Abstract
Salicylic acid (SA) is implicated in the induction of programmed cell death (PCD) associated with pathogen defense responses because SA levels increase in response to PCD-inducing infections, and PCD development can be inhibited by expression of salicylate hydroxylase encoded by the bacterial nahG gene. The acd11 mutant of Arabidopsis (Arabidopsis thaliana L. Heynh.) activates PCD and defense responses that are fully suppressed by nahG. To further study the role of SA in PCD induction, we compared phenotypes of acd11/nahG with those of acd11/eds5-1 and acd11/sid2-2 mutants deficient in a putative transporter and isochorismate synthase required for SA biosynthesis. We show that sid2-2 fully suppresses SA accumulation and cell death in acd11, although growth inhibition and premature leaf chlorosis still occur. In addition, application of exogenous SA to acd11/sid2-2 is insufficient to restore cell death. This indicates that isochorismate-derived compounds other than SA are required for induction of PCD in acd11 and that some acd11 phenotypes require NahG-degradable compounds not synthesized via isochorismate.
Plants possess an immune system to defend themselves against pathogen infection. An intensively studied inducible immune response occurs when a pathogen carrying an avirulence (avr) gene is recognized directly or indirectly by a cognate resistance (R) gene in the plant. This leads to activation of defenses that restrict pathogen growth in infected tissues and in noninfected tissues by a process referred to as systemic acquired resistance (SAR). These defense responses are typically accompanied by localized programmed cell death (PCD) around the site of infection in the hypersensitive response (HR; Nimchuk et al., 2003). In the absence of an R-avr interaction, basal resistance responses are also activated, although they may not successfully restrict pathogen growth, and disease symptoms may develop (Glazebrook et al., 1997).
The importance of salicylic acid (SA) in the induction of such resistance responses is supported by both gain- and loss-of-function evidence. SA levels increase upon many avirulent and some virulent infections (Malamy et al., 1990; Métraux et al., 1990; Heck et al., 2003), and application of exogenous SA, or generation of high endogenous SA levels by expression of bacterial SA synthesizing enzymes, is sufficient to induce resistance to many normally virulent pathogens (White, 1979; Ward et al., 1991; Verberne et al., 2000; Mauch et al., 2001). Loss-of-function analyses have relied upon SA depletion by transgenic expression of a bacterial SA hydroxylase encoded by nahG. NahG abrogates local R function elicited by a range of bacterial, oomycete, and viral pathogens (Delaney et al., 1994; Rairdan and Delaney, 2002) as well as SAR (Gaffney et al., 1993) and basal resistance responses to virulent bacteria, fungi, and oomycetes (Delaney et al., 1994; Reuber et al., 1998). These results have been supported by the characterization of two SA-deficient mutants of Arabidopsis (Arabidopsis thaliana L. Heynh.), sid2 and eds5 (Rogers and Ausubel, 1997; Nawrath and Métraux, 1999; Dewdney et al., 2000). Both sid2 and eds5 show enhanced susceptibility to many virulent infections and strongly reduced induction of SAR. However, only some R-mediated local resistance responses are affected in sid2 and eds5 (Nawrath and Métraux, 1999), and in most cases their hypersusceptibility is not as pronounced as in NahG plants. SID2 encodes a pathogen induced isochorismate synthase (ICS; Wildermuth et al., 2001), while EDS5 encodes an orphan multidrug and toxin extrusion transporter that may be involved in a positive feedback loop stimulating SA accumulation as EDS5 expression is induced by SA (Nawrath et al., 2002). The strong reduction of SA accumulation in sid2 mutants in response to infection establishes the isochorismate pathway as the major route to defense-associated SA. A second Phe ammonia lyase (PAL)-dependent SA biosynthesis pathway from cinnamic acid via benzoic acid has also been described (Yalpani et al., 1993; Ribnicky et al., 1998), although the functional relevance of this pathway is unclear.
The involvement of SA in activation of PCD in the HR is supported by similar lines of evidence but remains less clear. SA does not induce HR-like PCD on its own in whole plants, although it may induce PCD in cell culture (Kawai-Yamada et al., 2004). HR induced by two Peronospora parasitica isolates avirulent on Arabidopsis appears to depend on SA since nahG, eds5, and sid2 blocked the HR in response to infection, although trailing necrosis surrounding growing hyphae was still observed (Nawrath and Métraux, 1999). Similarly, nahG delays the HR of tobacco (Nicotiana tabacum) in response to tobacco mosaic virus (Mur et al., 1997). Consistent with these observations of PCD attenuation by SA removal, exogenous SA strongly accelerated HR cell death in soybean (Glycine max) suspension cells (Shirasu et al., 1997) and induced cell death in Arabidopsis lsd1 mutants and RPW8 enhanced transcription lines kept under conditions nonpermissive for spontaneous HR-like cell death development (Dietrich et al., 1994; Xiao et al., 2003). On the other hand, HR is normally induced in eds5 and sid2 in response to high doses of Pseudomonas syringae pv tomato DC3000 expressing the avr gene avrRpm1, avrRpt2, or avrRps4 (Nawrath and Métraux, 1999; Dewdney et al., 2000). Clearer evidence pointing to a role of SA in PCD comes from the analysis of Arabidopsis acd and lsd mutants that spontaneously activate PCD and defense responses. In many of these mutants, including acd6-1, acd11, ssi1, and lsd6, nahG expression completely suppresses PCD development, while this can be restored by application of SA agonists such as 2,6-dichloroisonicotinic acid (INA) and benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH; Weymann et al., 1995; Rate et al., 1999; Shah et al., 1999; Brodersen et al., 2002). Similarly, nahG inhibits PCD induction by the mycotoxin fumonisin B1 (Asai et al., 2000). Thus, the clearest links of SA to PCD induction are based on analysis of NahG plants.
