Abstract
Classical forward genetics, the identification of genes responsible for mutant phenotypes, remains an important part of functional characterization of the genome. With the advent of extensive genome sequence, phenotyping and genotyping remain the critical limiting variables in the process of map-based cloning. Here, we reduce the genotyping problem by hybridizing labeled genomic DNA to the Affymetrix Arabidopsis (Arabidopsis thaliana) ATH1 GeneChip. Genotyping was carried out on the scale of detecting greater than 8,000 single feature polymorphisms from over 200,000 loci in a single assay. By combining this technique with bulk segregant analysis, several high heritability development and circadian clock traits were mapped. The mapping accuracy using bulk pools of 26 to 100 F2 individuals ranged from 0.22 to 1.96 Mb of the mutations revealing mutant alleles of EARLY FLOWERING 3, EARLY FLOWERING 4, TIMING OF CAB EXPRESSION 1, and ASYMMETRIC LEAVES 1. While direct detection of small mutations, such as an ethyl-methane sulfonate derived single base substitutions, is limited by array coverage and sensitivity, large deletions such as those that can be caused by fast neutrons are easily detected. We demonstrate this by resolving two deletions, the 77-kb flavin-binding, kelch repeat, f-box 1 and the 7-kb cryptochrome2-1 deletions, via direct hybridization of mutant DNA to ATH1 expression arrays.
The properties that originally made organisms such as yeast (Saccharomyces cerevisiae), nematode (Caenorhabditis elegans), Arabidopsis (Arabidopsis thaliana), and fruit fly (Drosophila melanogaster) models for scientific research were their amenability to genetic studies: easily reared short life cycle, simple controlled mating, and fecundity. Only later was the serendipity of their small genomes realized and capitalized on by sequencing their entire genomes. A fully sequenced and annotated genome alone has limited value in revealing the functional relevance of genes. Forward genetics via mutagenesis is a traditional approach to assign gene function. This practice involves the identification of phenotypically divergent individuals and subsequent identification of the causal genetic difference, thereby connecting a gene with a phenotype and/or function. Advances in genome biology have facilitated this approach, thus increasing the usefulness of model systems for determining the role of genes in organismal physiology.
In Arabidopsis, genetic variation is commonly induced with the chemical mutagen ethyl-methane sulfonate (EMS), which alkalates guanine residues (Koornneef, 2002). Since alkalated guanine pairs to thymine instead of cytosine, EMS treatment produces conversions of GC to AT. The outcome of such nucleotide changes is either synonymous, missense, or stop codons. High-energy particles are also an effective mutagen. Bombarding seed or pollen with γ-rays, x-rays, or fast neutrons will create deletions ranging from a single base to greater than 100 kb. Genetic linkage mapping, i.e. cosegregation analysis, is one approach to locate these randomly created mutations.
Besides genetic variation created by mutagens, there is extensive naturally occurring variation among Arabidopsis accessions. That variation can be exploited within a segregating population to mark loci associated with a phenotype. Sequencing projects have identified several thousand polymorphisms that serve as anchored molecular markers (Schmid et al., 2003; Torjek et al., 2003). The complete genome sequence of the accession Columbia (Col) and approximately 70% genome coverage of the accession Landsberg erecta (Ler) revealed more than 50,000 polymorphisms that can be utilized as molecular markers (The Arabidopsis Genome Initiative, 2000; Jander et al., 2002). Cosegregation of anchored molecular markers and a phenotype, i.e. genetic linkage, is a strong indicator of the local region where the mutation lies.
High-density oligonucleotide arrays are an effective platform to measure numerous polymorphic loci in a single assay (Hazen and Kay, 2003). The Affymetrix Arabidopsis ATH1 GeneChip is designed to quantitatively measure the transcript abundance of more than 20,000 genes. The expression level of each gene is a function of the hybridization intensity of usually 11 25-mers, referred to as features. DNA genotyping uses each one of the features independently without regard to the gene or probeset to which they belong. Many sequence polymorphisms can be detected as a difference in hybridization intensity between two strains when randomly labeled genomic DNA is profiled. These are referred to as single feature polymorphisms (SFPs; Borevitz et al., 2003; Winzeler et al., 2003). An allele with a perfect match to an array feature may hybridize with a detectably greater affinity than one with a mismatch sequence; thus, the detectable sequence polymorphism functions as a molecular marker. Hybridization intensity to a microarray is a quantitative measurement; therefore, the difference between two measurements is quantitative as well. By comparing replicate samples of the certain accessions to replicates of the reference Col accessions, >8,000 SFPs were identified and reliably scored (Wolyn et al., 2004; Borevitz, 2005; Werner et al., 2005a, 2005b).
