Abstract
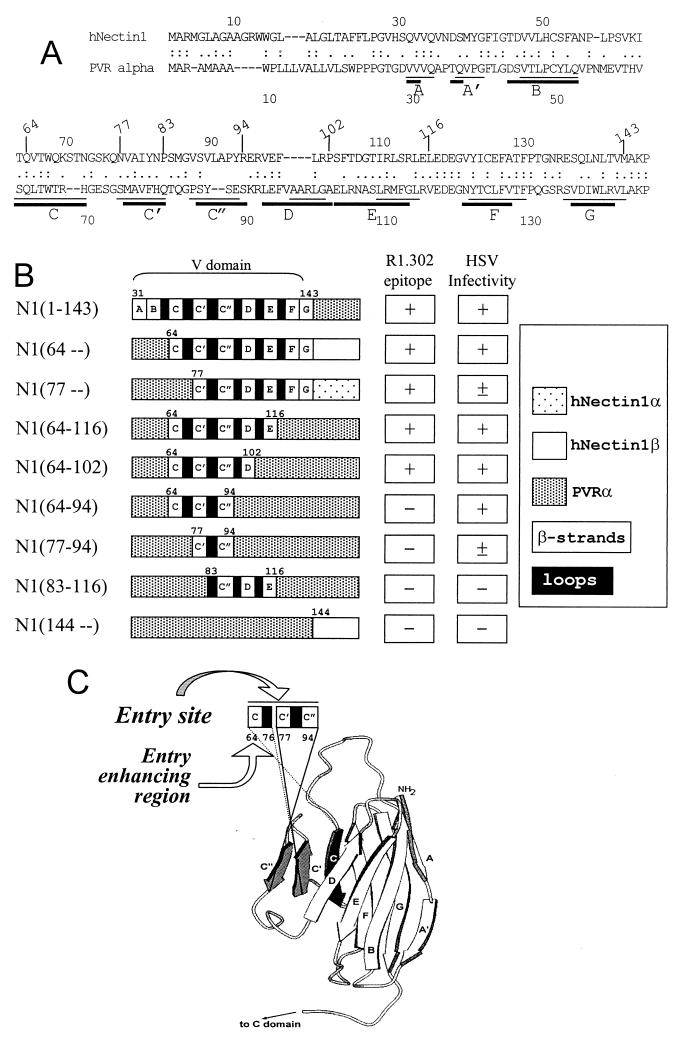
Human nectin1 (hNectin1), an adhesion molecule belonging to the nectin family of the immunoglobulin superfamily, mediates entry of herpes simplex virus (HSV) into cells. The hNectin1 domain that mediates virus entry into cells and also binds glycoprotein D (gD) has been localized to the first N-terminal V-type domain. The poliovirus receptor (PVR) is a structural homolog to nectins, but it cannot function as an HSV entry receptor. hNectin1-PVR chimeras were constructed to functionally locate the site on hNectin1 involved in HSV entry (HSV entry site). The epitope recognized by monoclonal antibody (MAb) R1.302, which is able to block HSV entry, was also located. The chimeric receptors were designed to preserve the overall structure of the V domain. The HSV entry activity mapped entirely to the hNectin1 portion located between residues 64 and 94 (64-94), likely to encode the C, C′, and C" β-strands and intervening loops. In turn, this site consisted of two portions: one with low-level basal activity for HSV entry (77-94), and one immediately upstream (residues 64 to 76) which greatly enhanced the HSV entry activity of the downstream region. The gD-binding site mapped substantially to the same site, whereas the MAb R1.302 epitope also required a further downstream portion (95-102). The involvement of the 64-76 portion is at difference with previous indirect mapping results that were based on competitive binding studies (C. Krummenacher et al., J. Virol. 74:10863–10872, 2000). The A, A′, B, D, E, F, and G β-strands and intervening loops did not appear to play any role in HSV entry. According to the predicted three-dimensional structure of PVR, the C C′ C" site is located peripherally in the V domain and very likely represents an accessible portion at the cell surface.
The receptors which mediate entry of herpes simplex virus (HSV) into cells belong to three structurally unrelated molecular families. They are nectin1 (CD111), a member of the immunoglobulin (Ig) superfamily, HveA, a member of the tumor necrosis factor receptor family, and modified heparan sulfate (4, 5, 9, 12, 20, 27, 35, 37). Of these, nectin1 is widespread in human cell cultures and is expressed in human organs and tissues targeted by HSV (9, 12). Nectin1 mediates both the entry of the virion into cells (9, 12) and cell-to-cell spread of virus (8), a relevant route of transmission given that in humans HSV spreads anterograde, i.e., from mucocutaneous tissues to nerve endings, and retrograde by direct cell-to-cell transmission. Of the 11 glycoproteins contained in the HSV virion, glycoprotein D (gD) is the one which interacts with the entry receptor (7, 9, 16), whereas gB and gC interact with the attachment receptor heparan sulfate proteoglycans (for reviews, see references 5 and 37). The site on nectin1 that mediates virus entry (hereafter called the HSV entry site) and also binds gD has been localized to the first N-terminal domain of the protein (7, 17). This is one of the three Ig-like domains that compose the ectodomain, and it has a V-like structure; the remaining two domains have a C-type structure. The V domain alone, engineered as soluble molecule, is able to compete with full-length transmembrane receptor and to block HSV entry, and it is also able to bind soluble forms of gD (7, 17). The V domain also carries the epitope recognized by monoclonal antibody (MAb) R1.302, which is able to block HSV infectivity (9) and therefore must overlap, at least in part, with the HSV entry site. Functional studies to locate the HSV entry site within the V domain have not been reported. In binding competition assays, gD interfered with the binding between nectin1 and a MAb directed to 80 to 104 (80-104) linear epitope of nectin1, a finding that was interpreted as evidence that this represents the gD-binding region (15).