However, recent reports comparing phenotypes of NahG plants with SA-deficient eds5 and sid2 mutants indicate that the effects of nahG expression are not as straightforward as previously thought. For example, the loss of nonhost resistance toward P. syringae pv phaseolica in Arabidopsis expressing nahG was not due to reduced SA levels but to the generation of SA breakdown products (van Wees and Glazebrook, 2003), whereas the failure of nahG-expressing plants to accumulate camalexin and ethylene in response to P. syringae pv tomato infections resulted from the action of salicylate hydroxylase on as yet unidentified compounds different from SA (Heck et al., 2003).
We previously characterized the lethal acd11 mutant in which PCD and defense responses activated at the seedling stage can be completely suppressed by nahG (Brodersen et al., 2002). Here, we use the acd11 mutant in combination with nahG, eds5, and sid2 to further examine the role of SA in PCD. To our knowledge, such a side-by-side comparison of the effect of nahG, eds5, and sid2 on PCD and SA accumulation using the same system has not been reported. We show that while SA may play a role in PCD initiation in this system, it is not the only isochorismate-derived compound required for PCD induction. In addition, some acd11 phenotypes are dependent on NahG degradable compounds not derived from isochorismate.
RESULTS
Penetrance of acd11 Phenotypes in Columbia and Landsberg erecta Accessions
Our previously isolated acd11-1 deletion mutant allele is in the Landsberg erecta (Ler) accession, while mutant alleles of EDS5 (eds5-1 to eds5-3) and SID2 (sid2-1 and sid2-2) were only available in Columbia (Col-0) at the beginning of this study. Since mixed genetic backgrounds arising from crosses of mutants in nonisogenic parental accessions may complicate interpretations of observed double mutant phenotypes, we first made use of a cross of acd11-1 to wild-type Col-0 to examine phenotypes conferred by acd11-1 in Col-0. In the F2 population, 19% of the progeny (18/95) showed a typical Acd− phenotype whose onset occurred at the same time as the acd11-1Ler control. None of the 77 phenotypically wild-type plants were homozygous for acd11-1, indicating that the slightly distorted segregation ratio was not due to the presence of a recessive acd11 suppressor in Col-0. In addition, we recently isolated an acd11 T-DNA insertion allele in Col-0 (acd11-2, SALK_018628) with an Acd− phenotype identical to that of acd11-1. We next analyzed total SA levels in acd11-1, acd11-2, and acd11-1LerxCol and found that they accumulated similar high levels of total SA compared with the corresponding ACD11 controls (data not shown). This indicates that the Acd− and SA accumulation phenotypes are not affected in a mixed Ler/Col-0 genetic background, so that acd11-1 crossed to eds5-1 and sid2-2 may be suitable for examining roles of SA in PCD induction.
Phenotypes of acd11/eds5-1 and acd11/sid2-2 Mutants
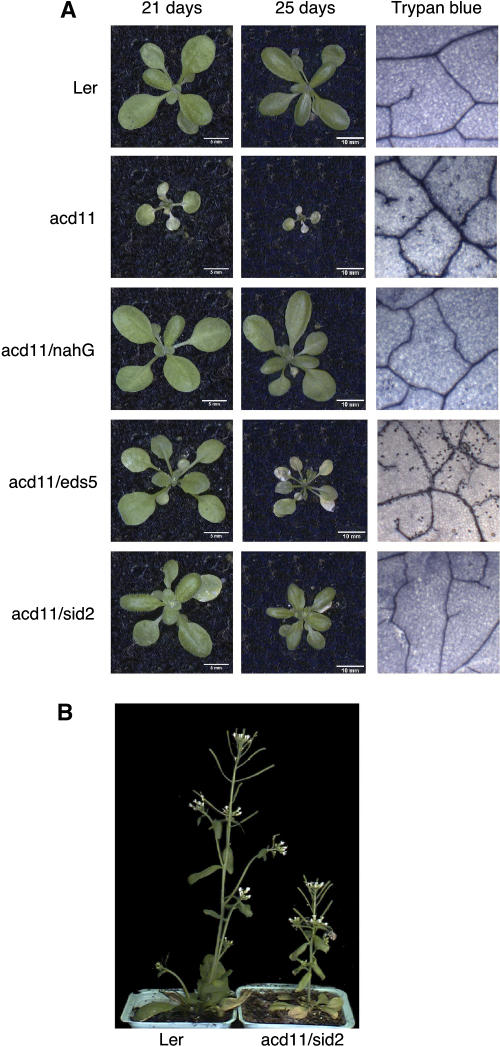
Families homozygous for eds5-1 and sid2-2 and heterozygous for acd11-1 were selected in the F3 generation of crosses of eds5-1 and sid2-2 to acd11-1. These lines were grown together with acd11-1 and acd11/nahG (Brodersen et al., 2002) for phenotypic comparison. In three independent F3 families, acd11/eds5-1 exhibited cell death symptoms 4 to 5 d later than acd11 and consequently grew much larger than acd11 (average leaf area over all leaves 32 mm2 versus 11 mm2 in acd11-1; Fig. 1A). However, once leaf yellowing initiated in acd11/eds5-1, it developed as quickly as in acd11 to engulf the rosette. Like acd11, acd11/eds5-1 exhibited strong shoot inhibition, and stem and inflorescence formation was rarely observed, although a few flowers and siliques typically formed directly at the rosette. Microscopic examination of trypan blue-stained leaves showed that they contained numerous dead cells similar to acd11 (Fig. 1A). Thus, except for being delayed, acd11/eds5-1 exhibited the same phenotypes as acd11.
Figure 1.
