the 219 sequences, the percent A+T distribution is shown in the top graph. Using the sequence with the lowest percent A+T from each patient as a normal reference, the percent G in the GA or GG context that was mutated to A was calculated for each sequence, and the distribution of percent G-to-A mutation was examined (bottom graph). Based on the bimodal distribution of these parameters, 149 sequences were classified as normal (less than or equal to 66% A+T and less than 20% G-to-A mutation) and 70 were classified as hypermutated (67% A+T and more than 21% G-to-A mutation).
The percent G-to-A mutation was then tabulated. The number of G residues susceptible to hypermutation (i.e., followed by G or A) in the normal reference sequence from each patient was compared to the number of these G's transitioned to A's in all other sequences from that patient (Fig. 4, bottom). Among the sequences with 60 to 64% A+T, the number of G-to-A mutations was less than 20% while in the sequences with A+T content greater than 66%, the range of G-to-A mutation was 21 to 100%. From these distributions we operationally defined normal sequences as those with 65% A+T or less and less than 20% G-to-A mutation, and hypermutated sequences as those with more than 66% A+T and more than 20% G-to-A mutation. By these criteria, 149 of the different patient sequences were normal and 70 were hypermutated.
The sequence data established that, among the 53 patients, 23 (43%) harbored a mixture of normal and hypermutated HIV-1 sequences, while from the remaining 30 patients, only normal sequences were recovered by the techniques used (Table 2). Hypermutated sequences were found in both asymptomatic and AIDS patients, and in patients with infections with four different HIV-1 subtypes.
Parameters of hypermutation.
Seventy hypermutated sequences were recovered directly from patient PBMC, together with normal reference sequences from each patient, by a specific and consistent method. This permitted, for the first time, a systematic evaluation of the parameters of HIV-1 hypermutation in vivo. The specificity of the hypermutations was extraordinary (Fig. 5, top). The sequences showed elevated A content and reduced G content compared to normal sequences, while their C and T content was completely unaltered. In the 23 patients from whom both normal and hypermutated sequences were recovered, the normal sequences had the same base composition as those of the 30 patients whose recovered sequences were entirely normal (data not shown).
FIG. 5.
Parameters of hypermutation in sequences from patient PBMC. One hundred forty-nine normal and 70 hypermutated sequences from patient PBMC were examined for base composition (top). The normal sequences had a narrow distribution of percent G and percent A, centered on 22 and 37%, respectively. Hypermutated sequences showed a broad range of G content from 5 to 20% and a range of A content from 40 to 54%. The distribution of percent C and percent T was the same in normal and hypermutated sequences. Hypermutated sequences were arranged in order of increasing hypermutation (bottom two graphs). While G-to-A mutations in the GA or GG context increased 10-fold, other mutations (G to A in the GC and GT context and all other mutations) showed no discernible increase. The percentages of available G replaced by A in the GA and GG contexts were calculated separately and compared over the range of G-to-A substitution (bottom). More of the G residues in the GA context than in the GG context were replaced by A at all levels of hypermutation.
Not all of the mutations in hypermutated sequences were G-to-A mutations. Other mutations included G to A in the context of GC or GT, and all other transitions and transversions. While the range of G-to-A substitution was 20 to 94% in hypermutants, other mutations ranged from 0 to 2.7%. We determined how other mutations were distributed with respect to the range of hypermutation (Fig. 5, bottom). There was no observed trend for increase in other mutations even at the highest levels of G-to-A saturation. Hypermutation appears to be entirely restricted to G-to-A substitution, generates a broad range of G-to-A substitution, and appears to occur completely independently of other mutations.
The dinucleotide context of hypermutation was then considered (Fig. 5, bottom). Although the GA and the GG context were both used in almost every hypermutated sequence, there was a consistent trend for preferential mutation of G in the GA context rather than the GG context. With respect to the saturation of the available GA and GG dinucleotides, at all levels of G-to-A replacement, the percentage of G replaced by A in the dinucleotide context GA was about twofold higher than the percentage of G replaced by A in the GG context. Although G-to-A substitutions in the GC and GT dinucleotide contexts were previously referred as hypermutation related (60), the observed number of these replacements was limited, ranging from 0 to 3 (mean, 0.6) per hypermutated sequence. In addition, no trend for increase in G-to-A mutations was evident in these contexts. At the highest levels of G-to-A mutation, virtually all of the GA and GG dinucleotides were mutated, while G-to-A mutations in GC or GT (other mutations; Fig. 5) were not elevated.
In summary, hypermutated HIV-1 protease sequences recovered from patient PBMC exhibited a broad range of G-to-A mutation that was entirely restricted to the GA or GG dinucleotide context, with a consistent preference for G-to-A mutation in the dinucleotide GA relative to GG at all levels of G-to-A saturation. Other mutations, including G-to-A substitutions in the GC or GT context, occurred at much lower levels and independently of hypermutations.
