Abstract
Comprehensive functional data on plant R2R3-MYB transcription factors is still scarce compared to the manifold of their occurrence. Here, we identified the Arabidopsis (Arabidopsis thaliana) R2R3-MYB transcription factor MYB12 as a flavonol-specific activator of flavonoid biosynthesis. Transient expression in Arabidopsis protoplasts revealed a high degree of functional similarity between MYB12 and the structurally closely related factor P from maize (Zea mays). Both displayed similar target gene specificity, and both activated target gene promoters only in the presence of a functional MYB recognition element. The genes encoding the flavonoid biosynthesis enzymes chalcone synthase, chalcone flavanone isomerase, flavanone 3-hydroxylase, and flavonol synthase were identified as target genes. Hence, our observations further add to the general notion of a close relationship between structure and function of R2R3-MYB factors. High-performance liquid chromatography analyses of myb12 mutant plants and MYB12 overexpression plants demonstrate a tight linkage between the expression level of functional MYB12 and the flavonol content of young seedlings. Quantitative real time reverse transcription-PCR using these mutant plants showed MYB12 to be a transcriptional regulator of CHALCONE SYNTHASE and FLAVONOL SYNTHASE in planta, the gene products of which are indispensable for the biosynthesis of flavonols.
Members of the MYB transcription factor superfamily are characterized by the presence of an amino acid motif structurally and functionally related to the DNA-binding domain of the product of the retroviral oncogene v-MYB and its animal cellular counterpart c-MYB. MYB proteins have been identified in a large number of eukaryotic organisms ranging from fungi (Stober-Grässer et al., 1992; Ohi et al., 1994; Tice-Baldwin et al., 1989) to plants (for review, see Martin and Paz-Ares, 1997; Jin and Martin, 1999) and to vertebrates (Gonda et al., 1985; Slamon et al., 1986; Nomura et al., 1988). While the MYB domain of c-MYB consists of three imperfect repeats (referred to as R1, R2, and R3), proteins with other numbers of MYB repeats have also been identified (Kranz et al., 2000; Riechmann et al., 2000; Stracke et al., 2001; Jiang et al., 2004). In contrast to the situation in animals, R2R3-MYB genes in plants comprise a large gene family. In Arabidopsis (Arabidopsis thaliana), 126 MYB genes of the R2R3 type have been described (Stracke et al., 2001; GenBank accession no. AF469468 for At3g60460). For Oryza sativa (Jia et al., 2004) and maize (Zea mays; Rabinowicz et al., 1999), a minimum of 80 R2R3-MYB genes has been reported.
Up to now, no or only few functional data are available for the overwhelming majority of plant MYB genes. The functional data available indicate that MYB transcription factors are involved in a wide array of cellular processes. These include development (e.g. AtMYB0/GL1 [Oppenheimer et al., 1991; Esch et al., 1994; Shikazono et al., 1998], AtMYB66/WER [Lee and Schiefelbein, 1999], and AtMYB103 [Higginson et al., 2003]), signal transduction (e.g. AtMYB2 [Urao et al., 1993] and AtMYB34/ATR1 [Bender and Fink, 1998]), plant disease resistance (e.g. AtMYB30 [Daniel et al., 1999]), cell division (e.g. AtCDC5, [Hirayama and Shinozaki, 1996]), and secondary metabolism (e.g. AtMYB75/PAP1, AtMYB90/PAP2 [Borevitz et al., 2000], AtMYB4 [Jin et al., 2000], AtMYB123/TT2 [Nesi et al., 2001; Baudry et al., 2004] and AtMYB61 [Penfield et al., 2001]).
