Abstract
ENHANCED DISEASE RESISTANCE 1 (EDR1) encodes a CTR1-like kinase and was previously reported to function as a negative regulator of disease resistance and ethylene-induced senescence. Here, we report that the edr1 mutant displays enhanced stress responses and spontaneous necrotic lesions under drought conditions in the absence of pathogen, suggesting that EDR1 is also involved in stress response signaling and cell death regulation. Double mutant analysis revealed that these drought-induced phenotypes require salicylic acid but not ethylene signaling pathways. In addition, the edr1-mediated ethylene-induced senescence phenotype was suppressed by mutations in EIN2, but not by mutations in SID2, PAD4, EDS1, or NPR1, suggesting that EDR1 functions at a point of cross talk between ethylene and salicylic acid signaling that impinges on senescence and cell death. Two edr1-associated phenotypes, drought-induced growth inhibition and ethylene-induced senescence, were suppressed by mutations in ORE9, implicating ubiquitin-mediated protein degradation in the regulation of these phenotypes. However, the ore9 mutation did not suppress edr1-mediated enhanced disease resistance to powdery mildew or spontaneous lesions, indicating that these phenotypes are controlled by separate signaling pathways. To investigate the function of the EDR1 kinase domain, we expressed the C-terminal third of EDR1 in wild-type Columbia and edr1 backgrounds under the control of a dexamethasone-inducible promoter. Overexpression of the EDR1 kinase domain in an edr1 background had no obvious effect on edr1-associated phenotypes. However, overexpression of the EDR1 kinase domain in a wild-type Columbia background caused dominant negative phenotypes, including enhanced disease resistance to powdery mildew and enhanced ethylene-induced senescence; thus, the overexpressed EDR1 kinase domain alone does not exert EDR1 function, but rather negatively affects the function of native EDR1 protein.
Plants defend themselves against pathogens by many different mechanisms (Dangl and Jones, 2001). To identify genes regulating plant disease responses, we previously screened for Arabidopsis (Arabidopsis thaliana) mutants that displayed enhanced disease resistance to virulent pathogens. The enhanced disease resistance 1 (edr1) mutant displays enhanced disease resistance to the bacterium Pseudomonas syringae and the ascomycete fungus Erysiphe cichoracearum (Frye and Innes, 1998). EDR1 encodes a protein kinase with similarity to CTR1 (Frye et al., 2001), a negative regulator of ethylene responses (Kieber et al., 1993; Cao et al., 1997). The original edr1 mutation causes a premature stop codon that eliminates the kinase domain, suggesting that EDR1 is a negative regulator of disease resistance (Frye et al., 2001). This resistance is suppressed by mutations that block salicylic acid (SA) perception (npr1/nim1; Cao et al., 1997; Ryals et al., 1997) or reduce SA production (eds1 and pad4; Zhou et al., 1998; Falk et al., 1999), and is also suppressed by the transgene NahG, which lowers endogenous SA levels (Frye et al., 2001). In contrast, the ein2 mutation, which disrupts all known ethylene responses (Alonso et al., 1999), does not suppress edr1-mediated enhanced disease resistance. These data demonstrate that the edr1-mediated powdery mildew resistance phenotype requires an intact SA signaling pathway, but not ethylene-induced pathways (Frye et al., 2001).
In addition to the enhanced disease resistance, edr1 also shows an enhanced ethylene-induced senescence phenotype (Frye et al., 2001). Although it has been shown that SA and ethylene signal transduction pathways may cooperate to control certain processes (Geffroy et al., 1999; Clarke et al., 2000; Greenberg et al., 2000; Wang et al., 2002), the contributions of SA and ethylene signaling pathways to the edr1-enhanced senescence phenotype have not been defined. Ethylene-induced senescence in Arabidopsis appears to be regulated at least in part by the F-box protein ORE9, as ore9 mutants display a delayed ethylene-induced senescence phenotype (Woo et al., 2001). ORE9 physically interacts with one of the SCF complex components ASK1 (Arabidopsis Skp1-like 1) in vitro, suggesting ORE9 may regulate the plant senescence process by protein ubiquitylation (Woo et al., 2001). Recently, the F-box proteins EBF1 and EBF2 were shown to regulate levels of the EIN3 protein, which regulates transcription of several ethylene response element binding proteins (Guo and Ecker, 2003; Potuschak et al., 2003). Because ctr1 mutations cause an increase in steady-state levels of EIN3 protein (Guo and Ecker, 2003; Potuschak et al., 2003), CTR1 may be functioning at least indirectly to regulate the rate of EIN3 ubiquitylation by EBF1 and EBF2. By analogy, EDR1 may regulate the ubiquitylation rate of a yet-to-be identified substrate of ORE9. To test this hypothesis, and to gain insight into the relationship between the ethylene-induced and pathogen-induced phenotypes in edr1 plants, we used double mutant analyses involving edr1, ore9, and various SA pathway mutants. These analyses revealed that edr1-mediated disease resistance and ethylene responses are independent pathways and that ORE9 is required for a subset of the edr1-mediated phenotypes.