Loss of informational content in hypermutated sequences.
Hypermutated and normal DNA sequences were translated into protein sequences and evaluated for the integrity of the genetic information. Virtually all of the hypermutated sequences from patient PBMC had lost their protein-coding potential (Table 3); 69 of 70 had an in-frame stop codon, compared to 3 of 149 normal sequences. In addition, there was a net accumulation of positively charged amino acids, mostly from decreased Gly, Asp, and Arg and increased Lys and Asn in hypermutants. The proteases encoded by hypermutated sequences had, on average, a fivefold increase in net charge (Table 3). It is noteworthy that hypermutated sequences with the lowest levels of G-to-A substitutions sustained inactivating in-frame stop codons and many nonsynonymous nucleotide substitutions. These combined effects make it highly unlikely that functional proteases, and, consequently, viable viruses can be derived from hypermutated sequences.
TABLE 3.
Genetic damage in hypermutated sequences
| Source | Sequence type | n | No. of in-frame stop codons | Mean (range) net charge of protein |
|---|---|---|---|---|
| Patient PBMC | Normal | 149 | 3 | 2.5 (0.0–9.0) |
| Hypermutated | 70 | 69 | 12.6 (3.0–23.0) | |
| Virus cultures | Normal | 73 | 3 | 1.9 (0.0–4.0) |
| Hypermutated | 58 | 48 | 10.9 (2.0–23.0) |
Relationship of hypermutation to stage of disease and genetic subtype.
The relationships between hypermutation, clinical stage, and HIV-1 subtype were investigated (Table 2). Hypermutated sequences were recovered at essentially the same rate from AIDS patients (42%) and early asymptomatic individuals (44%). Hypermutation was found in 57% of subtype A, 67% of subtype D, and 44% of subtype B infections, as well as in A+D dually infected and unclassified samples. Although subtype C was the most frequently represented subtype among AIDS patients, hypermutation was detected only in 3 of 14 (21%) patients infected with this subtype. Subtype C hypermutated sequences also tended to have lower levels of G-to-A mutation than the other subtypes, but in this small sample the differences did not reach statistical significance (data not shown).
Investigation of hypermutation in cell culture.
The link between unbalanced dNTP pools and hypermutation in vivo is probable but not yet conclusive. Experiments with cell-free systems have shown that unbalances in dNTP pools during reverse transcription can lead to hypermutation, but the specificity for the GA and GG dinucleotide context has been difficult to reproduce. On the other hand, hypermutants in the correct dinucleotide context have been recovered from HIV-1 cultures in PBMC, but the exact nature of the cell population that produced them and the status of the dNTP pools in this population have not been elucidated. In CD4+ T cells, which are a main target of HIV 1 replication in vivo, dNTP pools are elevated when the cells are activated and in cycle (20) but are unbalanced and fluctuating in resting T cells and during the transition from a resting to an activated state (5, 10, 20, 58). We designed an experiment in which the timing of HIV-1 infection was varied with respect to activation of resting CD4+ T cells by PHA; the outcome measure was recovery of hypermutated sequences whose characteristics corresponded exactly to those found in vivo.
The overall results are shown in Fig. 6. Proviral DNA was established in all culture conditions, as evidenced by its recovery by DNA PCR; a mock-infected culture was HIV-1 DNA PCR negative (data not shown). p24 antigen in the culture supernatant, indicative of virus production, was observed only when cultures were stimulated with PHA either before, or simultaneously with, virus infection.
The HIV-1 protease gene was amplified from all time points and culture conditions (Materials and Methods) and evaluated for the presence of normal and hypermutated sequences using the HA yellow gel system (Fig. 6). It was possible to visualize abundant hypermutated sequences in the bulk PCR product from some of the culture conditions (Fig. 6A, lanes a to e, g, and h). The richest source of hypermutants was culture II, in which infection and activation occurred simultaneously (Fig. 6A, lanes b through e). Hypermutated sequences could also be visualized in conditions III and IV (lanes g and h), which represented PHA activation for less than 8 h before infection. Stimulation with PHA for 12 h or more (conditions V, VI, VII, and VIII) yielded only normal sequences (Fig. 6B, lanes f and i through l). However, by pre-enrichment for hypermutants using ScrF1 and AvaII digestion (see Materials and Methods), rare hypermutants could be observed in late time points in most cultures (Fig. 6, lanes f∗, i∗, j∗, k∗, and l∗). Finally, hypermutated sequences were found in condition IX, where virus infection preceded PHA activation by 24 h. In this case, the sequences had very low levels of G-to-A mutation, mostly in the GG context (see below) and were not distinguished on HA yellow gels (Fig. 6A, lanes m and n), but they could be recovered by cloning of the bulk PCR product. Overall, these analyses establish that HIV-1 infection of resting CD4+ T cells either shortly before or shortly after activation generated abundant hypermutated sequences with high levels of G-to-A mutation, while infection after longer periods of activation yielded principally normal sequences.