In addition to nectin1, the human nectin family comprises the following structurally related molecules: nectin2 (CD112) (10), nectin3 (33), and the poliovirus receptor (PVR) (22). Nectin2 mediates entry of HSV gD mutants but not of wild-type HSV (19, 40). PVR (12) or nectin3 (unpublished observations) cannot function as an HSV receptor. For nectin1 and nectin2, three and two isoforms are known, respectively, which share the ectodomain (9, 10, 12, 18, 20). The designation of different isoforms of nectin1 by using Greek letters is discussed in reference 23.
Structurally, PVR may be considered the prototype member of the nectin family. Cryoelectron microscopy and X-ray crystallography have been used to define the structure of PVR and of the complex between the receptor and the poliovirus (2, 13). Each Ig-type domain has a β-barrel fold in which all β-strands (named A to G) run parallel or antiparallel to the long axis of the domain and are connected by more flexible interstrand loops. The V-type domain, relative to the C type, contains in addition the C′ and C" strands. A, B, E, and D strands form one face of the barrel. C, G, and F strands form the opposite surface of the barrel. C, C′, and C" strands are laterally located. For PVR, mutations in the predicted C-C′, C′-C", D-E, and E-F interstrand loops and in the C" and D strands disrupt poliovirus binding (1, 3, 28); the viral surface contacts mainly the C C′ C" surface (13).
The objective of the present study was to functionally locate the HSV entry site within the V domain of nectin1 and to fine-map the MAb R1.302 epitope. Inasmuch as the Ig V-like domain is a complex and rather rigid structure, deletions of entire β-strands, of portions or of groups of β-strands, are likely to alter the structure itself and to result in loss of interaction with HSV, not because the actual residues that interact with HSV have been deleted but because the overall structure of the V-like domain has been altered. We took advantage of the fact that PVR is structurally related to nectin1, but cannot function as an HSV entry receptor, to construct chimeric molecules where groups of β-strands and connecting loops of nectin1 replaced the corresponding regions of the V domain of PVR. This approach was meant to preserve the overall structure of the V domain: it lead to the locating of the HSV entry site at the portion of the molecule between residues 64 and 94, predicted to contain the C, C′, and C" strands and interstrand loops. Functionally, this site is divided into portions. The most upstream one (64-76) has enhancing activity over the 77-94 portion, which displays a low-level basal activity for HSV entry.
MATERIALS AND METHODS
Cells and viruses.
Cells were grown in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum. The J1.1-2 cell line, a derivative of BHKtk− cells highly resistant to HSV infection and a derivative thereof expressing human nectin1β (hNectin1β) are described elsewhere (9). The cell line stably expressing the N1(1-143) chimera, previously named V(HlgR-PVRα), was described previously (7). Stable cell lines expressing chimeric receptors were obtained by selection with neomycin G418, followed by sorting with a fluorescence-activated cell sorter (FACS). The HSV-1 recombinant R8102 carrying a lacZ gene inserted between the UL3 and UL4 genes under the control of the α27 promoter was described previously (9). Viruses were grown and titrated by plaque assay in Vero cells. Virus infectivity was detected by β-galactosidase (β-Gal) expression by reading the optical density at 405 nm (OD405) (9, 27) or by light microscopy observation of β-Gal-expressing cells.
Construction of hNectin1-PVR chimeric receptors.
Chimeric primers overlapping the hNectin1α or -β and PVR sequences to be joined were synthesized both as sense and antisense primers. The chimeric primers were used separately with appropriate external primers designed on either the hNectin1 sequence or PVR sequence to generate two fragments, one N-terminal and one C-terminal (see Table 1 for primer sequences). The N-terminal and the C-terminal fragments were then mixed in equimolar amounts and joined through a polymerization reaction (20 to 25 cycles of denaturation, annealing, and extension) which exploited the complementarity of the chimeric primer and the ability of the fragments to act both as primer and template for each other. The chimeras were cloned in pcDNA3.1 (Invitrogen) and sequenced for accuracy.
TABLE 1.
