Abstract
The breakdown of most nuclear and cytoplasmic proteins involves their partial cleavage by the 26S proteasome followed by further disassembly to free amino acids by the combined action of endo- and exopeptidases. In animals, one important intermediate exopeptidase is tripeptidyl peptidase (TPP)II, which digests peptide products of the 26S proteasome and other endopeptidases into tripeptides. Here, we describe the purification and characterization of TPPII from Arabidopsis (Arabidopsis thaliana). Like its animal counterparts, Arabidopsis TPPII exists as a soluble, approximately 5- to 9-MD complex. Two related species of 153 and 142 kD are present in the purified preparations that are derived from a single TPP2 gene. Sequencing by Edman degradation of the intact polypeptides and mass spectrometry of proteolytic fragments demonstrated that the 142-kD form mainly differs from the 153-kD form by a truncation at the C-terminal end. This serine protease is a member of the subtilisin superfamily and is sensitive to the inhibitors alanine-alanine-phenylalanine-chloromethylketone and butabindide, which are diagnostic for the TPPII subfamily. The Arabidopsis TPP2 gene is widely expressed in many tissue types with related genes evident in other plant genomes. Whereas the 26S proteasome is essential, TPPII appears not as important for plant physiology. An Arabidopsis T-DNA mutant defective in TPP2 expression displays no phenotypic abnormalities and is not hypersensitive to either amino acid analogs or the 26S proteasome inhibitor MG132. As a consequence, plants likely contain other intermediate exopeptidases that assist in amino acid recycling.
Proteolysis serves a variety of essential functions, including the elimination of misfolded or damaged proteins, the precise removal of regulatory proteins, and the maintenance of free amino acid pools needed for continual protein synthesis (Vierstra, 1996; Tomkinson, 1999). To facilitate this breakdown, plants and animals have evolved several proteolytic mechanisms for each subcellular compartment. In the vacuole/lysosome, proteins are catabolized by a variety of proteases and peptidases following their delivery to this hydrolytic compartment via endocytic and autophagic mechanisms (Thompson and Vierstra, 2005). For nuclear and cytoplasmic proteins, as well as abnormal polypeptides transported in a retrograde fashion from the endoplasmic reticulum to the cytoplasm, a major route involves ubiquitin (Ub) and the 26S proteasome (Smalle and Vierstra, 2004). Here, proteins destined for degradation are selectively tagged by the covalent attachment of multiple Ubs. These ubiquitinated proteins are then recognized and cleaved into smaller fragments by the 26S proteasome, a self-compartmentalized ATP-dependent protease complex with broad substrate and cleavage specificity.
Products of the 26S proteasome are predominantly peptides 6 to 12 amino acids in length (Wenzel et al., 1994; Kisselev et al., 1999; Voges et al., 1999). Complete recycling requires further cleavage of these peptides by intermediate endo/exopeptidases into shorter polymers, the products of which are then hydrolyzed to single amino acids by carboxyl- and amino-exopeptidases specific for tri- and dipeptides (Tomkinson, 1999). The intermediate peptidases are especially important for the rapid trimming of partial breakdown products that, if allowed to accumulate, could interfere with the functions and interactions of the parent proteins. They may also be important for generating bioactive peptides from larger precursors and for inactivating these peptides by further processing (e.g. Rose et al., 1996). Endo/exopeptidase activities also appear critical in mammalian cells for generating major histocompatibility complex class I antigens from proteasomal degradation products (Seifert et al., 2003; Reits et al., 2004). Whereas the plant 26S proteasome and several exopeptidases that release single amino acids have been described biochemically (e.g. Callis, 1995; Gu et al., 1996), the endo/exopeptidases that fulfill these intermediate roles are not well known.
In animals, tripeptidyl peptidase (TPP)II is an intermediate exopeptidase thought to be necessary for efficient protein turnover (EC 3.4.14.10; Tomkinson, 1999). This aminopeptidase was first identified as a Ser protease related to subtilisin that can, with broad sequence specificity, release tripeptides from the N terminus of oligopeptides (Balow et al., 1986). TPPII is composed of a single, approximately 140- to 150-kD polypeptide that oligomerizes into an approximately 5- to 9-MD complex. Electron microscopy (EM) shows that this oligomer assumes a twisted double-strand superstructure, which appears to create a central channel that may compartmentalize the active sites (Geier et al., 1999; Rockel et al., 2002). Assembly of this superstructure substantially enhances the peptidase activity of TPPII, indicating that the active sites may work cooperatively and possibly exploit assembly/disassembly of the oligomer to help regulate their activity (Osmulski and Gaczynska, 1998; Tomkinson, 2000). In combination with other endopeptides such as neurolysin, prolyl oligopeptidase, and thimet oligopeptidase (Saric et al., 2004), TPPII appears to play an essential role in amino acid recycling in animals (Tomkinson, 1999). Mammalian cultured cells adapted for growth on high concentrations of 26S proteasome inhibitors exhibit increased TPPII activity (Glas et al., 1998; Geier et al., 1999). Thus, under extreme conditions, TPPII may help replace the 26S proteasome in maintaining sufficient pools of free amino acids necessary for survival.
Even though genes encoding exopeptidases like TPPII are evident in various plant genomes, none of the encoded proteins have been characterized. Here, we describe the purification and biochemical characterization of TPPII from Arabidopsis (Arabidopsis thaliana). The purified protein behaves as a Ser protease and, like its animal counterparts, assembles into a large oligomeric complex. This complex contains two proteins of 153 and 142 kD that are derived from a single TPP2 gene, with the smaller version missing part of the C-terminal end. A T-DNA disruption mutant of the TPP2 locus is phenotypically indistinguishable from the wild type, indicating that this exopeptidase is not essential and suggesting that it works in concert with other endo/exopeptidases in plants to complete the digestion of large peptides to free amino acids.
RESULTS
Purification of TPPII from Arabidopsis
During our previous attempts to purify the 26S proteasome from Arabidopsis seedlings (Yang et al., 2004), we identified a large contaminating complex in the final preparations that contained two proteins of 153 and 142 kD. Following SDS-PAGE, these two species were analyzed by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF)-mass spectrometry (MS) fingerprinting following digestion of each with trypsin. The molecular masses of at least five peptides from each species matched the theoretical tryptic map of a single Arabidopsis open reading frame At4g20850 (now designated TPPII for the protein and TPP2 for the genomic locus), which has strong sequence similarity to animal and Drosophila TPPII (Renn et al., 1998). Even though two TPPII species of 153 and 142 kD were evident by SDS-PAGE, only one TPP2 coding sequence could be found in the near-complete Arabidopsis genomic sequence. This heterogeneity suggested either that two related polypeptides were generated by alternate splicing of the TPP2 transcript or that the initial translation product of TPP2 was modified, either in vivo or following homogenization, to alter its SDS-PAGE mobility.
Given the potential connection of TPPII with the Ub/26S proteasome pathway in other eukaryotes (Glas et al., 1998; Geier et al., 1999; Princiotta et al., 2001) and the predicted importance of intermediate exopeptidases such as TPPII in recycling amino acids (Tomkinson, 1999), we attempted to further characterize this Arabidopsis enzyme. To isolate TPPII specifically, we modified the purification protocol for the 26S proteasome to selectively enrich for this exopeptidase from whole seedlings. The separation behavior of both the 26S proteasome and TPPII indicated that their copurification resulted mainly from the similar native size of their superstructures. To avoid this overlap, we omitted ATP from all buffers to promote dissociation of the 26S complex into the smaller 19S regulatory particle (RP) and 20S core protease (CP) subcomplexes. At first we also included the protease inhibitor phenylmethylsulfonyl fluoride (PMSF) in the extraction buffer to minimize possible proteolytic breakdown of TPPII from 153 kD to the smaller 142-kD species. Because this inhibitor did not diminish the relative abundance of the 142-kD species, it was omitted from subsequent extractions to avoid inhibiting the peptidase activity of TPPII. Throughout the purification, levels of TPPII were monitored by a peptidase assay using the tripeptide Ala-Ala-Phe (AAF)-7-amido-4-methylcoumarin (AMC), an effective substrate of its animal orthologs (Balow et al., 1986).