These combined approaches provided 131 different HIV-1 protease sequences from virus cultures for analysis. The distribution of percent A+T was evaluated, and results are shown in Figure 7A. Seventy-three sequences were classified as normal and 58 as hypermutated. Hypermutants from culture, like those from patient PBMC (Fig. 4), had a broad range of AT content, with some showing as high as 78% A+T. The normal sequences were narrowly distributed around 62% A+T, that of the NL4-3 virus innoculum. The different culture conditions were examined for the range of hypermutants that they produced (Fig. 7B). Conditions II, III, and IV, in which abundant hypermutation was detected in the bulk PCR product, also yielded a full range of G-to-A mutation. The hypermutants from cultures IX and I showed only lower levels of G-to-A mutation. Based on abundance and the range of G-to-A substitution, the hypermutants recovered from cultures II to IV most closely resembled those found in patient PBMC.
FIG. 7.
Parameters of hypermutation in virus cultures. A total of 227 HIV-1 protease sequences, 131 of which were unique, were obtained from virus cultures. The unique sequences were compared for their distribution of percent A+T (A). Seventy-three were normal and 58 were hypermutated. (B) Distribution of G-to-A mutation in sequences from different culture conditions. Roman numerals refer to the culture conditions described in the legend to Fig. 6. Filled symbols, normal sequences; open symbols, hypermutated sequences. The cultures where abundant hypermutants covering the full range of G-to-A replacement were found are shaded. Below, the average ratio of percent G-to-A substitutions in the GA versus GG context is plotted for the sequences (all except six) in which both contexts were used. (C) Base composition of normal and hypermutated sequences from virus cultures. (D) Specificity for G-to-A substitutions in the GA or GG context in contrast to all other mutations, and the preferential use of GA over GG over the range of hypermutation.
The hypermutants from culture were further examined for their correspondence to hypermutants generated in vivo by analysis of the base composition, accumulation of other mutations, and context preference (Fig. 7C and D). Once again, there was no alteration in percent C or T, the mutations were entirely restricted to G-to-A substitutions in the GA or GG context without significant accumulation of other mutations, and the GA context was generally used in preference to GG, essentially over the full range of G-to-A substitution. However, when sequences from each culture condition were analyzed separately, an interesting pattern of dinucleotide context utilization was noted (Fig. 7B, lower graph). In cultures IX, I, and II, where PHA either was not added or was added simultaneously with or after infection with the virus, the preference for GA over GG was not strong. With prior PHA stimulation for 3 h or more (cultures III to VIII), the saturation of available GA dinucleotides occurred at 4 to 6 times that of available GG dinucleotides.
Finally, hypermutated and normal protease gene sequences recovered from cell culture were examined for the loss of informational content (Table 3). Twenty-seven out of 36 hypermutated sequences (75%) were associated with in-frame stop codons compared to 3 of 73 (4%) normal sequences. Like those from patients, hypermutants from culture had at least a fivefold increase in the net charge of the encoded protein.
DISCUSSION
This study represents the first systematic evaluation of patient PBMC and virus cultures with a consistently applied methodology optimized for detection of hypermutated HIV-1 sequences. Hypermutated sequences could be detected in 43% of patients and in virus cultures when T-cell stimulation was initiated at the time of, or shortly before, virus infection. The nearly identical quantitative parameters of the hypermutants recovered from patients and from virus cultures suggest that T cells in the process of becoming activated are also the main source of hypermutation in vivo.
Some quantitative parameters of hypermutation have been confirmed. The excessive accumulation of nonsynonymous substitutions and stop codon formation observed in hypermutants reinforces the perception that they can no longer generate viable viruses. However, hypermutation is not accompanied by a general increase in mutation rate; it is completely selective for G residues followed by A or G. We examined the question of whether hypermutation affects GC and GT dinucleotides, although at lower levels, in addition to the preferred GA and GG contexts. Our data are most consistent with the idea that G-to-A substitutions in GC and GT contexts are not part of the hypermutation process, but are only background mutations, for the following reasons: (i) the utilization of GC and GT did not increase proportionally as the saturation of GA and GG increased from 10% to more than 90% (other mutations; Fig. 5) and (ii) the levels of G-to-A substitution in GC and GT were never significantly higher than those of other types of mutations. However, our observations are limited to the HIV-1 protease gene, and examination of full HIV-1 genomes for the context of hypermutation would be required to fortify this conclusion. A consistent preference for saturation of available GA over GG dinucleotides within hypermutated sequences was also noted.
