Abstract
Plant reproductive development is dependent on successful pollen-pistil interactions. In crucifers, the pollen tube must breach the stigma surface and burrow through the extracellular matrix of the stigma epidermal cells and transmitting tract cells before reaching its ovule targets. The high degree of specificity in pollen-pistil interactions and the precision of directional pollen tube growth suggest that signals are continually being exchanged between pollen/pollen tubes and cells of the pistil that line their path. However, with few exceptions, little is known about the genes that control these interactions. The specialized functions of stigma epidermal cells and transmitting tract cells are likely to depend on the activity of genes expressed specifically in these cells. In order to identify these genes, we used the Arabidopsis (Arabidopsis thaliana) ATH1 microarray to compare the whole-genome transcriptional profiles of stigmas and ovaries isolated from wild-type Arabidopsis and from transgenic plants in which cells of the stigma epidermis and transmitting tract were specifically ablated by expression of a cellular toxin. Among the 23,000 genes represented on the array, we identified 115 and 34 genes predicted to be expressed specifically in the stigma epidermis and transmitting tract, respectively. Both gene sets were significantly enriched in predicted secreted proteins, including potential signaling components and proteins that might contribute to reinforcing, modifying, or remodeling the structure of the extracellular matrix during pollination. The possible role of these genes in compatible and incompatible pollen-pistil interactions is discussed.
The stigma and transmitting tract of the style play critical roles in triggering, promoting, and guiding the growth of pollen tubes toward their ovule targets. In crucifers, the stigma epidermis is also the major site for intraspecific and interspecific pollen recognition. In Arabidopsis (Arabidopsis thaliana), the stigma is capped by approximately 150 finger-like cells, called papillar cells (Bowman et al., 1999). Analysis of the dry stigmas of other crucifers (Heslop-Harrison and Shivanna, 1977) has indicated that the surface of each papillar cell is covered by an interrupted layer of cutin, called the cuticle, and a superficial proteinaceous pellicle layer of poorly defined molecular composition (Gaude and Dumas, 1986; Hiscock et al., 2002), which may function in the adhesion and hydration of pollen grains through interactions with components of the pollen surface (Stead et al., 1980; Gaude and Dumas, 1986; Zinkl et al., 1999). For their part, cells of the transmitting tract that line the path of pollen tube growth are thought to produce molecules that contribute to the rapid rate and directionality of tube elongation (Sanchez et al., 2004). Despite the importance of these cells, however, the mechanisms that underlie successful pollen-pistil interactions are poorly understood.
Only a few genes have been identified in crucifers, which are expressed specifically in the stigma epidermis or transmitting tract, and the best-characterized genes are those that function in the recognition or response phases of self-incompatibility (SI) in obligate out-crossing species, such as Brassica and Arabidopsis lyrata (for review, see Kachroo et al., 2002). As a prelude to gain a molecular understanding of the function of stigma and transmitting tract cells, it is important to generate a near-complete catalog of genes expressed specifically in these cells. However, both types of cells are difficult to isolate by conventional microdissection methods in the numbers required for gene expression profiling. Several physical or biochemical methods for the isolation of specific cell or tissue types for RNA analysis have been reported recently (Karrer et al., 1995; Brandt et al., 1999; Birnbaum et al., 2003; Kerk et al., 2003). However, these methods typically produce only small numbers of cells and require amplification of the extracted RNA prior to microarray hybridization (Klur et al., 2004), a process that results in significant loss of information with only approximately 37% of the RNA being amplified.
An alternative to the direct isolation of specific cell types, one that avoids the use of specialized equipment as well as RNA amplification, is the use of strains that differ in the presence/absence of specific cell or tissue types. In one example of this approach, Arabidopsis homeotic mutants were used to enrich for genes expressed in specific floral organs (Scutt et al., 2003; Zik and Irish, 2003; Wellmer et al., 2004). Another example is the use of transgenic plants in which the specific cell type of interest is genetically ablated by expression of a cell-autonomous cytotoxin, such as diphtheria toxin subunit A (DT-A) or RNase under the control of a tightly regulated cell type-specific promoter. Genetic ablation has been shown to be an effective tool for killing or ablating specific cell types in plants (Mariani et al., 1990; Thorsness et al., 1991, 1993; Day and Irish, 1997). The rationale underlying the use of genetic ablation for the identification of cell type-specific transcripts is that such transcripts will be missing or very much reduced in samples ablated for this cell type relative to wild-type samples. In contrast, the levels of transcripts that are expressed in both ablated and neighboring cells will be equivalent in the two samples, because the number of ablated cells is usually small relative to other cells in the tissue samples. It should be noted, however, that genes identified by this approach might also be expressed in cell types or tissues not included in the analysis. Despite this caveat, the approach is promising and should be increasingly applicable to a variety of cell types as more tissue-specific promoters are identified.
We had previously used genetic ablation of Brassica stigma epidermal cells in conjunction with differential mRNA display and identified a few stigma epidermal cell-specific genes (Kang and Nasrallah, 2001). The availability of the Affymetrix Arabidopsis whole-genome ATH1 array, which contains probe sets corresponding to 23,000 Arabidopsis genes, allowed us to extend this approach in a genome-wide search for genes expressed specifically along the path of pollen tube growth. Here, we report on the generation and analysis of Arabidopsis pistils in which epidermal cells and transmitting tract cells were genetically ablated by expression of DT-A under the control of a promoter expressed specifically in these cells. Gene expression profiling of ablated and wild-type tissues identified 115 and 34 genes predicted to be expressed specifically in the stigma epidermis and transmitting tract, respectively. Both gene sets were significantly enriched in predicted secreted proteins that could function during pollination in the remodeling of the extracellular matrix (ECM) or as small signaling molecules. In addition, the stigma dataset included candidate orthologs of some, but not all, genes previously implicated in compatible or incompatible pollinations in other species.
RESULTS AND DISCUSSION
Genetic Ablation of the Stigma Epidermis and Transmitting Tract of the Style and Ovary
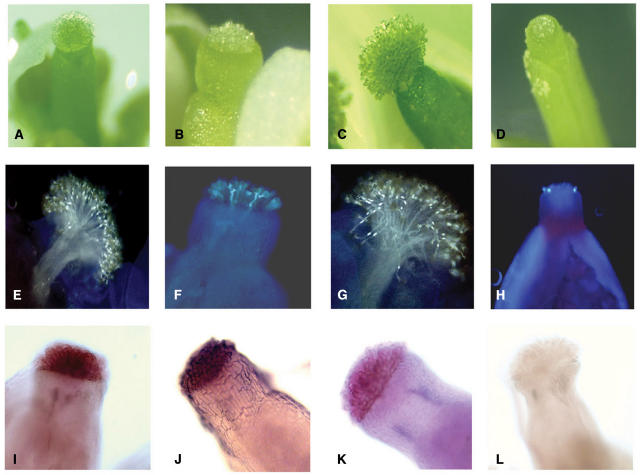
The promoter of ψSRK (At4g21370), the nonfunctional, but transcribed, Arabidopsis ortholog of the S-locus receptor kinase gene (SRK), which is the determinant of SI specificity in the stigma, was previously shown to be active exclusively in the stigma epidermis and transmitting tract of the style and ovary (Kusaba et al., 2001). Transformation of Arabidopsis with ψSRKpr::uidA::nos, a chimeric gene containing 1.6 kb of sequence 5′ of the ψSRK coding region (Kusaba et al., 2001), produced intense GUS staining in these cells and in no other cells of transformants (Kusaba et al., 2001). The same promoter fragment was fused to DT-A and used to transform Arabidopsis. The resulting transgenic plants developed normally, except that their stigmatic papillar cells and the transmitting tract cells of their stigmas, styles, and ovaries (i.e. the cells in which the ψSRK promoter is active) were ablated and nonfunctional. The most obvious defect was that the papillar cells of transgenic stigmas failed to elongate normally. At stage 13 of floral bud development (approximately 1 d before flower opening), ablated papillar cells were 3- to 5-fold shorter than wild-type papillar cells (compare Fig. 1, A and B), and the size difference was magnified by stage 14, when wild-type cells had elongated appreciably and were approximately 50 μm in length (compare Fig. 1, C and D). Unlike wild-type stigmas (Fig. 1, E and G), ablated stigmas failed to support normal pollen tube growth (Fig. 1, F and H). Pollen grains from untransformed plants or from DT-A transformants only occasionally hydrated and produced a pollen tube on the stunted stage 13 papillar cells (Fig. 1F), but they were never observed to germinate on stage 14 stigmas (Fig. 1H).