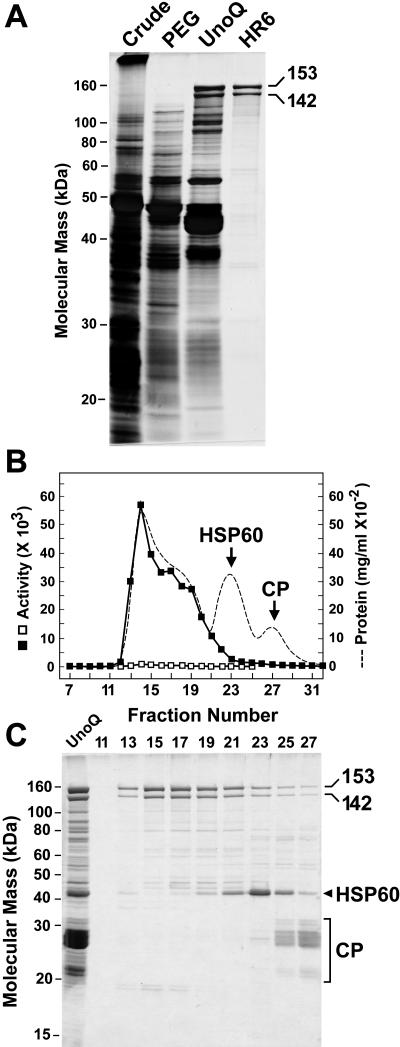
Purification of the TPPII to near homogeneity was achieved by differential polyethylene glycol (PEG) 8000 precipitation and sequential FPLC using a UnoQ anion-exchange column and a Superose HR6 size exclusion column (Fig. 1). While little purification was achieved by the PEG precipitation steps, they did remove most of the contaminating activity toward the AAF-AMC substrate (butabindide resistant; Table I). Subsequent MALDI-TOF analysis of tryptic and Glu-C peptides confirmed that the 153- and 142-kD forms were TPPII (see below). A minor contaminant of approximately 45 kD was also detected in the final preparations that eluted from the HR6 column as a 3-MD complex (Fig. 1, B and C). MALDI-TOF-MS fingerprinting of this polypeptide identified it as a chloroplast precursor chaperonin orthologous in sequence to human and yeast (Saccharomyces cerevisiae) heat shock protein-60 (At3g13470). The 20S CP of the 26S proteasome was also detected after HR6 FPLC, but eluted as a much smaller particle than TPPII (Fig. 1, B and C). We typically obtained a 500-fold purification of TPPII with an approximately 5% overall yield (based on the butabindide-inhibitable peptidase activity; Table I). Approximately 50 μg of TPPII were isolated from 200 g fresh weight of green seedlings. The Km of 0.11 mm and the Vmax of 4.8 nmol/min were determined for the AAF-AMC substrate; the former value is close to those determined for the rat and human orthologs (Balow et al., 1986; Rose et al., 1996).
Figure 1.
Purification of TPPII from Arabidopsis seedlings. A, SDS-PAGE analysis of the TPPII purification steps. Samples from the crude extract, 2% to 8% PEG precipitation, pooled fractions from the UnoQ anion-exchange column, and the peak fraction from the Superose HR6 column were subjected to SDS-PAGE and silver staining. Positions of the 153- and 142-kD TPPII species are indicated. B and C, Elution profile of TPPII during size exclusion chromatography. The TPPII-containing fractions from UnoQ FPLC (lane C, left) were subjected to FPLC on a HR6 Superose column. B, TPPII peptidase activity using AAF-AMC as the substrate of the fractions with (□) and without (▪) the addition of 50 μm butabindide. Elution profile of total protein (A280) is included. C, SDS-PAGE of the column fractions surrounding the peak of TPPII peptidase activity. The positions of the 153- and 142-kD forms of TPPII are shown. The SDS-PAGE migration and elution position of the heat shock protein-60 chaperonin and the 20S CP of the 26S proteasome are indicated in B and C.
Table I.
Purification of TPPII from Arabidopsis
| Total Protein
|
TPPII Activity
|
Purification
|
Yield
|
|||
|---|---|---|---|---|---|---|
| Amount | Percentage | Totala | % Butabindide Sensitiveb | |||
| mg | % | μmol L−1 min−1 mg−1 | % | -fold | % | |
| Crude extract | 535 | 100 | 18 | 19 | 1 | 100 |
| 2% PEG | 300 | 56 | 20 | 19 | 1 | 62 |
| 10% PEG | 59 | 11 | 36 | 93 | 1 | 23 |
| UnoQ6 | 7.6 | 1.4 | 44 | 98 | 12 | 17 |
| Superose HR6 | 0.05 | 0.01 | 1,672 | 99 | 464 | 5 |
μmol L−1 min−1 mg−1 AMC released using AAF-AMC as the substrate.
Percent total activity inhibited by 50 μm butabindide.
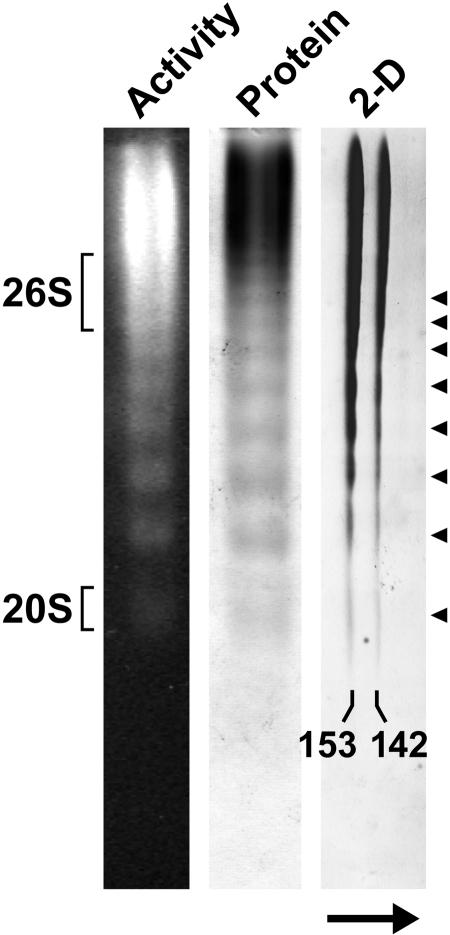
The 153- and 142-kD TPPII species eluted from the size exclusion column as oligomers of 5 to 9 MD, indicating that Arabidopsis TPPII, like its animal counterparts (Geier et al., 1999; Rockel et al., 2002), assembles into higher order complexes (Fig. 1, B and C). The higher mass complexes appeared to have more peptidase activity, suggesting that the individual subunits are more active when assembled. A similarly large complex could be seen by native PAGE as detected by both protein and activity staining (Fig. 2). Prolonged electrophoresis also revealed a uniform laddering of discrete subpopulations with smaller native sizes that retained peptidase activity. Although the precise masses of these species were not determined, they appeared to differ by much more than the mass of a single 153- or 142-kD polypeptide, suggesting that this laddering was caused by the loss of discrete segments containing multiples of these polypeptides. Native PAGE coupled with SDS-PAGE of the complex showed that both the 153- and 142-kD polypeptides had a coincident electrophoresis pattern during native PAGE (Fig. 2). A similar coincident elution was evident by size exclusion chromatography (Fig. 1, B and C). Consequently, it appears that both polypeptides were present together at a constant ratio within the purified TPPII superstructure.
Figure 2.
Native PAGE of Arabidopsis TPPII. The peak fraction from the Superose HR6 FPLC was subjected to native PAGE and TPPII was either detected by fluorescence following overlay of the gel with the substrate AAF-AMC or by staining with silver nitrate. Migration positions of the 26S proteasome and its 20S CP in an adjacent lane are indicated. For two-dimensional PAGE, TPPII was first subjected to native PAGE followed by SDS-PAGE in the second dimension and the gel was then stained with silver nitrate. The arrow indicates the electrophoretic direction of the SDS-PAGE. The positions of the 153- and 142-kD forms of TPPII are shown. Arrowheads locate the series of discrete size species of TPPII observed by native PAGE.