Efficient detection of hypermutated sequences requires the proper combination of PCR variables, including DNA sampling, primer design, annealing temperatures, and a physical method for separation and enrichment of hypermutants. While far more hypermutated sequences were detected here than in other studies, they still may have been underrepresented in our analyses. PCR primers matching all possible hypermutants were not included, in part because of their low annealing temperature. The detection of hypermutants on HA yellow gels, while efficient, may be influenced by the context of the G-to-A transitions and by their distribution within the sequence. Indeed, the DNA-binding preferences of bisbenzamide for runs of AT, particularly AAAA and AATT, would suggest that GA to AA would be more efficiently detected than GG to AG, but the fact that the vast majority of hypermutants we recovered used both contexts suggests that this was not a major obstacle to recovery. Finally, we have examined 297 bp of a 9,200-bp genome. Many hypermutated proviruses may have been missed by our procedure. The common perception that hypermutated sequences are rare in vivo should be revised in light of these considerations.
Careful definition of the quantitative parameters of hypermutation is necessary in order to define its biochemical basis. Here we show that the process is completely independent of the normal accumulation of mutations in HIV-1 and covers a much broader dynamic range. Figure 5 shows that the rate of accumulation of all other mutations, including G-to-A substitution in the GT and GC contexts, remained stable while G-to-A substitutions reached virtual saturation. The preferential utilization of available GA over GG dinucleotides over the whole dynamic range, and the fact that the vast majority of hypermutants utilized both GA and GG contexts, suggests that both contexts are recognized during a single reverse transcription event. It could be that the efficiency of hypermutation in the GA context is intrinsically, but not overwhelmingly, higher, or the frequency of extension past the mismatch could be higher for the GA context. The high error rate of HIV-1 RT is apparently necessary to permit both “normal” mutation and hypermutation, but hypermutation should not be thought of as an elevated error rate for RT; it is clearly an independent process with separate biochemical requirements which must vary over a wide range during HIV-1 replication in vivo.
Could hypermutation be influenced by viral variation, or is it essentially a passive function of the condition of the cell population in which HIV-1 replicates? Evidence that there may be a viral component appears in Table 2. Hypermutated sequences were recovered from a smaller percentage of patients infected with subtype C, compared to other subtypes. While itself not significant in this small sample (P = 0.078), the percent G-to-A substitution was also skewed to the lower end of the observed range in subtype C sequences. Stronger evidence would be needed, including larger numbers of patients with a broad range of clinical stages, to establish a definitive relationship between hypermutation and HIV-1 subtype.
The experiments with virus cultures are illuminating with respect to the conditions under which hypermutation occurs. First, the evidence that true hypermutants were generated in CD4+ T cells in culture should be reiterated. The target cell populations were free of HIV-1 because DNA extracted from the cells of mock-infected cultures carried in parallel were HIV-1 PCR negative. The virus stock was molecularly cloned and verified to be free of hypermutation by sequencing of the viral RNA by RT-PCR and by DNase treatment to remove any contaminating proviral DNA remaining from virus propagation. DNA PCR of the culture supernatants after infection was negative. The identity of the target cell population was carefully established; the culture contained principally, if not exclusively, CD4+ T cells. The major contaminant was B cells, which do not support HIV-1 replication. Most, if not all, CD4+ T cells were quiescent. It has been previously documented that infection of quiescent cells results in the absence of virus progeny (68) and that viral DNA can be detected after infection of unstimulated cells although it is not associated with virus production (69, 70). In our experiments, proviral DNA was established but there was no detectable virus production in the absence of PHA stimulation, which is consistent with the infection of quiescent cells. Most of the cells were HLA-DR negative. It is highly probable that those cells that were HLA-DR+, ranging from 15 to 18% in different experiments, represented recently, but not currently, activated cells.
Hypermutants were recovered principally from virus cultures in which HIV-1 infection was simultaneous with, or briefly preceded by, PHA stimulation (Fig. 6). The parameters of the hypermutants were quantitatively identical to those recovered from patient PBMC (Fig. 7), establishing a direct link between the circumstances established in culture and those leading to hypermutation in vivo. Our results suggest that hypermutation may be the direct and predictable outcome of certain biochemical stages during early PHA stimulation of T cells. Even the dinucleotide context preference is partially parsed out by these experiments; we note that in culture conditions XI, I, and II, where PHA either was not added or was added along with or after the virus, utilization of available GA and GG was almost equivalent. The strong bias for GA appeared once PHA preceded HIV-1 infection by only 3 h (Figure 7B). It is tempting to speculate that GG preference hypermutants derive from resting cells while those with the strong GA preference are generated in cells that are in the process of becoming activated.