Phenotypes of acd11/eds5, acd11/sid2, and acd11/nahG. A, Phenotypes and trypan blue stainings of leaves at the rosette stage. Trypan blue stainings were performed at day 25. B, Stature of mature acd11/sid2 relative to wild-type Ler.
The effect of the sid2-2 mutation was much more pronounced. In four independent F3 families, rosettes of acd11/sid2 were found to develop normally, although they were reduced in size compared with wild type or acd11/nahG (average leaf areas over all leaves 45 mm2 in acd11-1/sid2-2; 76 mm2 in acd11/nahG and 76 mm2 in Ler; Fig. 1A). Around the time of bolting, some leaf chlorosis occurred, but flower and silique bearing bolts were produced. Although smaller than wild type and acd11/nahG plants, acd11/sid2-2 did not show the strong shoot inhibition observed in acd11 and acd11/eds5-1 (Fig. 1B). Neither the young green nor the older yellowing acd11/sid2-2 leaves contained dead cells, as revealed by trypan blue staining. Thus, most but not all acd11 phenotypes were suppressed by sid2-2. As previously described, no differences were observed between the acd11-1/nahGLerxCol line and the wild type. We also constructed an acd11-1/nahG line in Ler that showed complete suppression of the Acd− phenotype (Cui et al., 2002; data not shown). These results demonstrate marked differences between the effects of nahG expression and sid2-2 and eds5-1 mutations on the acd11 phenotype in which the strength of suppression decreases in the order nahG > sid2-2 > eds5-1. The observation that double mutant phenotypes were the same in independent families of the acd11-1/sid2-2 and acd11-1/eds5-1 crosses confirms that the mixed genetic background has little, if any, influence on the double mutant analysis. For this reason, experiments described below were performed with a single double mutant family from each cross.
SA Accumulation in Double Mutants
Due to the proposed key role of SA in PCD and defense induction, the double mutant results may be explained by different effects of nahG, sid2-2, and eds5-1 on SA accumulation in acd11. To test this, we compared total (the sum of free and Glc conjugated) SA levels in the different genetic backgrounds (Table I). This showed that acd11/eds5-1 accumulated SA to approximately 10% of the levels observed in acd11, whereas only basal SA levels could be detected in acd11/nahG or acd11/sid2-2. Thus, the weaker suppression of acd11 by eds5-1 might be due to incomplete reduction of SA accumulation, whereas this cannot explain the differences observed between acd11/sid2-2 and acd11/nahG. More specifically, these findings appear to exclude the possibility that SA synthesized via the SID2-independent PAL pathway could cause the phenotypic differences between acd11/sid2 and acd11/nahG because sid2 mutation alone leads to full suppression of SA accumulation in acd11.
Table I.
Accumulation of total SA in single and double mutants
SA and SA-glucoside levels determined by HPLC in the wild type, in single mutants and nahG, and in the acd11/eds5, acd11/sid2, and acd11/nahG double mutants as shown. “Young” refers to 18-d-old rosettes (third pair of true leaves emerging; all samples except “acd11/sid2 old”), and “old” refers to yellowing leaves on 34-d-old bolting plants. Each measurement is an average of triplicate samples, and the entire experiment was repeated twice with similar results.
| Genotype | Total SA |
|---|---|
| μg/g FW | |
| acd11 | 82 ± 14 |
| Ler | 1.3 ± 0.48 |
| acd11/sid2 young | 1.5 ± 0.52 |
| acd11/sid2 old | 1.7 ± 0.38 |
| sid2 | 1.2 ± 0.52 |
| acd11/eds5 | 8.5 ± 2.9 |
| eds5 | 1.3 ± 0.35 |
| acd11/nahG | 0.87 ± 0.14 |
| nahG | 1.01 ± 0.82 |
Expression of Cell Death and Defense Marker Genes
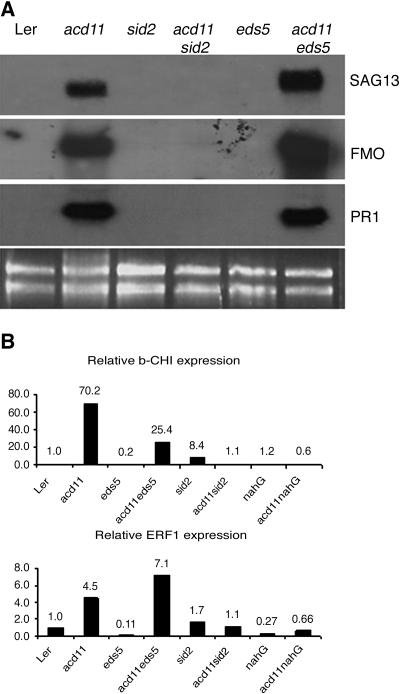
To characterize the phenotypes of the double mutants at the molecular level, we first monitored the expression of molecular markers for cell death and SA-dependent defense. We chose the SA-dependent defense marker PR1 (At2g14610) and the cell death-associated genes SAG13 (At2g29350) and FMO (At1g19250), encoding a putative short-chain alcohol dehydrogenase and a putative flavin containing monooxygenase, respectively. These genes are all strongly expressed in acd11 (Brodersen et al., 2002). RNA-blot analysis showed that these three markers were strongly expressed in acd11/eds5-1, while their expression was almost completely suppressed in acd11/sid2-2 and acd11/nahG (Fig. 2A; Brodersen et al., 2002). Thus, although these markers do not distinguish between acd11/nahG and acd11/sid2-2, this molecular analysis supports the whole plant and cellular phenotypes described above.
Figure 2.