Sequence Analysis of the Arabidopsis TPP2 Gene
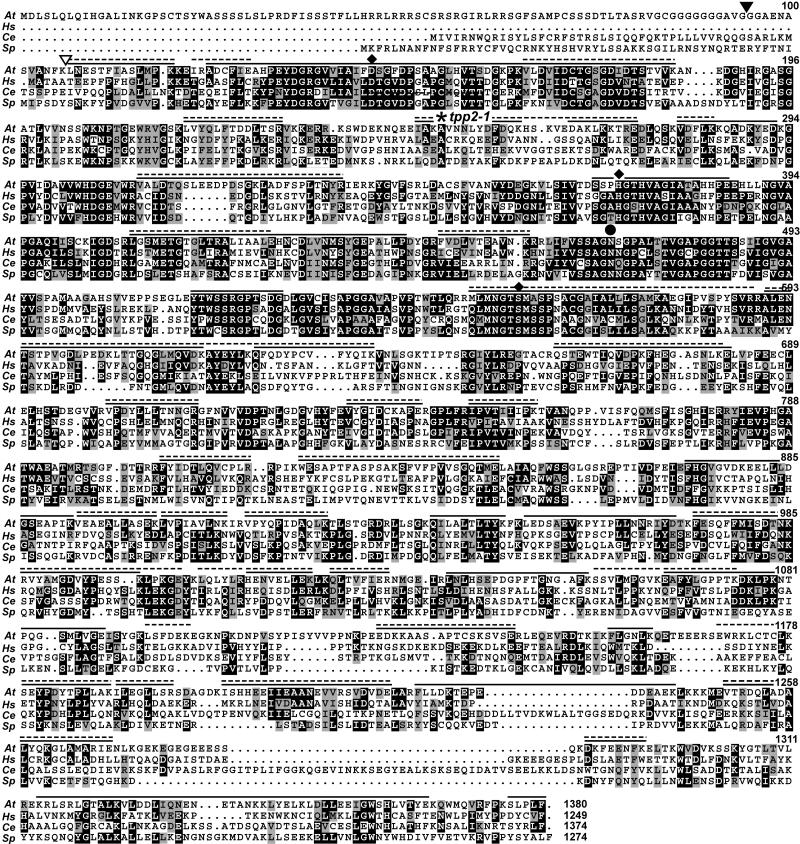
Alignment of the Arabidopsis TPP2 genomic sequence with those of several cDNAs present in the expressed sequence tag (EST) database allowed us to assemble the complete open reading frame encoding this protease. The predicted initial transcript of 4,143 bp encodes a 1,380-amino acid protein with a calculated mass of 154 kD, which is close to the apparent molecular mass of the larger 153-kD species as determined by SDS-PAGE. Overall, Arabidopsis TPPII is 36%/53% and 32%/51% identical/similar to its human and Schizosaccharomyces pombe orthologs, respectively. Alignment of the Arabidopsis TPPII with related sequences from yeast, Caenorhabditis elegans, and humans confirmed its inclusion within the subtilisin superfamily (Fig. 3). Like other subtilisin-type Ser proteases, Arabidopsis TPPII has the positionally conserved Asp, His, and Ser residues (positions 147, 372, and 558, respectively) that form the catalytic triad as well as the Asn (residue 469) that stabilizes the tetrahedral intermediate (Hilbi et al., 2002). Consistent with their importance, the sequence blocks around these catalytic sites are some of the most conserved regions within this protein family. Similar to other TPPII proteins, the Arabidopsis form also has an approximately 200-amino acid insertion (residues 152–369) between the catalytic Asp and His residues that appears to distinguish these peptidases from other members of the subtilisin superfamily (Renn et al., 1998; Tomkinson et al., 2002). Like its close relatives in other species, Arabidopsis TPPII also has an extended C-terminal domain (residues 1,195–1,380) that is highly variable with numerous alignment gaps as compared to its orthologs (Fig. 3).
Figure 3.
Amino acid sequence comparison of Arabidopsis TPPII with orthologs from other species. Sequences from Arabidopsis (At), Homo sapiens (Hs), C. elegans (Ce), and S. pombe (Sp) were aligned by ClustalW and viewed with MACBOXSHADE. Identical and similar amino acids are shown in black and gray boxes, respectively. The diamonds identify the conserved Asp, His, and Ser residues that form the catalytic triad. The circle indicates the Asn that stabilizes the tetrahedral intermediate (Renn et al., 1998). The site of the T-DNA insertion for the tpp2-1 mutant is indicated by the asterisk. The solid and dashed lines indicate the composite coverage of the regions from the 153- and 142-kD species, respectively, as identified by MS sequencing of trypsin and Glu-C peptides (see Table II). The black and white triangles locate the N-terminal amino acid of the 153- and 142-kD species, respectively, as determined by Edman degradation of the full-length proteins. GenBank accession numbers for H. sapiens, C. elegans, and S. pombe sequences are NP_003282, NP_495221, and NP_594951, respectively.
The larger predicted size of Arabidopsis TPPII relative to its animal orthologs is mainly due to a long N-terminal extension of 102 amino acids (Fig. 3). Although both the C. elegans and S. pombe versions are predicted to have N-terminal extensions, these extensions are shorter than that for Arabidopsis TPPII and appear unrelated in sequence. Reverse transcription (RT)-PCR analysis confirmed that the nucleotide sequence for this extension is present in the Arabidopsis TPP2 transcript (data not shown). Whether this entire sequence is translated is unclear because a second Met codon can be found at codon 69. The full extension is notably enriched in Ser and Gly residues and displays some homology with Arabidopsis transit peptides that direct the transport of proteins into chloroplasts (Psort software; http://psort.nibb.ac.jp). We consider this predicted localization to be unlikely based on data with its mammalian and yeast counterparts showing that TPPII is a cytosolic protein (Rose et al., 1996; Geier et al., 1999).
Searches of the near-complete Arabidopsis genomic sequence with TPP2 failed to find related loci. We also failed to detect variant cDNAs among the 28 ESTs present in the Arabidopsis database (http://www.arabidopsis.org) that would imply alternate splicing of the TPP2 transcript. Related genes were evident in rice (Oryza sativa [AK067099]) and maize (Zea mays [AY105477]), suggesting that TPPII-like activities are present in all plants. Analysis of the Arabidopsis EST and massively parallel signature sequences (http://mpss.udel.edu/at) databases indicated that TPP2 is widely expressed in most tissues (data not shown). TPP2 mRNA levels were highest in callus as compared to differentiated tissues such as leaves, roots, and siliques.
Inhibitor Sensitivity of TPPII
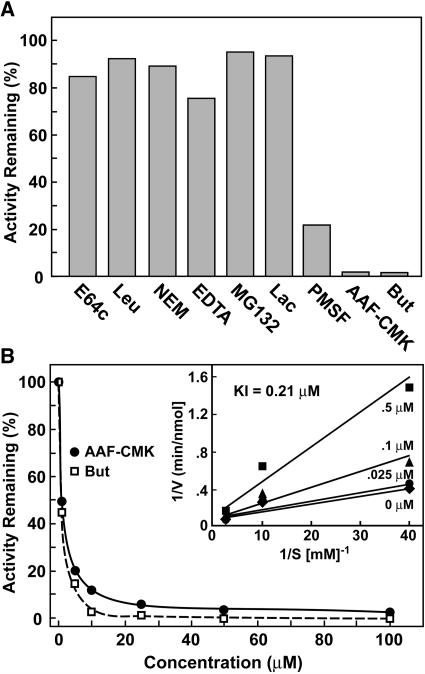
Sensitivity of Arabidopsis TPPII to various protease inhibitors confirmed the expectation, based on sequence comparisons, that it is a member of the TPPII subfamily of subtilisin-type proteases (Fig. 4A). Whereas the Ser protease inhibitor PMSF attenuated its activity, the Cys (E64c, leupeptin) and metalloprotease inhibitors (EDTA) were without an effect. Mammalian and Drosophila TPPIIs are sensitive to thiol-reactive compounds, mainly through their reaction with a positionally conserved Cys (residue 760 in Drosophila TPPII; Renn et al., 1998). This Cys has been substituted for an Ala in Arabidopsis TPPII, potentially explaining why this ortholog appears insensitive to N-ethylmaleimide (NEM; Fig. 4A). The peptidase activity of the preparations was also insensitive to the 26S proteasome inhibitors, MG132, and lactacystin, confirming that the activity was derived from TPPII and not the 26S proteasome CP that may have contaminated the final preparations.