To gain additional insight into how EDR1 regulates disease resistance and ethylene-induced senescence, we transiently overexpressed the EDR1 kinase domain in planta. EDR1 consists of an N-terminal putative regulatory domain that comprises the first two-thirds of the protein and a C-terminal kinase domain that comprises the last one-third. Previously, we showed that overexpression of a kinase deficient form of the full-length EDR1 gene enhances powdery mildew resistance and ethylene-induced senescence in Arabidopsis (Tang and Innes, 2002). However, we could not detect any EDR1 protein, although the transgene was highly transcribed, suggesting that these phenotypes resulted from inhibition of translation (Tang and Innes, 2002). The C terminus of EDR1 alone displays kinase activity (Tang and Innes, 2002). We thus attempted to complement an edr1 mutant with the kinase domain alone (kdEDR1). We first attempted to overexpress kdEDR1 under control of the cauliflower mosaic virus (CaMV) 35S promoter. However, we were unable to recover plants that overexpressed kdEDR1 at the RNA or protein level. From this, we hypothesized that the EDR1 kinase domain may be toxic to the plant (Tang and Innes, 2002). In this paper, we overexpress kdEDR1 in both the ecotype Columbia (Col-0) and edr1 backgrounds under the control of a dexamethasone-inducible promoter. The kdEDR1 protein did not rescue edr1 phenotypes and surprisingly, it disrupted the native EDR1 function in wild-type Col-0 plants.
RESULTS
Drought Stress Induces Hypersensitive Response-Like Lesions on edr1 Mutant Plants
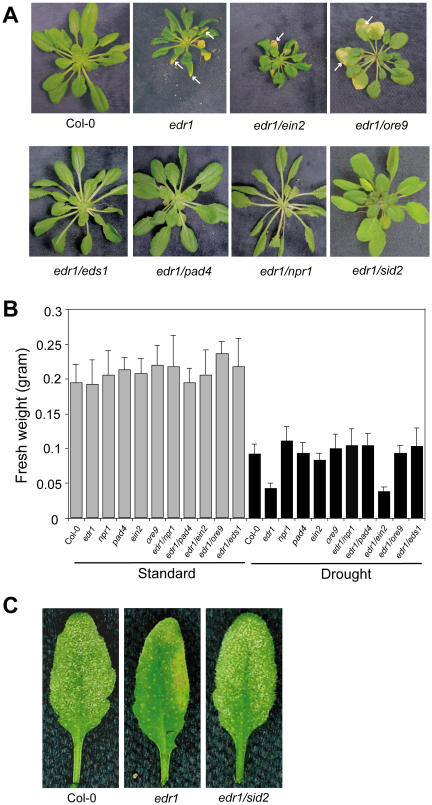
Previously, we reported that the edr1 mutant is phenotypically normal in the absence of pathogen when grown under optimal conditions, displaying neither microscopic nor macroscopic lesions (Frye and Innes, 1998). However, we observed that edr1 plants appeared more sensitive than wild-type plants to underwatering and overwatering, often growing slower than wild-type plants. To characterize edr1-mediated stress response phenotypes, we grew plants under standard growth conditions (see “Materials and Methods”) for 3 weeks and then stopped watering the plants for 2 weeks. The edr1 plants were significantly smaller than wild-type Col-0 plants at the end of the 2-week-drought period, despite being nearly identical in size at the start (Fig. 1A). Control edr1 plants grown with the standard watering regime did not significantly differ in size from Col-0 at 5 weeks (Fig. 1B). In addition, edr1 plants also developed spontaneous yellow-brown necrotic lesions under drought conditions that were similar in appearance to those induced by powdery mildew infection (Fig. 1C). To quantify the edr1-mediated drought-induced growth phenotype, we weighed the individual plants (fresh weight) grown under standard or drought conditions. Figure 1B shows that edr1 mutant plants weighed almost the same as Col-0 wild-type plants when grown under standard conditions but weighed significantly less than the Col-0 wild-type plants when grown under drought conditions. These data suggest that EDR1 may also regulate plant growth and cell death in response to drought stress, in addition to its previously reported roles in disease resistance and senescence.
Figure 1.
The edr1 mutant displays drought-induced growth inhibition and cell death phenotypes. A, Plants were grown under standard growth conditions for 3 weeks, and then watering was withheld for 2 weeks. Compared to Col-0 wild-type plants, edr1 mutant plants were smaller and displayed spontaneous yellow-brown necrotic patches (arrows). The edr1-mediated drought-induced growth and cell death phenotypes were suppressed by mutations that block SA-mediated defense responses (eds1, pad4, npr1, and sid2), but not by a mutation that blocks ethylene response (ein2). The ore9 mutation suppressed edr1-mediated drought-induced growth inhibition, but not spontaneous lesions. B, Quantification of drought-induced growth inhibition. Aerial portions were weighed immediately after removal from soil. Bars represent the mean and sd of values from 10 plants. C, SID2 is required for edr1-mediated resistance to powdery mildew infection. Leaves were removed from plants for photography 8 d after infection. Abundant white powder visible on the leaves in the left and right sections indicates asexual sporulation, thus a susceptible response. The middle leaf displays yellow-brown necrotic patches and little powder, typical of an edr1-mediated resistance response.