Expression of defense and cell death markers in single and double mutants. A, RNA-blot analysis of steady-state levels of FMO, SAG13, and PR1 mRNA in the wild type, in single mutants, and in the acd11/eds5 and acd11/sid2 double mutants. rRNA controls for equal loading are shown at the bottom. In some experiments, weak FMO, SAG13, and PR1 signals were detected in acd11/sid2-2. B, Real-time RT-PCR analysis of mRNA accumulation of ethylene response genes PR3 and ERF1 in single and double mutants. Expression of ubiquitin (UBQ10) was used to normalize cDNA input from the different genetic backgrounds.
In contrast to sid2, nahG has been reported to suppress ethylene accumulation following virulent and avirulent bacterial infections (Heck et al., 2003). This leaves open the possibility that the phenotypic differences between acd11/sid2 and acd11/nahG may be due to differential ethylene accumulation and/or signaling in the two backgrounds. To test this hypothesis, we used real-time reverse transcription (RT)-PCR to monitor the expression of the ethylene-induced genes PDF1.2 (At5g44420), PR3 (At3g12500), ERF1 (At3g23240), and EBP (At3g16770), encoding plant defensin, basic chitinase, and two ethylene response element binding transcription factors, respectively (Buttner and Singh, 1997; Solano et al., 1998). No significant induction of PDF1.2 and EBP was observed in acd11 (data not shown), whereas PR3 and ERF1 were clearly induced in acd11 and acd11/eds5. Importantly, expression of both genes was equally strongly suppressed in acd11/sid2 and acd11/nahG (Fig. 2B). Thus, this analysis does not support differences in ethylene accumulation and/or signaling as a cause of the phenotypic differences between acd11/sid2 and acd11/nahG. This result is consistent with the fact that the ein2 mutation conferring complete insensitivity to ethylene does not appreciably influence the acd11 phenotype (Brodersen et al., 2002).
nahG Is Epistatic to sid2-2
Two models may explain why nahG expression results in stronger acd11 suppression than sid2-2. First, catechol produced by NahG-catalyzed decarboxylative hydroxylation of SA may act to fully suppress acd11 phenotypes in concert with SA depletion. Alternatively, NahG acts to modify cellular metabolites other than SA (or other isochorismate-derived metabolites) that are required for development of the residual chlorosis and growth phenotypes observed in acd11/sid2-2.
Analysis of the epistatic relationship between sid2 and nahG may distinguish between these possibilities (Heck et al., 2003; van Wees and Glazebrook, 2003). In the first model noted above, sid2 would be epistatic to nahG since catechol cannot be produced in the SA-deficient sid2 background. In the second model, nahG would be epistatic to sid2 since NahG could still inactivate the additional set of putative, active metabolites in a sid2 background. To examine this, acd11/nahG was crossed to acd11/sid2-2. In the segregating F2 generation, none of 10 yellowing plants typical of the acd11/sid2-2 phenotype contained the nahG transgene, whereas several completely green plants were found by PCR to be sid2-2 homozygous and to contain the nahG transgene. F3 progeny from many of these plants segregated 3:1 for the fully green:yellowing phenotypes, demonstrating that the effects of nahG are epistatic to sid2-2.
SA Does Not Restore Cell Death in acd11/sid2-2
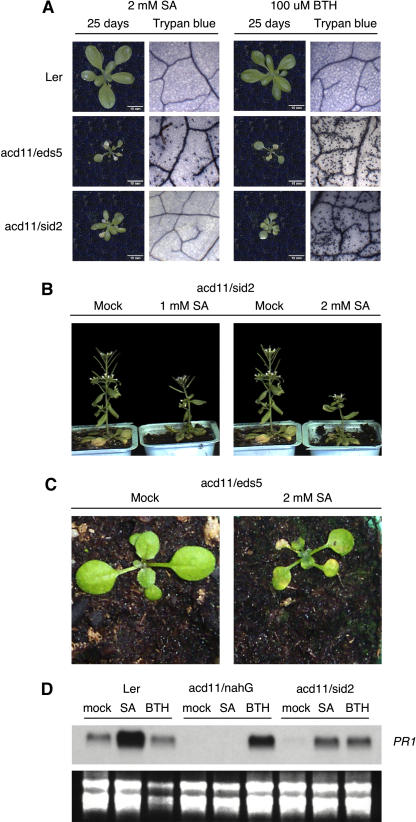
To more directly study the role of SA in cell death induction in acd11, we analyzed the effect of application of exogenous SA to acd11/eds5-1 and acd11/sid2-2 mutants. Treatment with BTH that completely restores cell death in acd11/nahG was used as a control. Both application of 1 and 2 mm SA and 100 μm BTH significantly accelerated cell death in acd11/eds5-1, suggesting that the delayed death phenotype of acd11/eds5-1, relative to acd11, was due to reduced SA levels (Fig. 3C). In contrast, trypan blue staining showed that neither 1 nor 2 mm SA was sufficient to restore leaf cell death in acd11/sid2-2, although a dose-dependent growth reduction was observed. However, cell death in acd11/sid2-2 was fully restored by treatment with 100 μm BTH (Fig. 3A) or 325 μm INA (data not shown). When SA concentrations were elevated to 5 mm, some acd11/sid2-2 individuals wilted quickly but did not exhibit the phenotype typical of acd11 after BTH application. Notably, some sid2-2 single mutant plants also wilted following this treatment with 5 mm SA, indicating that this level of application affects plants lacking the acd11 mutation.
Figure 3.