Figure 1.
Effects of genetic ablation and cell type-specific expression in Arabidopsis pistils. Images of stigmas at stage 13 of floral bud development in wild-type (A) and ablated (B) pistils. Images of stigmas at stage 14 of floral bud development in wild-type (C) and ablated (D) pistils. Fluorescent micrographs of pollinated stigmas from stage 13 wild-type (E) and ablated (F) floral buds, and from stage 14 wild-type (G) and ablated (H) floral buds. Stigmas were pollinated with wild-type pollen grains, and pollen tubes were allowed to develop overnight. Nonradioactive whole-mount in situ hybridizations of wild-type stage 13 pistils using antisense riboprobes specific for selected genes in the stigma dataset: At5g59810 (I), At2g02850 (J), and At5g19880 (K). A sense riboprobe negative control is shown in L.
Comparative Whole-Genome Transcriptional Profiling of Ablated and Wild-Type Tissues
Stigmas and ovaries were hand dissected from ablated and wild-type pistils before anther dehiscence from stage 13 floral buds to avoid contamination with pollen grains. Stigmas were excised by cutting the pistil just below the junction of stigma and style, and the ovary samples were obtained by cutting the pistil just above the junction of style and ovary. The stigma samples thus contained papillar cells and transmitting tract cells, while the ovary samples contained transmitting tract cells. Approximately 150 stigmas and 75 ovaries typically yielded approximately 10 μg of total RNA, an amount sufficient for microarray hybridization.
The RNA was used as a template for synthesis of biotin-labeled cRNA probes, which were subsequently hybridized to ATH1 whole-genome oligonucleotide arrays (Affymetrix, Santa Clara, CA). Two biological replicates of the hybridizations were performed, which permitted two pairwise comparisons of ablated and wild-type stigma or ovary samples and allowed evaluation of result reproducibility. The hybridization signals were highly reproducible, except for a few genes (see “Materials and Methods”), and correlation coefficients in the two stigma and ovary replicates were 0.83 and 0.70, respectively. Because gene expression in wild-type tissue was used as the baseline for comparisons to ablated tissue, a signal log ratio (SLR; a measure of the change in hybridization signal intensity between samples) value of 1 indicates a gene that shows a 2-fold increased transcript level in ablated samples relative to wild type, whereas a SLR of −1 indicates a gene that shows a 2-fold decreased transcript level in ablated samples relative to wild type. Genes that exhibited negative SLR values of at least −1 in the two biological replicates were considered to be candidate papillar cell-specific genes (stigma samples) or transmitting tract-specific genes (ovary samples).
By using the parameters outlined in “Materials and Methods,” approximately 63% of the probe sets, i.e. 14,000 genes, were called present in wild-type stigmas, and 161 genes reproducibly exhibited >2-fold decrease in hybridization signal only in ablated stigmas relative to wild-type stigmas, with 10 genes exhibiting reductions of 10-fold or greater (SLR > −3.4; Supplemental Table I). In the ovary samples, approximately 64% of the genes produced a hybridization signal in wild type, and 61 were found to exhibit reductions of 2-fold or greater only in ablated ovary samples (Supplemental Table II). Sixteen genes were also scored as being reduced in both the stigma and ovary samples, some of which might be expressed specifically in the transmitting tract. A total of 343 genes reproducibly gave hybridization signals that were >2-fold higher in the ablated stigma samples relative to wild-type stigma samples, and 62 genes produced a similar hybridization pattern in ovary samples (data not shown). These genes include several genes previously shown to be induced in programmed cell death, such as genes that function in the generation of reactive oxygen species, calmodulin- and calcium-dependent protein kinases, and genes of the ethylene biosynthetic or signaling pathways (Swidzinski et al., 2002). Therefore, it is likely that many of these genes are related to the programmed cell death response triggered by expression of the DT-A toxin (Day and Irish, 1997). It should be noted that the use of protoplasting, which is often used for isolation of specific cell types, also induces several hundred transcripts (Birnbaum et al., 2003). However, while the inclusion of several controls is essential for microarray data interpretation when protoplasting is used, an advantage of the genetic ablation strategy is that the artifactual gene set induced by expression of the toxin is readily distinguished from the candidate cell-specific gene set because of opposite directions of change in hybridization signal exhibited by the two datasets.
Candidate Stigma Epidermis- and Transmitting Tract-Specific Genes
After eliminating artifactual genes from consideration, we are left with 115 genes in the candidate stigma-specific gene set (hereafter designated the stigma dataset), and 34 genes in the candidate transmitting tract-specific gene set (hereafter designated the transmitting-tract dataset; Supplemental Table II). These genes are scattered throughout the Arabidopsis genome and do not generally occur in physical clusters, as is sometimes observed for coordinately regulated genes (Williams and Bowles, 2004), even in cases where two or more members of a gene family are predicted to have papillar cell- or transmitting tract-specific expression. Seventeen gene families were found in the stigma dataset, the largest being lipid transfer protein (LTP), peroxidase, and protein disulfide isomerase families, each of which includes three or four putative papillar cell-specific genes.
Tight physical linkage was observed only in At3g26450 and At3g26460, which encode proteins with similarity to major latex proteins. These two genes share 92% sequence similarity, suggesting that they arose by relatively recent gene duplication or were homogenized by gene conversion. Interestingly, with the exception of these major latex protein-related genes and three protein disulfide isomerase-like genes that shared >85% sequence similarity, gene family members in the stigma dataset were generally quite diverged from each other, suggesting that their products might have distinct activities or functions. Together with the absence of gene families in the transmitting tract dataset, this observation suggests a relatively low level of genetic redundancy among genes expressed specifically in the stigma epidermis and transmitting tract.
Validation of Microarray Results
The efficacy of the genetic ablation approach used here was initially confirmed by the fact that our datasets included 36 genes previously identified as being enriched in the carpels of floral homeotic mutants (Scutt et al., 2003; Wellmer et al., 2004). Furthermore, several genes or homologs of genes previously shown to be expressed specifically in subsets of pistil cells were correctly assigned in this study. Thus, the stigma dataset contained probable orthologs of Brassica papillar cell-specific genes, including At3g12000 (AtS1), At2g33850 (Brassica Pis63-1), At2g47550 (Brassica PPme), and At5g61790 (Brassica Pcnx; Dwyer et al., 1992; Robert et al., 1994; Kang and Nasrallah, 2001). The ovary dataset contained At1g72290, a gene previously identified as expressed specifically in the transmitting tract (Scutt et al., 2003). Notably, At1g69870, another gene previously reported to be expressed in all floral tissues with a relatively higher expression level in the transmitting tract and stigma epidermis (Scutt et al., 2003), did not exhibit differential hybridization signals between our wild-type and ablated ovary samples. It did show, however, reduced hybridization signals in ablated stigmas relative to wild-type stigmas, with a level of reduction (1.8-fold; SLR = −0.9) just below the cutoff used in this study. This result underscores the robustness of the genetic ablation strategy and of the criteria we used for identifying genes expressed specifically in stigma papillar cells or transmitting tract cells. It also indicates that the stringent criterion of a SLR = 1 cutoff value likely led to omission from our datasets of genes that, although not specifically expressed in papillar or transmitting tract cells, nevertheless exhibit increased expression in these cells relative to other cells within the pistil.