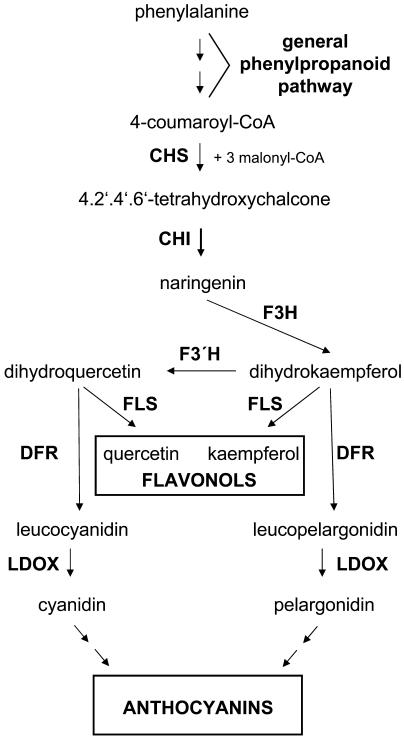
The well-established flavonoid biosynthetic pathway (Fig. 1) serves as a useful model for studying metabolic regulation. More than 8,000 different flavonoids have been identified in vascular plants (Pietta, 2000). This multitude of structures arises from a wide array of diverse modifications of the basic flavonoid scaffold, including methylations, acetylations, and glycosylations. Corresponding to the large variety of existing flavonoid compounds, their functions within the plant kingdom are diversified too, e.g. as pigments in flowers and fruit, as UV screens, as phytoalexins, or as regulators of auxin transport (for review, see Harborne and Williams, 2000). The structural genes of the enzymes involved in flavonoid formation are predominantly regulated at the level of transcription (for review, see Weisshaar and Jenkins, 1998). The transcriptional regulation of the first enzyme in the flavonoid-specific part of phenylpropanoid biosynthesis, chalcone synthase (CHS), has been investigated in detail in Arabidopsis. Within an operationally defined minimal AtCHS promoter, three different cis-acting elements have been identified (Hartmann et al., 1998, 2005). An ACGT containing element (ACE) and a MYB recognition element (MRE) comprise the light regulatory unit (LRUAtCHS), which confers responsiveness to UV-containing white light to the CHS promoter. The ACE and the MRE have been shown to be bound by basic region Leu zipper and MYB transcription factors, respectively (Armstrong et al., 1992; Feldbrügge et al., 1997; Hartmann et al., 1998). A third cis-acting element has recently been identified between the ACE and the MRE in the CHS promoter. This element, designated R response element (RRE), is required for the action of basic region helix-loop-helix (BHLH)-type transcription factors on the CHS promoter and might be involved in the regulation of the tissue-specific expression of CHS (Hartmann et al., 2005).
Figure 1.
Simplified, schematic representation of the biosynthesis of flavonols and anthocyanidins. Abbreviations of the enzyme designations are CHS, chalcone synthase; CHI, chalcone flavanone isomerase; F3H, flavanone 3-hydroxylase; FLS, flavonol synthase; F3′H, flavonoid 3′-hydroxylase; DFR, dihydroflavonol 4-reductase. Note that for the biosynthesis of flavonols the action of FLS is required while for the formation of anthocyanins DFR is necessary.
Many R2R3-MYB proteins share an extended degree of sequence similarity, especially within the highly conserved MYB domain. However, it remains unclear if in general this apparent structural redundancy also accounts for functional similarity. For example, the structurally closely related proteins ZmC1/Pl (Cone et al., 1993) from maize, PhMYBAN2 from Petunia hybrida (Quattrocchio et al., 1993), and AmMYBROSEA from Antirrhinum majus (Schwinn et al., 2001) belong to a subfamily of R2R3-MYB proteins controlling anthocyanin biosynthesis. Although the loss-of-function phenotype (reduction or loss of pigmentation) is the same in all these cases, the three factors display slightly different, though overlapping target gene specificities (Martin and Paz-Ares, 1997), indicating no complete functional homology between these closely related MYB proteins. An example for the existence of true functional homology between closely related MYB proteins are the two Arabidopsis factors MYB66/WER and MYB0/GL1, which are involved in the regulation of trichome and root hair development, respectively. By reciprocal complementation experiments, it was shown that MYB66/WER and MYB0/GL1, although expressed in different tissues, encode functionally fully equivalent proteins. Hence, their unique roles in plant development are entirely due to differences in their expression patterns (Lee and Schiefelbein, 2001).
Because of the strong amino acid sequence similarity of the Arabidopsis R2R3-MYB factor MYB12 to the maize MYB protein ZmP (84% identity within the MYB domain, 67% overall similarity) that controls phlobaphene synthesis in floral organs (Styles and Ceska, 1977; Grotewold et al., 1994; Chopra et al., 1996), we selected MYB12 to investigate its role as a part of the regulatory network controlling phenylpropanoid metabolism in Arabidopsis. We studied MYB12 function by transient coexpression using protoplasts of cultured Arabidopsis cells. Knockout mutants and ectopic overexpression plants were generated and investigated with respect to a flavonoid phenotype. MYB12 was found to be a flavonol-specific activator of flavonoid biosynthesis with the two flavonoid biosynthesis genes CHS and FLS (flavonol synthase) as its primary targets. Transcriptional activation by MYB12 is coactivator-independent but requires the presence of functional MREs within target promoters.