Responses of acd11/eds5 and acd11/sid2 to SA and BTH. The compounds were sprayed onto leaves at concentrations of 2 mm (SA) and 100 μm (BTH). Treatments were done at day 18 and pictures taken at day 25. Trypan blue stainings were also performed at day 25. Nontreated control plants are shown in Figure 1. A, Visible phenotypes and microscopic analysis of trypan blue-stained leaves after treatments. Mock-treated controls behaved as plants shown in Figure 1A. B, Dose-dependent growth inhibition of acd11/sid2 by SA. Plants were allowed to continue growth for 14 d after a single treatment with SA. C, Acceleration of the Acd− phenotype in acd11/eds5 by exogenous SA. Plants were treated at day 12 and photographed 5 d later. D, Accumulation of PR1 mRNA in Ler, acd11/nahG, and acd11/sid2 24 h after treatments with either 2 mm SA or 100 μm BTH. sid2 and nahG PR1 inductions were tested on a separate blot and were similar to the double mutants shown.
In addition to growth inhibition (Fig. 3B), SA treatments efficiently induced accumulation of PR1 mRNA in Ler and acd11/sid2, confirming that gene expression typical of SA application was induced by the treatments (Fig. 3D).
DISCUSSION
NahG Substrates Different from SA May Be Involved in acd11 Phenotypes
The failure to restore PCD in acd11/sid2 by exogenous SA could have a trivial cause, such as reduced SA uptake or responsiveness of acd11/sid2. Two lines of evidence suggest that this is not the case. First, the SA treatment led to a similar fold induction of PR1 in Ler and acd11/sid2, demonstrating that responsiveness to SA is intact in acd11/sid2. Second, the same treatment was sufficient to accelerate cell death in acd11/eds5-1. This indicates that there is no technical problem precluding cell death induction by SA in our treatments.
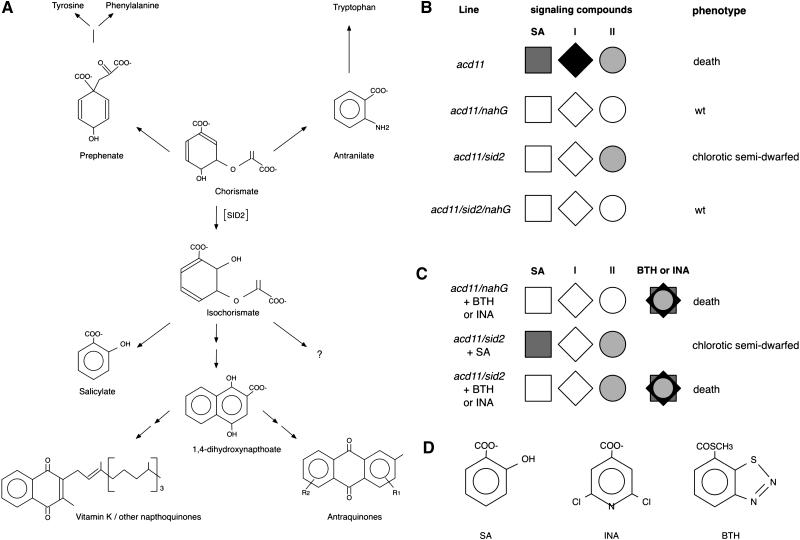
Assuming that the only function of SID2 is to convert chorismate to isochorismate, two models may explain the lack of cell death induction by SA in acd11/sid2. In the first model, ICS mutation leads to accumulation of PCD inhibitory compounds, while in the second model, it impairs accumulation of PCD activating compounds. We do not find accumulation of PCD inhibitory compounds a likely explanation because of the central position of chorismate, the ICS substrate, in primary metabolic pathways (Fig. 4A). As a precursor to Phe, Tyr, and Trp, chorismate is a branchpoint in aromatic amino acid biosynthesis, and given the high fluxes of chorismate to metabolites other than isochorismate, it is not certain that it would even accumulate in acd11/sid2-2. Rather, it is likely that SA is not the only isochorismate-derived compound required for PCD induction. We refer to this other group of PCD-promoting isochorismate-derived compounds as group 1 (Fig. 4, B and C).
Figure 4.
Overview of classes of metabolites required for acd11 phenotypes. A, Elements of biochemical pathways around isochorismate. The ensemble of isochorismate-derived metabolites different from SA is candidate group 1 compounds. The naphthoquinone/anthraquinone intermediate 1,4-dihydroxynaphtoic acid is highlighted due to its structural similarity with NahG substrates. The question mark indicates that there may be other, as yet unknown, biochemical pathways that use isochorismate as a precursor for synthesis of group 1 compounds. B and C, A black box model illustrating contributions to acd11 phenotypes of the two classes of compounds discussed in the text. It is assumed that the two classes of compounds are necessary but not sufficient for cell death and chlorosis and that BTH and INA are able to imitate the actions of both classes of compounds. D, Comparison of the chemical structures of SA, BTH, and INA.
Although we cannot exclude leakiness of the eds5-1 splice acceptor site mutant allele, the accumulation of group 1 compounds appears to be independent of EDS5 since PCD was only delayed in acd11/eds5-1 and could be accelerated by exogenous SA. Importantly, group 1 compounds or their precursors must be substrates of the NahG salicylate hydroxylase because PCD was completely blocked in acd11/nahG. In addition, the action of group 1 compounds must be mimicked by BTH and INA since PCD was fully restored by BTH and INA in both acd11/sid2-2 and acd11/nahG. Based on these observations, it is likely that group 1 compounds are structurally related to SA because a test of the substrate specificity of salicylate hydroxylase using 15 differently substituted benzoic and naphtoic acids suggested a requirement for a free carboxyl group and hydroxyl groups in the ortho-position (Yamamoto et al., 1965). Nonetheless, since BTH is not particularly closely related to SA structurally (Fig. 4D), we cannot exclude the possibility that NahG acts to deplete an SA-related, inactive precursor for a PCD-promoting compound unrelated to SA.