The Spontaneous Hypersensitive Response-Like Lesions in edr1 Are SA Dependent But Ethylene Independent
To gain insight into how EDR1 regulates plant growth and cell death, we analyzed whether mutations in the SA and ethylene signal transduction pathways suppress the edr1-associated growth and spontaneous hypersensitive response (HR)-like phenotypes. Through double mutant analyses, we found that eds1-1, nim1-1 (an allele of npr1), pad4-2, and sid2-2 all suppressed both the edr1-mediated drought-induced growth and the spontaneous HR-like phenotypes, while ein2-1 did not. These data indicate that edr1-mediated growth inhibition and cell death phenotypes are also SA dependent and ethylene independent (Fig. 1). This was consistent with our previous finding that edr1-mediated enhanced disease resistance requires SA, but not ethylene (Frye et al., 2001). We confirmed the SA requirement for edr1-mediated resistance by assaying resistance in the edr1/sid2-2 double mutant, which we had not tested previously. As expected, the sid2 mutation, which dramatically reduces SA levels in Arabidopsis (Nawrath and Metraux, 1999), suppressed the edr1-mediated resistance to powdery mildew (Fig. 1C). Taken together, these observations suggest that the edr1-mediated enhanced disease resistance, drought-induced growth, and cell death phenotypes may share the same signal transduction pathways.
edr1-Mediated Ethylene-Induced Enhanced Senescence Is EIN2 Dependent, But NPR1, PAD4, and SID2 Independent
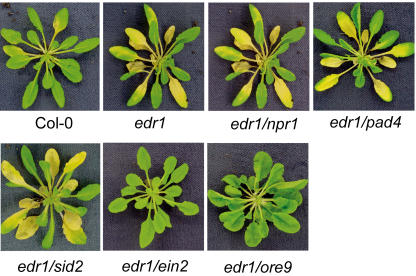
To understand why edr1 mutant plants display ethylene-induced enhanced senescence and to gain insight into the cross talk between the ethylene and SA pathways, we tested the ethylene phenotypes of double mutants edr1/ein2-1, edr1/npr1(nim1-1), edr1/pad4-2, and edr1/sid2-2. As shown in Figure 2, the edr1-mediated ethylene-induced senescence phenotype was suppressed by the ein2 mutation, but not by the npr1, pad4, or sid2 mutations. The edr1/ein2 double mutant displayed a delayed senescence phenotype similar to the ein2 single mutant when plants were treated with 100 ppm ethylene for 3 d. While older leaves in Col-0 plants yellowed, whole plants of edr1/ein2 stayed green. These data suggest that the edr1-mediated ethylene-induced senescence phenotype functions independent of SA signaling.
Figure 2.
The enhanced ethylene-induced senescence phenotype of edr1 mutants is EIN2 and ORE9 dependent, but NPR1, PAD4, and SID2 independent. Five-week-old plants grown under standard conditions were placed in a sealed chamber containing 100 ppm ethylene for 3 d and then photographed. The ein2 and ore9 mutations blocked the edr1-mediated ethylene-induced senescence phenotype, but the pad4, npr1, and sid2 mutations had no effect.
A Subset of the edr1-Mediated Stress-Induced and Ethylene-Induced Phenotypes Require the F-Box Gene ORE9
To investigate whether ORE9 plays a role in edr1-mediated phenotypes, we crossed edr1 with ore9-1 and tested the phenotypes of edr1/ore9 double mutants. Interestingly, the ore9 mutation suppressed the edr1-mediated ethylene-induced senescence (Fig. 2) and drought stress response (growth inhibition; Fig. 1), but did not affect edr1-mediated spontaneous lesions (Fig. 1) and the enhanced powdery mildew resistance phenotype (data not shown). These data indicate that ORE9 is involved in at least a subset of edr1-mediated phenotypes and that protein degradation may play a role in EDR1 regulation of drought stress responses and ethylene-induced senescence. ORE9 also controls shoot lateral branching, as ore9 mutations show an enhanced shoot branching phenotype (Stirnberg et al., 2002; ORE9 is called MAX2 in this publication). The edr1 mutation had no effect on the ore9-mediated shoot branching phenotype (data not shown).
edr1-Mediated Disease Resistance Does Not Require NDR1 or COI1
To gain more insight into edr1-mediated disease resistance signaling, we also tested two other signaling components, NDR1 and COI1, in the edr1-mediated disease resistance pathway. NDR1 encodes a small, highly basic, putative membrane protein, and is required for disease resistance mediated by some CC-NBS-LRR type R genes (Century et al., 1997). COI1 encodes a protein containing an F-box motif and is a central component of the jasmonic acid response pathway (Xie et al., 1998). Loss-of-function coi1 mutants also display a male sterile phenotype (Xie et al., 1998). The edr1 mutation had no effect on the coi1-associated male sterile phenotype (data not shown). Neither ndr1 nor coi1 suppressed the edr1-mediated enhanced powdery mildew resistance (data not shown). The failure of coi1 to suppress edr1-mediated resistance is consistent with our earlier observation that ein2 mutations also do not suppress this phenotype (Frye et al., 2001); thus, we conclude that this resistance phenotype functions independent of the jasmonic acid pathway. Table I summarizes the data obtained from the double mutant analyses.