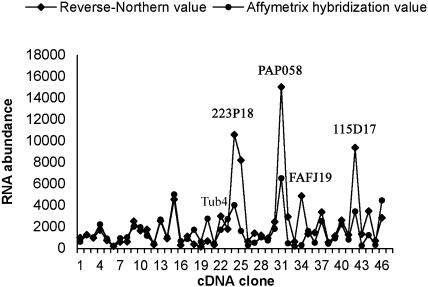
We then used the relatively high-throughput method of reverse northern-blot analysis (see “Materials and Methods”) as an independent means to examine the relative steady-state levels of transcripts in wild-type stigmas. Forty-five genes for which cDNAs are available from the Arabidopsis Biological Resource Center (ABRC) were amplified and subjected to gel-blot analysis using a cDNA probe synthesized from wild-type stigma RNA. After normalization of the signals relative to the β-tubulin4 gene, the reverse northern-blot data were found to be in agreement with the microarray data for most genes (Fig. 2). Three exceptions were genes encoding a peroxidase, a LTP, and a Pro-rich protein, all of which belong to gene families with several members expressed in stigmas. Thus, the discrepancy observed for these genes between the reverse northern-blot and microarray data is most likely due to cross-hybridization of closely related gene family members in the reverse northern-blot analysis.
Figure 2.
Comparison of hybridization signals obtained on the ATH1 array and by reverse northern-blot analysis for 45 genes from the stigma dataset. cDNA clones were amplified and arrayed as described in “Materials and Methods.” Each data point represents the hybridization signal produced by one cDNA clone. RNA abundance is shown in arbitrary numbers. The β-tubulin4 (Tub4) signal was used to normalize the signals. Accession numbers are shown for cDNAs that produced higher signals on the reverse northern-blot analysis than on the ATH1 array.
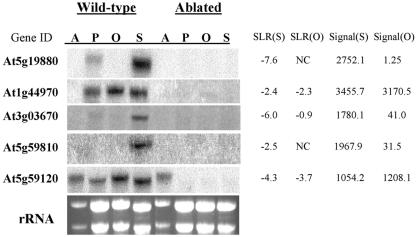
Direct validation of our results was obtained by RNA gel-blot analysis for five previously uncharacterized genes. These five genes, which belong to the peroxidase gene family (At5g19880, At1g44970, and At3g03670) and the subtilisin gene family (At5g59810 and At5g59120), were representative of the genes identified in this study. They produced different hybridization signals in wild-type stigmas (ranging from 1,054–3,455), and they also exhibited different levels of reduction in hybridization signal in ablated stigmas relative to wild-type stigmas (ranging from 5-fold [SLR = −2.4 for At1g44970] to 194-fold [SLR = −7.6 for At5g19880]). As shown in Figure 3, RNA gel-blot analysis with gene-specific probes (see “Materials and Methods”) detected transcripts derived from these genes in wild-type tissue, but not in ablated tissue, as expected. Importantly, transcripts of At5g19880, At3g03670, and At5g59810, which were identified only in the stigma dataset, were detected in stigma, but not ovary, RNA. In contrast, At1g44970 and At5g59120 were identified in both the stigma and ovary datasets, and their transcripts were detected at relatively high levels in both stigma and ovary RNA (Fig. 3). Furthermore, for four of the five genes, transcripts were detected only in pistils and not in anthers. At5g59120 transcripts, on the other hand, were also detected in anthers of ψSRK pr::DT-A transformants and wild-type plants, underscoring the fact that the genetic ablation strategy can only inform on specificity of gene expression within the pistil.
Figure 3.
RNA gel-blot analysis of selected genes from the stigma dataset. RNA was extracted from anthers (A), whole pistils (P), ovaries (O), and stigmas (S) of wild-type and ablated plants. Ten micrograms of total RNA were loaded in each lane and blotted RNA was hybridized with probes specific for each of the listed genes. The intensity of ethidium bromide staining of ribosomal RNA served as a loading control. To the right of each image are the array hybridization signals (averages of two replicates) obtained with wild-type stigma (signal S) and ovary (signal O), and the SLR values (averages of two replicates) calculated for the stigma [SLR(S)] and ovary [SLR(O)] wild-type versus ablated comparisons. NC, No change. The SLR(O) of −0.9 scored for At3g03670 is not reflected in differential hybridization signals on gel blots of wild-type and ablated ovary RNA because the low transcript abundance (signal of 41.0 in wild-type ovary) is below the limits of detection of the RNA gel blot.
Indeed, a search of the Arabidopsis UniGene database, a compilation of all known expressed sequence tag (EST) data containing clusters of all EST sequences originating from individual genes along with the tissue from which the EST sequences were derived, showed that many of the genes in our datasets are expressed in other plant tissues. However, for 19 of the 115 putative papillar cell-specific genes and 7 of the 34 putative transmitting tract-specific genes, ESTs were found only in floral organ libraries (Supplemental Tables I and II). Within the limits of this analysis, these genes are therefore likely to function exclusively in the reproductive phase of plant development, possibly only in the stigma or transmitting tract. Twenty-nine other genes were not represented in any UniGene tissue library (Supplemental Tables I and II). These genes produced hybridization signals that ranged from 64 to 3,602, with six genes producing average signals greater than 400, a signal typically produced by relatively abundant transcripts. For these six genes at least, a likely explanation for their absence from the UniGene database is that they are expressed exclusively in stigma epidermis or transmitting tract cells, since these cells would represent only a small fraction of cells included in available floral organ cDNA libraries.
Cell Type-Specific Expression Patterns Confirmed by Whole-Mount In Situ Hybridization
As further confirmation that our inferences relative to cell type-specific expression were valid, three-dimensional whole-mount in situ hybridization was performed for a subset of genes to determine the actual cellular localization of their transcripts. Three genes from the stigma dataset were selected that exhibited relatively high hybridization signals, ensuring that their transcripts could be detected by the in situ hybridization method. Additionally, one gene (At2g02850) is the probable ortholog of a gene previously implicated in pollination in lily (Lilium longiflorum; see below). As illustrated in Figure 1, I to K, the transcripts of At5g59810, At2g02850, and At5g19880, all of which produced negative SLRs in stigma samples only, were restricted to the stigma papillar cells.
These results confirm our predictions of cell type-specific expression for the genes in the stigma and transmitting tract datasets. Within each dataset, the genes are expected to be coordinately regulated and they may therefore share promoter motifs responsible for their cell type-specific expression. However, analysis of 1,000-bp regions located upstream of the start codons of the genes using motif analysis (http://www.arabidopis.org) and PLACE (http://www.dna.affrc.go/PLACE) failed to identify conserved cis-acting elements, suggesting that different regulatory elements can direct cell type-specific expression in the pistil.
Functional Classification of Candidate Stigma Epidermis- and Transmitting Tract-Specific Genes
The genes identified in this study may be grouped into nine functional categories (Fig. 4) based on gene annotations in the Munich Information Center for Protein Sequences Arabidopsis DataBase (MIPS AtDB) and The Arabidopsis Information Resource (TAIR), and, in some cases, on published empirical studies. As with other genome-wide transcript-profiling studies (e.g. Zhu et al., 2001), the largest category in all three datasets consisted of genes encoding unknown proteins and the next largest category consisted of genes for various metabolic pathways. Defense-related genes also comprised a major category in our datasets, consistent with previous studies that documented the increased resistance of floral organs to pathogen invasion and the constitutive expression of several defense-associated proteins in pistils (Kononowicz et al., 1992; de Oliveira et al., 1993; Delp and Palva, 1999; Scutt et al., 2003; Lan et al., 2004). It should be noted, however, that these defense-related genes might function not only in defense against pathogens, but also in response to pollination. Incompatible pollen-pistil interactions are sometimes associated with the accumulation of intermediates of the phenylpropanoid pathway (Elleman and Dickinson, 1999), which also occurs in the plant's response to pathogens and stress (Schuler and Werck-Reichhart, 2003). Consistent with this observation, the stigma dataset included a cytochrome P450 gene (CYP75B1; At5g07990) that encodes a flavonoid 3′-hydroxylase, a key enzyme in the phenylpropanoid pathway, and two genes (At1g65060 and At3g21230) for 4-coumarate:CoA ligase, which catalyzes the last step of the phenylpropanoid pathway.