A previous study demonstrated that hypermutants were recovered, although at low frequencies, after infection of unstimulated (2%) and stimulated (0.9%) bulk PBMC (62). Such recovery of hypermutants was ascribed to virus infection of a small proportion of PBMC with distorted dNTP pools, reinforcing the concept of an occasional and sporadic event. Because the composition of dNTP pools fluctuates as cells progress through the cell cycle (5, 20, 45), we investigated if the timing of HIV-1 infection with respect to T-cell stimulation could influence the generation of hypermutation. Our results define a window of susceptibility to hypermutation during initial events of T-cell stimulation. The target cell population for hypermutation must be continually renewed in vivo, commensurate with the proportion of resting T cells that become newly activated during active virus replication. Once PHA had been present for 8 h or more, the recovery of hypermutants plummeted. They may be no longer generated once T cells proceed far enough along the activation pathway. Nonetheless, it appears that the loss of informational content observed in hypermutants would preclude their further expansion except as passive passengers in the genomes of T cells. Such sequences should be included in conceptualizations of the latent reservoir of HIV-1 in patients.
Is hypermutation related to the observation that the genomes of HIV-1 and other lentiviruses are AT rich? The total and unrecoverable loss of coding potential of hypermutated HIV sequences (Table 3) would suggest that they are incapable of generating progeny virions and cannot contribute to the HIV-1 gene pool. However, it is important to consider the dynamic range of G-to-A substitution. For purposes of analysis we classified sequences as either normal or hypermutated, but, of course, the frequency of G-to-A substitution is a continuum, and those sequences with the lowest levels of hypermutation may sometimes be viable.
We have shown that hypermutation is much more abundant in patient PBMC than previously recognized. It will be important to determine what fraction of the genome in hypermutants remains unaffected, as well as the potential for hypermutated proviruses to generate viable viruses through recombination. Hypermutated proviruses could represent a difficult-to-eradicate remnant of HIV-1 during highly active antiretroviral therapy. Another aspect is the fact that the vast majority of hypermutated protease sequences present in this study were associated with a disrupted coding potential. Since protease has a crucial role during the HIV-1 replication cycle, the loss of its coding potential integrity should lead to abortive infections. Our data demonstrated that hypermutation may reduce the pool of viable replicating genomes in an infecting HIV-1 population. Detection of HIV-1 hypermutated sequences in at least 43% of patients studied suggests that hypermutation may happen in a systematic way in HIV-1-infected individuals. Based on these two observations one can speculate that hypermutation may be seen as a result of a host-associated mechanism which may decrease virus replication.
A better understanding of the metabolic profile of early proliferating T cells in relation to their expanding dNTP pools would lead to a better interpretation of the correlation between temporal aspects of cell cycle metabolism and HIV-1 hypermutation. This interpretation could develop into new strategies of promoting a hypermutation-inducible state in HIV-1 target cells, leading to irreversible mutagenesis of viral genomes. A better comprehension of the cellular mechanisms involved in HIV-1 hypermutation could open new avenues for virus control.
ACKNOWLEDGMENTS
We thank Casey Vibbard, Mark Louder, and Phil Ehrenberg for critical technical assistance. We are grateful to John Mascola for the support provided during virus propagation procedures. We are indebted to Michael Hoelscher for providing clinical samples from Tanzanian patients. We also thank Mary Marovich and Jean Carr for careful review of the manuscript.
The present work was supported by a cooperative agreement between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Department of Defense.
REFERENCES
- 1.Abu-Daya A, Brown P M, Fox K R. DNA sequence preferences of several AT-selective minor groove binding ligands. Nucleic Acids Res. 1995;23:3385–3392. doi: 10.1093/nar/23.17.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 3.Bianchi V, Borella S, Rampazzo C, Ferraro P, Calderazzo F, Bianchi L C, Skog S, Reichard P. Cell cycle-dependent metabolism of pyrimidine deoxynucleoside triphosphates in CEM cells. J Biol Chem. 1997;272:16118–16124. doi: 10.1074/jbc.272.26.16118. [DOI] [PubMed] [Google Scholar]