Table I.
Suppression of edr1-mediated phenotypes by other characterized Arabidopsis mutations
−, Did not suppress the edr1-mediated phenotype; +, suppressed the edr1-mediated phenotype; NA, not analyzed.
| Mutation | Ethylene-Induced Senescence | Powdery Mildew Resistance | Drought-Induced Lesions | Drought-Induced Growth Inhibition |
|---|---|---|---|---|
| npr1 | − | + | + | + |
| pad4 | − | + | + | + |
| eds1 | − | + | + | + |
| ore9 | + | − | − | + |
| ein2 | + | − | − | − |
| ndr1 | − | − | − | − |
| coi1 | NA | − | NA | NA |
| sid2 | − | + | + | + |
Overexpression of the EDR1 C Terminus in Transgenic Plants
The EDR1 C terminus alone displays kinase activity in vitro (Tang and Innes, 2002), suggesting that the edr1 C terminus (kdEDR1) may be responsible for EDR1 function and that the N-terminal portion of EDR1 might function as a regulator to control kinase activity. To investigate the function of the EDR1 kinase domain in planta, we overexpressed kdEDR1 in both the Col-0 and edr1 backgrounds. Previously, we attempted to overexpress kdEDR1 using a CaMV 35S promoter, but we were unable to detect any expression at either the RNA or protein levels in all examined Col-0 and edr1 transgenic lines, suggesting that the EDR1 kinase domain may be toxic to plants. To overcome this problem, we used a dexamethasone-inducible system to overexpress kdEDR1. The dexamethasone-inducible promoter was fused to the C-terminal domain of EDR1, and a start codon was added in frame (see “Materials and Methods”). The construct was then transformed into Col-0 and edr1. Thirty and 35 T2 lines were recovered from the Col-0 and edr1 backgrounds, respectively. The number of T-DNA insertion sites was estimated by the segregation of hygromycin resistance in T2 lines. Transgenic lines containing only one site of insertion were selected and used for further characterization. Neither Col-0 nor edr1 transgenic plants showed any morphologic or developmental defects prior to treatment with dexamethasone.
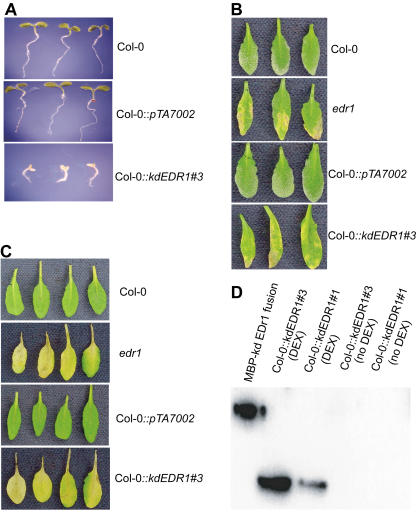
To investigate whether overexpression of the EDR1 kinase domain is toxic, we treated transgenic plants with dexamethasone at different developmental stages. Eight Col-0::kdEDR1 lines were sown and grown on one-half Murashige and Skoog medium with or without dexamethasone. Seeds from these transgenic lines all germinated and grew normally on plates without dexamethasone. In dexamethasone containing plates (200 nm), the growth of all eight Col-0::kdEDR1 transgenic lines was inhibited to various degrees. Three of these eight lines displayed severe growth inhibition. The seedlings of these three lines became yellow and died 5 d after germination, while control nontransgenic Col-0 and vector control plants were still green and viable (Fig. 3A), demonstrating that overexpression of the EDR1 kinase domain is toxic to plants. This observation is consistent with our previous results, in which we failed to recover any stable transgenic lines overexpressing the kinase domain under the control of a CaMV 35S promoter. However, transient overexpression of the EDR1 kinase domain had no obvious effects on older plants. Five-week-old Col-0::kdEDR1 transgenic plants were sprayed with 50 μm dexamethasone and no obvious phenotypes were observed, although the EDR1 kinase domain was highly expressed at the protein level (data not shown).
Figure 3.
Overexpression of the EDR1 kinase domain. A, Overexpression of the EDR1 kinase domain is toxic to plants in the early development stage. Seeds from wild-type Col-0, Col-0::kdEDR1#3, and Col-0::pTA7002 vector control plants were sown and grown on one-half Murashige and Skoog medium containing 200 nm dexamethasone. Three seedlings from each line were removed from the plates and photographed 5 d after germination. B, Overexpression of the EDR1 kinase domain enhances powdery mildew resistance. Transgenic plants carrying the dexamethasone-inducible EDR1 kinase domain were inoculated with E. cichoracearum and then sprayed with 50 μm dexamethasone. The phenotype was scored 8 d after inoculation. Wild-type and vector control plants displayed abundant spores (visible as a white powder) and lacked the yellow-brown necrotic patches observed in the edr1 mutant and transgenic Col::kdEDR1 plants. C, Overexpression of the EDR1 kinase domain enhances ethylene-induced senescence. Transgenic plants were sprayed with 50 μm dexamethasone and placed in a sealed chamber containing 100 ppm ethylene for 3 d. D, Immunoblot analysis of EDR1 protein content. Five-week-old plants were sprayed with 50 μm dexamethasone and total protein was extracted from rosette leaves 3 d after spraying. As a positive control, a purified recombinant EDR1 kinase domain-maltose-binding protein fusion was included in the blot. EDR1 was detected using antiserum raised against the C-terminal 16 amino acids of EDR1 (Tang and Innes, 2002).