As shown in Figure 4A, isochorismate is a precursor for a range of plant metabolites, including phylloquinone (vitamin K1; Poulsen and Verpoorte, 1991), other naphthoquinones (Müller and Leistner, 1978), and anthraquinones (Inoue et al., 1984; Sieweke and Leistner, 1992; Stalman et al., 2003). While phylloquinone has no apoptosis-inducing activity in animal cells, such activity has been found for both the related menaquinone (vitamin K2) and several anthraquinones (Miyazawa et al., 2001; Lin et al., 2003; Yeh et al., 2003). The synthetic, water-soluble menadione (vitamin K3) is also a potent inducer of PCD in tobacco protoplasts (Sun et al., 1999), and menadione bisulphite induces rapid and extensive leaf necrosis when sprayed onto Arabidopsis plants at 1 to 3 mm concentration (P. Brodersen and F.G. Malinovsky, unpublished data). This raises the possibility that isochorismate-derived naphthoquinones and/or anthraquinones may be used as endogenous PCD regulatory signals in plants such that inhibition of their accumulation by ICS mutation leads to defective PCD induction. In this regard, it is interesting to note that the anthraquinone/naphthoquinone biosynthesis intermediate 1,4-dihydroxynaphtoic acid (Fig. 4A) is a likely NahG substrate, as 1-hydroxynapthoic acid is a NahG substrate and 2,5-dihydroxybenzoic acid is hydroxylated more efficiently than SA by NahG (Yamamoto et al., 1965).
The observation that nahG acts as a stronger acd11 suppressor than sid2-2 may be explained by three scenarios: (1) by protective action of SA degradation products in acd11/nahG, (2) by accumulation of isochorismate precursors promoting chlorosis and growth inhibition in acd11/sid2-2 due to ICS mutation, and (3) by NahG degradation of chlorosis-promoting and growth-inhibiting compounds that accumulate in acd11/sid2-2 because they are synthesized independently of ICS. The first scenario of a significant contribution of SA degradation products to acd11 suppression may be excluded because nahG was found to be epistatic to SA-deficient sid2-2. The second scenario of an accumulation of isochorismate precursors is unlikely because they would have to be NahG substrates whose minimal structural requirements are not met by isochorismate precursors. Thus, the third scenario appears likely. If so, then a second group of compounds (group 2), distinct from the isochorismate-derived group 1 described above, is involved in promoting growth inhibition and chlorosis (Fig. 4, B and C). Like group 1, group 2 may be, or may be derived from, compounds closely related to SA. The PAL pathway is a possible source of such compounds since both benzoic acid and SA can be synthesized via this pathway in several species (Métraux, 2002). However, repeated treatments starting at the seedling stage with the PAL inhibitor 2-aminoindane-2-phosphonic acid (AIP; Zön and Amrhein, 1992) did not reduce or delay leaf chlorosis in acd11/sid2 rosettes, although these treatments consistently resulted in sterility, indicating that they were effective (data not shown). These preliminary results do not support involvement of the PAL pathway in the generation of group 2 compounds.
We note that while our results suggest that compounds possibly related to SA act to promote cell death, they do not exclude a role for SA itself in promoting this process. On the contrary, the acceleration of PCD by exogenous SA in acd11/eds5-1 confirms that SA does potentiate PCD and suggests that it can be observed in acd11/eds5-1 because other PCD-promoting compounds are already present in this background. If so, comparative metabolite profiling in acd11/eds5-1 versus acd11/sid2-2 could identify the group 1 compounds of isochorismate-derived PCD-promoting signals in an unbiased manner.
Pathways of SA Biosynthesis in PCD Induction
The biosynthetic pathway of SA potentiating cell death is unclear at present. PAL-dependent SA biosynthesis has been shown to operate during tobacco mosaic virus-induced resistance in tobacco (Yalpani et al., 1993; Ribnicky et al., 1998). A study using AIP for PAL inhibition concluded that HR and resistance mediated by RPP4 in Arabidopsis also requires PAL-dependent SA synthesis (Mauch-Mani and Slusarenko, 1996). Nonetheless, the characterization of SID2 has established that the isochorismate pathway is the major source of SA in resistance responses, at least in Arabidopsis (Nawrath and Métraux, 1999; Wildermuth et al., 2001). To reconcile these findings, Wildermuth et al. (2001) proposed that while SID2-dependent SA synthesis is required in cells expressing defense responses, the PAL pathway may be important for SA synthesis in cells about to undergo PCD.
Our results provide an example that an SA pool potentiating PCD may be derived entirely from isochorismate synthesized by SID2. However, this does not exclude participation of the PAL pathway in SA synthesis for PCD in other contexts. Indeed, it is reasonable to question the general applicability of our results with a PCD/resistance system based solely upon the acd11 mutation. Nonetheless, acd11 cell death and resistance activation share genetic requirements with responses controlled by some R genes of the Toll/Interleukin-1 receptor-NBS-LRR class, such as dependence on NPR1 for PR gene expression, full dependence on EDS1 and PAD4, as well as complete independence of NDR1, PBS3, RAR1, and components of ethylene and jasmonate signaling (Brodersen et al., 2002; P. Brodersen and N.H.T. Petersen, unpublished data). This suggests that our results may extend to PCD controlled by some Toll/Interleukin-1 receptor-NBS-LRR class R genes.
BTH and INA as SA Agonists
BTH and INA have been widely used as SA agonists in the plant defense field because they induce typical SA target genes and provide resistance to the same set of pathogens as SA (Uknes et al., 1992; Lawton et al., 1996). Our study provides an example that BTH and INA may have physiological effects in addition to those that overlap with SA effects, suggesting that results of experiments using these SA analogs should be interpreted with caution with respect to roles of endogenous SA.