A very practical, effective, and rapid approach to using SFPs for mapping mutations is in combination with bulk segregant analysis (BSA; Michelmore et al., 1991; Wolyn et al., 2004). For example, the array genotype of a pool of mutant F2 individuals is compared to a pool of wild-type F2 individuals from the same segregating population. The pools are expected to have equal mixtures of both parent genotypes at loci unlinked to the mutation and therefore exhibit hybridization intensity intermediate to that of the parent genotypes. In the region of the mutation, the mutant pool is enriched for mutant genotype alleles and the wild-type phenotype pool enriched for the wild-type parent alleles. In a test case using the first generation Affymetrix Arabidopsis AtGenome1 GeneChip, the discrete qualitative erecta mutation was mapped to within a few centimorgans of the actual mutation by comparing pools of 15 wild-type and erecta F2 individuals (Borevitz et al., 2003). Using the ATH1 GeneChip, in three F2 populations segregating for mutations in LUX ARRHYTHMO (LUX), which confers an arrhythmic circadian clock in constant light, the mutation was mapped to within 0.723, 2.295, and 1.235 Mb by comparing pools of 50 individuals (S.P. Hazen, T.F. Schultz, J.L. Pruneda-Paz, J.O. Borevitz, J.R. Ecker, and S.A. Kay, unpublished data).
Here, we report the results of several empirical studies mapping EMS mutations in genes involved in development (asymmetric leaves 1 [as1]), and the circadian clock (early flowering 3 [elf3], early flowering 4 [elf4], and timing of cab expression 1 [toc1]). In each case, BSA mapping with SFP genotyping rapidly mapped the mutation to a rough interval suitable for fine mapping or direct sequencing of candidate genes. We also demonstrate an approach to identify mutant loci without having to make a mapping cross, by directly delineating the fast neutron derived deletions responsible for flowering time mutations flavin-binding, kelch repeat, f-box 1 (fkf1) and cryptochrome2-1 (cry2-1).
RESULTS AND DISCUSSION
Isolating and Mapping Circadian Clock Mutants
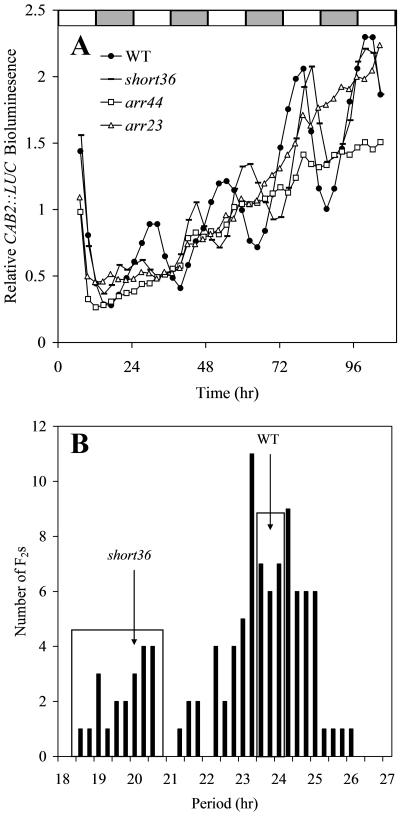
The circadian clock allows an organism to anticipate environmental changes and time specific physiological events to occur at certain times of day. A powerful laboratory tool used as an indicator of the clock is LUCIFERASE (LUC) fused with the promoter of a circadian regulated gene, namely, CHLOROPHYLL A-B BINDING PROTEIN 2 (CAB2; Millar et al., 1995). To isolate mutants of the circadian clock, M2 seedlings from EMS mutagenized CAB2::LUC reporter line in the Col background were screened for long hypocotyl, a common characteristic of clock mutants (Dowson-Day and Millar, 1999). The rhythms produced by the circadian clock were then monitored with the CAB2::LUC reporter. A circadian process is often described in the form of a wave, which has three basic properties: period, phase, and amplitude. From this screen, mutants with short period (short36) and no amplitude, i.e. arrhythmic (arr23 and arr44), were identified (Fig. 1, A and B).