To assess the function of the EDR1 kinase domain in powdery mildew resistance, Col-0 and edr1 transgenic lines containing the dexamethasone-inducible kdEDR1 construct were inoculated with E. cichoracearum. To induce the expression of the EDR1 kinase domain, the plants were treated with 50 μm dexamethasone after inoculation with E. cichoracearum. Without dexamethasone treatment, the Col-0 and edr1 transgenic plants displayed normal powdery mildew disease responses, which were indistinguishable from untransformed Col-0 and edr1 plants. Surprisingly, transient overexpression of the EDR1 kinase domain enhanced powdery mildew resistance on Col-0 wild-type plants, phenocopying the edr1 mutant (Fig. 3B). Transgenic Col-0 plants carrying the vector alone were not altered in susceptibility to powdery mildew (Fig. 3B). The edr1::kdEDR1 lines did not show any phenotype changes. These results conflict with the hypothesis that removal of the N-terminal non-kinase domain will produce a constitutively active kinase that can exert the function of the full-length protein. Rather, these results indicate that the EDR1 N-terminal domain is required for EDR1 function and that the overexpressed EDR1 N-terminal deletion form might interfere with native EDR1.
Previously, we observed that edr1 plants display an enhanced ethylene-induced senescence (Frye et al., 2001). When 5-week-old plants are treated with ethylene for 3 d, the oldest two leaves become yellow, however, in the edr1 mutant, this yellowing occurs on much younger leaves. To investigate whether overexpression of the C-terminal domain of EDR1 can affect Col-0 and edr1 ethylene responses, we exposed 5-week-old Col-0 wild-type, edr1 and Col-0::kdEDR1 transgenic lines to 100 ppm ethylene for 3 d under 9 h light. The oldest two leaves in wild-type Col-0 yellowed. However, in edr1, this yellowing was visible in much younger leaves as described previously (Frye et al., 2001). Without dexamethasone treatment, the Col-0 and edr1 kdEDR1 transgenic plants displayed ethylene responses similar to untransformed Col-0 and edr1 plants. Dexamethasone-treated transgenic plants, however, showed an ethylene response similar to the edr1 plants (Fig. 3C), while Col-0 lines carrying the vector only showed no significant difference from non-transgenic Col-0 plants (Fig. 3C). The dexamethasone-inducible kdEDR1 construct did not complement the edr1-mediated ethylene-induced senescence phenotype. These data are consistent with the results we obtained from powdery mildew tests and indicate that transient overexpression of the EDR1 kinase domain causes a dominant negative phenotype.
To investigate the mechanisms underlying the dominant negative phenotypes of the Col-0::kdEDR1 lines, we assessed EDR1 protein levels in the transgenic lines using immunoblot analyses. Figure 3D shows that the EDR1 kinase domain was highly expressed at the protein level in the kdEDR1 transgenic plants treated with dexamethasone, suggesting that the dominant negative phenotypes of these lines are caused by the kinase domain interfering with full-length EDR1 function. In the absence of dexamethasone treatment, the EDR1 KD protein was not detected, demonstrating the efficacy of the EDR1 induction by dexamethasone (Fig. 3D).
DISCUSSION
While the edr1 mutant is phenotypically wild-type in the absence of pathogen under optimal growth conditions, we observed that edr1 mutants were stunted and displayed spontaneous lesions under drought conditions. This observation suggests that edr1 mutants are more sensitive to environmental conditions than wild-type Col-0 plants and that EDR1 might be involved in regulating stress induced growth and cell death in addition to the previously reported disease resistance and ethylene responses.
To date, a number of mutants that display enhanced disease resistance to pathogens have been identified. Many of them constitutively display defense response phenotypes, including spontaneous lesions and defense gene expression. Several of these are also smaller in size, indicating a correlation between defense induction and growth inhibition (Dietrich et al., 1995; Rate et al., 1999). Several mutants conditionally display microscopic lesions. These pleiotropic phenotypes also suggest a connection between defense responses and stress responses.