- 4.Borman A M, Quillent C, Charneau P, Kean K M, Clavel F. A highly defective HIV-1 group O provirus: evidence for the role of local sequence determinants in G→A hypermutation during negative-strand viral DNA synthesis. Virology. 1995;208:601–609. doi: 10.1006/viro.1995.1191. [DOI] [PubMed] [Google Scholar]
- 5.Bray G, Brent T P. Deoxyribonucleoside 5′-triphosphate pool fluctuations during the mammalian cell cycle. Biochim Biophys Acta. 1972;269:184–191. doi: 10.1016/0005-2787(72)90425-x. [DOI] [PubMed] [Google Scholar]
- 6.Brodine S K, Shaffer R A, Starkey M J, Tasker S A, Gilcrest J L, Louder M K, Barile A, VanCott T C, Vahey M T, McCutchan F E, Birx D L, Richman D D, Mascola J R. Drug resistance patterns, genetic subtypes, clinical features, and risk factors in military personnel with HIV-1 seroconversion. Ann Intern Med. 1999;131:502–506. doi: 10.7326/0003-4819-131-7-199910050-00004. [DOI] [PubMed] [Google Scholar]
- 7.Bronson E C, Anderson J N. Nucleotide composition as a driving force in the evolution of retroviruses. J Mol Evol. 1994;38:506–532. doi: 10.1007/BF00178851. [DOI] [PubMed] [Google Scholar]
- 8.Cattaneo R. Biased (A→I) hypermutation of animal RNA virus genomes. Curr Opin Genet Dev. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 9.Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter M A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen A, Barankiewicz J, Lederman H M, Gelfand E W. Purine and pyrimidine metabolism in human T lymphocytes. Regulation of deoxyribonucleotide metabolism. J Biol Chem. 1983;258:12334–12340. [PubMed] [Google Scholar]
- 11.Delassus S, Cheynier R, Wain-Hobson S. Evolution of human immunodeficiency virus type 1 nef and long terminal repeat sequences over 4 years in vivo and in vitro. J Virol. 1991;65:225–231. doi: 10.1128/jvi.65.1.225-231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domingo E, Martinez-Salas E, Sobrino F, de la Torre J C, Portela A, Ortin J, Lopez-Galindez C, Perez-Brena P, Villanueva N, Najera R, VandePol S, Steinhauer D, DePolo N, Holland J J. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 13.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake J W. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci USA. 1993;90:4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eigen M, Schuster P. The hypercycle. A principle of natural self-organization. A. Emergence of the hypercycle. Naturwissenschaften. 1977;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 16.Felder M P, Laugier D, Yatsula B, Dezelee P, Calothy G, Marx M. Functional and biological properties of an avian variant long terminal repeat containing multiple A-to-G conversions in the U3 sequence. J Virol. 1994;68:4759–4767. doi: 10.1128/jvi.68.8.4759-4767.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 18.Fitzgibbon J E, Mazar S, Dubin D T. A new type of G→A hypermutation affecting human immunodeficiency virus. AIDS Res Hum Retrovir. 1993;9:833–838. doi: 10.1089/aid.1993.9.833. [DOI] [PubMed] [Google Scholar]
- 19.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 20.Gao W Y, Cara A, Gallo R C, Lori F. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc Natl Acad Sci USA. 1993;90:8925–8928. doi: 10.1073/pnas.90.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodenow M, Huet T, Saurin W, Kwok S, Sninsky J, Wain-Hobson S. HIV-1 isolates are rapidly evolving quasispecies: evidence for viral mixtures and preferred nucleotide substitutions. J Acquir Immune Defic Syndr. 1989;2:344–352. [PubMed] [Google Scholar]
- 22.Gunther S, Sommer G, Plikat U, Iwanska A, Wain-Hobson S, Will H, Meyerhans A. Naturally occurring hepatitis B virus genomes bearing the hallmarks of retroviral G→A hypermutation. Virology. 1997;235:104–108. doi: 10.1006/viro.1997.8676. [DOI] [PubMed] [Google Scholar]
- 23.Hajjar A M, Linial M L. Modification of retroviral RNA by double-stranded RNA adenosine deaminase. J Virol. 1995;69:5878–5882. doi: 10.1128/jvi.69.9.5878-5882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hecker K H, Roux K H. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques. 1996;20:478–485. doi: 10.2144/19962003478. [DOI] [PubMed] [Google Scholar]
- 25.Hoelscher M, Hanker S, Barin F, Cheingsong-Popov R, Dietrich U, Jordan-Harder B, Olaleye D, Nagele E, Markuzzi A, Mwakagile D, Minja F, Weber J, Gurtler L, Von Sonnenburg F. HIV type 1 V3 serotyping of Tanzanian samples: probable reasons for mismatching with genetic subtyping. AIDS Res Hum Retrovir. 1998;14:139–149. doi: 10.1089/aid.1998.14.139. [DOI] [PubMed] [Google Scholar]
- 26.Holland J J, De La Torre J C, Steinhauer D A. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 27.Holland J J, Domingo E, de la Torre J C, Steinhauer D A. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J Virol. 1990;64:3960–3962. doi: 10.1128/jvi.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janini L M, Pieniazek D, Peralta J M, Schechter M, Tanuri A, Vicente A C, de la Torre N, Pieniazek N J, Luo C C, Kalish M L, Schochetman G, Rayfield M A. Identification of single and dual infections with distinct subtypes of human immunodeficiency virus type 1 by using restriction fragment length polymorphism analysis. Virus Genes. 1996;13:69–81. doi: 10.1007/BF00576981. [DOI] [PubMed] [Google Scholar]