RESULTS
Cotransfection Analyses Reveal MYB12 and ZmP As Flavonol-Specific Activators of Flavonoid Biosynthesis in the At7 Cell Culture System
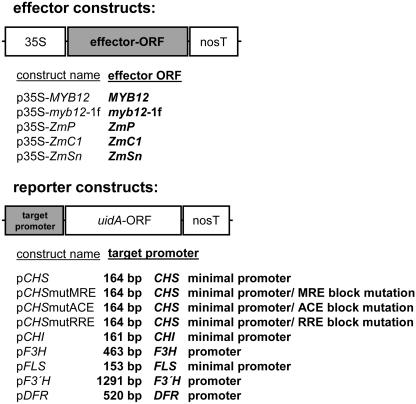
To investigate if the structural similarity of MYB12 and ZmP equivalents functional similarity of the two factors, a transient expression system using protoplasts of cultured At7 cells was utilized. In this system, the cotransfection of effector and reporter plasmid constructs (Fig. 2) allows the quantification of promoter activity as well as the determination of the transactivation potential of a transcription factor by measuring uidA (β-glucuronidase [GUS]) reporter activity.
Figure 2.
Schematic representations of the effector and reporter constructs used for transient expression analyses. Effector constructs contain the cauliflower mosaic virus 35S promoter fused to the respective effector ORFs followed by the nopaline synthase terminator (nosT). Reporter constructs contain the respective target promoters fused to the uidA-ORF followed by nosT. The length of the individual promoter fragments used is indicated. The three mutated variants of the CHS minimal promoter are named according to the mutated cis-element, respectively.
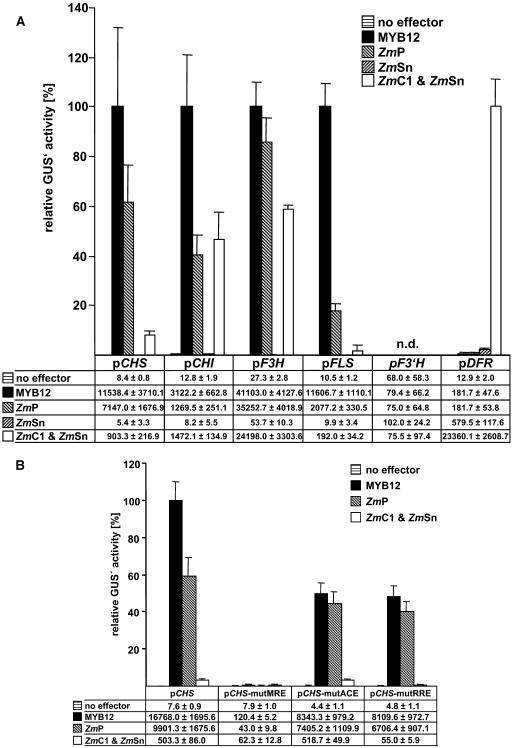
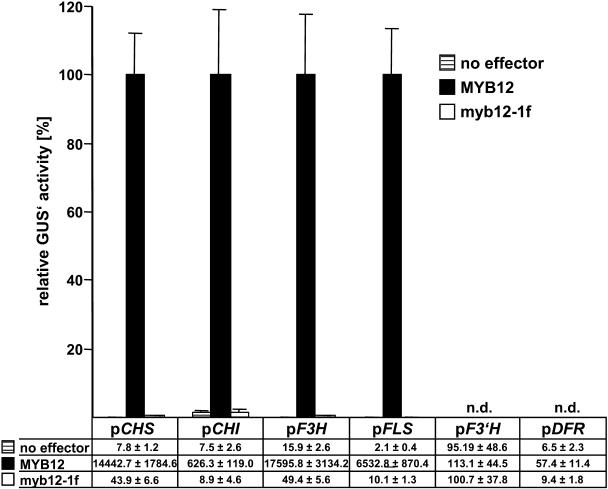
Figure 3A summarizes transfection experiments carried out to compare the transactivation capacities of MYB12 and ZmP within the flavonoid biosynthetic pathway (see Fig. 1). The promoters of the flavonoid biosynthesis genes CHS, CHI (chalcone flavanone isomerase), F3H (flavanone 3-hydroxylase), FLS, F3′H (flavonoid 3′-hydroxylase), and DFR were tested as potential targets for MYB12 and ZmP. MYB12 strongly activated the promoters of CHS, F3H, FLS, and, to some lesser extent, the CHI promoter. A similar activation pattern is displayed by ZmP, although the overall relative induction of promoter activity is generally lower than for MYB12; notably, FLS induction reached only 18% of that observed for MYB12. Neither MYB12 nor ZmP were able to activate the F3′H or DFR promoters to a significant extent. A combination of two factors from maize, the R2R3-MYB factor ZmC1 and the BHLH factor ZmSn, served as positive control for the functionality of the DFR promoter construct that was strongly activated by the combination of ZmC1/ZmSn. The factor ZmSn, when used in a control experiment as a single effector, showed marginal activation potential for the DFR promoter but no activation of the other promoters (Fig. 3A). MYB12 and ZmP did, in contrast to ZmC1, activate the target promoters independently of the presence of the BHLH factor ZmSn. In summary, the transactivation properties of MYB12 and ZmP in the transient expression system were almost identical with the exception of quantitative differences in the responsiveness of the FLS promoter to these two R2R3-MYB factors.
Figure 3.