Sequences of primers employed to generate the chimeric receptors
| Chimera | N-ter primer | Chimeric primer (sense) | C-ter primer |
|---|---|---|---|
| N1(1-143) | PRR1V5, TAATAAGCTTATGGCTCGGATGGGGCTTGCGGGC (hNectin1)a | R1VRV3, CAATCTCACGGTGATGGCCAAGCCCCAGAACAC | R1VR-CC3, GTTAGGATCCTCACCTTGTGCCCTCTGTCTG (PVRα) |
| N1(64–) | PVR5, GAATAAGCTTATGGCCCGAGCCATGGCCGCCGCGTG (PVRα) | HIMUT4, CAACATGGAGGTGACGCATGTGTCACAGGTCACATGGCAGAAGTCCACC | VRHICC3, GTTAGGATCCCTAGGGGCTCTCTCCTCGAGG (hNectin1β) |
| N1(77–) | PVR5Bam, GAATGGATCCATGGCCCGAGCCATGGCCGCCGCGTG (PVRα) | Mut5forw, TGGGCGCGGCATGGTGAATCTGGCAACGTGGCCATCTACAACCCATCC | HveCHind, ATTAAAGCTTCTACACGTACCACTCCTTCTTGG (hNectin1α) |
| N1(64-116) | PVR5Nhe, GATTGCTAGCATGGCCCGAGCCATGGCCGCCGCGTG [N1(64–)] | Mut6forw CGCCTCTCCCGCCTGGAGCTGGAGGATGAAGGCAACTACACCTGC | PVR3Hind, ATTAAAGCTTCACCTTGTGCCCTCTGTCTGTGG (PVRα) |
| N1(64-102) | PVR5Nhe, GATTGCTAGCATGGCCCGAGCCATGGCCGCCGCGTG [N1(64–)] | Mut10forw, GAGCGTGTGGAATTCCTGCGGCCCGAGCTGCGGAATGCCTCGCTGAGG | PVR3Hind, ATTAAAGCTTCACCTTGTGCCCTCTGTCTGTGG (PVRα) |
| N1(64-94) | PVR5Nhe, GATTGCTAGCATGGCCCGAGCCATGGCCGCCGCGTG [N1(64–)] | Mut9forw, GTGTCCGTGCTGGCTCCCTACCGCCTGGAATTCGTGGCAGCCAGACT | PVR3Hind, ATTAAAGCTTCACCTTGTGCCCTCTGTCTGTGG (PVRα) |
| N1(77-94) | PVR5Nhe, GATTGCTAGCATGGCCCGAGCCATGGCCGCCGCGTG [N1(77–)] | mut9forw, GTGTCCGTGCTGGCTCCCTACCGCCTGGAATTCGTGGCAGCCAGACT | PVR3Hind, ATTAAAGCTTCACCTTGTGCCCTCTGTCTGTGG (PVRα) |
| N1(83-116) | PVR5Nhe, GATTGCTAGCATGGCCCGAGCCATGGCCGCCGCGTG (PVRα) | Mut7forw, GGCAGCATGGCCGTCTTCCACCAACCATCCATGGGCGTGTCCGTGCTG | PVR3Hind, ATTAAAGCTTCACCTTGTGCCCTCTGTCTGTGG [N1(64-116)] |
| N1(144–) | PVR5, GAATAAGCTTATGGCCCGAGCCATGGCCGCCGCGTG (PVRα) | VRHI-CC5, GATATCTGGCTCCGAGTGCTTGCCAAACCCACCAATTGGATAG | VRHICC3, GTTAGGATCCCTAGGGGCTCTCTCCTCGAGG (hNectin1β) |
Shown in parentheses is the template used to obtain the fragment.
Antibodies.
The R1.302 MAb directed to the V domain of hNectin1 was described previously (7, 21). The anti-PVR antibodies P242, 280, D171, PV.404, and P44 have been described elsewhere (1, 3, 14, 21, 25).
IFA.
For FACS analysis of immunoreactivity to R1.302 and anti-PVR antibodies and analysis of gD binding, live cells were incubated with the following antibodies: P44 and P242 hybridoma cell supernatants at 1:2 dilution, MAb 280 ascitic fluid at 1:1,000 dilution, purified D171 and PV.404 at 10 μg/ml, or 0.5 μg of recombinant gD(Δ290-299t)/ml and mouse anti-gD MAb H170 (Goodwin Cancer Research Institute, Plantation, Fla.) (1:1,000), followed by a 1:50 goat anti-mouse phycoerythrin antibody (Beckman-Coulter-Immunotech, France). All steps were done in phosphate-buffered saline supplemented with 5% fetal colf serum for 1 h at 4°C. Cells were subsequently analyzed in a FACScan flow cytometer (Becton Dickinson). For immunofluorescence assays (IFAs), J1.1-2 cells stably expressing chimeric receptors or pcDNA3.1 as a negative control were grown on glass coverslips, fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 10 min, and then reacted with MAb R1.302 (1:100), followed by anti-mouse antibodies (1:100) conjugated to fluorescein isothiocyanate (Jackson Immunoresearch Laboratories).
Infectivity assay.
J1.1-2 cells were transfected with the chimeric constructs by means of Lipofectamine reagent (Life Technologies) according to the manufacturer's instructions and infected at 30 h after transfection with R8102 at 10 PFU/cell. After 16 h, virus infectivity was detected by β-Gal expression by staining with o-nitrophenyl-β-d-galactopyranoside (ONPG) and reading the OD405 (9, 27) or by light microscopy observation of β-Gal-expressing cells after staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
RESULTS
Construction of chimeric hNectin1-PVR receptors.
The structure of nectin1 has not been solved. By contrast, the structure of PVR has been the object of numerous studies (for recent studies, see references 2, 13, and 32). In order to proceed with the substitution of groups of strands and interstrand loops of hNectin1 with the corresponding regions of its structural homolog PVR, we performed an alignment of hNectin1 and PVR sequences (Fig. 1A). As illustrated in Fig. 1A, while there is an agreement on the overall structure of the PVR V domain, there is no universal agreement on the boundaries of the single β-strands. Fig. 1B shows a schematic representation of the chimeric hNectin1-PVR constructs generated in this study. They are designated as N1 (for nectin1) followed by numerals in parentheses which define the nectin1 portion present in the chimera. For each construct to be generated, two fragments were derived by PCR, one N-terminal and one C-terminal, and then joined by PCR, and the chimeras were cloned in pcDNA3.1. Table 1 reports the sequences of the primers and the respective DNA employed as template. J1.1-2 cells, which are negative for HSV receptors and resistant to HSV infection (9), were transfected with the plasmids encoding the chimeric receptors. They were either employed 30 h after transfection (transient expression) or selected for acquired neomycin G418 resistance (stable transformants) and subsequently employed as such and subjected to single-cell cloning or enriched by FACS sorting, as specified below.
FIG. 1.
(A) Alignment of hNectin1 (CD111) and PVR (CD155) V domain sequences (31). The bars define the location of the predicted PVR β-strands according to reference 13 (thin bars) and reference 2 (thick bars). (B) Schematic linear representation of the composition of the V domain of chimeric hNectin1-PVR receptors and summary of their properties. The chimeric receptors were constructed by means of the primers described in Table 1. The first 30 residues of hNectin1 are predicted to encode the signal sequence and to be cleaved off in the mature form of the molecule. The N1(1-143) chimera was described previously and designated as V(HIgR-PVRα) (7). (C) Simplified view of the structural model of PVR, adapted from reference 32. The region encompassing the C, C′, and C" β-strands, identified as the HSV entry site on nectin1, is shaded in black and gray.