Figure 4.
Effect of various protease inhibitors on Arabidopsis TPPII activity. A, Peptidase activity against the AAF-AMC substrate (100 μm) was measured with or without inhibitors and expressed as a percent of uninhibited activity. Concentrations used were E-64, 100 μm; leupeptin (Leu), 25 μm; NEM, 1 mm; EDTA, 1 mm; MG132, 100 μm; lactacystin (Lac), 5 μm; PMSF, 1 mm; AAF-CMK, 100 μm; and butabindide (But), 25 μm. B, Effect of various AAF-CMK and butabindide concentrations on TPPII peptidase activity using 100 μm AAF-AMC. Inset shows a double reciprocal Lineweaver-Burk plot for various concentrations of AAF-AMC and butabindide that was used to calculate a Ki of 0.21 μm.
Like TPPII preparations from animals, the Arabidopsis peptidase was effectively quenched by the inhibitors AAF-chloromethylketone (CMK) and butabindide (Fig. 4, A and B), thus placing the enzyme in the TPPII clade of the subtilisin superfamily (Siezen and Leunissen, 1997; Renn et al., 1998). Butabindide was specifically designed as a competitive inhibitor of TPPII, using the rat peptidase for the assay (Rose et al., 1996). The concentration needed for 50% inhibition of initial activity for butabindide and AAF-CMK (using 100 μm AAF-AMC) was approximately 5 μm, with 100 μm of each effectively inhibiting >97% of the peptidase activity of TPPII (Fig. 4B). For butabindide, assays with a range of substrate concentrations provided a Ki of 0.21 μm (Fig. 4B), which is close to the value determined for Drosophila TPPII (Renn et al., 1998). Of the AAF-AMC degrading activity in crude seedling extracts, only 19% was inhibited by butabindide (Table I), suggesting that peptidases other than TPPII can also break down this substrate in crude plant extracts and likely in vivo. Possible candidates include TPPI, a structurally distinct but functionally related tripeptidase that may be located in the vacuole (Tomkinson, 1999), and various aminopeptidases working in concert.
Sequence Analysis of the TPPII Proteins
In the absence of either additional Arabidopsis TPP2 genes or evidence for alternative splicing of a single TPP2 transcript, synthesis of the 142-kD form of TPPII could occur by either differential translation of the TPP2 mRNA, processing of the initial translation product, and/or by posthomogenization proteolysis of the 153-kD species. Calculations of apparent molecular masses suggested that the two forms differ by approximately 100 amino acids. To help determine which regions in the 153-kD species account for this difference, we subjected both species to thorough sequencing by MS. The 153- and 142-kD polypeptides were separated by SDS-PAGE, individually excised from the gels, and digested with either trypsin or Glu-C. The resulting peptides were then subjected to online liquid chromatography followed by electron spray ionization (ESI) ion trap tandem MS (MS/MS) analysis. As can be seen in Table II and Figure 3, this sequencing identified a large array of peptides from both the 142- and 153-kD forms (56% coverage for the 153-kD species). All these peptides could be located in the derived amino acid sequence from the TPP2 gene, indicating that no genes other than TPP2 were responsible for their synthesis. Both sets of peptides showed similar coverage at the N-terminal end of the TPPII protein, with each set failing to contain fragments upstream of Lys-107. Several peptides corresponding to the C-terminal end could be found for the 153-kD species, including a pentapeptide that covered the predicted C-terminal Phe residue. However, for the 142-kD species, the most C-terminal peptide we identified ended at Lys-1297, with no coverage for the last 84 residues (Table II; Fig. 3).
Table II.
MS analysis of TPPII peptides
| Start | Tryptic Peptide Sequencea | Start | Glu-C Peptide Sequencea | ||
|---|---|---|---|---|---|
| 107 | LNESTFIASLMPK | B | 122 | IRADCFIE | L |
| 124 | ADCFIEAHPEYDGR | B | 134 | YDGRGVVIAIFD | U |
| 138 | GVVIAIFDSGFDPSAAGLHVTSDGKPK | U | 247 | IAKAVNNLYD | B |
| 165 | VLDVIDCTGSGDIDTSTVVK | B | 247 | IAKAVNNLYDFDQKHSKVE | L |
| 218 | LVYQLFTDDLTSR | B | 266 | DAKLKKTRE | B |
| 250 | AVNNLYDFDQK | B | 282 | FLKKQADKYE | U |
| 280 | VDFLK | B | 321 | PDSGKLAD | U |
| 310 | VALDTQSLEEDPDSGK | B | 369 | SSPHGTHVAGIATAHHPE | L |
| 326 | LADFSPLTNYR | B | 414 | TGTGLTRALIAALE | B |
| 409 | LGSMETGTGLTR | B | 428 | HNCDLVNMSYGEPALLPD | U |
| 449 | FVDLVTEAVNK | B | 516 | YTWSSRGPTSDGD | U |
| 552 | MLMNGTSMASPSACGAIALLLSAMK | B | 604 | DKLTTGQGLMQVDKAYE | L |
| 577 | AEGIPVSPYSVR | L | 675 | GASNLKE | U |
| 590 | ALENTSTPVGDLPEDK | B | 732 | VYGIDCKAPE | B |
| 606 | LTTGQGLMQVDK | B | 824 | SAPTFASPSAKSFVFPVVSGQTME | B |
| 618 | AYEYLK | L | 872 | FHGVGVDKE | U |
| 624 | QFQDYPCVFYQIK | B | 883 | LLDGSEAPIKVE | U |
| 660 | QSTEWTIQVDPK | B | 957 | VKPYIPLLNNRIYD | U |
| 701 | VPDYLLLTNNGR | B | 975 | SQFFMISDTNKRVYAMGDVYPE | U |
| 748 | IPVTIIIPK | B | 983 | TNKRVYAMGDVYPE | L |
| 806 | FYIDTLQVCPLR | B | 997 | SSKLPKGE | L |
| 822 | WESAPTFASPSAK | B | 1,066 | AFYLGPPTKDKLPKNTPQGSMLVGE | U |
| 880 | EELLLDGSEAPIK | U | 1,126 | EDKKAASAPTCSKSVSE | B |
| 893 | VEAEALLASEK | B | 1,170 | WRKLCTCLKSE | L |
| 904 | LVPIAVLNK | B | 1,181 | YPDYTPLLAKILE | B |
| 915 | VPYQPIDAQLK | B | 1,218 | VVRSVDVDE | B |
| 939 | QILALTLTYK | U | 1,251 | VTRDQLADALYQKGLAMARIE | B |
| 951 | LEDSAEVKPYIPLLNNR | U | 1,318 | KRLSRLGTALKVLDDLIQNE | U |
| 973 | FESQFFMISDTNK | B | 1,340 | TANKKLYE | U |
| 987 | VYAMGDVYPESSK | U | 1,348 | LKLDLLEE | U |
| 1,007 | LQLYLR | L | 1,356 | IGWSHLVTYE | U |
| 1,013 | HENVELLEK | B | |||
| 1,024 | QLTVFIER | B | |||
| 1,038 | LNLHSEPDGPFTGNGAFK | U | |||
| 1,056 | SSVLMPGVK | L | |||
| 1,065 | EAFYLGPPTK | B | |||
| 1,080 | NTPQGSMLVGEISYGK | B | |||
| 1,096 | LSFDEK | L | |||
| 1,155 | FLGNLK | U | |||
| 1,179 | SEYPDYTPLLAK | B | |||
| 1,191 | ILEGLLSR | B | |||
| 1,205 | ISHHEEIIEAANEVVR | B | |||
| 1,221 | SVDVDELAR | B | |||
| 1,230 | FLLDK | U | |||
| 1,235 | TEPEDDEAEKLK | U | |||
| 1,254 | DQLADALYQK | B | |||
| 1,289 | DKFEENFK | B | |||
| 1,329 | VLDDLIQNENETANKK | U | |||
| 1,376 | SLPLF | U |
U, L, and B, respectively, denote peptides detected only in upper (153-kD), only in lower (142-kD), or in both polypeptides present in purified preparations of TPPII (see Fig. 1).