Cotransfection analysis of target gene specificities and cis-element requirements in At7 protoplasts. A, Cotransfection assays to determine the activation of the promoters of flavonoid biosynthesis genes by MYB12, ZmP, ZmSn, and the combination of ZmC1/ZmSn. The mean value of luciferase activities was 6,800 RLU μg−1 s−1. Samples were incubated for 20 h in the dark at 26°C prior to harvesting. B, Identification of cis-elements required for the activation of the CHS minimal promoter by MYB12, ZmP, and ZmC1+ZmSn. The mean value of luciferase activities was 7,600 RLU μg−1 s−1. Samples were incubated for 8 h in the dark at 26°C prior to harvesting. A and B, The reporter/effector combinations used are indicated. Columns represent the relative GUS′ activity in cotransfection experiments using discrete reporter/effector combinations. The highest occurring GUS′ activity measured for each individual reporter construct was set to 100%. Each column represents the mean of six independent measurements. Error bars illustrate the sd of mean values. The data tables below the graphs give the respective absolute GUS′ activity values measured. n.d., Not determined.
MYB12 Requires a Functional MYB Recognition Element for Transactivation
To identify functional cis-acting elements required for MYB12 action, we assayed CHS promoter variants containing block mutations in the MRE, ACE, or the RRE (Fig. 3B). A functional MRE was shown to be required for activation by MYB12. Again, ZmP behaves similarly to MYB12, while the RRE was not necessary. By using ZmC1 and ZmSn in a control experiment, the functionality of the RRE was confirmed. Mutation of the ACE of the LRUAtCHS did not interfere with action of the R2R3-MYB factors tested.
Isolation of a myb12-ko Line and Generation of a MYB12-OX Line
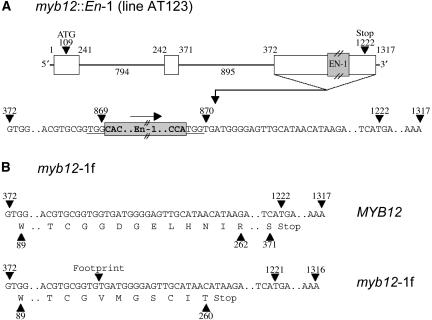
The isolation of a myb12∷En-1 line (designated AT123, Columbia [Col-0] background) from the En/Spm mutagenized AMAZE2 population allowed the in planta analysis of MYB12 function and the validation of the activity of MYB12 in regulating flavonoid biosynthesis. Figure 4A depicts the genomic structure of the wild-type MYB12 gene and of the myb12∷En-1 allele. The presence of approximately 12 En-1 elements in this line (data not shown) rendered it genetically unstable, impeding functional analyses. Repeated backcrossing to wild-type plants (accession Col-0) led to the isolation of an En-1-free, homozygous myb12-ko line carrying the myb12-1f footprint allele that is derived from myb12∷En-1 by excision of the En-1 transposon. The myb12-1f allele has a single base pair deleted at the original insertion site (Fig. 4B), which causes a frameshift that results in a premature termination of translation. The myb12-1f-encoded protein lacks 111 C-terminal amino acids of the wild-type MYB12 protein. The myb12-ko line was complemented with a 4.5-kb genomic fragment containing the MYB12 coding region under the control of the native MYB12 promoter. The resulting transgenic line was designated MYB12-COMP. In addition to the loss-of-function myb12-ko line, a homozygous 35S-MYB12 ectopic overexpression line (MYB12-OX) was constructed. The plants of both the myb12-ko and the MYB12-OX line showed no obvious phenotypic changes when compared to wild type under greenhouse conditions.