Figure 1.
Three circadian clock mutants with either short circadian period or arrhythmic expression of the CAB2::LUC reporter. A, Transgenic seedlings carrying the CAB2::LUC reporter were entrained under 12-h-light:12-h-dark cycles for 7 d. Bioluminescence was then monitored under constant light conditions in wild type (black circles), short36 (dash), arr44 (white square), and arr23 (white triangles). B, The distribution of period lengths in the short36 × Ler F2 population. DNA from plants in each box was pooled for array hybridization.
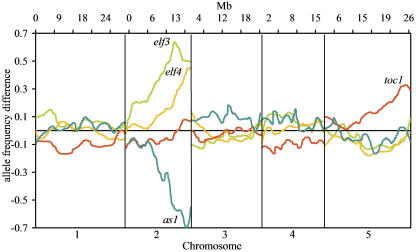
The F2 populations segregating for arrhythmic phenotype (Fig. 1A) derived from a cross with Ler were classified into discrete categories, either rhythmic or not in segregants that contained the CAB2::LUC reporter. Segregation was consistent with a single recessive mutation for each of the three populations (data not shown). As the mutations are in the Col background, at the mutant locus and linked regions, the arrhythmic group was homozygous Col and the rhythmic (wild-type) group a 2:1 mixture of heterozygotes and homozygous Ler. Unlinked loci are of roughly equal proportion of Col and Ler alleles in both bulk pools. Thus, the largest difference in allele frequency between the pools is the predicted location of the mutation. The greatest difference in allele frequency between the arr23×Ler arrhythmic F2s and the rhythmic F2s was near the bottom of chromosome 2 at 11.288 Mb, very near the circadian clock component, ELF3 (Fig. 2). The mutant phenotype of elf3 is similar to arr23 making ELF3 a candidate gene (Hicks et al., 1996; Zagotta et al., 1996). Sequencing arr23 revealed a missense mutation in ELF3 at a codon (P667L) conserved across several species (Hicks et al., 2001). This nucleotide conversion is consistent with the type of mutation caused by EMS (GC to AT) and the phenotype is consistent with many known mutant alleles of ELF3 strongly suggesting that this is the cause of the arr23 mutant phenotype. The predicted location of arr23 is 222 kb from this mutation (Table I).
Figure 2.
Bulk segregant array mapping of four mutants involved in development or the circadian clock. Horizontal axis represents the five Arabidopsis chromosomes. Vertical axis represents allele frequency differences between mutant and wild-type pools and is positive for mutants in the Col background and negative for mutants in the Ler background.
Table I.
Accuracy of array mapping for circadian clock and developmental mutations (this paper or corresponding reference)
na, Not applicable.
| Gene
|
AGI
|
Mapping Accuracy
|
Pool Sizes
|
Inheritance
|
Mutagen
|
Mutation
|
Background
|
Phenotype
|
|
|---|---|---|---|---|---|---|---|---|---|
| kb | cM | ||||||||
| ERECTA | AT2G26330 | 569 | 2.28 | 15 | Recessive | X-ray | I > K | Ler | Short internodes (Torii et al., 1996; Borevitz et al., 2003) |
| ELF3 (ARR23) | AT2G25930 | 222 | 0.89 | 73 | Recessive | EMS | P > L | Col | Early flowering, arrhythmic circadian clock |
| ELF4 (ARR44) | AT2G40080 | 1,960 | 7.84 | 50 | Recessive | EMS | Q > stop | Col | Early flowering, arrhythmic circadian clock |
| TOC1 (SHORT36) | AT5G61380 | 224 | 0.90 | 26 | Recessive | EMS | Q > stop | Col | Short circadian period |
| AS1 (BIBB) | AT2G37630 | 1,679 | 6.72 | 100 | Recessive | EMS | W > stop | Ler | Rumpled leaves with a characteristic asymmetry |
| LUXa | AT3G46640 | 726 | 2.90 | 50 | Recessive | EMS | R > stop | Col | Early flowering, arrhythmic circadian clock (S.P. Hazen, T.F. Schultz, J.L. Pruneda-Paz, J.O. Borevitz, J.R. Ecker, and S.A. Kay, unpublished data) |
| 1,234 | 4.94 | 50 | R > stop | ||||||
| 2,396 | 9.58 | 50 | Q > stop | ||||||
| FKF1 | AT1G68050 | na | na | na | Recessive | Fast neutron | 77-kb deletion | Col | Late flowering |
| CRY2 | AT1G04400 | na | na | na | Recessive | Fast neutron | 7-kb deletion | Col | Late flowering |
S.P. Hazen, T.F. Schultz, J.L. Pruneda-Paz, J.O. Borevitz, J.R. Ecker, and S.A. Kay (unpublished data).