The link between stress and defense responses is also supported by analysis of ozone-induced responses in Arabidopsis (Rao and Davis, 1999; Overmyer et al., 2000; Rao et al., 2002). Ozone induces HR-like lesions in Arabidopsis, along with ethylene and SA biosynthesis. Significantly, ozone-induced lesions are attenuated by mutations in either the SA or ethylene signal transduction pathways, and blocks in the SA signaling pathway block ozone-induced ethylene production (Rao and Davis, 1999; Overmyer et al., 2000; Rao et al., 2002). Thus, SA appears to potentiate lesion formation directly and by enhancing ethylene biosynthesis. Drought treatment in Arabidopsis induces ethylene biosynthesis (Rao and Davis, 1999; Rao et al., 2002), but the failure of the ein2-1 mutation to suppress drought-induced phenotypes in edr1 plants indicates that ethylene is not causally related to these phenotypes.
edr1-Mediated Enhanced Disease Resistance and Ethylene-Induced Senescence Are Controlled by Separate Pathways
The high similarity between the CTR1 and EDR1 protein sequences led us to investigate whether EDR1 directly regulates ethylene responses. Unlike ctr1 mutants, edr1 plants display a normal triple response and the ethylene-inducible genes CHIB and HEL display normal ethylene inducibility (Frye et al., 2001). However, edr1 plants do display an enhanced ethylene-induced senescence phenotype. In addition, the defense gene PR-1 is highly induced by ethylene in edr1 mutant plants (Frye et al., 2001). These data suggest there is an overlap between the genetic control of ethylene-induced senescence and SA-induced defense responses. However, the ein2 mutation has no effect on edr1-mediated disease resistance, although it suppresses the edr1-mediated ethylene-induced senescence. Furthermore, the ore9 mutation has no effect on edr1-mediated disease resistance, but abolishes the enhanced ethylene-induced senescence phenotype. These data demonstrate that the edr1-mediated enhanced senescence and enhanced disease resistance are separate pathways requiring different components. However, it is unclear whether EIN2 and ORE9 function in the same pathway; the ein2 mutation affects only the edr1-mediated ethylene phenotype, while the ore9 mutation also affects edr1-mediated drought-induced growth phenotypes. This observation suggests that EIN2 may only function in the ethylene signal transduction pathway, while ORE9 may also function in other developmental processes involving EDR1.
The edr1-mediated disease resistance and drought-induced spontaneous lesion phenotypes appear to share the same signal transduction pathway. The npr1, pad4, eds1, and sid2 mutations all suppressed these two phenotypes, while ein2, ore9, and ndr1 mutations did not. However, the drought-induced growth phenotype was separable from the drought-induced HR-like phenotype, because the growth phenotype, but not the lesion phenotype, was suppressed by the ore9 mutation. This observation suggests that inhibition of growth requires the function of the ORE9 protein and is a response that occurs downstream, or independent from, the cell death phenotype. Because disease resistance is maintained in the edr1/ore9 double mutant, it appears that growth inhibition is not an obligatory cost of enhanced disease resistance.
Protein Degradation May Be Involved in edr1-Associated Stress Responses and Ethylene Responses
In the double mutant analysis, we demonstrated that an F-box protein, ORE9, is involved in the drought stress responses and ethylene-induced enhanced senescence. This suggests that protein degradation may play a role in the EDR1-mediated signal transduction pathway since ORE9 has been shown to interact with ASK1, a component of the SCF ubiquitin ligase complex, in vitro (Woo et al., 2001). EDR1 may negatively regulate both stress and ethylene responses by affecting the ORE9 dependent protein degradation pathways. In this scenario, edr1 plants can respond to stress and ethylene more readily, and these processes require ORE9 to degrade regulators involved in stress and ethylene responses. In the simplest model, wild-type EDR1 protein would function normally to phosphorylate ORE9, keeping it inactive. Loss of EDR1 function would activate ORE9, causing degradation of negative regulators of senescence, making plants more prone to environmental signals that promote senescence. Removal of ORE9 then stabilizes these negative regulators, preventing premature senescence. Alternatively, EDR1 may phosphorylate these negative regulators directly, which then prevents their binding to ORE9 and subsequent degradation. Loss of EDR1 function would thus promote binding of the regulators to ORE9 and their subsequent degradation. Loss of ORE9 would then restore stability to these negative regulators. This latter scenario is somewhat at odds with data on F-box proteins in yeast and animals, in which phosphorylation of substrates promotes binding to F-box proteins, rather than inhibiting binding (Reed, 2003).
EDR1 Function Requires More Than Just the Kinase Domain
EDR1 has significant similarity to the CTR1 protein, which encodes a negative regulator of the ethylene response pathway. CTR1 and EDR1 belong to the B3 subgroup of plant Raf-like kinases (Ichimura et al., 2002). Like the EDR1 protein, CTR1 displays a Ser/Thr protein kinase activity in vitro. However, unlike Raf-1, deletion of the N-terminal domain of CTR1 does not elevate the kinase activity, indicating that the N terminus does not inhibit kinase function (Huang et al., 2003). In addition, overexpression of the CTR1 N terminus causes a dominant negative phenotype. Further analysis of several mutant alleles demonstrates that the CTR1 kinase activity is required for CTR1 function (Huang et al., 2003).