Figure 4.
Genomic structure of myb12 alleles. A, Schematic overview and partial structure of the third exon of the originally isolated myb12∷En-1 line (AT123). White boxes symbolize exons and the connecting lines introns. Numerical values above exons indicate nucleotide positions relative to the transcription start site. Intron lengths are indicated. The En-1 transposon insertion took place between nucleotides at positions 869 and 870. The nucleotides of the 3-bp target sequence duplication of the En-1 transposon are highlighted by underscore. The arrow upside the transposon marks its orientation. B, Comparison of nucleotide and protein sequence of wild-type (MYB12) and myb12-1f alleles. Only the third exon is shown. In the genetically stable myb12-1f line the deletion of the guanidine nucleotide at position 869 leads to the premature termination of translation. The myb12-1f protein comprises only 260 amino acids compared to 370 in the wild-type case. In addition the sequence of the last seven amino acids is altered at six positions.
The myb12-1f Derived Protein Is Not Functional in Cotransfection Assays
To address the question if the myb12-1f allele is still functional in activating the MYB12 target promoters identified above, we used the myb12-1f allele to generate an effector plasmid for cotransfection analyses. Figure 5 summarizes the results of the corresponding cotransfection experiments, illustrating that neither of the tested promoters showed any response to the myb12-1f protein. Thus, the truncated protein seems to be devoid of any activation potential indicating that the myb12-1f allele is not functional.
Figure 5.
The myb12-1f allele is not functional. The reporter/effector combinations used are indicated. Columns represent the relative GUS′ activity in cotransfection experiments using discrete reporter/effector combinations. The highest occurring GUS′ activity measured for each reporter construct was set to 100%. Each column represents the mean of six independent measurements. Error bars illustrate the sd of mean values. The data table below the graph gives the respective absolute GUS′ activity values measured. The mean value of luciferase activities was 4,500 RLU μg−1 s−1. Samples were incubated for 8 h in the dark at 26°C prior to harvesting.
MYB12-OX and myb12-ko Plants Display a Flavonol Accumulation Phenotype
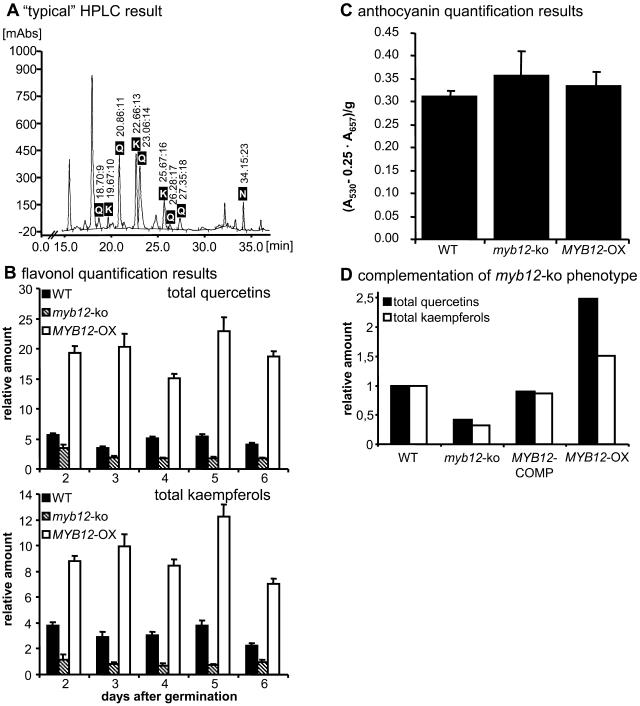
Plants of the myb12-ko and MYB12-OX lines were analyzed for their flavonol contents by HPLC. Pilot experiments showed that young myb12-ko seedlings contained reduced amounts of flavonoids, while seedlings as well as leaves of MYB12-OX plants displayed an increased flavonoid content. For the following analyses, we concentrated on a series of 2- to 6-d-old developing seedlings grown under continuous white light that were sampled at 1-d intervals. In wild-type seedlings, five different