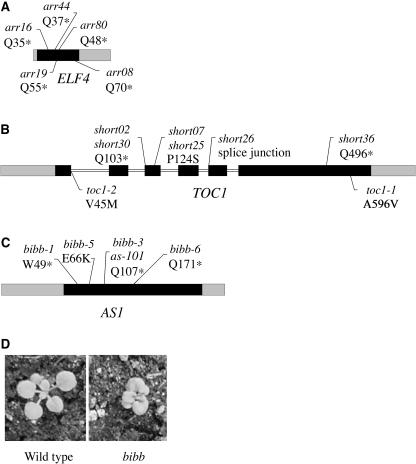
Following the same procedure, the arr44 mutation was quickly mapped to the bottom of chromosome 2 at 18.701 Mb, well below ELF3 (Fig. 2). Besides elf3, the only other gene in the region known to cause CAB2::LUC arrhythmia and long hypocotyl when mutated is ELF4 (Doyle et al., 2002). The predicted location of arr44 is 1.96 Mb from ELF4. Sequencing ELF4 in arr44 revealed the conversion of codon 37 (CAA) to a stop codon (TAA; Table I). As observed in arr23, this nucleotide conversion is consistent with the type of mutation caused by EMS (Koornneef, 2002). In addition, we directly sequenced ELF4 in other arr mutant lines displaying a similar phenotype and identified four different ELF4 mutant alleles, all of which introduce stop codons (Fig. 3A).
Figure 3.
A to C, Intron/exon structures of ELF4 (A), TOC1 (B), and AS1 (C). Exons are indicated by black boxes, UTRs by gray boxes, and introns by white boxes. Molecular changes in elf4, toc1, and as1 alleles are shown neighboring the position. D, The cabbage-like rosette leaves of the bib-1 mutant.
As opposed to the discrete classification of rhythmic versus arrhythmic phenotype, circadian period is a quantitative measurement. Such period mutants could be mapped as quantitative trait loci using extreme array mapping, by selecting pools of extreme phenotype plants from an F2 distribution (Borevitz et al., 2003; Wolyn et al., 2004). The period of short36 is considerably shorter (approximately 20 h) than wild type (approximately 24 h) and the period distribution in the F2 is bimodal (Fig. 1, A and B). The short period pool was constructed of 26 individuals with a period length less than 22 h and the wild-type pool of 26 F2 plants with a period ranging from 23.75 to 24.74 h. The difference in allele frequency between short36 mutant and wild-type pools was greatest at the bottom of chromosome 5 at 24.92 Mb, 224 kb from a candidate gene TOC1 (Fig. 2). Sequencing revealed a nucleotide conversion from cytosine to thymine changing Gln to a stop codon in the last exon of TOC1 (Table I). Five other mutants with toc1-like phenotypes were isolated from the same screen as short36, each one with either a missense, splice junction mutation, or a premature stop codon in TOC1 (Fig. 3B).
Isolating and Mapping a Developmental Mutant
In addition to analyzing mutants defective in controlling their circadian clocks, we successfully used array-based bulk segregant mapping to position mutations that affect leaf and flower development. To isolate floral organ shedding mutants in Arabidopsis, mature M2 plants from an EMS-mutagenized Ler population were screened for defects in this process. An isolated mutant with defects in both flower and leaf development was named bibb (bib) due to the resemblance of its rumpled leaves with short petioles to Bibb lettuce (Fig. 3D). Three additional mutants with a bib-like appearance were isolated from this and a previous screen (Liljegren et al., 2000).