MS analyses implied that the difference between the 153- and 142-kD species was generated by loss of the ≤85 residues from the C-terminal end. However, given the absence of any peptides from both species matching the first 105 residues, we could not rule out the possibility that the N-terminal end was also affected. The unusually long N-terminal extension predicted for Arabidopsis TPPII also raised the possibility that the internal Met at position 69 and not the predicted Met-1 is the first amino acid. To resolve these ambiguities, we determined the N-terminal sequence of both the 153- and 142-kD forms by Edman degradation. (MS/MS analysis of tryptic fragments failed to find any acetylated amino acids, suggesting that neither species contains a block N terminus.) The N-terminal sequence of the 142-kD species was determined to be Leu-Asn-Glu-Ser, indicating that this protein started at Leu-107 (Fig. 3). For the 153-kD species, two sequences were detected. The predominant one was Gly-Gly-Ala-Glu, with the minor one being Leu-Asn-Glu-Ser, indicating that the 153-kD protein most often began at Gly-95, with a minor form beginning 12 residues later at Leu-107 (Fig. 3). Taken together, the 153-kD form of TPPII appeared to have arisen by N-terminal cleavage of a larger form (possible beginning at Met-1 or Met-69) to generate a dominant 1,285-residue protein (actual mass of 142 kD) beginning at Gly-95. The 142-kD species was created by N-terminal cleavage before Leu-107. It was also missing ≤85 residues at the C terminus to generate a predicted protein with an actual mass of ≤131 kD. Whether this processing/cleavage happens in vivo or in vitro remains unclear.
Genetic Analysis of the TPP2 Gene
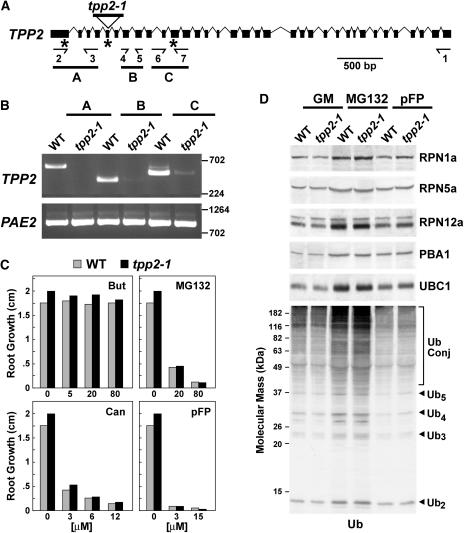
To help assess the phenotypic functions of TPPII in planta, we analyzed an Arabidopsis mutant in the Columbia (Col-0) ecotype background that harbored a T-DNA disruption (SALK_085776) of the corresponding gene. The TPP2 gene encompasses 8,988 bp and contains 34 exons and 33 introns (Fig. 5A). Sequence analysis of the tpp2-1 locus indicated that the T-DNA inserted within the fifth exon upstream of nucleotide 248 (Fig. 5A). Kanamycin resistance associated with the T-DNA segregated in a 3:1 pattern, indicating that the T-DNA inserted at a single site. Both the right- and left-border T-DNA primers, in combination with gene-specific primers, amplified the upstream and downstream regions of TPP2, respectively. Sequence analysis of these products showed that insertion of the T-DNA did not induce any secondary effects on the TPP2 locus. RT-PCR analysis of total RNA isolated from wild-type and homozygous tpp2-1 seedlings demonstrated that the mutation altered expression of the TPP2 gene. Whereas we could detect the expected TPP2 PCR product from wild-type RNA using primers upstream of the T-DNA (primers 2 and 3), no products were evident from the tpp2-1 RNA (Fig. 5B). When primers downstream of the T-DNA were used (primers 4 and 5 or 6 and 7), we could reproducibly detect a low amount of the expected PCR products from the tpp2-1 RNA, indicating that the region downstream of the T-DNA was transcribed, albeit at low levels (Fig. 5B). Since this shorter transcript would lack the codon for the essential Asp (residue 152) of the catalytic triad, the resulting truncated protein would be enzymatically inactive even if translated (Figs. 3 and 5). Consequently, we predict that this mutant represents a null allele.
Figure 5.
Genomic and phenotypic descriptions of the Arabidopsis tpp2-1 mutant. A, Gene structure of Arabidopsis TPP2. Coding region and introns are indicated by boxes and lines, respectively. The positions of the T-DNA in tpp2-1 and the locations of the primer pairs used for RT-PCR are indicated. B, RT-PCR analysis of the tpp2-1 insertion mutants. Total seedling RNA was reverse transcribed with primer 1 (see “Materials and Methods”). The reverse transcribed products were then PCR amplified using the A (primers 2 and 3), B (primers 4 and 5), and C (primers 6 and 7) reactions. RT-PCR of PAE2 was included as a control. C, Root growth of wild-type (gray bars) and homozygous tpp2-1 (black bars) seedlings on various concentrations of butabindide (But), MG132, canavanine (Can), and pFP. D, Immunoblot analysis of crude extracts from wild-type and tpp2-1 seedlings with antibodies against Ub, subunits of the CP (PBA1), RP base (RPN1a), and RP lid (RPN5a and RPN12a) from the 26S proteasome, and UBC1, a Ub-conjugating enzyme. Seven-day-old seedlings were exposed for 7 d on solid GM without or with 50 μm MG132 or 50 μm pFP. The migration position of free Ub, Ub chains containing 2 to 5 Ubs, and Ub conjugates are indicated.
Phenotypic analysis of tpp2-1 seedlings indicated that the corresponding protein is not essential in Arabidopsis. Homozygous mutant seeds germinated normally and the plants developed, flowered, and set seeds indistinguishable to wild type under standard growth conditions (data not shown; Fig. 5C). It has been shown that mammalian TPPII can partially substitute for the 26S proteasome when the activity of the latter is blocked by the 26S proteasome inhibitors such as MG132 (Glas et al., 1998; Geier et al., 1999; Princiotta et al., 2001). To test for a similar connection in plants, we examined the effects of several inhibitors of the 26S proteasome and TPPII using root growth as the assay. Predicting that Arabidopsis tpp2-1 seedlings might then be hypersensitive to MG132, we examined their growth response to a range of MG132 concentrations. However, as can be seen in Figure 5C, both wild-type and tpp2-1 seedlings were similarly sensitive. Numerous Ub/26S proteasome pathway mutants are hypersensitive to amino acid analogs, presumably because they cannot efficiently remove the resulting abnormal proteins that incorporate these analogs (e.g. Girod et al., 1999; Yan et al., 2000). When tpp2-1 plants were tested similarly, no hypersensitivity was seen for canavanine or p-fluorophenylalanine (pFP), analogs of Arg and Phe, respectively (Fig. 5C). The specific inhibitor of animal TPPII, butabindide, was shown to hyperactivate at least one neuropeptide hormone response in mice, presumably via its ability to prevent TPPII from degrading the peptide (Rose et al., 1996). To test whether butabindide might have a pharmacological effect on Arabidopsis, we exposed roots to a range of butabindide concentrations. Even at a butabindide concentration approximately 400 times that of the Ki for TPPII in vitro (80 μm), no detectable effect on growth was observed (Fig. 5C). In a similar fashion, both wild-type and tpp2-1 seedlings were insensitive to another TPP-type peptidase inhibitor, AAF-CMK (data not shown).