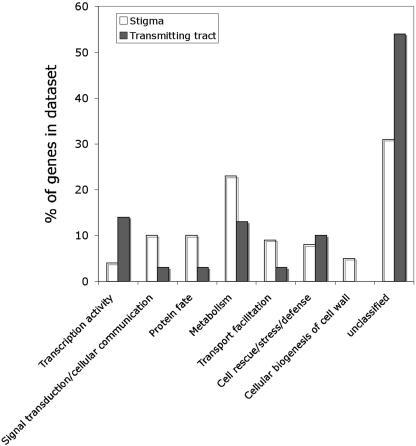
Figure 4.
Comparison of functional gene categories in the stigma and transmitting tract datasets. The bars indicate the number of genes assigned to each functional category as a percentage of the total number of genes in each dataset. The transmitting tract dataset contained no genes in the “cellular biogenesis of cell wall” category.
Comparison of the gene complements in the stigma and transmitting tract datasets reveals significant differences in the representation of genes within the various functional categories (Fig. 4). The stigma dataset is enriched for genes predicted to function in signal transduction, cell communication, protein fate, and transport relative to the transmitting tract dataset, but it contains a significantly lower proportion of transcription-related genes both relative to the transmitting tract (4% versus 14%) and the Arabidopsis genome as a whole (4% versus approximately 17%). Furthermore, both datasets are enriched for genes with unclassified function (especially the transmitting tract dataset) and relatively poorly populated with genes predicted to function in photosynthesis, the cell cycle, and nucleic acid biosynthesis.
The ECM of the Stigma and Transmitting Tract, Signaling, and Pollen-Pistil Interactions
One group of genes that is overrepresented in our datasets consists of genes predicted to encode proteins with N-terminal signal peptides, which would enter the secretory pathway. While only 17.6% of all genes in the Arabidopsis genome are predicted to code for proteins with signal peptides (Arabidopsis Genome Initiative, 2000), TargetP predictions (Emanuelsson et al., 2000) showed that such genes comprised 36% (41 genes) and 29% (10 genes) of genes in the stigma and transmitting-tract datasets, respectively (Tables I and II). Among these, three genes encode proteins containing both a signal sequence and a glycosylphosphatidylinositol (GPI) anchor and are potentially located in the cell wall in close association with the plasma membrane. Additionally, two genes encode proteins containing a GPI anchor, but no signal peptide, indicative of a possible association with the cytoplasmic face of the plasma membrane. Thus, a significant proportion of genes expressed in cells along the path of pollen tube growth potentially function in the endoplasmic reticulum or cell surface, encoding either proteins targeted to the cell surface (plasma membrane or ECM) or proteins that function in the processing of secreted proteins (such as protein disulfide isomerases and calnexin). The number of these genes is likely to be even larger because signal peptide-predicting programs can fail to identify a significant number of secreted proteins. Interestingly, a similar enrichment in genes for secreted proteins was also observed among a subset of genes predicted to function specifically in reproductive development (Hennig et al., 2004).
Table I.
Papillar cell-specific genes predicted to encode secreted proteins
| Affy ID | AGI | SLRa | Gene Descriptionb | Length of Small (<200 Amino Acid) Proteinsc |
|---|---|---|---|---|
| 247494_at | At5g61790 | −1.3 | Calnexin-like protein | |
| 251840_at | At3g54960 | −1.3 | Protein disulfide isomerase | |
| 262504_at | At1g21750 | −1.5 | Putative protein disulfide isomerase | |
| 251182_at | At3g62600 | −1.3 | Putative DnaJ-like protein | |
| 245956_at | At5g28540 | −1.2 | Luminal binding protein | |
| 255690_at | At4g00360 | −2.4 | Putative cytochrome P450/CYP86A2 | |
| 246145_at | At5g19880 | −7.6 | Putative peroxidase | |
| 254914_at | At4g11290 | −3.3 | Putative peroxidase | |
| 259197_at | At3g03670 | −6.0 | Putative peroxidase | |
| 245151_at | At2g47550 | −1.9 | Putative pectinesterase | |
| 252971_at | At4g38770 | −2.0 | Extensin-like | |
| 258805_at | At3g04010 | −1.7 | Putative β-1,3-glucanase | |
| 255822_at | At2g40610 | −2.2 | Putative expansin/AtEXP8 | |
| 261226_at | At1g20190 | −4.5 | Putative expansin/AtEXP11 | |
| 254403_at | At4g21323 | −2.5 | Subtilisin-like protease | |
| 249856_at | At5g22980 | −1.3 | Putative Ser carboxypeptidase III | |
| 267253_at | At2g22960 | −2.3 | Putative Ser carboxypeptidase I | 184 |
| 251838_at | At3g54940 | −1.9 | Putative Cys proteinase | |
| 247686_at | At5g59700 | −1.5 | Putative receptor-like protein kinases | |
| 245080_at | At2g23300 | −1.3 | Putative receptor-like protein kinases | |
| 250437_at | At5g10430 | −1.5 | AGP (GPI anchor) | 135 |
| 251065_at | At5g01870 | −1.4 | Putative LTP | 116 |
| 256145_at | At1g48750 | −1.6 | Putative LTP | 94 |
| 261385_at | At1g05450 | −4.1 | Putative LTP | |
| 267472_at | At2g02850 | −1.2 | Putative plantacyanin | 129 |
| 249748_at | At5g24620 | −1.4 | Putative thaumatin-like | |
| 254226_at | At4g23690 | −2.5 | Disease resistance protein | 187 |
| 251746_at | At3g56060 | −2.8 | Mandelonitrile lyase-like protein | |
| 261485_at | At1g14360 | −1.7 | Putative UDP-galactose transporter | |
| 256636_at | At3g12000 | −4.4 | S-locus related protein SLR1 homolog/AtS1 | |
| 249073_at | At5g44020 | −3.2 | Vegetative storage protein-like | |
| 267483_at | At2g02810 | −2.0 | Unknown protein | |
| 262503_at | At1g21670 | −1.8 | Hypothetical protein | |
| 264382_at | At2g25110 | −1.3 | Unknown protein | |
| 253437_at | At4g32460 | −1.7 | Putative protein | |
| 253687_at | At4g29520 | −1.2 | Putative protein | |
| 254372_at | At4g21620 | −1.4 | Putative protein | 131 |
| 260553_at | At2g41800 | −2.4 | Unknown protein | |
| 262750_at | At1g28710 | −2.1 | Unknown protein | |
| 267459_at | At2g33850 | −2.6 | Unknown protein | |
| 264774_at | At1g22890 | −1.3 | Unknown protein | 73 |
SLR, Average SLR of two independent replicates.
Gene description according to TAIR and the Affymetrix analysis center.
Protein length in amino acids is shown only for proteins predicted to be <200 amino acids in length.
Table II.
Transmitting tract-specific genes predicted to encode secreted proteins
| Affy ID | AGI | SLRa | Gene Descriptionb | Length of Small (<200 Amino Acid) Proteinsc |
|---|---|---|---|---|
| 255054_at | At4g09740 | −3.8 | Endo-β-1,4-glucanase; cellulase | |
| 254543_at | At4g19810 | −2.7 | Putative chitinase | |
| 258918_at | At3g10560 | −2.1 | Putative cytochrome P450/CYP77A3 | |
| 252897_at | At4g39480 | −1.4 | Cytochrome P450/CYP96A1 | |
| 259799_at | At1g72290 | −2.0 | Putative drought-induced protein | |
| 262454_at | At1g11190 | −3.5 | bfn1 like protein | |
| 263482_at | At2g03980 | −1.6 | Putative lipase/hydrolase | |
| 255575_at | At4g01430 | −2.2 | Nodulin/MtN21 | |
| 254781_at | At4g12840 | −1.5 | Putative protein | |
| 264636_at | At1g65490 | −1.3 | Hypothetical protein | 88 |
SLR, Average SLR of two independent replicates.