- 29.Ji J P, Loeb L A. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992;31:954–958. doi: 10.1021/bi00119a002. [DOI] [PubMed] [Google Scholar]
- 30.Johnson P R, Hamm T E, Goldstein S, Kitov S, Hirsch V M. The genetic fate of molecularly cloned simian immunodeficiency virus in experimentally infected macaques. Virology. 1991;185:217–228. doi: 10.1016/0042-6822(91)90769-8. [DOI] [PubMed] [Google Scholar]
- 31.Julias J G, Kim T, Arnold G, Pathak V K. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J Virol. 1997;71:4254–4263. doi: 10.1128/jvi.71.6.4254-4263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Julias J G, Pathak V K. Deoxyribonucleoside triphosphate pool imbalances in vivo are associated with an increased retroviral mutation rate. J Virol. 1998;72:7941–7949. doi: 10.1128/jvi.72.10.7941-7949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim T, Mudry R A, Jr, Rexrode II C A, Pathak V K. Retroviral mutation rates and A-to-G hypermutations during different stages of retroviral replication. J Virol. 1996;70:7594–7602. doi: 10.1128/jvi.70.11.7594-7602.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunkel T A. Biological asymmetries and the fidelity of eukaryotic DNA replication. Bioessays. 1992;14:303–308. doi: 10.1002/bies.950140503. [DOI] [PubMed] [Google Scholar]
- 35.Kunz B A, Kohalmi S E. Modulation of mutagenesis by deoxyribonucleotide levels. Annu Rev Genet. 1991;25:339–359. doi: 10.1146/annurev.ge.25.120191.002011. [DOI] [PubMed] [Google Scholar]
- 36.Kunz B A, Kohalmi S E, Kunkel T A, Mathews C K, McIntosh E M, Reidy J A. International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat Res. 1994;318:1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 37.Kurath G, Rey M E, Dodds J A. Analysis of genetic heterogeneity within the type strain of satellite tobacco mosaic virus reveals variants and a strong bias for G to A substitution mutations. Virology. 1992;189:233–244. doi: 10.1016/0042-6822(92)90699-p. [DOI] [PubMed] [Google Scholar]
- 38.Lang W, Anderson R E, Perkins H, Grant R M, Lyman D, Winkelstein W, Royce R, Levy J A. Clinical, immunologic, and serologic findings in men at risk for acquired immunodeficiency syndrome. The San Francisco Men's Health Study. JAMA. 1987;257:326–330. [PubMed] [Google Scholar]
- 39.Li Y, Kappes J C, Conway J A, Price R W, Shaw G M, Hahn B H. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol. 1991;65:3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Tang X P, McArthur J C, Scott J, Gartner S. Analysis of human immunodeficiency virus type 1 gp160 sequences from a patient with HIV dementia: evidence for monocyte trafficking into brain. J Neurovirol. 2000;6(Suppl. 1):S70–S81. [PubMed] [Google Scholar]
- 41.Loucks E, Chaconas G, Blakesley R W, Wells R D, van de Sande J H. Antibiotic induced electrophoretic mobility shifts of DNA restriction fragments. Nucleic Acids Res. 1979;6:1869–1879. doi: 10.1093/nar/6.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansky L M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology. 1996;222:391–400. doi: 10.1006/viro.1996.0436. [DOI] [PubMed] [Google Scholar]
- 43.Martinez M A, Sala M, Vartanian J P, Wain-Hobson S. Reverse transcriptase and substrate dependence of the RNA hypermutagenesis reaction. Nucleic Acids Res. 1995;23:2573–2578. doi: 10.1093/nar/23.14.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez M A, Vartanian J P, Wain-Hobson S. Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc Natl Acad Sci USA. 1994;91:11787–11791. doi: 10.1073/pnas.91.25.11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathews C K, Ji J. DNA precursor asymmetries, replication fidelity, and variable genome evolution. Bioessays. 1992;14:295–301. doi: 10.1002/bies.950140502. [DOI] [PubMed] [Google Scholar]
- 46.Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp Cell Res. 1989;181:305–316. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- 47.Monken C E, Wu B, Srinivasan A. High resolution analysis of HIV-1 quasispecies in the brain. AIDS. 1995;9:345–349. [PubMed] [Google Scholar]
- 48.Muller W, Hattesohl I, Schuetz H J, Meyer G. Polyethylene glycol derivatives of base and sequence specific DNA ligands: DNA interaction and application for base specific separation of DNA fragments by gel electrophoresis. Nucleic Acids Res. 1981;9:95–119. doi: 10.1093/nar/9.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Hara P J, Nichol S T, Horodyski F M, Holland J J. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell. 1984;36:915–924. doi: 10.1016/0092-8674(84)90041-2. [DOI] [PubMed] [Google Scholar]