Attenuation of 26S proteasome activity in Arabidopsis by mutation of specific subunits leads to a coordinated transcriptional up-regulation of most, if not all, genes encoding components of the complex, indicating that plants like yeast contain a feedback loop to adjust 26S proteasome levels to meet demand (Smalle et al., 2003; Yang et al., 2004). Given the potential connection of TPPII to the 26S proteasome, a similar up-regulation might occur when TPPII levels are diminished. However, immunoblot analyses of a battery of 26S proteasome subunits for the CP (PBA1), and RP base (RPN1a) and lid (RPN5a and RPN12a) subcomplexes failed to detect such coregulation. The levels of all five subunits in the tpp2-1 mutant remained similar to wild type when grown either on Gamborg G5 medium (GM) or on GM containing concentrations of MG132 or pFP that inhibit growth (Fig. 5D). The basal level of Ub conjugates as well as the levels increased by MG132 were also unaffected by the tpp2-1 mutation (Fig. 5D). Taken together, it appears that removal of TPPII does not detectably affect abnormal protein removal, amino acid recycling, and the Ub/26S proteasome pathway, suggesting that other intermediate peptidases assume its functions when absent.
DISCUSSION
Recycling of amino acids appears to require the coordinated action of endopeptidases, like the 26S proteasome, which selects targets and makes the initial cleavages, followed by intermediate endo/exopeptidases that further process the oligopeptides into di- and tripeptides, and finally various carboxy- and aminopeptidases that digest the short peptides into free amino acids (Tomkinson, 1999). Among other possible functions, TPPII has been proposed to be a key intermediate exopeptidase responsible for the generation of tripeptides from longer peptides. Several previous studies have characterized Drosophila and mammalian TPPII at the biochemical and structural levels. Here we report the purification and preliminary genetic characterization of the Arabidopsis version. Like its counterparts from other species, Arabidopsis TPPII is a Ser protease with sequence similarity to subtilisin-type peptidases. Although we have not confirmed that TPPII is a tripeptidyl peptidase, the fact that it digests AAF-AMC, a peptide substrate preferred by TPPII orthologs from other eukaryotes, and that it is effectively blocked by micromolar concentrations of AAF-CMK and butabindide, both specific inhibitors of TPPII-type activities, strongly support this mechanism of action. Preliminary genomic analyses suggest a wide distribution of TPPII in both monocotyledonous and dicotyledonous plants. Although the TPP2 gene has not yet been identified in soybeans, a >1-MD protease with related enzymatic properties has been described (Shimotsuura et al., 1992).
Purified Arabidopsis TPPII consists of two related polypeptides of 153 and 142 kD, which are derived from a single TPP2 gene. Both polypeptides are processed from a larger precursor by cleavage at the N-terminal end. MS fingerprinting demonstrated that the interior sequence of the two proteins appears to be contiguous. However, we could not identify peptides beyond Lys-1297 in the 142-kD species, implying that it is missing as much as 10 kD from the C-terminal end. Why the C-terminal end is absent from the 142-kD species remains unclear. Among the numerous ESTs from the TPP2 gene, there is no evidence for alternative transcripts, thus precluding differential RNA processing as the mechanism. Another possible scenario is that proteolytic processing of the 153-kD polypeptide, either in vivo or in vitro, generates the 142-kD species. It is notable that the purified TPPII complex from both Drosophila and mouse lymphoma cells also contains two species, the larger of which is typically more abundant (Renn et al., 1998; Geier et al., 1999; Rockel et al., 2002). For Drosophila TPPII, the smaller species was even present in recombinant preparations from human embryonic kidney cells, with prolonged storage above 4°C promoting its conversion from the larger species (Renn et al., 1998). Taken together, a likely scenario is that the C-terminal end of the intact TPPII protein is exposed in the assembled complex and thus it is intrinsically susceptible to proteolytic cleavage. Given the likelihood that such cleavage occurs following tissue homogenization, either by self-processing of TPPII using its reported endopeptidase activity (Geier et al., 1999) or by another protease, we attempted to block the cleavage of Arabidopsis TPPII by including PMSF in the purification protocol. Unfortunately, no change in the relative abundance of the 142-kD species was observed. To resolve this issue, antibodies against Arabidopsis TPPII are clearly needed to identify the protein in crude extracts prior to purification.
A striking feature of animal TPPIIs is their ability to assemble into a large oligomeric complex of approximately 5 to 9 MD, with EM pictures of the mammalian and Drosophila particles showing that it resembles a twisted double-strand ribbon (Geier et al., 1999; Rockel et al., 2002). Both size exclusion chromatography and native PAGE indicate that a similarly large complex is assembled in Arabidopsis. The reason for this structure is unclear; it could either help orient the active sites such that they work cooperatively or compartmentalize the active sites to shield the cytoplasm from inadvertent proteolysis. Although archaebacteria do not contain obvious TPPII orthologs, they contain tricorn protease, which appears to have an analogous intermediate role in processing oligopeptides (Tamura et al., 1996). Of interest here is that tricorn protease also assembles into a distinctly shaped 14-MD complex with an obvious channel. Consequently, it is possible that such higher order structures are critical for the roles of these exopeptidases in selectively and efficiently processing long peptides into discrete di- and tripeptide fragments.
The EM pictures of TPPIIs also show that the strands are composed of discrete segments that may represent individual or multimers of the TPPII polypeptide (Geier et al., 1999; Rockel et al., 2002). While our native PAGE of Arabidopsis TPPII indicated that most of the activity was present in a large complex, a uniform laddering of smaller enzymatically active particles was also seen that appears to represent partially disassembled species missing increasing numbers of repeated segments. It was possible that the dissociated segments were enriched in the smaller 142-kD species and possibly even generated upon formation of this smaller polypeptide. However, SDS-PAGE following native PAGE showed that both the larger complexes and these smaller species contained a similar ratio of the 153 and 142 polypeptides.
Even though TPPII has been proposed to participate in numerous processes, final confirmation awaits genetic analyses. A role for TPPII in antigen presentation by human cells was inferred by the ability of AAF-CMK to block the production of antigenic peptides (Seifert et al., 2003). However, genome-wide RNA interference of TPP2 showed that reduction of the transcript did not result in any obvious phenotypes in C. elegans, even though the gene is normally well expressed at all stages of development (Kamath et al., 2003). This nonessential nature of TPP2 is also supported by the pilot study searching for essential genes in S. pombe (Decottignies et al., 2003). In S. cerevesiae, a TPP2-type gene is notably absent from the genome. With the analysis of the Arabidopsis tpp2-1 mutation, we show here that TPPII is also not essential in plants. Homozygous tpp2-1 mutant plants germinated, developed, flowered, and produced seed indistinguishable from wild type. Taken together, it is likely that other exopeptides can assume the roles of TPPII in amino acid recycling when absent. TPPII has also been proposed to assist in forming and/or degrading peptide hormones in mammals, a scenario supported by both peptidase assays on prohormones and the phenotypic effects of the TPPII inhibitor butabindide (Rose et al., 1996). The absence of any developmental defects for the tpp2-1 mutant or for wild-type seedlings when exposed to high concentrations of butabindide or AAF-CMK implies that TPPII does not have a similar role in Arabidopsis.
The observations that TPPII is up-regulated in mammalian cells adapted to high concentrations of 26S proteasome inhibitors have been used to suggest that TPPII and the 26S proteasome work cooperatively in degrading proteins to amino acids (Glas et al., 1998; Geier et al., 1999). However, more recent studies by Princiotta et al. (2001) suggest that these adapted cells still have residual 26S proteasome activity and thus may not obligatorily need TPPII for survival. Similarly, our analysis of Arabidopsis seedlings missing TPPII found no evidence for a connection between TPPII and the 26S proteasome in plants. tpp2-1 plants were not hypersensitive to the 26S proteasome inhibitor MG132, nor were they hypersensitive to amino acid analogs, which, when incorporated into abnormal proteins, require the 26S proteasome for removal. Attenuated proteolysis generated by 26S proteasome mutations has been shown to coordinately enhance accumulation of both mRNA and protein for a battery of 26S proteasome subunits in yeast, animals, and Arabidopsis (Xie and Varshavsky, 2001; Meiners et al., 2003; Yang et al., 2004). Our data showed that a similar up-regulation does not occur upon loss of TPPII.
While the 26S proteasome is clearly a key protease in the removal of abnormal polypeptides and the breakdown of important regulatory proteins, the entire process of amino acid recycling likely requires a host of additional endo/exopeptidases to complete the task. Our discovery of Arabidopsis TPPII hopefully identifies one such additional component important to this process in plants. While the functions of TPPII remain unclear, the coupling of the tpp2 mutant with those in other amino acid recycling systems (e.g. Ub/26S proteasome and autophagy) may eventually reveal its roles in plant biology.