quercetin and three different kaempferol derivatives were detected that originate from different glycosylation patterns of the aglycone (Fig. 6A). Under the growth conditions used, all plants contained higher amounts of total quercetin than total kaempferol. For Col-0 wild-type seedlings, the average ratio between quercetins and kaempferols was about 1.5. At all time points both the quercetin and the kaempferol content of myb12-ko seedlings was clearly reduced compared to the wild-type reference. For MYB12-OX seedlings, the opposite effect was observed; quercetin and kaempferol content were significantly increased (Fig. 6B). To check whether the biochemical alterations also extend to anthocyanidins, the anthocyanidin content of 6-d-old seedlings of wild-type, myb12-ko, and MYB12-OX seedlings was determined. No significant differences in anthocyanin content were detected (Fig. 6C), indicating that MYB12 does not control anthocyanin accumulation. Taken together, a clear and specific correlation between MYB12 activity and the flavonol content in developing seedlings was observed.
Figure 6.
Biochemical myb12-ko phenotype. A, Representation of a selected HPLC result. The example shows the chromatogram obtained from a methanolic extract of 2-d-old wild-type seedlings. Peaks identified as corresponding to quercetin or kaempferol derivatives and the internal standard naringenin are labeled. Retention times are indicated above the peaks. K, Kaempferol; N, naringenin; Q, quercetin. B, Relative quantification of flavonols (total quercetins and total kaempferols) in methanolic extracts of developing Arabidopsis seedlings by HPLC analysis. The age and the genotype of the seedlings (WT, wild type) analyzed are indicated. Naringenin was used as an internal standard (relative amount arbitrarily set as one). Error bars indicate the sd of the average of relative quercetin/kaempferol amounts determined as triplicates in two independent biological replicates. C, Photometric determination of anthocyanin content in methanolic extracts of 6-d-old Arabidopsis seedlings. A530, Absorption at 530 nm; A657, absorption at 657 nm. Error bars represent the sd of the average from a total of six measurements using two independent biological replicates. D, Complementation of the myb12-ko phenotype by transformation of myb12-ko plants with a 4.5-kb MYB12 genomic fragment. Relative flavonol contents in methanolic extracts of 6-d-old wild-type, myb12-ko, MYB12-COMP, and MYB12-OX plants were determined by HPLC.
MYB12-COMP Plants Show Normal Flavonol Accumulation
Successful complementation of the myb12-ko line was demonstrated by HPLC analysis of 6-d-old MYB12-COMP seedlings. As shown in Figure 6D, relative quercetin and kaempferol amounts in MYB12-COMP and wild-type plants were very similar. Both quercetin and kaempferol levels in the MYB12-COMP line reached about 90% of wild-type levels.