- 50.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perrino F W, Preston B D, Sandell L L, Loeb L A. Extension of mismatched 3′ termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci USA. 1989;86:8343–8347. doi: 10.1073/pnas.86.21.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perry S T, Flaherty M T, Kelley M J, Clabough D L, Tronick S R, Coggins L, Whetter L, Lengel C R, Fuller F. The surface envelope protein gene region of equine infectious anemia virus is not an important determinant of tropism in vitro. J Virol. 1992;66:4085–4097. doi: 10.1128/jvi.66.7.4085-4097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Pieniazek D, Peralta J M, Ferreira J A, Krebs J W, Owen S M, Sion F S, Filho C F, Sereno A B, de Sa C A, Weniger B G, Heyward W L, Ou C Y, Pieniazek N J, Schochetman G, Rayfield M. Identification of mixed HIV-1/HIV-2 infections in Brazil by polymerase chain reactioin. AIDS. 1991;5:1293–1299. doi: 10.1097/00002030-199111000-00002. [DOI] [PubMed] [Google Scholar]
- 53.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 54.Rose P P, Korber B T. Detecting hypermutations in viral sequences with an emphasis on G→A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- 55.Roux K H. Using mismatched primer-template pairs in touchdown PCR. Biotechniques. 1994;16:812–814. [PubMed] [Google Scholar]
- 56.Salminen M O, Ehrenberg P K, Mascola J R, Dayhoff D E, Merling R, Blake B, Louder M, Hegerich S, Polonis V R, Birx D L, Robb M L, McCutchan F E, Michael N L. Construction and biological characterization of infectious molecular clones of HIV-1 subtypes B and E (CRF01-AE) generated by the polymerase chain reaction. Virology. 2000;278:103–110. doi: 10.1006/viro.2000.0640. [DOI] [PubMed] [Google Scholar]
- 57.Salminen M O, Koch C, Sanders-Buell E, Ehrenberg P K, Michael N L, Carr J K, Burke D S, McCutchan F E. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 58.Tyrsted G. Effect of hydroxyurea and 5-fluorodeoxyuridine on deoxyribonucleoside triphosphate pools early in phytohemagglutinin-stimulated human lymphocytes. Biochem Pharmacol. 1982;31:3107–3113. doi: 10.1016/0006-2952(82)90087-9. [DOI] [PubMed] [Google Scholar]
- 59.Vartanian J P, Henry M, Wain-Hobson S. Hypermutagenic PCR involving all four transitions and a sizeable proportion of transversions. Nucleic Acids Res. 1996;24:2627–2631. doi: 10.1093/nar/24.14.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vartanian J P, Meyerhans A, Asjo B, Wain-Hobson S. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J Virol. 1991;65:1779–1788. doi: 10.1128/jvi.65.4.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vartanian J P, Meyerhans A, Sala M, Wain-Hobson S. G→A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci USA. 1994;91:3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vartanian J P, Plikat U, Henry M, Mahieux R, Guillemot L, Meyerhans A, Wain-Hobson S. HIV genetic variation is directed and restricted by DNA precursor availability. J Mol Biol. 1997;270:139–151. doi: 10.1006/jmbi.1997.1104. [DOI] [PubMed] [Google Scholar]
- 63.Wain-Hobson S. Retroviral G to A hypermutation, p. III-57–III-63. In: Myers G, Korber B T, Foley B T, Jeang K-T, Mellors J M, Wain-Hobson S, editors. Human retroviruses and AIDS—a compilation and analysis of nucleic acid and amino acid sequences. Vol. 1996. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1996. [Google Scholar]
- 64.Wain-Hobson S, Sonigo P, Guyader M, Gazit A, Henry M. Erratic G→A hypermutation within a complete caprine arthritis-encephalitis virus (CAEV) provirus. Virology. 1995;209:297–303. doi: 10.1006/viro.1995.1261. [DOI] [PubMed] [Google Scholar]
- 65.Wawer C, Ruggeberg H, Meyer G, Muyzer G. A simple and rapid electrophoresis method to detect sequence variation in PCR-amplified DNA fragments. Nucleic Acids Res. 1995;23:4928–4929. doi: 10.1093/nar/23.23.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkelstein W, Lyman D M, Padian N, Grant R, Samuel M, Wiley J A, Anderson R E, Lang W, Riggs J, Levy J A. Sexual practices and risk of infection by the human immunodeficiency virus. The San Francisco Men's Health Study. JAMA. 1987;257:321–325. [PubMed] [Google Scholar]
- 67.Yuste E, Lopez-Galindez C, Domingo E. Unusual distribution of mutations associated with serial bottleneck passages of human immunodeficiency virus type 1. J Virol. 2000;74:9546–9552. doi: 10.1128/jvi.74.20.9546-9552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zack J A. The role of the cell cycle in HIV-1 infection. Adv Exp Med Biol. 1995;374:27–31. doi: 10.1007/978-1-4615-1995-9_3. [DOI] [PubMed] [Google Scholar]
- 69.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 70.Zack J A, Haislip A M, Krogstad P, Chen I S. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zoubak S, Rynditch A, Bernardi G. Compositional bimodality and evolution of retroviral genomes. Gene. 1992;119:207–213. doi: 10.1016/0378-1119(92)90273-r. [DOI] [PubMed] [Google Scholar]