Reactivity of chimeric receptors to MAbs to nectin1 and to PVR.
As a first characterization of the constructs, we ascertained that the encoded proteins were expressed in the transfected cells and were able to reach the cell surface. To this end, we measured reactivity to a panel of MAbs to PVR, which constitutes the C-terminal portion of all constructs, except N1(64–) and N1(77–). The anti-PVR antibodies were addressed either to the V domain (P242, 280, D171) (1, 3, 14, 25) or to the C-C domains (PV.404, P44) (3, 14, 21). We also measured reactivity to MAb R1.302 directed to nectin1. Stable transformants of J1.1-2 cells transfected with the plasmids encoding the chimeric receptors and selected for G418 resistance were assayed by IFA microscopy or FACS analysis. Typical results are shown in Fig. 2 (IFA with MAb R1.302) and Fig. 3 (FACS with MAbs R1.302 and anti-PVR) and are summarized in Table 2 (FACS with PVR MAbs) and Fig. 1B. PVR antibodies showed that all the constructs whose C terminus consisted of PVR sequence were able to reach the cell surface. As expected, the constructs carrying the C domains of PVR, namely N1(1-143), N1(64-116), N1(64-102), N1(64-94), N1(77-94), and N1(83-116), were only detected by PV.404 and P44 MAbs, whereas the chimera carrying the entire V domain of PVR, N1(144–), was only detected by P242, 280, and D171 MAbs (Fig. 3 and Table 2). No reactivity to anti-PVR MAbs was detected with the N1(64–) and N1(77–) chimeras carrying the cytoplasmic tail of nectin1α or -β, but these chimeras were positive with MAb R1.302. Concerning MAb R1.302 (Fig. 2 and 3), N-terminus reactivity was still present in the construct containing nectin1 downstream from residue 77 [N1(77–)] and absent in the construct containing nectin1 downstream from residue 83. At the C terminus, reactivity was present in the construct containing the nectin1 portion upstream of residue 102 [N1(64-102)] but absent from the constructs containing nectin1 upstream of residue 94 [N1(64-94) and N1(77-94)]. These results define an essential part of the MAb R1.302 epitope as being located between residues 77 and 102, which likely includes the C′, C", and D strands and the intervening loops.
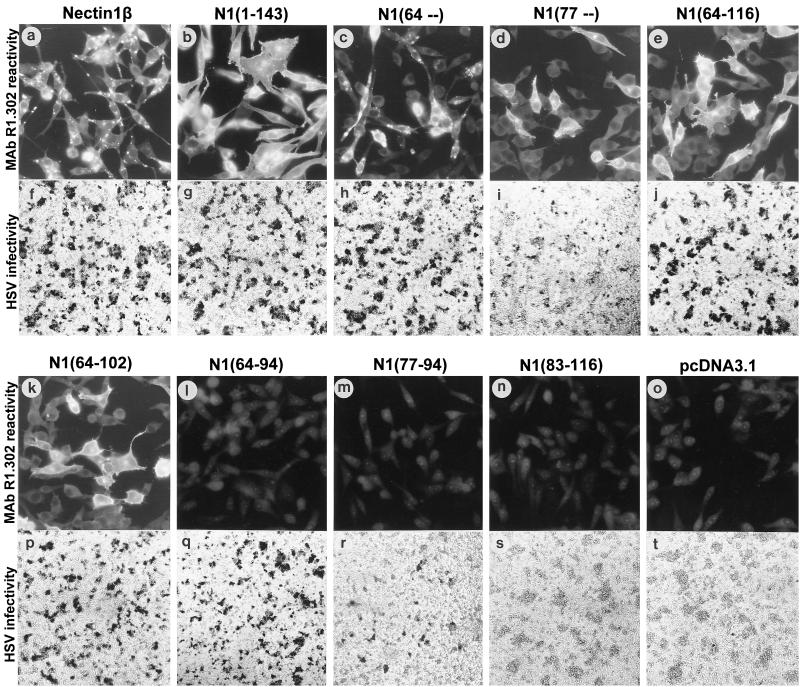
FIG. 2.
Representative gallery of cells expressing chimeric hNectin1-PVR receptors. In each row, the upper panels show IFA reactivity with MAb R1.302, whereas the bottom panels show HSV R8102 infection, detected as β-Gal activity (9). The positive and negative controls consisted of full-length nectin1β (a, f) and of cells transfected with pcDNA3.1 alone (o, t). For IFA, nectin1β was determined in a cloned cell line and N1(1-143) was studied in G418-selected cells sorted by FACS; the remaining panels show G418-selected cells that were not subjected to FACS sorting. For the infectivity assay, all cultures were assayed after transient expression.
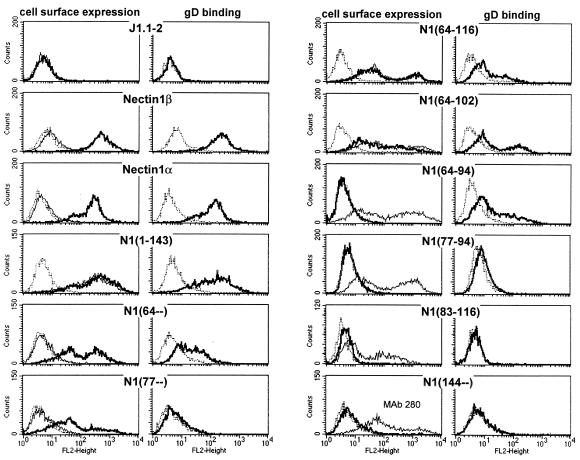
FIG. 3.