An F2 mapping population derived from a cross of bib-1 to Col segregated in the expected ratio for a single recessive mutation; mutants were readily distinguished from wild type at the rosette stage. Bulk segregant array genotyping positioned the bib mutation on chromosome 2 (Fig. 2) within 1.68 Mb of ASYMMETRIC LEAVES1 (AS1). AS1 encodes a Myb-domain transcription factor that negatively regulates expression of KNOX homeobox genes in developing lateral organ primordia (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001). Like bib, as1 mutants produce rumpled, cabbage-like leaves, although defects in floral organ shedding have not previously been observed. Sequencing bib-1 revealed a single nucleotide change in AS1 that would convert a highly conserved Trp (codon 49) to a stop codon in the Myb DNA-binding domain (Table I). Three of the other bib-like mutants also contained mutations in AS1 (Fig. 3C).
Delineating Large Deletions
Using replicate samples and initially only a partial genome array, 105 potential natural deletions were detected between Col and Ler (Borevitz et al., 2003), while 542 potential natural deletions were found between Col-gl1 and Kas using a full genome expression array (Wolyn et al., 2004). Two hundred fifty-three and 286 potential deletions were found in Bur and Lz accessions compared to Col (Werner et al., 2005a), while 210 and 325 potential natural deletions were found in Nd and Ler compared to the reference Col (S.P. Hazen, T.F. Schultz, J.L. Pruneda-Paz, J.O. Borevitz, J.R. Ecker, and S.A. Kay, unpublished data; Werner et al., 2005b). Potential deletions were restricted to a significance threshold, were greater than 100 bp, and contained at least four adjacent features to avoid excessive false positives. Less conservative parameters would increase the number of deletions detected. Large deletions created by fast neutrons should also be easily detected. This offers the opportunity to identify an induced mutation by directly comparing hybridization patterns of mutant and wild-type strains to delineate the deletion(s). We verified this using the flowering time deletion mutants fkf1 and cry2-1.
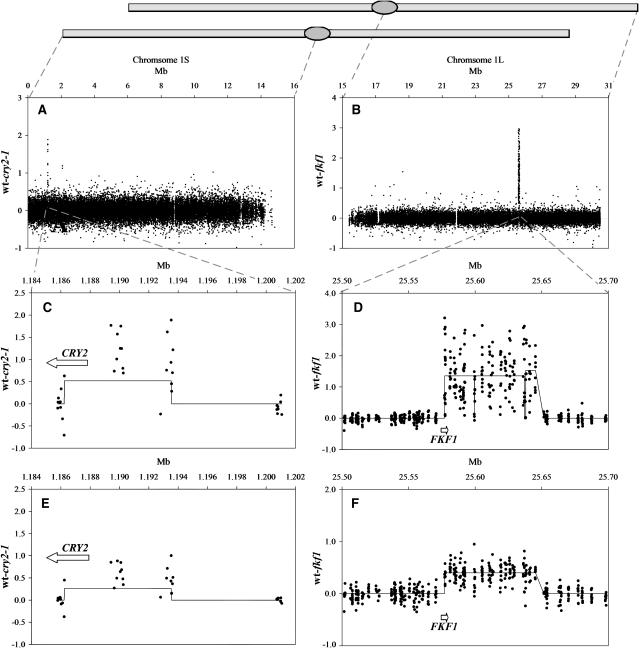
The fkf1 fast neutron induced deletion was estimated to be 65 to 80 kb (Nelson et al., 2000), which represented the loss of several genes. The cause of the phenotype was deduced by complementation with subclones from the missing region. To more accurately characterize the deleted region, single replicates genomic DNA from fkf1 and wild-type Col was hybridized to the ATH1 array. Of the 26,213 unique features that correspond to the long arm of chromosome 1, a large block hybridized with much greater intensity in wild type than in fkf1 (Fig. 4B). The missing 185 features suggest the deletion is at least 77 kb and contains 22 genes (Fig. 4D). This deletion could also be detected in a heterozygous form when an equal mixture of genomic DNA from fkf1 and Col was labeled and hybridized to an array. Upon comparison of this synthetic F1 with Col, the deletion was again clearly visible (Fig. 4F).
Figure 4.