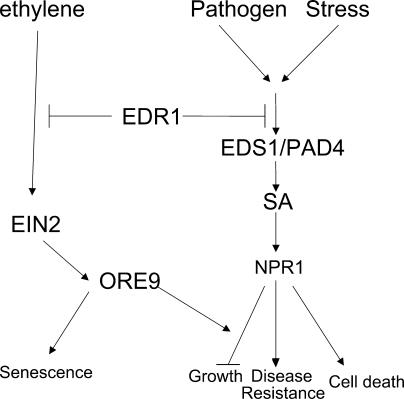
Although the EDR1 C-terminal domain is highly similar to the CTR1 kinase domain and the EDR1 C-terminal domain displays protein kinase activity in vitro (Tang and Innes, 2002), it is not known whether kinase activity is required in the EDR1-mediated signal transduction pathway. The EDR1 C terminus is similar to the catalytic domain of the animal Raf protein; however, this similarity is too low to infer any substrate specificity (Tang and Innes, 2002). To investigate the function of the EDR1 kinase domain, we overexpressed the EDR1 C terminus in the wild-type and edr1 mutant backgrounds. The EDR1 kinase domain does not complement the edr1 mutant phenotype. Rather, it negatively affects native EDR1 function, indicating that EDR1 function is dependent upon the N-terminal regulatory region. Alternatively, the EDR1 kinase domain may compete with the EDR1 full-length protein by sequestering substrates in planta. Our results support a model for edr1-mediated disease resistance, ethylene-induced senescence, and drought-induced spontaneous lesion and growth phenotypes (Fig. 4). In this model, E. cichoracearum and stress both induce SA accumulation, which lead to formation of lesions and inhibition of growth in the absence of EDR1. Thus, the normal function of EDR1 would be to prevent inappropriate initiation of cell death, growth inhibition, and senescence.
Figure 4.
A model summarizing EDR1's role in programmed cell death, drought tolerance, and senescence. E. cichoracearum and drought induce SA accumulation, which promotes HR-like lesions. This process is negatively regulated by EDR1 to prevent inappropriate initiation of stress responses. These stress responses inhibit growth, and such growth inhibition requires both NPR1 and ORE9 function. EDR1 also negatively regulates ethylene-induced senescence, which requires the function of EIN2 and ORE9, but not NPR1, EDS1, or PAD4.
MATERIALS AND METHODS
Plants Growth and Inoculation with Erysiphe cichoracearum
Arabidopsis (Arabidopsis thaliana) seeds were sown in pots filled with vegetable plug mix (Scotts, Marysville, OH). Seeded pots were held at 4°C for 3 d before moving to a growth room maintained at 23°C and 70% relative humidity. Plants were subjected to a 9-h-light/15-h-dark cycle with an average light intensity of 150 μE m−2 s−1 at plant level. E. cichoracearum was maintained on hyper-susceptible pad4 plants and passed to fresh plants every 8 to 10 d. To inoculate plants, diseased plants were used to brush healthy 4- to 6-week-old plants to pass asexual spores (conidia) onto the new plants. Disease phenotypes were scored 8 d after inoculation.
Drought Stress Assay
Plants were grown in growth rooms as described above for 3 weeks, then watering was withheld for 2 weeks. The aerial portions of plants were then weighed and photographed. Ten plants of each line were weighed and the mean weight was used to represent the growth phenotype.
Ethylene-Induced Senescence Assay
Six-week-old plants were placed in a sealed chamber containing 100 ppm ethylene for 3 d. The plants were scored and photographed as previously described (Frye et al., 2001).
Double Mutant Analysis
Isolation of edr1/ein2-1, edr1/eds1-1, edr1/pad4-2, and edr1/npr1(nim1-1) double mutants has been described previously (Frye et al., 2001). The edr1/ore9-1, edr1/ndr1-3, edr1/coi1-1, and edr1/sid2-2 double mutants were created by standard genetic crosses. To identify double mutants, we used PCR to amplify the respective genes, which were then sequenced to identify plants that were homozygous for the mutations. All double mutant analyses was conducted on F3 homozygous lines except for edr1/coi1-1, which is male sterile for the double mutant. F2 individuals homozygous for edr1/coi1-1 were identified and assessed by PCR followed by direct sequencing.
Plant Transformation Constructs
A full-length EDR1 cDNA carried in the pGEM-T Easy vector (Promega, Madison, WI) was used as a PCR template. Primers (5′-AAGGCTCGAGATGGATGTTGGTGAATGTGAAATTC-3′ and 5′-AAGGTCTAGACTATTGTGGTGTAGGAAGTACAAGC-3′) were used to amplify the kinase domain. Restriction sites for XhoI and XbaI and a start codon ATG were incorporated in the primer. The PCR products were cloned into the pTA7002 vector, which contains a dexamethasone-inducible promoter (Aoyama and Chua, 1997). The clones were verified by sequencing and transformed into Agrobacterium tumefaciens strain GV3101 by electroporation.
Plant Transformation
Arabidopsis plants were transformed by using a floral dip method (Clough and Bent, 1998). Transgenic plants (T1 generation) were selected by growing on 0.5× Murashige and Skoog salts (Life Technologies, Grand Island, NY), plus 0.8% agar and 50 μg/mL hygromycin. T1 lines were allowed to self-fertilize and lines with single insertion loci were selected based on 3:1 segregation of hygromycin resistance in the T2 generation.