FACS analysis of immunoreactivity to R1.302 and anti-PVR MAbs and of gD binding. Cells were incubated either with R1.302 or PV.404 MAbs or with gD(Δ290-299t) followed by anti-gD MAb H170. Except for nectin1α and -β, all cultures were subjected to G418 selection followed by FACS sorting. After FACS sorting and culturing, the cells exhibited heterogeneous fluorescence profiles. This most likely reflects cell populations with heterogeneity in the extent of expression. Cell surface expression analysis: dashed line, negative control for MAb reactivity consisting of isotype-matched irrelevant MAb, followed by secondary antibody; thin line, anti-PVR MAb PV.404; thick line, anti-Nectin1 MAb R1.302. gD-binding analysis: dashed line, negative control consisting of no gD, followed by MAb H170; thick line, gD at 0.5 μg/ml.
TABLE 2.
Summary of FACS reactivity to PVR MAbs of cells expressing the chimeric receptorsa
| Receptor | Reactivity to anti-PVR MAb
|
||||
|---|---|---|---|---|---|
| PV.404 | P44 | P242 | 280 | D171 | |
| Nectin1β | − | − | − | − | − |
| N1(1-143) | + | + | − | − | − |
| N1(64–) | − | − | − | − | − |
| N1(64-116) | + | + | − | − | − |
| N1(64-102) | + | + | − | − | NDb |
| N1(64-94) | + | + | − | − | − |
| N1(77–) | − | − | − | − | − |
| N1(77-94) | + | + | − | − | ND |
| N1(83-116) | + | + | − | − | − |
| N1(144–) | − | − | + | + | + |
| PVR (Cos cells) | + | + | + | + | + |
For experimental details, see the legend to Fig. 3.
ND, not done.
While all constructs exhibited cell surface expression, the intracellular distribution of the MAb R1.302-reactive compartment displayed two somewhat different patterns (Fig. 2). Thus, all the chimeras carrying the C-terminal portion of PVR exhibited a common pattern characterized by a rather diffuse cytoplasmic staining (Fig. 2b, e, and k). The pattern for N1(77–) construct, carrying the cytoplasmic tail of nectin1α, was similar (panel d). By contrast, the construct carrying the cytoplasmic tail of nectin1β [N1(64–)], and nectin1β itself, exhibited a predominantly vesicular-like distribution (panels c and a, respectively). The intracellular distribution of the gD(Δ290-299t)-binding compartment paralleled closely that of MAb R1.302 (data not shown). Nectin1α, but not nectin1β, carries at the C terminus the consensus sequence A/ExYV that binds the PDZ domain of afadin (38) and enables localization at adherens junctions in polarized epithelial cells. While these differences are not relevant to the function of hNectin1α and -β as HSV receptors, as the two isoforms cannot be differentiated with respect to ability to act as HSV entry receptors or degree of cell surface expression (reference 9 and Fig. 3), it is possible that, in nonpolarized cells, the nectin1β cytoplasmic sequence may favor a preferential accumulation in vesicular-like structures. The PVRα does not carry a typical PDZ-domain binding site. Its localization at adherens junctions remains to be investigated, although its cellular distribution clearly included a diffuse cytoplasmic pattern in addition to a high level of cell surface expression.
HSV infectivity mediated by chimeric hNectin1-PVR receptors.
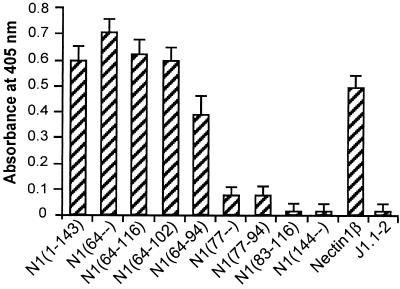
Having ascertained that all chimeric receptors are able to reach the cell surface, we then compared the levels of HSV infectivity. J1.1-2 cells transiently expressing the chimeric receptors were infected with the HSV-1 recombinant R8102, which carries a LacZ reporter gene under the immediate early α27 promoter (9). Infection was monitored and quantified as β-Gal activity. Typical results are shown in Fig. 2 and Fig. 4 and are summarized in Fig. 1B. Chimeric molecules containing hNectin1 residues downstream of residue 64 enabled expression of the reporter gene to a level comparable to that seen in cells expressing the entire nectin1 V domain [N1(1-143)] or full-length hNectin1β (Fig. 2h, j, p, and q). Cells expressing the two constructs containing nectin1 downstream from residue 77 [N1(77–) and N1(77-94)] were infected with HSV at reduced efficiency (compare Fig. 2i with r; Fig. 4). Cells expressing the construct containing nectin1 downstream from residue 83 [N1(83-116)] were not infected at all (Fig. 2s). For the C terminus, infectivity was present in cells expressing the construct containing the nectin1 portion upstream of residue 94 (Fig. 2h, i, j, and p to r). The key constructs were therefore N1(64-94) and N1(77-94), which exhibit similar levels of cell surface expression (Fig. 3). We infer from their properties that (i) the minimal region that mediated HSV entry with an efficiency similar to that of full-length nectin1 is located between residues 64 and 94, predicted to include the C, C′, and C" strands and intervening loops. (ii) Within this region, two portions were differentiated. The 77-94 portion displayed a low-level, basal HSV entry activity. The adjacent upstream portion (64-76) greatly enhanced the HSV entry activity.
FIG. 4.