Gene description according to TAIR and the Affymetrix analysis center.
Protein length in amino acids is shown only for proteins predicted to be <200 amino acids in length.
Three classes of signal peptide-containing genes that might function in the development or pollination-related functions of pistil cells are discussed here.
The Stigma Cuticle
The stigma cuticle presents a barrier to pollen hydration and germination, and this barrier must be breached by the action of cutinases (esterases) from the stigma pellicle and pollen grain surface (Hiscock et al., 2002). Removal of the stigmatic pellicle or treatment of stigmas with a cutinase inhibitor inhibits pollen tube penetration through the stigma cell wall (Heslop-Harrison and Heslop-Harrison, 1975; Hiscock et al., 2002). Furthermore, the heavily cutinated surface of leaf epidermal cells cannot support pollen tube growth, except in cuticle-defective mutants and cutinase-expressing transgenic Arabidopsis (Lolle and Cheung, 1993; Sieber et al., 2000).
Several genes that might function in the biogenesis of the stigma cuticle were identified in the stigma dataset. These include LTPs, of which one proposed function is the transport of cutin monomers to the cell surface (Kader, 1996), and two genes previously implicated in the biosynthesis of cutin in epidermal cells of Arabidopsis: the At1g49430 gene, whose product, LACS2, exhibits long-chain acyl-CoA synthetase activity (Schnurr et al., 2004), and At4g00360 (CYP86A2), which shares 87% amino acid sequence similarity with the fatty acid ω-hydroxylase LACERTA (LCR; CYP86A8; Wellesen et al., 2001).
ECM Modification and Remodeling
Successful pollinations in crucifers are associated with the expansion or loosening of the stigmatic papillar cell wall and the transmitting tract ECM to accommodate the growing pollen tube. However, little is known about the mechanism of wall expansion and about the nature and origin (pollen coat, stigma epidermis, or both) of the signals that trigger ECM remodeling during pollination or stigma maturation. Transmitting electron microscopic images have shown that pollen tubes invade the expanded papillar cell wall by growing between its outer and inner layers (Kandasamy et al., 1994), and that application of compatible pollen coating initiates wall expansion and clustering of endoplasmic reticulum stacks and Golgi bodies beneath the site of papillar wall expansion (Robert et al., 1984).
Our datasets contained several genes for putative cell wall-localized enzymes that might contribute to ECM modification during pollination. The stigma epidermis appears to be particularly enriched in gene products that might function in regulating pollen tube penetration through the stigma papillar cell wall (Table I). Among these proteins are two α-expansins, a pectinesterase, three peroxidases, and several proteases (a subtilisin-like Ser protease, two Ser carboxypeptidases, and a Cys-type peptidase). Because of the documented activity of their better studied relatives in the disassembly of ECM structural components, some of these proteins (e.g. the expansins and proteases) may be assumed to facilitate pollen tube growth by loosening the ECM or by generating or modifying precursors or signals for directional tube growth.
In the case of the putative pectinesterase and peroxidases, however, roles in either wall loosening or rigidification are possible. For example, cell wall-localized peroxidases can cause an increase in wall extensibility by generating hydroxyl radicals that degrade cell wall polysaccharides (Schopfer, 2001) or they can increase wall rigidity by mediating rapid cross-linking (insolubilization) of the structural cell wall Pro-rich extensins, as occurs upon physical damage, treatment with fungal elicitors, and pathogen infection (Bradley et al., 1992; Brownleader et al., 2000; Jackson et al., 2001). The presence of stigma surface peroxidases has been used as an indication of stigma receptivity for pollination in several species (e.g. see Galen and Plowright, 1987), implicating peroxidases in wall loosening during pollination or stigma maturation. However, it is tempting to speculate that a papillar cell-specific peroxidase activity might function to strengthen the epidermal cell wall, possibly by cross-linking the extensin-like At4g38700 protein product, which is the most highly expressed of the structural cell wall proteins in the stigma dataset.
Cell-Cell Communication and Signal Transduction
Genes predicted to function in cell-cell communication and signal transduction are of particular interest in the context of pollen-pistil interactions. Consistent with the primary recognition role of the stigma in discriminating between pollen grains, the stigma dataset contained a significant proportion of genes (10%) in this category. Most noteworthy are genes encoding putative receptor-like protein kinases (RLK), which, by virtue of their predicted plasma membrane localization, might interact with ligands held on the pollen coat or pollen tube surface. Two RLKs were identified in the stigma dataset, At2g23300 and At5g59700, which belong to the Leu-rich repeat and Cys-rich protein kinase subfamilies (Shiu and Bleecker, 2003), respectively. A third RLK gene, At1g66880 (a member of the LRK10L subfamily; Shiu and Bleecker, 2003), was identified in the transmitting tract dataset. However, this RLK gene may function at least partly in processes unrelated to pollination because it has been shown to be a stress-responsive gene in previous microarray experiments (Zipfel et al., 2004).
Also of interest are genes that encode small secreted proteins that might function as signaling molecules, as precursors for peptide hormones, or as ligands for receptors located on the surfaces of pollen grains or pistil cells. The importance of small proteins has been documented in several systems, for example, the self-incompatibility response of crucifers (Kachroo et al., 2002; Takayama et al., 2001), meristem development (Gross-Hardt and Laux, 2003), and defense (Pearce and Ryan, 2003). In addition, several small secreted pistil proteins have recently been shown to interact with pollen-localized RLKs or to function in pollen adhesion or tube growth and guidance. In tomato, LeSTIG1, a small Cys-rich protein located in the style interacts with pollen RLKs (Tang et al., 2004). In lily, chemocyanin, a small basic protein related to plantacyanins, functions as a pollen tube attractant in the stigma (Kim et al., 2003), and a LTP, stigma/style Cys-rich adhesin, facilitates adhesion of pollen tubes to the stylar transmitting tract cells and also enhances the action of chemocyanin in the stigma (Park et al., 2000; Kim et al., 2003).
Thirty of the 115 genes in the stigma dataset are predicted to encode proteins smaller than 200 amino acids in length, eight of which contain signal peptides (Table I). Included in this set of genes is AtAGP4 (At5g10430), a member of the arabinogalactan protein (AGP) gene family, some members of which are thought to function in cell-cell interactions (Majewska-Sawka and Nothnagel, 2000). Interestingly, AGPs are enriched in the exudates of stigmas from several species, where they might function to enhance pollen tube growth (Wu et al., 2000). We also identified a plantacyanin gene and four LTP genes (Table III), but not a candidate ortholog of LeSTIG1. While the mechanisms underlying pollen-pistil interactions might not be conserved between the highly diverged lily and Arabidopsis taxa, it is possible that plantacyanin and at least one of the LTP genes have functions similar to those fulfilled by their counterparts in the lily stigma. This is particularly likely for plantacyanin (At2g02850), a gene with no related paralogs in the Arabidopsis genome, which is expressed both at high levels (based on microarray hybridization signal) and specifically (see Fig. 1J, in situ hybridization) in Arabidopsis stigma epidermal cells. Of the four putative secreted LTPs identified in this study, At2g38530 (LTP2) and At5g01870 (LTP6-like) are most similar in sequence to lily stigma/style Cys-rich adhesin. However, the sequence similarity between the Arabidopsis and lily genes is only approximately 40%, and it remains to be determined whether either one or both of these genes function as a stigma adhesin.
Table III.