Precise mapping of induced deletions by directly hybridizing fast neutron mutant and wild type to arrays. The vertical axis is hybridization intensity difference between Col and either cry2-1 (A, C, and E) or fkf1 (B, D, and F). Horizontal axis is Mb on chromosome 1. Line through the data points represents clusters that identify groups of 25-mer features with differential hybridization indicative of the deletions.
The fast neutron mutant cry2-1 is photoperiod insensitive and late flowering (Guo et al., 1998). The deletion is known to include the 5′ region of CRY2, while the precise upstream location was unknown. Both cry2-1 and wild-type DNA were hybridized to single ATH1 arrays. Of the 26,684 unique features corresponding to the short arm of chromosome 1, there was a group of features missing in the apical region (Fig. 4A). The missing 19 features suggest the deletion is at least 7 kb disrupting at least two genes (Fig. 4C). The CRY2 gene is also partially deleted. This deletion could also be detected in a heterozygous form when comparing Col with the average of the cry2-1 and Col array genotype. Upon comparison of this in silico F1 with Col, the deletion was again clearly visible (Fig. 4E). Fine mapping of the deletion with nested PCR primers confirmed that the left border of the deletion is 480 bp from the 3′ end of CRY2 removing a total of 8.1 kb in the cry2-1 mutant.
CONCLUSION
High-Density Genome Coverage
Map-based cloning first relies on detecting an association between a marker genotype and a phenotype. Subsequently, mapping resolution is a function of marker density and number of recombination events. With the 8,000 confident SFPs used here, marker resolution was on average every 15 kb. With new whole genome tiling arrays, we expect an SFP on average every 700 bp; thus, marker density is no longer limiting. By using pools of recombinant lines, we increase the number of recombination events assayed in a single hybridization; however, the rare close recombination events are diluted. An important advance toward the fine mapping of novel loci will take advantage of pools of preselected recombinant lines for array genotyping when clear candidates are not identified. Here, a fine recombination event can be fully exploited with high-density SFPs.
Accuracy and Heritability
If our candidate genes are all correct, the array mapping accuracy of the examples described ranged from 0.222 to 2.396 Mb. This corresponds well with the 7 cM 95% credible interval determined by simulations (Borevitz, 2005). Thus, the mutation will most likely be within 2 Mb of the BSA peak, producing a list of approximately 400 candidate genes. If there is contamination of incorrect genotypes in the bulk pools due to misscoring low heritability traits, this reduces the mapping accuracy since the signal (allele frequency difference between pools) is also reduced. Practically, larger pools will not be greatly affected and the approach is robust to moderate levels of contamination as evidence by the mapping of large effect quantitative trait loci (Wolyn et al., 2004).
Flexibility
SFP discovery was conducted via the comparison of replicate arrays of Col and Ler. Beyond the sequence necessary to design the oligonucleotide array, sequencing other accessions is not necessary for array-based genotyping and mapping. Any two accessions can be used in a cross to BSA map with SFPs following replicate hybridizations of both accessions and the identification of polymorphic features.
Deletions
We have shown that induced deletions the size of a gene size or greater can be readily identified by direct hybridization of DNA from mutant lines in comparison with wild-type controls. Future generations of whole genome tiling arrays (Borevitz and Ecker, 2004; Mockler et al., 2005) will be even more powerful to localize small deletions and replicate hybridizations can be used to improve signal to noise if needed. This approach is feasible with mutations in other backgrounds provided there is synteny with the Col reference genome. Proof that a predicted deletion is causative will rely on cosegregation analysis and complementation, but certainly a short list of candidates is quickly generated. With this approach to quickly identify deletion mutations, fast neutron mutagenesis is sure to regain popularity for future mutant screens. In addition, when T-DNA induced mutations do not cosegregate with the mutation (untagged), the phenotypic cause could be identified as an unlinked deletion with the array hybridization approach shown here. Also, it is possible to combine BSA with deletion mapping to show that a particular lesion is associated with the phenotype; however, small deletions of less than approximately four features are unlikely to be directly identified without candidate genes in mind (Gong et al., 2004).