Comparison of eds5 and sid2
The SA-deficient mutants eds5-1 and sid2-2 behaved differently in two important ways in this study. First, acd11/sid2-2 exhibited complete suppression of SA accumulation, while approximately 10% of the total SA level in acd11 remained in acd11/eds5-1. This compares well with analyses of total SA levels induced by ozone and P. syringae pv tomato DC3000(avrRpt2) infection in Col-0, sid2, and eds5, which showed that sid2 total SA remained at basal levels, while eds5 accumulated approximately 10% of the wild-type induced level (Nawrath and Métraux, 1999; Heck et al., 2003). Second, sid2-2, but not eds5-1, appeared to affect other as yet unidentified compounds required for PCD induction. Thus, as has been shown for nahG, care should be taken when evaluating the importance of SA solely based on analyses of the sid2 mutant.
MATERIALS AND METHODS
Construction of Double and Triple Mutants
Crosses of ACD11/acd11 (pollen donor) to eds5-1 and sid2-2 were confirmed by selection of kanamycin-resistant F1 progeny, transferred to soil, and allowed to self-pollinate. Forty F2 plants from each cross were again allowed to self-pollinate, and 25 F3 families segregating for kanamycin resistance were screened for homozygosity of eds5-1 by DNA sequencing (Nawrath et al., 2002) or for sid2-2 by PCR with primers detecting the approximately 50-bp deletion in exon IX of the SID2 gene (Wildermuth et al., 2001). Three ACD11/acd11;eds5-1/eds5-1 and four ACD11/acd11;sid2-2/sid2-2 families were identified and used for initial phenotypic analysis, and a single family from each cross was used for subsequent SA measurements, gene expression, and phenotypic analyses. acd11/sid2-2 was later kept as a double homozygote isolated from the segregating F3 family.
acd11/sid2-2/nahG was constructed by crossing acd11/nahG (pollen donor) to acd11/sid2-2. F1 progeny were phenotypically wild type and contained nahG detected by PCR. In F2, plants with typical acd11/sid2-2 phenotypes were checked for nahG by PCR and were in all cases shown to lack the nahG transgene. Wild type-looking plants were screened for sid2-2 homozygotes by PCR. Most of these families segregated roughly 3:1 for green versus yellowing plants, but our numbers were not high enough to deduce statistically significant ratios for segregating (nahG heterozygous) to nonsegregating (nahG homozygous) families.
Sequences of primers used to detect acd11-1, eds5-1, sid2-2, and nahG as well as all other novel materials developed in this study are available from the authors upon request.
RNA Analyses
Total RNA was extracted by Trizol reagent (Invitrogen, Carlsbad, CA). Northern blotting and synthesis of radiolabeled probes was done according to standard protocols. cDNA templates for PR1, SAG13, and FMO were amplified by PCR as described (Brodersen et al., 2002). For RT-PCR and quantitative (Q)-PCR analysis, RNA samples were first treated with RQ1 DNase (Promega, Madison, WI). RT reactions were done with 1 μg of RNA and 0.5 μg of (dT)21 primer at 42°C with 0.1 unit of RT (Promega) and 2 units of RNasin (Promega) for 1 h in 20-μL reactions. Q-PCR was performed using the SYBR Green protocol (Applied Biosystems, Foster City, CA) with 10 pmol of each primer and 0.5-μL aliquot of RT reaction product in a 25-μL reaction. Q-PCR reactions were in triplicate and averaged for each line individually. Quantification of the threshold cycle (CT) values obtained by Q-PCR analysis was done by the 2−ΔΔCT method (Livak and Schmittgen, 2001) after verifying that CT(ubiquitin) − CT(target) remained constant for each of the target genes tested over a 100-fold cDNA dilution series.
SA Measurements
Extraction and quantitation of total SA levels were done on 2- to 3-week-old leaf tissue by the procedure described by Newman et al. (2001).
Trypan Blue Staining
Trypan blue staining was carried out as described by Bowling et al. (1997).
Plant Treatments
Plants were grown in Percival Scientific growth chambers (Perry, IA) at 16 h light/8 h darkness with a day temperature of 21°C and a night temperature of 16°C. A 1 m stock of SA (Sigma-Aldrich, St. Louis) dissolved in ethanol was diluted into aqueous solution at 1 or 2 mm final concentration and sprayed onto the leaves. A 50:50 weight granular formulation (Novartis Crop Protection, Copenhagen) of BTH was dispersed in water (100 μm BTH) and sprayed onto the leaves.
Acknowledgments
We thank the Arabidopsis Biological Resource Center (Ohio State University, Columbus) and Frederick M. Ausubel for providing eds5-1 and sid2-2 seeds, Jerzy Zön for providing AIP, Urs Neuenschwander for providing BTH, participants of the 2002 molecular plant biology course at Copenhagen University for performing the crosses of acd11 to eds5-1 and sid2-2, Ole Mattson for help with microscopy, and Lubna Ghanem for technical assistance.
This work was supported by the Faculty of Science, University of Copenhagen (P.B.), and by the Royal Veterinary and Agricultural University (M.-A.N.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.059303.