Genes previously implicated in the pollen-pistil interactions of Brassica, Arabidopsis, and lily
*, LTP.
| Gene
|
Arabidopsis Gene ID
|
Hybridization Signala
|
SLRb
|
Unigene Librariesc
|
||
|---|---|---|---|---|---|---|
| Ovary | Stigma | Ovary | Stigma | |||
| POP2 | At3g22200 | 791 | 770 | −0.25 | −0.25 | |
| SCA | At2g38540* | 5115 | 6444 | 0.2 | −0.3 | |
| At5g59320* | 4000 | 1042 | 0.2 | 0.35 | ||
| At5g59310* | 983 | 505 | 0.0 | 0.1 | ||
| Chemocyanin | At2g02850 | 4977 | 2735 | 0.3 | −1.2 | |
| ARC1 | At1g29340 | 316 | 500 | 0.2 | −0.2 | R, W, L, SF |
| MLPK | At2g28930 | 343 | 347 | 0.0 | −0.75 | R, W, SF, FB |
| THL1 | At5g42890 | 512 | 1853 | −0.2 | −0.8 | R, L, I, SF |
| THL2 | At1g19730 | 581 | 380 | −0.1 | 0.15 | SF, FB |
| KAPP | At5g19280 | 132 | 138 | −0.3 | −0.15 | R, W, S, G |
| SNX1 | At5g06140 | 4762 | 321 | 0.0 | −0.15 | R, W, L, S, SF |
| AtS1d | At3g12000 | 17 | 5122 | −0.2 | −4.4 | I |
Values are averages of array hybridization signals obtained in two independent replicates of stigma and ovary samples.
The SLRs shown are averages of two replicate experiments.
Unigene cDNA libraries containing ESTs for the genes listed: R, Roots; W, whole plants including stages from germination to seed; L, leaf or rosette; SF, mix of silique and fowers; FB, flower buds; S, seed; G, green silique; I, inflorescence.
AtS1 is shown as an example of a gene known to exhibit papillar cell-specific expression.
Pistil Genes Previously Implicated in the Pollen-Pistil Interactions of Crucifers
As stated earlier, very few pistil genes that function in pollination have been identified in crucifers. One gene, POP2 (At3g22200), which has been implicated in pollen tube guidance, encodes a transaminase that might degrade γ-amino butyric acid (GABA), thus creating a GABA gradient that increases from stigma to micropyle and guides the pollen tube to the ovules (Palanivelu et al., 2003). While the GABA gradient must be perceived by the pollen tube as it grows within the transmitting tract, the POP2 protein was immunolocalized to all cells of the pistil (Palanivelu et al., 2003). Consistent with this expression pattern, POP2 exhibited a <1-fold reduced hybridization signal in ablated tissues relative to wild-type tissues (Table III), and it was therefore excluded from our stigma and transmitting tract datasets.
Other than POP2, all other pistil genes with proven or suggested roles in the pollen-pistil interactions of crucifers are related to the recognition or signaling phases of the SI response. The best studied of these genes is SRK. The highly polymorphic SRK is activated upon self-pollination by allele-specific interaction with its cognate pollen coat-localized ligand, the S-locus Cys-rich protein (SCR; for review, see Kachroo et al., 2002). This interaction triggers a signaling cascade within the stigma papillar cells, which culminates in the inhibition of pollen germination or pollen tube growth into the epidermal cell wall. Arabidopsis is a self-fertile species with no known occurrence of SI and it contains nonfunctional versions of SRK and SCR (Kusaba et al., 2001). However, we had previously introduced the SI trait into this species by expression of an SRK/SCR gene pair isolated from self-incompatible A. lyrata (Nasrallah et al., 2002, 2004). This result demonstrates that downstream components of the SRK signal transduction pathway are functional in Arabidopsis. It was of interest, therefore, to determine whether the probable Arabidopsis orthologs of genes previously shown or suggested to have SI-related functions were represented in our stigma dataset.
The Arabidopsis ψSRK gene (At4g21370), which provided the promoter for our genetic ablation study, is not represented on the ATH1 array. The array did contain elements corresponding to genes that produced the best hits in BLAST searches with several Brassica SI-associated genes. Among these are M-locus protein kinase (MLPK; Murase et al., 2004) and arm repeat containing (ARC1; Gu et al., 1998; Stone et al., 1999), both of which are predominantly expressed in the stigma and are reportedly required for full expression of the SI trait, albeit by unknown mechanisms. In addition, several genes were identified as SRK kinase-domain interactors in yeast (Saccharomyces cerevisiae), including thioredoxin-like (THL1 and THL2), kinase-associated protein phosphatase (KAPP), and Bo SNX1 (nexin), all of which are expressed in a variety of plant tissues and have no proven role in SI (Bower et al., 1996; Vanoosthuyse et al., 2003). As shown in Table III, none of the Arabidopsis genes exhibited significantly reduced signals in ablated stigmas relative to wild-type stigmas, suggesting that these genes are not specific to the stigma epidermis within the pistil. Consistent with this conclusion, the genes exhibited equivalent hybridization signals in stigma and ovary samples, except for the THL1-like gene, which produced a higher signal in stigma relative to ovary samples. The SNX1-like gene is even expressed at significantly higher levels in ovaries than in stigmas (Table III). Furthermore, the occurrence of ESTs for these genes in cDNA libraries derived from vegetative tissues are indicative of ubiquitous expression (Table III). In the case of At1g29340 and At2g28930, which exhibit, respectively, 77% and only 52% overall amino acid identity to Brassica MLPK and ARC1, these observations are not consistent with the stigma-predominant expression of their Brassica counterparts. It is possible that At1g29340 and At2g28930 may have assumed or retained pollination-unrelated functions since the divergence of the Arabidopsis and Brassica lineages. Alternatively, these genes may not be the functional orthologs of Brassica ARC1 and MLPK, in which case other genes expressed in the Arabidopsis stigma epidermis must fulfill whatever role ARC1 and MLPK have in the Brassica SI response.
CONCLUSION
The genetic ablation approach used here successfully identified genes expressed specifically along the path of pollen tubes. Most of these genes were not previously studied or reported to be expressed in stigma or transmitting tract cells, and their potential involvement in pollination remains to be determined. The number of predicted papillar cell-specific genes is more than 3 times larger than that of predicted transmitting tract-specific genes, suggesting a higher degree of functional specialization in the stigma epidermis than in the transmitting tract. While the biological roles of these genes have yet to be determined, it is likely that at least some of the papillar cell-specific genes have functions related to the development of the stigma epidermis, in pollen recognition, or in the promotion of adhesion, hydration, and germination of pollen grain. The transmitting tract-specific genes might function in the development of transmitting tract cells or in the promotion and guidance of the pollen tube. Future functional characterization of these genes, using the molecular and genetic resources available in Arabidopsis, promises to elucidate mechanisms underlying compatible pollen-pistil interactions, which so far have been recalcitrant to genetic and molecular approaches. Based on this study and several other recent genome-wide expression studies of Arabidopsis floral tissues, a detailed knowledge of transcription in reproductive tissues is emerging. The increased resolution provided by studies in which specific reproductive cell types are targeted for analysis, such as the one described here, will be required to extend this knowledge and elucidate the transcriptional programs of each of the highly specialized cell types that together contribute to the success of plant reproduction.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants of the C24 ecotype were transformed with ψSRK::DT-A, a chimeric gene consisting of the promoter of the ψSRK (At4g21370; Kusaba et al., 2001) fused to DT-A by Agrobacterium tumefaciens-mediated transformation. Transgenic plants were selected on media containing kanamycin and examined microscopically to determine the efficiency of cell ablation. Arabidopsis C24 wild-type plants and ψSRK::DT-A transformants were grown under the same conditions for tissue collection.