Comparison of R8102 infectivity in cells expressing the chimeric hNectin1-PVR receptors. Cells transiently expressing the receptors were infected with R8102 (10 PFU/cell) at 30 h after transfection. Sixteen hours later, cells were solubilized with 0.5% Nonidet P-40 and β-Gal activity was quantified with ONPG (9, 27). Each bar represents the average of three replicate samples.
Binding activity to gD.
The localization of the gD-binding site on nectin1 may be determined essentially by two types of approaches, each with advantages and disadvantages. Enzyme-linked immunosorbent assays and biosensor assays are very sensitive techniques that rely on soluble recombinant proteins (7, 17, 41). These assays may not fully preserve the same conformation and oligomerization displayed by the full-length molecule in its natural context. By contrast, cells which express nectin1 can bind recombinant soluble gD (9) and can therefore indicate a biologically significant location of the gD-binding site on nectin1 when the receptor is expressed at the cell surface, in its natural conformation and context. Two forms of recombinant gD have been reported to bind in a detectable manner to cells expressing gD. They are (i) gD(Δ290-299t), a soluble gD in which the residues 290 to 299 were replaced with an unrelated sequence (29); and (ii) gD-Fc, a chimeric gD fused to the Fc portion of rabbit IgG (11). gD(306t), a form of gD truncated about 20 residues upstream from the transmembrane region, does not appear to produce detectable binding, at least under the experimental conditions tested in our laboratory. Previously, it was reported that gD(Δ290-299t) binds to soluble nectin1 with a 100-fold-higher affinity than gD(306t) (16). The higher affinity is consistent with the finding that binding to cells is detectable with gD(Δ290-299t) but not with gD(306t). In enzyme-linked immunosorbent assays, wild-type gD, affinity-purified from infected cells, bound soluble nectin1-Fc molecules significantly more strongly than gD(Δ290-299t) (23). Thus, different forms of gD display different binding strengths, a phenomenon currently not well understood.
Here, the binding of gD(Δ290-299t) to cells expressing the chimeric receptors was detected by FACS analysis, following reactivity with anti-gD MAb H170 (30). Figure 3 shows that the gD(Δ290-299t) binding activity to cells expressing the full-length nectin1α and -β, or a chimera containing the entire nectin1 V domain [N1(1-143)], was readily detectable and of high intensity. The binding to cells expressing hNectin1 constructs downstream of residue 64 could be detected with a 5- to 10-fold-reduced efficiency; the binding to cells expressing hNectin1 downstream from residue 77 was barely detectable. The binding to cells expressing hNectin1 downstream from residue 83 was null.
DISCUSSION
In the studies reported here, we mapped the site of interaction of nectin1 responsible for entry of HSV-1 into susceptible cells and for the binding to gD. The donor sequences were components of the V domain of nectin1. The recipient was PVR, a member of the same family of proteins. The mapping of active domains by exchange of amino acid stretches between two structurally related proteins is a powerful technique for mapping functional domains, provided the degree of homology of the two related proteins is sufficient to ensure that the basic structure is similar. Sequences should be distant enough so that transfer of positionally corresponding stretches complements existing similarities in structure to fully restore function. The extent of homology between PVR and nectin1 is 31% in the V domain, which is low enough to ensure that the extent of basal interaction with HSV gD is nonexistent but high enough to ensure some structural conservation. Our results are consistent with these objectives and show the following:
(i) The HSV entry site was localized to the portion of hNectin1 between residues 64 and 94, which is likely to encode the C, C′, and C" β-strands and intervening loops.
(ii) This site consisted of two functional portions: the 77-94 portion (C′-C"), which mediated a low-level, basal extent of entry, and the upstream portion (64-76) (C), which augmented the HSV entry activity to a level comparable to that seen with full-length nectin1. The latter portion may participate in the entry process directly, i.e., by contributing residues that interact with HSV gD, or indirectly, i.e., by stabilizing, or contributing to, the structure of the receptor. The higher extent of gD binding exhibited by N1(64-94) relative to that of N1(77-94) favors the first possibility.
(iii) Our constructs ruled out the direct participation of the A, A′, and B and D, E, F, and G portions in the entry process.
(iv) The epitope recognized by MAb R1.302 is localized at the 77-102 portion. Inasmuch as the epitope included the 95-102 portion (D), which is not involved in HSV entry, there was only a partial overlap between the MAb R1.302 epitope and the HSV entry site. Of note, blocking the 77-94 region appears to be sufficient in order to completely prevent virus entry.
(v) There was an overall parallel between the gD-binding activity and the HSV entry site. The key constructs were N1(64-94), N1(77-94), and N1(83-116), which exhibited high, low, and null HSV entry, respectively, as well as gD-binding activity. Thus, the gD-binding activity localized to the region on nectin1 where the HSV entry site was located.
Recently, the localization of the gD-binding site and of the MAb R1.302 epitope on nectin1 were determined by an indirect approach based on competition with MAbs directed to linear epitopes of the V domain (15). Thus, (i) soluble gD [gD(285t)] interfered with the interaction of a soluble recombinant nectin1 with anti-nectin1 MAbs directed to the 80-104 linear epitopes. (ii) The MAb R1.302 epitope was mapped to the 70-104 amino acid region. (iii) The contributions of the 70-80 region to the gD-binding site were not investigated, nor were the contributions of the regions upstream of residue 70 and downstream of residue 104. Clearly this approach was more indirect relative to the one applied in our study, and the reduction in MAb binding may have resulted from steric hindrance or from conformational changes induced by binding of the antibodies. For instance, such conformational changes were observed with HveA following binding to gD and to its additional ligands (34).