References
- Asai T, Stone JM, Heard JE, Kovtun Y, Yorgey P, Sheen J, Ausubel FM (2000) Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12: 1823–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Pike HC, Olszak B, Skov S, Odum N, Jørgensen LB, Brown RE, Mundy J (2002) Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev 16: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M, Singh KB (1997) Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA 94: 5961–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Jander G, Racki LR, Kim PD, Pierce NE, Ausubel FM (2002) Signals involved in Arabidopsis resistance to Trichoplusia ni caterpillars induced by virulent and avirulent strains of the phytopathogen Pseudomonas syringae. Plant Physiol 129: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24: 205–216 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77: 565–577 [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM (1997) Use of Arabidopsis for genetic dissection of plant defense responses. Annu Rev Genet 31: 547–569 [DOI] [PubMed] [Google Scholar]
- Heck S, Grau T, Buchala A, Métraux J-P, Nawrath C (2003) Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J 36: 342–352 [DOI] [PubMed] [Google Scholar]
- Inoue K, Shiobara Y, Nayeshiro H, Inouye H, Wilson G, Zenk MH (1984) Biosynthesis of anthraquinones and related compounds in Gallium mollugo cell suspension cultures. Phytochemistry 23: 307–311 [Google Scholar]
- Kawai-Yamada M, Ohori Y, Uchimiya H (2004) Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell 16: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10: 71–82 [DOI] [PubMed] [Google Scholar]
- Lin S, Fujii M, Hou DX (2003) Rhein induces apoptosis in HL-60 cells via reactive oxygen species-independent mitochondrial death pathway. Arch Biochem Biophys 418: 99–107 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002–1004 [DOI] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Gaille C, Kull B, Haas D, Reimmann C (2001) Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J 25: 67–77 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Slusarenko AJ (1996) Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métraux J-P (2002) Recent breakthroughs in the study of salicylic acid biosynthesis. Trends Plant Sci 7: 332–334 [DOI] [PubMed] [Google Scholar]
- Métraux J-P, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250: 1004–1006 [DOI] [PubMed] [Google Scholar]
- Miyazawa K, Yaguchi M, Funato K, Gotoh A, Kwanishi Y, Nishizawa Y, You A, Ohyashiki K (2001) Apoptosis/differentiation-inducing effects of vitamin K2 on HL-60 cells: dichotomous nature of vitamin K2 in leukemia cells. Leukemia 15: 1111–1117 [DOI] [PubMed] [Google Scholar]
- Müller WU, Leistner E (1978) Metabolic relation between naphtalene derivatives in Juglans. Phytochemistry 17: 1735–1738 [Google Scholar]
- Mur AJ, Bi Y-M, Darby RM, Firek S, Draper J (1997) Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. Plant J 12: 1113–1126 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux J-P (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux J-P (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, von Roepenack-Lahaye E, Parr A, Daniels MJ, Dow JM (2001) Induction of hydroxycinnamoyl-tyramine conjugates in pepper by Xanthomonas campestris: a plant defense response activated by hrp gene-dependent and -independent mechanisms. Mol Plant Microbe Interact 14: 785–792 [DOI] [PubMed] [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF, Dangl JL (2003) Recognition and response in the plant immune system. Annu Rev Genet 37: 579–609 [DOI] [PubMed] [Google Scholar]
- Poulsen C, Verpoorte R (1991) Roles of chorismate mutase, isochorismate synthase and anthranilate synthase in plants. Phytochemistry 30: 2873–2878 [Google Scholar]
- Rairdan GJ, Delaney TP (2002) Role of salicylic acid and NIM1/NPR1 in race-specific resistance in Arabidopsis. Genetics 161: 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT (1999) The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber TL, Plotnikova JM, Dewdney J, Rogers EE, Wood W, Ausubel FM (1998) Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J 16: 473–485 [DOI] [PubMed] [Google Scholar]
- Ribnicky DM, Shulaev V, Raskin I (1998) Intermediates of salicylic acid biosynthesis in tobacco. Plant Physiol 118: 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM (1997) Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9: 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Kachroo P, Klessig DF (1999) The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11: 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Krishnamachari Rajasekhar V, Dixon RA, Lamb C (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9: 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieweke H-J, Leistner E (1992) o-Succinylbenzoate: coenzyme A ligase from anthraquinone producing cell suspension cultures of Gallium mollugo. Phytochemistry 31: 2329–2335 [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalman M, Koskamp AM, Luderer R, Vernooy JH, Wind JC, Wullems GJ, Croes AF (2003) Regulation of anthraquinone biosynthesis in cell cultures of Morinda citrifolia. J Plant Physiol 160: 607–614 [DOI] [PubMed] [Google Scholar]
- Sun Y-L, Zhao Y, Hong X, Zhai Z-H (1999) Cytochrome c release and caspase activation during menadione-induced apoptosis in plants. FEBS Lett 462: 317–321 [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees SC, Glazebrook J (2003) Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolica is due to degradation products of salicylic acid. Plant J 33: 733–742 [DOI] [PubMed] [Google Scholar]
- Verberne MC, Verpoorte R, Bol JF, Mercado-Blanco J, Linthorst HJB (2000) Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat Biotechnol 18: 779–783 [DOI] [PubMed] [Google Scholar]
- Ward E, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Métraux J-P, Ryals JA (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymann K, Hunt M, Uknes S, Neuenschwander U, Lawton K, Steiner HY, Ryals J (1995) Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7: 2013–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RF (1979) Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99: 410–412 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xiao S, Brown S, Patrick E, Brearley C, Turner JG (2003) Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid–dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15: 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani N, León J, Lawton MA, Raskin I (1993) Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol 103: 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Katagiri M, Maeno H, Hayaishi O (1965) Salicylate hydroxylase, a monooxygenase requiring flavin adenine dinucleotide. J Biol Chem 240: 3408–3413 [PubMed] [Google Scholar]
- Yeh FT, Wu CH, Lee HZ (2003) Signaling pathway for aloe-emodin-induced apoptosis in human H460 lung nonsmall carcinoma cell. Int J Cancer 106: 26–33 [DOI] [PubMed] [Google Scholar]
- Zön J, Amrhein N (1992) Inhibitors of phenylalanine ammonia-lyase: 2-aminoindan-2-phosphonic acid and related compounds. Liebigs Ann Chem 625–628