MATERIALS AND METHODS
TPPII Purification
TPPII was purified from Arabidopsis (Arabidopsis thaliana) ecotype Col-0 seedlings using a protocol similar to that for the 26S proteasome (Yang et al., 2004), with the exception that ATP was omitted from all solutions. Seedlings were grown for 10 d in liquid GM (Gibco BRL, Gaithersburg, MD) at 24°C under continuous light, washed, frozen to liquid nitrogen temperatures, and stored at −80°C. The frozen tissue was pulverized and homogenized in 1.25 mL/g fresh weight of extraction buffer (50 mm potassium phosphate, pH 6, 2 mm MgCl2, 5% [v/v] glycerol, and 5 mm 2-mercaptoethanol) supplemented with 5% polyvinylpyrrolidone and 0.6% sodium metabisulfite just before use. All subsequent steps were performed at 0°C to 4°C.
The extract was filtered through 4 layers of cheesecloth and 2 layers of Miracloth and clarified at 30,000g for 15 min. The supernatant was made 2% (w/v) PEG 8000 (Sigma, St. Louis), stirred for 30 min, and reclarified at 30,000g for 30 min. The supernatant was made 8% (w/v) PEG 8000 and stirred for 30 min, and the TPPII-containing precipitate was collected by centrifugation at 12,000g for 15 min. The pellet was resuspended in extraction buffer, clarified, and fractionated by FPLC on a 6-mL UnoQ6 (Bio-Rad, Richmond, CA) anion-exchange column using a 120-mL 0 to 1 m KCl gradient in extraction buffer. Fractions (3 mL) containing TPPII activity (as determined by peptidase assay) eluted between 220 and 280 mm KCl; these fractions were pooled and reprecipitated with 10% (w/v) PEG 8000. The pellet was resuspended in the extraction buffer containing 20% glycerol and further resolved by size exclusion FPLC using a 24-mL Superose HR6 10/30 column (Pharmacia, Piscataway, NJ) with a flow rate of 0.1 mL/min. Peak fractions (0.5 mL) of TPPII activity were pooled and stored at −80°C.
TPPII Aminopeptidase Assay
Fractions were incubated for various times at 37°C with 0.01 to 1 mm AAF-AMC in 15 mm Tris, pH 7.0, and 5 mm MgCl2 (Balow et al., 1986), and then quenched by adding 0.9 mL of 80 mm sodium acetate, pH 4.3. Released AMC was monitored by fluorescence using excitation and emission wavelengths of 380 and 440 nm, respectively. Data were plotted in arbitrary fluorescence units using free AMC as a standard. MG132, (2S)-[1-[(2S)-2-amino-1-oxobutyl]-N-butyl]-2,3-dihydro-1H-indole-2-carboxamide oxalate (butabindide oxalate; Tocris, Ellisville, MO), PMSF, AAF-CMK (BioMol, Swedesboro, NJ), NEM, clasto-lactacystin β-lactone (lactacystin), (+)-(2S,3S)-3-[[(S)-3-methyl-1-[(3-methylbutyl) carbamoyl]butyl]carbamoyl]-2-oxiranecarboxylic acid (E-64c), leupeptin, and Na2EDTA were used in the inhibitor studies.
ESI Ion Trap-MS/MS Analysis and N-Terminal Sequencing
The in-gel tryptic or Glu-C digestion of TPPII followed the protocol as described (http://biotech.wisc.edu/ServicesResearch/MassSpec/ingel.htm). Briefly, sections of SDS-PAGE gels containing TPPII (visualized by Coomassie Blue staining) were excised, washed, treated with 50 mm DTT and 55 mm iodoacetamide, and incubated at 37°C overnight with 20 ng/μL trypsin (Promega, Madison, WI) or 25 ng/μL Glu-C (Roche, Indianapolis) in 100 mm ammonium bicarbonate, pH 8.0. The release peptides were extracted in 0.1% trifluoroacetic acid followed by 5% trifluoroacetic acid and 75% acetonitrile, and vacuum dried. MS sequencing employed capillary liquid chromatography-MS/MS system consisting of an HPLC connected to an ESI ion-trap MS (Surveyor HPLC and LCQ deca XPplus; ThermoFinnigan, San Jose, CA). A fritless 100- × 365-μm fused silica capillary microcolumn was prepared by pulling the tip of the capillary to approximately 2 μm with a P-2000 laser puller (Sutter Instruments, Novato, CA) and packing the capillary with 10 cm of 5 μm-diameter C18 beads (Western Analytical Products, Murrieta, CA). The vacuum-dried digests of TPPII were reconstituted in 10 μL of 95% water, 0.1% formic acid, and 5% acetonitrile. One-half of the sample was loaded onto the fused-silica capillary microcolumn at a flow rate of 1 μL/min for 30 min. The peptides were eluted over 150 min at a flow rate of 300 nL/min with a gradient of 5% to 80% acetonitrile in 0.1% formic acid. A full-mass scan was performed between 400 to 2,000 m/z, followed by 3 MS/MS scans of the 3 highest intensity parent ions at 45% relative collision energy. The acquired MS/MS spectra were searched against an Arabidopsis protein database derived from the National Center for Biotechnology Information (NCBI) nonredundant database (http://www.ncbi.nlm.nih.gov) using the SEQUEST program (ThermoFinnigan).
For N-terminal sequencing of TPPII, the 153- and 142-kD species were separated by SDS-PAGE and electrophoretically transferred onto a PVDF membrane (Millipore, Bedford, MA). Regions of the membrane containing the two species were excised, washed extensively with water, and subjected to 8 cycles of Edman degradation using a 494 Procise protein sequencer followed by a 140C analyzer (Applied Biosystems, Foster City, CA).
PAGE and Native/SDS-PAGE Two-Dimensional Electrophoresis
Nondenaturing PAGE followed the protocol as described in Yang et al. (2004). Peptidase activity of TPPII was detected by overlaying the gel with 100 μm AAF-AMC in 15 mm Tris, pH 7.0, and 5 mm MgCl2 and incubating the gel at 37°C. The released AMC product was visualized by fluorescence under UV light. The 26S proteasome and its 20S CP were purified as described (Yang et al., 2004). For two-dimensional native/SDS-PAGE, gel lanes from native PAGE were incubated with 1% SDS and 1% 2-mecaptoethanol for 30 min and subjected to SDS-PAGE in the second dimension.
Identification and Isolation of the Arabidopsis TPP2 Gene
The Arabidopsis genomic and EST databases (http://genome-www.stanford.edu/Arabidopsis) were searched by BLAST (Altschul et al., 1997) for TPP2 sequences using the yeast (Saccharomyces cerevisiae) and human proteins as queries. The full-length cDNA for TPP2 (GenBank accession no. AY096651) was aligned with the genomic region (At4g20850) to define the intron/exon boundaries. Amino acid sequence comparisons and phylogenetic analyses were performed using MACBOXSHADE (Institute of Animal Health, Pirbright, UK) and ClustalW (http://www.ebi.ac.uk/clustalw), respectively.
Identification and Analysis of the tpp2-1 Mutation
The tpp2-1 insertion mutant (SALK-085776) in the Arabidopsis ecotype Col-0 background was identified in the SIGNAL T-DNA collection (Alonso et al., 2003) and obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, OH). The mutation was tracked by PCR using the TPP2-specific 5′ (primer 2) or 3′ (primer 5) primers (see below), in combination with a T-DNA-specific primer (Alonso et al., 2003). The presence of the T-DNA was followed in subsequent generations by PCR and by kanamycin resistance conferred by the T-DNA. The heterozygous tpp2-1 plants were backcrossed 3 times to wild-type Col-0 Arabidopsis and selfed to obtain homozygous plants.