Irrespective of the slight differences, and of different sensitivities of the assays, the two studies point to the region at C′-C" as critical for the HSV entry site, the gD-binding activity, and the MAb R1.302 epitope. In addition, our studies extend the mapping data, as they rule out a contribution of the sequences upstream and downstream of amino acids 64 and 94, respectively. Interestingly, comparison between human and murine nectin1 sequences highlights in the 70-95 amino acid region (C′-C") a 28% amino acid difference, as compared to 9 and 8% for the V domain and the entire ectodomain, respectively. This region overlaps with the HSV entry site identified in this study and may represent a site of variations for nectins from different species. Whether this region fully accounts for the observed differences in gD-binding activity between the human and the murine receptors remains to be determined (23, 24).
According to the predicted structure of PVR and several other Igs, the C, C′, and C" β-strands and the intervening loops are peripherally exposed portions in the V domain, and the C′ and C" strands form a lateral loop (2) (Fig. 1C). For nectin1 expressed at the cell surface, this is consistent with accessibility of this site to HSV and virion gD and to the blocking MAb, R1.302. It is relevant that also the site recognized by poliovirus on PVR, and the site recognized by human immunodeficiency virus gp120 on CD4 map to the corresponding regions of their respective receptors (2, 13, 26). It is very likely that this reflects the location and accessibility of this site at the cell surface.
Finally, it should be noted that the objective of conducting mapping studies is not only to define the regions of interaction between the receptor and the virus. These studies may be relevant for the control of HSV infection in humans. The emergence of HSV strains resistant to the commonly used anti-HSV drug acyclovir (6) creates a need for novel drugs that are addressed to different targets. For human immunodeficiency virus as well as for influenza virus, drugs designed to disrupt the interaction of the virus with the cognate receptor are under study or are already commercially available (36, 39). The definition of the HSV entry site in nectin1—narrowed in this study to 31 residues—may lead to the design of novel therapeutics able to block the interaction of HSV with nectin1.
ACKNOWLEDGMENTS
We thank Elisabetta Romagnoli and Stephanie Fabre for invaluable assistance with cell cultures and FACS analyses. We thank G. H. Cohen and R. Eisenberg (Philadelphia, Pa.) for the gift of gD(Δ290-299t), B. Roizman (Chicago, Ill.) for the gift of R8102, Akio Nomoto (Tokyo, Japan) for the gift of P242 and P44 MAbs, and Philip Minor (Hertfordshire, United Kingdom) for the gift of MAb 280.
The work done at the University of Bologna was supported by grants from Telethon (grant A141), Target Project in Biotechnology/CNR, MURST (40%), University of Bologna (60%), and pluriannual plan. The studies at INSERM U.119, Marseille, were aided by INSERM, the Association pour la Recherche Contre le Cancer, and the Ligue Nationale Française Contre le Cancer.
REFERENCES
- 1.Aoki J, Koike S, Ise I, Sato-Yoshida Y, Nomoto A. Amino acid residues on human poliovirus receptor involved in interaction with poliovirus. J Biol Chem. 1994;269:8431–8438. [PubMed] [Google Scholar]
- 2.Belnap D M, McDermott B M, Jr, Filman D J, Cheng N, Trus B L, Zuccola H J, Racaniello V R, Hogle J M, Steven A C. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc Natl Acad Sci USA. 2000;97:73–78. doi: 10.1073/pnas.97.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhardt G, Harber J, Zibert A, deCrombrugghe M, Wimmer E. The poliovirus receptor: identification of domains and amino acid residues critical for virus binding. Virology. 1994;203:344–356. doi: 10.1006/viro.1994.1493. [DOI] [PubMed] [Google Scholar]
- 4.Campadelli-Fiume G. Virus receptor arrays, CD46 and human herpesvirus 6. Trends Microbiol. 2000;8:436–438. doi: 10.1016/s0966-842x(00)01804-7. [DOI] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev Med Virol. 2000;10:305–319. doi: 10.1002/1099-1654(200009/10)10:5<305::aid-rmv286>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Scieux C, Garrait V, Socie G, Rocha V, Molina J M, Thouvenot D, Morfin F, Hocqueloux L, Garderet L, Esperou H, Selimi F, Devergie A, Leleu G, Aymard M, Morinet F, Gluckman E, Ribaud P. Resistant herpes simplex virus type 1 infection: an emerging concern after allogeneic stem cell transplantation. Clin Infect Dis. 2000;31:927–935. doi: 10.1086/314052. [DOI] [PubMed] [Google Scholar]
- 7.Cocchi F, Lopez M, Menotti L, Aoubala M, Dubreuil P, Campadelli-Fiume G. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc Natl Acad Sci USA. 1998;95:15700–15705. doi: 10.1073/pnas.95.26.15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocchi F, Menotti L, Dubreuil P, Lopez M, Campadelli Fiume G. Cell-to-cell spread of wild-type herpes simplex virus 1, but not of syncytial strains, is mediated by the immunoglobulin-like receptors that mediate virion entry, nectin 1 (HveC/HIgR/PRR1) and nectin2 (PRR2) J Virol. 2000;74:3909–3917. doi: 10.1128/jvi.74.8.3909-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bonafide receptor for herpes simplex viruses 1 and 2 in human cells. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberlé F, Dubreuil P, Mattei M G, Devilard E, Lopez M. The human PRR2 gene, related to the human poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 11.Geraghty R J, Jogger C R, Spear P G. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology. 2000;268:147–158. doi: 10.1006/viro.1999.0157. [DOI] [PubMed] [Google Scholar]