Transient Overexpression of the EDR1 Kinase Domain
The toxicity of the EDR1 kinase domain was examined in seedling and adult plants, respectively. Transgenic lines carrying DEX::kdEDR1 were sown on 0.5× Murashige and Skoog salts plus 0.8% agar and 200 nm dexamethasone. Seedlings were scored 5 d after germination. Five-week-old transgenic plants were sprayed with 50 μm dexamethasone and the phenotype observed for 1 week. To test the effect of the EDR1 kinase domain on powdery mildew disease resistance, transgenic plants were inoculated with E. cichoracearum and then immediately sprayed with 50 μm dexamethasone. The phenotype was scored 8 d after inoculation. In the ethylene-induced senescence assay, 5-week-old plants were sprayed with 50 μm dexamethasone and placed in a sealed chamber containing 100 ppm ethylene immediately for 3 d. To prepare protein extract, plant leaves were collected 3 d after spraying with dexamethasone.
Antibody Production
A peptide containing the C-terminal 16 amino acids of EDR1, showing the least similarity to homologs of EDR1 in Arabidopsis, was synthesized and conjugated to keyhole limpet hemocyanin (Tang and Innes, 2002). A polyclonal antiserum was raised in rabbits against this synthetic peptide as previously described. (Tang and Innes, 2002). Crude serum was used for immunoblot analyses.
Preparation of Protein Extracts
Preparation of protein extracts was conducted as previously described (Tang and Innes, 2002). Briefly, leaf material was ground in liquid nitrogen and extracted in two volumes of extraction buffer (50 mm HEPES, pH 7.4, 5 mm EDTA, 5 mm EGTA, 5 mm dithiothreitol, 10 mm NaF, 10 mm Na3VO4, 50 mm beta-glycerophosphate, 1 mm phenylmethylsulfonyl fluoride, 2 μg/mL antipain, 2 μg/mL aprotinin, and 2 μg/mL leupeptin, 10% glycerol). The extract was centrifuged at 4,000 rpm for 30 min at 4°C, and the supernatant was used as total protein. The protein concentration was determined using a protein assay kit (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as the standard.
Immunoblotting
Immunoblotting was conducted as described previously (Tang and Innes, 2002). Briefly, protein samples (20 μg total protein per lane) were separated by electrophoresis through a 10% SDS-polyacrylamide gel and protein was transferred to nitrocellulose membranes by wet electroblotting in transfer buffer (10 mm Tris, 192 mm Gly, and 20% methanol). The membrane was blocked using 5% nonfat dry milk in Tris-buffered saline (TBS) buffer (10 mm Tris-HCl, pH7.5, 100 mm NaCl) for 1 h, incubated with anti-EDR1 antisera at a dilution of 1:5,000 for 2 h at room temperature, and then washed three times with TBS buffer. The first two washes contained 0.05% Tween 20. The membrane was then incubated with anti-rabbit IgG antibody at a dilution of 1:10,000 for 1 h at room temperature. After three washes with TBS, the antibody-antigen was visualized using SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) following the manufacturer's protocol.
Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for non-commercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank H.G. Nam for providing ore9-1 mutant seeds, J.G. Turner for providing coi1-1 mutant seeds, and F.M. Ausubel for providing sid2-2 seeds, J. Parker for providing F2 seeds from a cross between edr1 and pad4-2 and between edr1 and eds1-1, and the Arabidopsis Biological Resource Center at Ohio State University for providing ein2-1 seeds. We also thank J. Glazebrook for critical reading of the manuscript.
This work was supported by the National Institutes of Health (grant no. R01 GM063761).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060400.
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1995) Arabidopsis mutants simulating disease resistance response. Cell 77: 565–577 [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Innes RW (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK Kinase. Proc Natl Acad Sci USA 98: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffroy V, Sicard D, de Oliveira JC, Sevignac M, Cohen S, Gepts P, Neema C, Langin T, Dron M (1999) Identification of an ancestral resistance gene cluster involved in the coevolution process between Phaseolus vulgaris and its fungal pathogen Colletotrichum lindemuthianum. Mol Plant Microbe Interact 12: 774–784 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Silverman FP, Liang H (2000) Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Tena G, Henry Y, Zhang S, Hirt H, Ellis BE, Morris PC, Wilson C, Champion A, Innes RW, et al (2002) Mitogen-activated protein kinase cascades in plants. Nomenclature of Arabidopsis MAP kinases (MPKs) and MAP kinase kinases (MKKs). Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Metraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H Jr, Kangasjarvi J (2000) Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12: 1849–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Rao MV, Davis KR (1999) Ozone-induced cell death occurs via two distinct mechanisms in Arabidopsis: the role of salicylic acid. Plant J 17: 603–614 [DOI] [PubMed] [Google Scholar]
- Rao MV, Hyung-il Lee H, Davis KR (2002) Ozone-induced ethylene production is dependent on salicylic acid, and both salicylic acid and ethylene act in concert to regulate ozone-induced cell death. Plant J 32: 447–456 [DOI] [PubMed] [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT (1999) The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI (2003) Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol 4: 855–864 [DOI] [PubMed] [Google Scholar]
- Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner HY, Johnson J, Delaney TP, Jesse T, Vos P, et al (1997) The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Tang D, Innes RW (2002) Overexpression of a kinase-deficient form of the EDR1 gene enhances powdery mildew resistance and ethylene-induced senescence in Arabidopsis. Plant J 32: 975–983 [DOI] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell (Suppl) 14: S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]