MATERIALS AND METHODS
Isolation and Analysis Mutants
The isolation and analysis of circadian mutants has been previously described (S.P. Hazen, T.F. Schultz, J.L. Pruneda-Paz, J.O. Borevitz, J.R. Ecker, and S.A. Kay, unpublished data). The isolation of developmental mutants has also been described (Liljegren et al., 2000).
Bulk Segregant Mapping with Array Genotyping
This method has been described in detail (Borevitz, 2005). Example data and analysis scripts are available http://naturalvariation.org/methods. Raw hybridization data for mapping and deletion experiments are available here http://naturalvariation.org/methods. The allele frequency differences between pools for each mapping cross were output as text files and plotted together in excel.
Deletion Mapping
Deletion mapping for fast neutron mutations was performed similarly to the prediction of potential natural deletions (Borevitz, 2005) except that the hybridization difference between the wild-type and mutant arrays was used rather than the d-statistic of relative difference between accessions as replicates were not used. The clustering criteria were also modified to find larger deletions with 10 clusters per chromosome.
Acknowledgments
We thank H. Bird Richardson for expert assistance with figure formatting.
This work was supported by the National Institutes of Health (grant nos. GM56006 and GM67837 to S.A.K. and a Ruth L. Kirschstein National Research Service Award postdoctoral fellowship [GM071225] to S.P.H.), by the Department of Energy (grant no. DE–FG03–00ER15113), by the National Science Foundation (grant no. MCB–0213154 to J.R.E.), by the U.S. Department of Agriculture (National Research Initiative Competitive Grants Program postdoctoral fellowship to S.J.L.), and by the Helen Hay Whitney Foundation (fellowship to J.O.B.). F.G.H. is a Department of Energy-Energy Biosciences Fellow of the Life Sciences Research Foundation. This is manuscript number 17134–CB of the Scripps Research Institute.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.061408.
References
- Borevitz JO (2005) Array genotyping genotyping and mapping. In J Salinas, JJ Sanchez-Serrano, eds, Arabidopsis Protocols, Ed 2. Humana Press, Totowa, NJ
- Borevitz JO, Ecker JR (2004) Plant genomics: the third wave. Annu Rev Genomics Hum Genet 5: 443–477 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, Weigel D, Berry CC, Winzeler E, Chory J (2003) Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res 13: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17: 63–71 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Gong JM, Waner DA, Horie T, Li SL, Horie R, Abid KB, Schroeder JI (2004) Microarray-based rapid cloning of an ion accumulation deletion mutant in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HW, Yang WY, Mockler TC, Lin CT (1998) Regulations of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Kay SA (2003) Gene arrays are not just for measuring gene expression. Trends Plant Sci 8: 413–416 [DOI] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR (2001) EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M (2002) Classical mutagenesis in higher plants. In PM Gilmartin, C Bowler, eds, Molecular Plant Biology, Ed Vol 1. Oxford University Press, pp 1–11
- Liljegren SJ, Ditta GS, Eshed HY, Savidge B, Bowman JL, Yanofsky MF (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Carre IA, Strayer CA, Chua NH, Kay SA (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Chan S, Sundareson A, Chen H, Jacobsen SE, Ecker JR (2005) Applications of DNA tiling arrays for whole-genome analysis. Genomics 85: 1–15 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Sorensen TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B (2003) Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res 13: 1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y (2001) The asymmetric leaves2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torjek O, Berger D, Meyer RC, Mussig C, Schmid KJ, Sorensen TR, Weisshaar B, Mitchell-Olds T, Altmann T (2003) Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. Plant J 36: 122–140 [DOI] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D (2005. a) FRIGIDA-independent variation in flowering time of natural A. thaliana accessions. Genetics (in press) [DOI] [PMC free article] [PubMed]
- Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D (2005. b) Natural variation in flowering time of A. thaliana associated with a deletion in the FLC homolog FLM. Proc Natl Acad Sci USA 102: 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Castillo-Davis CI, Oshiro G, Liang D, Richards DR, Zhou Y, Hartl DL (2003) Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 163: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyn DJ, Borevitz WO, Loudet O, Schwartz C, Maloof J, Ecker JR, Berry CC, Chory J (2004) Light-response quantitative trait loci identified with composite interval and eXtreme array mapping in Arabidopsis thaliana. Genetics 167: 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR (1996) The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J 10: 691–702 [DOI] [PubMed] [Google Scholar]