- 12.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 13.He Y, Bowman V D, Mueller S, Bator C M, Bella J, Peng X, Baker T S, Wimmer E, Kuhn R J, Rossmann M G. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci USA. 2000;97:79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koike S, Ise I, Nomoto A. Functional domains of the poliovirus receptor. Proc Natl Acad Sci USA. 1991;88:4104–4108. doi: 10.1073/pnas.88.10.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krummenacher C, Baribaud I, Ponce De Leon M, Whitbeck J C, Lou H, Cohen G H, Eisenberg R J. Localization of a binding site for herpes simplex virus glycoprotein D on herpesvirus entry mediator C by using antireceptor monoclonal antibodies. J Virol. 2000;74:10863–10872. doi: 10.1128/jvi.74.23.10863-10872.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krummenacher C, Rux A H, Whitbeck J C, Ponce-de-Leon M, Lou H, Baribaud I, Hou W, Zou C, Geraghty R J, Spear P G, Eisenberg R J, Cohen G H. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J Virol. 1999;73:8127–8137. doi: 10.1128/jvi.73.10.8127-8137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez M, Cocchi F, Avitabile E, Leclerc A, Adelaide J, Campadelli-Fiume G, Dubreuil P. Novel, soluble isoform of the herpes simplex virus (HSV) receptor nectin1 (or PRR1-HIgR-HveC) modulates positively and negatively susceptibility to HSV infection. J Virol. 2001;75:5684–5691. doi: 10.1128/JVI.75.12.5684-5691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez M, Cocchi F, Menotti L, Avitabile E, Dubreuil P, Campadelli-Fiume G. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J Virol. 2000;74:1267–1274. doi: 10.1128/jvi.74.3.1267-1274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez M, Eberlé F, Mattei M G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 21.Lopez M, Jordier F, Bardin F, Coulombel L, Chabannon C, Dubreuil P. CD155 Workshop. Identification of a new class of IgG superfamily antigens expressed in hemopoiesis. In: Kishimoto T, et al., editors. Leukocyte typing VI, White cell differentiation antigens. New York, N.Y: Garland Publishing; 1997. pp. 1081–1083. [Google Scholar]
- 22.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 23.Menotti L, Avitabile E, Dubreuil P, Lopez M, Campadelli-Fiume G. Comparison of murine and human nectin1 binding to herpes simplex virus glycoprotein D (gD) reveals a weak interaction of murine nectin1 to gD and a gD-dependent pathway of entry. Virology. 2001;282:256–266. doi: 10.1006/viro.2001.0850. [DOI] [PubMed] [Google Scholar]
- 24.Menotti L, Lopez M, Avitabile E, Stefan A, Cocchi F, Adelaide J, Lecocq E, Dubreuil P, Campadelli Fiume G. The murine homolog of human-Nectin1δ serves as a species non-specific mediator for entry of human and animal αherpesviruses in a pathway independent of a detectable binding to gD. Proc Natl Acad Sci USA. 2000;97:4867–4872. doi: 10.1073/pnas.97.9.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minor P D, Pipkin P A, Hockley D, Schild G C, Almond J W. Monoclonal antibodies which block cellular receptors of poliovirus. Virus Res. 1984;1:203–212. doi: 10.1016/0168-1702(84)90039-x. [DOI] [PubMed] [Google Scholar]
- 26.Moebius U, Clayton L K, Abraham S, Harrison S C, Reinherz E L. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic binding studies of mutants in conjunction with a high-resolution CD4 atomic structure. J Exp Med. 1992;176:507–517. doi: 10.1084/jem.176.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 28.Morrison M E, He Y J, Wien M W, Hogle J M, Racaniello V R. Homolog-scanning mutagenesis reveals poliovirus receptor residues important for virus binding and replication. J Virol. 1994;68:2578–2588. doi: 10.1128/jvi.68.4.2578-2588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira L, Dondero D V, Gallo D, Devlin V, Woodie J D. Serological analysis of herpes simplex virus types 1 and 2 with monoclonal antibodies. Infect Immun. 1982;35:363–367. doi: 10.1128/iai.35.1.363-367.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Person W R, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- 32.Racaniello V R. Early events in poliovirus infection: virus-receptor interactions. Proc Natl Acad Sci USA. 1996;93:11378–11381. doi: 10.1073/pnas.93.21.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reymond N, Borg J, Lecocq E, Adelaide J, Campadelli-Fiume G, Dubreuil P, Lopez M. Human nectin3/PRR3: a novel member of the PVR/PRR/nectin family that interacts with afadin. Gene. 2000;255:347–355. doi: 10.1016/s0378-1119(00)00316-4. [DOI] [PubMed] [Google Scholar]
- 34.Sarrias M R, Whitbeck J C, Rooney I, Ware C F, Eisenberg R J, Cohen G H, Lambris J D. The three HveA receptor ligands, gD, LT-alpha and LIGHT bind to distinct sites on HveA. Mol Immunol. 2000;37:665–673. doi: 10.1016/s0161-5890(00)00089-4. [DOI] [PubMed] [Google Scholar]
- 35.Shukla D, Liu J, Blaiklock P, Shworak N W, Bai X, Esko J D, Cohen G H, Eisenberg R J, Rosenberg R D, Spear P G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 36.Sodroski J G. HIV-1 entry inhibitors in the side pocket. Cell. 1999;99:243–246. doi: 10.1016/s0092-8674(00)81655-4. [DOI] [PubMed] [Google Scholar]
- 37.Spear P G, Eisenberg R J, Cohen G H. Three classes of cell surface receptors for alphaherpesvirus entry. Virology. 2000;275:1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Von Itzstein M, Wu W Y, Kok G B, Pegg M S, Dyason J C, Jin B, Van Phan T, Smythe M L, White H F, Oliver S W, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 40.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 41.Whitbeck J C, Muggeridge M I, Rux A H, Hou W, Krummenacher C, Lou H, van Geelen A, Eisenberg R J, Cohen G H. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J Virol. 1999;73:9879–9890. doi: 10.1128/jvi.73.12.9879-9890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]