For RT-PCR, total RNA was isolated from 10-d-old seedlings using the Trizol protocol (Invitrogen, Carlsbad, CA). Each first-strand cDNA reaction was performed using a TPP2- specific primer (primer 1, GCCAGTTGATCACGTGTCACCTCCAT), a PAE2-specific primer, 1 μg of RNA, and Moloney murine leukemia virus reverse transcriptase (Promega). The first-strand cDNA was then PCR amplified using three sets of primer pairs for TPP2: set A (primer 2, TGCAGATTCACGGCGCGTTAATCAACAAAG and primer 3, CCAACTTGGAACCTACACGCCACTCTCC), set B (primer 4, GTGTGGCATGATGGAGAAGTATGGAGGGTTG and primer 5, TCTCCATAGCTCATGTTGACAAGATCACAG), and set C (primer 6, CAGACTATGGGCGCTTTGTTGATCTTGTTAC and primer 7, TATCAACTTGCATAAGCCCTTGTCCAGTAG); the position of each is identified left to right in Figure 2. The PAE2 primers are as described (Downes et al., 2003).
Phenotypic Analyses of tpp2-1
The wild-type and homozygous mutant seeds were vapor-phase sterilized for 4 h, stratified at 4°C for 3 d in the dark, and germinated on solid GM containing 0.8% agar. The plants were grown under a long-day photoperiod (16-h light/8-h dark). Effects of various inhibitors on root growth were measured by transferring 4-d-old seedlings of equal size to solid GM containing the inhibitors and growing the seedlings vertically for an additional 7 d. For immunoblot analyses, seedlings were frozen to liquid nitrogen temperatures and homogenized in SDS-PAGE sample buffer. The clarified extracts were subjected to SDS-PAGE, transferred to PVDF membranes, and probed with antibodies against PBA1, RPN12a, RPN5a, RPN1a, UBC1, and Ub as described (Smalle et al., 2002; Yang et al., 2004).
Acknowledgments
We thank Drs. Michael Westphall and Derek Gingerich for help with the MS analyses and genotyping, respectively.
This work was supported by the National Science Foundation Arabidopsis 2010 Initiative (grant no. MCB–0115870), by the U.S. Department of Energy Basic Energy Sciences program (grant no. DE–FG02–88ER13968 to R.D.V.), by the National Heart, Lung and Blood Institute-National Institutes of Health (grant no. N01–HV–28182 to L.M.S.), and by a Louis and Elsa Thomsen Wisconsin Distinguished Predoctoral Fellowship (to P.Y.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.057406.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balow R, Tomkinson B, Ragnarsson U, Zetterqvist O (1986) Purification, substrate specificity, and classification of tripeptidyl peptidase II. J Biol Chem 261: 2409–2417 [PubMed] [Google Scholar]
- Callis J (1995) Regulation of protein degradation. Plant Cell 7: 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A, Sanchez-Perez I, Nurse P (2003) Schizosaccharomyces pombe essential genes: a pilot study. Genome Res 13: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes B, Stupar R, Gingerich D, Vierstra R (2003) The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J 35: 729–742 [DOI] [PubMed] [Google Scholar]
- Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, Niedermann G (1999) A giant protease with potential to substitute for some functions of the proteasome. Science 283: 978–981 [DOI] [PubMed] [Google Scholar]
- Girod PA, Fu H, Zryd JP, Vierstra RD (1999) Multiubiquitin chain binding subunit MCB1 (RPN10) of the 26S proteasome is essential for developmental progression in Physcomitrella patens. Plant Cell 11: 1457–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glas R, Bogyo M, McMaster J, Gaczynska M, Ploegh H (1998) A proteolytic system that compensates for loss of proteasome function. Nature 392: 618–622 [DOI] [PubMed] [Google Scholar]
- Gu YQ, Chao WS, Walling LL (1996) Localization and post-translational processing of the wound-induced leucine aminopeptidase proteins of tomato. J Biol Chem 271: 25880–25887 [DOI] [PubMed] [Google Scholar]
- Hilbi H, Jozsa E, Tomkinson B (2002) Identification of the catalytic triad in tripeptidyl-peptidase II through site-directed mutagenesis. Biochim Biophys Acta 1601: 149–154 [DOI] [PubMed] [Google Scholar]
- Kamath R, Fraser A, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Akopian TN, Woo KM, Goldberg AL (1999) The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes: implications for understanding the degradative mechanism and antigen presentation. J Biol Chem 274: 3363–3371 [DOI] [PubMed] [Google Scholar]
- Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, Kruger E (2003) Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J Biol Chem 278: 21517–21525 [DOI] [PubMed] [Google Scholar]
- Osmulski PA, Gaczynska M (1998) A new large proteolytic complex distinct from the proteasome is present in the cytosol of fission yeast. Curr Biol 8: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Princiotta M, Schubert U, Chen W, Bennink J, Myung J, Crews C, Yewdell J (2001) Cells adapted to the proteasome inhibitor 4-hydroxy5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone require enzymatically active proteasomes for continued survival. Proc Natl Acad Sci USA 98: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reits E, Neijssen J, Herberts C, Benckhuijsen W, Janssen L, Drijfhout J, Neefjes J (2004) A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity 20: 495–506 [DOI] [PubMed] [Google Scholar]
- Renn S, Tomkinson B, Taghert P (1998) Characterization and cloning of tripeptidyl peptidase II from the fruit fly, Drosophila melanogaster. J Biol Chem 273: 19173–19182 [DOI] [PubMed] [Google Scholar]
- Rockel B, Peters J, Kuhlmorgen B, Glaeser R, Baumeister W (2002) A giant protease with a twist: the TPP II complex from Drosophila studied by electron microscopy. EMBO J 21: 5979–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C, Vargas F, Facchinetti P, Bourgeat P, Bambal RB, Bishop PB, Chan SMT, Moore ANJ, Ganellin CR, Schwartz JC (1996) Characterization and inhibition of a cholecystokinin-inactivating serine peptidase. Nature 380: 403–409 [DOI] [PubMed] [Google Scholar]
- Saric T, Graef C, Goldberg A (2004) Pathway for degradation of peptides generated by proteasomes: a key role for thimet oligopeptidase and other metallopeptidases. J Biol Chem 279: 46723–46732 [DOI] [PubMed] [Google Scholar]
- Seifert U, Maranon C, Shmueli A, Desoutter JF, Wesoloski L, Janek K, Henklein P, Diescher S, Andrieu M, de la Salle H, et al (2003) An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat Immunol 4: 375–379 [DOI] [PubMed] [Google Scholar]
- Shimotsuura I, Yoshida N, Ogata F, Ito A (1992) Evidence for and properties of a tripeptidyl peptidase in soybean (Glycine max Merr.) extract. Plant Sci 87: 1–7 [Google Scholar]
- Siezen R, Leunissen J (1997) Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci 6: 501–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra R (2002) Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babyichuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis thaliana growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra R (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Tamura T, Tamura N, Cejka Z, Hegerl R, Lottspeich F, Baumeister W (1996) Tricorn protease: the core of a modular proteolytic system. Science 274: 1385–1389 [DOI] [PubMed] [Google Scholar]
- Thompson A, Vierstra R (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Tomkinson B (1999) Tripeptidyl peptidases: enzymes that count. Trends Biochem Sci 24: 355–359 [DOI] [PubMed] [Google Scholar]
- Tomkinson B (2000) Association and dissociation of the tripeptidyl-peptidase II complex as a way of regulating the enzyme activity. Arch Biochem Biophys 376: 275–280 [DOI] [PubMed] [Google Scholar]
- Tomkinson B, Laoi B, Wellington K (2002) The insert within the catalytic domain of tripeptidyl-peptidase II is important for the formation of the active complex. Eur J Biochem 269: 1438–1443 [DOI] [PubMed] [Google Scholar]
- Vierstra R (1996) Proteolysis in plants: mechanisms and functions. Plant Mol Biol 32: 275–302 [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68: 1015–1068 [DOI] [PubMed] [Google Scholar]
- Wenzel T, Eckerskorn C, Lottspeich F, Baumeister W (1994) Existence of a molecular ruler in proteasomes suggested by analysis of degradation products. FEBS Lett 349: 205–209 [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A (2001) RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci USA 98: 3056–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD (2000) The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol 124: 1828–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Fu H, Walker J, Papa C, Smalle J, Ju Y-M, Vierstra R (2004) Purification of the Arabidopsis 26S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem 279: 6401–6413 [DOI] [PubMed] [Google Scholar]