RNA Isolation and Probe Labeling
Tissue for microarray analysis was collected from floral buds of wild-type and ablated plants at 1 d before anther dehiscence to avoid pollen contamination. The stigma samples were collected by cutting the pistil just below the base of the stigma, and the remainder of the pistil was used for the ovary samples. Total RNA was extracted using the DNA/RNA isolation kit (Qiagen, Valencia, CA). The yield and purity of total RNA were determined spectrophotometrically and visualized by denaturing formaldehyde gel electrophoresis. Five micrograms of total RNA were used for cDNA synthesis with the SuperScript double-stranded synthesis kit (Invitrogen, Carlsbad, CA) and an HPLC-purified poly (T)-nucleotide primer containing a sequence recognized by T7 RNA polymerase (GeneSet Oligos, La Jolla, CA). The double-stranded cDNA was purified according to the recommendations found in the “GeneChip Sample Cleanup Module” (Affymetrix, Santa Clara, CA) and used to generate biotinylated cRNA by in vitro transcription using T7 RNA polymerase from the BioArray high yield RNA transcript labeling kit (ENZO, Farmingdale, NY). All labeled cRNAs were purified, randomly fragmented to molecules of 200 nucleotides or less, and checked for degradation by agarose gel electrophoresis.
Affymetrix GeneChip Hybridization and Data Analysis
Hybridization to Affymetrix Arabidopsis ATH1 full-genome arrays using the biotinylated and fragmented cRNAs was performed by the Microarray Core Facility at the University of Pennsylvania (Philadelphia). The conditions for hybridization, washing, and staining with streptavidin-phycoerythrin were as described in the “Affymetrix GeneChip Expression Analysis” technical manual.
Affymetrix Microarray Suite 5.0 software was used to measure expression levels for target genes and the default values provided by Affymetrix were applied to all analysis parameters. The signal value, a relative measure of expression level, was computed for each assayed gene. Global scaling was applied to allow comparison of gene signals across multiple microarrays. The average total chip signal was calculated and used to determine what scaling factor was required to adjust the chip average to an arbitrary target of 150. All signal values from one microarray were then multiplied by the appropriate scaling factor. For each pairwise comparison, a quantitative estimate of the change in hybridization signal intensity for each probe set in ablated samples relative to wild-type samples was generated in the form of a SLR, which also estimates the direction of change of a signal (increase or decrease). Only the probe sets called present and marginal from wild-type tissue hybridization were considered as expressed in these tissues. Average values of SLRs and signal levels from duplicate arrays were used to evaluate the data.
The extent of reduction in hybridization signal was reproducible between the biological replicates for most genes, except that 30 genes in the stigma dataset and 11 genes in the ovary dataset produced erratic results, with greater than 2-fold fluctuations in the SLRs calculated for the two replicates. These genes, which included all nine of the heat shock proteins represented in the dataset, are all potentially involved in stress or wounding responses, suggesting that subtle differences in the growth or handling of tissues produced different levels of stress in the replicate experiments.
Genes were grouped into functional categories based on the annotation from TAIR and MIPS.
Generation of an EST Blot and Reverse Northern-Blot Hybridization
EST inserts were amplified from cDNA clones obtained from the ABRC using M13 forward with M13 reverse or T7 primers. β-Tubulin4 was used as expression control. An EST blot was generated by running equal amounts of amplified products on an agarose gel followed by transfer to GeneScreen Plus (DuPont, New England Nuclear, Boston) membrane. For reverse northern-blot analysis, the membrane was hybridized with a stigma cDNA probe prepared from 10 μg of total stigma RNA by first-strand cDNA synthesis in a 30-μL reaction containing 750 ng oligo(dT) primer, 0.75 mm each dT/G/CTP, 250 μCi radiolabeled 32P-dATP, and SuperScript II reverse transcriptase (Invitrogen).
After heating the cDNA probe at 94°C for 5 min, hybridization was allowed to proceed for at least 16 h. Hybridization conditions, posthybridization washes, and visualization and analysis of hybridization patterns were as described for RNA gel-blot analysis. Hybridization signals were normalized using β-tubulin4.
Total RNA Extraction, RNA Gel-Blot Analysis, and Whole-Mount In Situ Hybridization
Total RNA was extracted using the TRIzol Reagent (Invitrogen) from stigmas and ovaries 1 d before flower opening, as well as from anthers, leaves, and roots. Total RNA was quantified spectrophotometrically, separated on 1.2% (w/v) denaturing formaldehyde agarose gels, and transferred to GeneScreen Plus membrane (DuPont). The blots were prehybridized and hybridized at 65°C in 10% (w/v) dextran sulfate, 330 mm sodium phosphate, pH 7.0, 10 mm EDTA, and 5% (w/v) SDS. The probes used for hybridization were PCR products amplified with gene-specific primers, after confirming their specificity by BLAST searches of the Arabidopsis genome sequence. Washing was performed at low stringency in 2× SSC (1× SSC [0.15 m NaCl and 0.015 m sodium citrate]), 0.1% (w/v) SDS at 65°C (two washes of 15 min each) and high stringency in 0.2× SSC, 0.1% (w/v) SDS at 65°C (two washes of 10 min each). The hybridized membranes were exposed to storage phosphor screens (Molecular Dynamics, Sunnyvale, CA), scanned in a Storm 860 scanner (Molecular Dynamics), and the images were analyzed using ImageQuant software (Molecular Dynamics).
Whole-mount in situ hybridization was performed essentially according to Zachgo et al. (2000).
Supplementary Material
This work was supported by a grant from the U.S. Department of Agriculture.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060558.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bower MS, Matias DD, Fernandes-Carvalho E, Mazzurco M, Gu T, Rothstein SJ, Goring DR (1996) Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 8: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Baum SF, Eshed Y, Putterill J, Alvarez J (1999) Molecular genetics of gynoecium development in Arabidopsis. Curr Top Dev Biol 45: 155–205 [DOI] [PubMed] [Google Scholar]
- Brownleader MD, Hopkins J, Mobasheri A, Dey PM, Jackson P, Trevan M (2000) Role of extensin peroxidase in tomato (Lycopersicon esculentum Mill.) seedling growth. Planta 210: 668–676 [DOI] [PubMed] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ (1992) Elicitor- and wound- induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid, defense response. Cell 70: 21–30 [DOI] [PubMed] [Google Scholar]
- Brandt S, Kehr J, Walz C, Imlau A, Willmitzer L, Fisahn J (1999) A rapid method for detection of plant gene transcripts from single epidermal, mesophyll and companion cells of intact leaves. Plant J 20: 245–250 [DOI] [PubMed] [Google Scholar]
- Day CD, Irish VF (1997) Cell ablation and the analysis of plant development. Trends Plant Sci 2: 106–111 [Google Scholar]
- Delp G, Palva ET (1999) A novel flower-specific Arabidopsis gene related to both pathogen-induced and developmentally regulated plant β-1,3-glucanase genes. Plant Mol Biol 39: 565–575 [DOI] [PubMed] [Google Scholar]
- de Oliveira DE, Franco LO, Simoens C, Seurinck J, Coppieters J, Botterman J, Van Montagu M (1993) Inflorescence-specific genes from Arabidopsis thaliana encoding glycine-rich proteins. Plant J 3: 495–507 [DOI] [PubMed] [Google Scholar]
- Dwyer KG, Lalonde BA, Nasrallah JB, Nasrallah ME (1992) Structure and expression of AtS1, and Arabidopsis thaliana gene homologous to the S-locus related genes of Brassica. Mol Gen Genet 231: 442–448 [DOI] [PubMed] [Google Scholar]
- Elleman CJ, Dickinson HG (1999) Commonalities between pollen/stigma and host/pathogen interactions: calcium accumulation during stigmatic penetration by Brassica oleracea pollen tubes. Sex Plant Reprod 12: 194–202 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Galen C, Plowright RC (1987) Testing the accuracy of using peroxidase activity to indicate stigma receptivity. Can J Bot 65: 107–111 [Google Scholar]
- Gaude T, Dumas C (1986) Organization of stigma surface components in Brassica: a cytochemical study. J Cell Sci 82: 203–216 [DOI] [PubMed] [Google Scholar]
- Gross-Hardt R, Laux T (2003) Stem cell regulation in the shoot meristem. J Cell Sci 116: 1659–1666 [DOI] [PubMed] [Google Scholar]
- Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR (1998) Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci USA 95: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U, Kohler C (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol 135: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J, Heslop-Harrison Y (1975) Enzymic removal of the proteinaceous pellicle of the stigmatic papillae prevents pollen tube entry in the Caryophyllaceae. Ann Bot (Lond) 39: 163–165 [Google Scholar]
- Heslop-Harrison Y, Shivanna KR (1977) The receptive surface of the angiosperm stigma. Ann Bot (Lond) 41: 1233–1258 [Google Scholar]
- Hiscock SJ, Bown D, Gurr SJ, Dickinson HG (2002) Serine esterases are required for pollen tube penetration of the stigma in Brassica. Sex Plant Reprod 15: 65–74 [Google Scholar]
- Jackson PAP, Galinha CIR, Pereira CS, Fortunato A, Soares NC, Amancio SBQ, Ricardo CPP (2001) Rapid deposition of extensin during the elicitation of grapevine callus cultures is specifically catalyzed by a 40-kilodalton peroxidase. Plant Physiol 127: 1065–1076 [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Nasrallah ME, Nasrallah JB (2002) Self-incompatibility in the Brassicaceae: receptor-ligand signaling and cell-to-cell communication. Plant Cell (Suppl) 14: S227–S238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader J-C (1996) Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 627–654 [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Nasrallah JB, Nasrallah ME (1994) Pollen-pistil interactions and developmental regulation of pollen tube growth in Arabidopsis. Development 12: 3405–3418 [Google Scholar]
- Kang Y, Nasrallah JB (2001) Use of genetically ablated stigmas for the isolation of genes expressed specifically in the stigma epidermis. Sex Plant Reprod 14: 85–94 [Google Scholar]
- Karrer EE, Lincoln JE, Hogenhout S, Bennett AB, Bostock RM, Martineau B, Lucas WJ, Gilchrist DG, Alexander D (1995) In situ isolation of mRNA from individual plant cells: creation of cell-specific cDNA libraries. Proc Natl Acad Sci USA 92: 3814–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk NM, Ceserani T, Tausta L, Sussex IM, Nelson TM (2003) Laser capture microdissection of cells from plant tissues. Plant Physiol 132: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Mollet J-D, Dong J, Zhang K, Park S-Y, Lord EM (2003) Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc Natl Acad Sci USA 100: 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klur S, Toy K, Williams MP, Certa U (2004) Evaluation of procedures for amplification of small-size samples for hybridization on microarrays. Genomics 83: 508–517 [DOI] [PubMed] [Google Scholar]
- Kononowicz AK, Nelson DE, Singh NK, Hasegawa PM, Bressan RA (1992) Regulation of the osmotin gene promoter. Plant Cell 4: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Dwyer K, Hendershot J, Nasrallah JB, Nasrallah ME (2001) Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643 [PMC free article] [PubMed] [Google Scholar]
- Lan L, Chen W, Lai Y, Suo J, Kong Z, Li C, Lu Y, Zhang Y, Zhao X, Zhang X, et al (2004) Monitoring of gene expression profiles and isolation of candidate genes involved in pollination and fertilization in rice (Oryza sativa L.) with a 10K cDNA microarray. Plant Mol Biol 54: 471–487 [DOI] [PubMed] [Google Scholar]
- Lolle S, Cheung AY (1993) Promiscuous germination and growth of wild type pollen from Arabidopsis and related species on the shoot of the Arabidopsis mutant, fiddlehead. Dev Biol 155: 250–258 [DOI] [PubMed] [Google Scholar]
- Majewska-Sawka A, Nothnagel EA (2000) The multiple roles of arabinogalactan proteins in plant development. Plant Physiol 122: 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani C, De Beuckeleer M, Truettner J, Leemans J, Goldberg RB (1990) Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347: 737–741 [Google Scholar]
- Murase K, Shiba H, Iwano M, Che F-S, Watanabe M, Isogai A, Takayama S (2004) A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303: 1516–1519 [DOI] [PubMed] [Google Scholar]
- Nasrallah ME, Liu P, Nasrallah JB (2002) Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297: 247–249 [DOI] [PubMed] [Google Scholar]
- Nasrallah ME, Liu P, Sherman-Broyles S, Boggs NA, Nasrallah JB (2004) Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc Natl Acad Sci USA 101: 16070–16074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D (2003) Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59 [DOI] [PubMed] [Google Scholar]
- Park S-Y, Jauh G-Y, Mollet J-C, Eckard KJ, Nothnagel EA, Walling LL, Lord EM (2000) A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Ryan CA (2003) Systemic signaling in tomato plants for defense against herbivores. Isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. J Biol Chem 278: 30044–30050 [DOI] [PubMed] [Google Scholar]
- Robert L, Allard S, Gerster JL, Cass L, Simmonds J (1994) Molecular analysis of two Brassica napus genes expressed in the stigma. Plant Mol Biol 26: 1217–1222 [DOI] [PubMed] [Google Scholar]
- Sanchez AM, Bosch M, Bots M, Nieuwland J, Feron R, Mariani C (2004) Pistil factors controlling pollination. Plant Cell 16: S98–S106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P (2001) Hydroxyl radical-induced cell wall loosening in vitro and in vivo: implications for the control of elongation growth. Plant J 28: 679–688 [DOI] [PubMed] [Google Scholar]
- Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54: 629–667 [DOI] [PubMed] [Google Scholar]
- Scutt CP, Vinauger-Douard M, Fourquin C, Ailhas J, Kuno N, Uchida K, Gaude T, Furuya M, Dumas C (2003) The identification of candidate genes for a reverse genetic analysis of development and function in the Arabidospis gynoecium. Plant Physiol 132: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S-H, Bleecker AB (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Metraux J-P, Nawrath C (2000) Transgenic Arabidopsis plants expressing a fungal cutinse show alternations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12: 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead AD, Roberts IN, Dickinson HG (1980) Pollen-stigma interaction in Brassica oleracea: the role of stigmatic proteins in pollen grain adhesion. J Cell Sci 42: 417–423 [DOI] [PubMed] [Google Scholar]
- Stone SL, Arnoldo M, Goring DR (1999) A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Swidzinski JA, Sweetlove LJ, Leaver CJ (2002) A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J 30: 431–446 [DOI] [PubMed] [Google Scholar]
- Takayama S, Shimosato H, Shiba H, Funato M, Che FS, Watanabe M, Iwano M, Isogai A (2001) Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413: 534–538 [DOI] [PubMed] [Google Scholar]
- Tang W, Kelley D, Ezcurra I, Cotter R, McCormick S (2004) LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro. Plant J 39: 343–353 [DOI] [PubMed] [Google Scholar]
- Thorsness MK, Kandasamy MK, Nasrallah ME, Nasrallah JB (1993) Genetic ablation of floral cells in Arabidopsis. Plant Cell 5: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Tichtinsky G, Dumas C, Gaude T, Cock JM (2003) Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphate with the Brassica oleracea S locus receptor kinase. Plant Physiol 133: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A (2001) Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proc Natl Acad Sci USA 98: 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM (2004) Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EJB, Bowles DJ (2004) Coexpression of neighboring genes in the genome of Arabidopsis. Genome Res 14: 1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-M, Wong E, Ogdahl J, Cheung AY (2000) A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. Plant J 22: 165–176 [DOI] [PubMed] [Google Scholar]
- Zachgo S, Perbal M-S, Saedler H, Schwarz-Sommer Z (2000) In situ analysis of RNA and protein expression in whole mounts facilitates detection of floral gene expression dynamics. Plant J 23: 697–702 [DOI] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Han B, Brown D, Chang H-S, Zou G, Wang X (2001) Toward elucidating the global gene expression patterns of developing Arabidopsis: parallel analysis of 8300 genes by a high-density oligonucleotide probe array. Plant Physiol Biochem 39: 221–242 [Google Scholar]
- Zik M, Irish VF (2003) Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15: 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D (1999) Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126: 5431–5440 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.