The Expression Levels of Biosynthetic Genes in the Flavonoid Biosynthetic Pathway Is Affected by MYB12
To analyze the effect of MYB12 on the expression of flavonol biosynthesis genes in planta, quantitative real time reverse transcription (RT)-PCR analyses were carried out using double labeled fluorogenic hybridization probes (TaqMan probes). The PCR efficiencies of all TaqMan systems, i.e. primer and probe combinations, proved to be identical within the boundaries of the error of measurement; efficiency values reached 100% (Fig. 7A). Figure 7B depicts the results of the quantitative RT-PCR analyses. The MYB12 expression level in wild-type seedlings showed only little variation over the course of the kinetic, reaching a maximum at day four or five. The values for myb12-ko seedlings were virtually identical, which is explained by the fact that the chosen TaqMan probe for MYB12 was not able to differentiate between transcripts from the MYB12 wild type and the myb12-1f allele. MYB12 was strongly (over) expressed in MYB12-OX seedlings with relative expression levels ranging from 50- to 80-fold above that of 2-d-old wild-type seedlings. The biosynthesis genes of the flavonoid pathway can be grouped into four categories in terms of their responsiveness to MYB12; the expression of the members of the first category, CHS and FLS, showed a clear correlation to the MYB12 expression level. In myb12-ko seedlings, the expression of both genes was reduced, while overexpression of MYB12 resulted in increased expression of both genes. CHI and F3H constitute the second category. No effect of the lack of MYB12 activity (myb12-1f plants) was observed for CHI and F3H. However, the expression level of both genes was in general increased in response to elevated MYB12 expression. The effect was more pronounced for CHI (5- to 17-fold increase) than for F3H (2- to 5-fold increase). The third category solely consists of the F3′H gene whose expression was more or less unaffected by changes in MYB12 expression. The differences in F3′H expression were always less than 2-fold and not consistent in the myb12-ko and MYB12-OX plants, although a slight trend of increased expression is observed in some of the MYB12-OX samples. The single member of the fourth category is the DFR gene whose expression under the experimental conditions chosen was extremely low in all samples analyzed. In general, the expression values of DFR relative to the CHS calibrator were below 0.05 (data not shown). Taken together, the data from quantitative real time RT-PCR analyses showed that only the two genes of the first category, CHS and FLS, were fully MYB12 responsive. No myb12-1f effect was detected on the other flavonoid genes analyzed (CHI, DFR, F3H, and F3′H). However, CHI and F3H displayed elevated expression levels in response to increased MYB12 expression.
Figure 7.
CHS and FLS are primary target genes of MYB12. A, Determination of PCR efficiencies in quantitative real time PCR. The example shows the determination for MYB12 as PCR target. The plot of the natural logarithm of template concentration against the threshold cycle number results in a linear standard curve. The efficiency of the PCR system used can be calculated from the slope of the graph. The equation and the corresponding correlation coefficient of the linear regression line is shown. The calculated efficiency value is also indicated. For details of the method see “Materials and Methods.” E, PCR efficiency; ln, natural logarithm; R2, correlation coefficient of linear regression line. B, Results of quantitative real time RT-PCR analyses of RNA from developing Arabidopsis seedlings. Columns represent the relative expression levels of the distinct target genes tested. The mRNA amount in 2-d-old wild-type (WT) seedlings served as a calibrator for the calculation of relative expression levels in all cases (relative expression arbitrarily set to one). Relative expression was determined in triplicate measurements in two independent biological replicates. The age and the genotype of the seedlings analyzed are indicated.
DISCUSSION
MYB12 and ZmP Share Fundamental Functional Characteristics
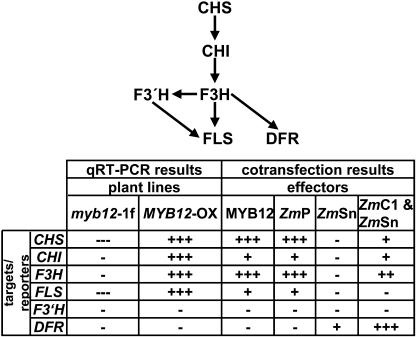
Cotransfection analyses revealed substantial functional similarities between the plant R2R3-MYB transcription factors AtMYB12 and ZmP. MYB12 acts via a MRE (Feldbrügge et al., 1997) and independently of a BHLH coactivator, similar to ZmP that was shown to bind the MREs in several flavonoid biosynthesis genes (Hartmann et al., 2005). Both factors show identical activation patterns in our test system by activating the CHS, CHI, F3H, and FLS promoters; all these genes are required for flavonol biosynthesis. AtF3′H and AtDFR are neither activated by MYB12 nor by ZmP (summarized in Fig. 8). In maize, the C2 (=CHS), CHI, and A1 (=DFR) genes that are required for the formation of 3-deoxy-flavonoids and phlobaphenes are regulated by ZmP (Grotewold et al., 1994). Although not formally proven, ZmP is probably no activator of ZmF3H, otherwise its activity would lead to the formation of anthocyanins rather than phlobaphenes in maize kernels. This difference, i.e. the responsiveness of AtF3H but not AtDFR to MYB12 in contrast to the responsiveness of ZmDFR but not ZmF3H to ZmP, probably reflects the divergent evolution of the promoter sequences of these two genes, constituting the basis for the observed metabolic difference between maize and Arabidopsis. A similar differential regulation has also been described for the CHS promoter between maize and P. hybrida (Quattrocchio et al., 1998).
Figure 8.
Overview of MYB12 effects on flavonoid gene expression. The top of the figure shows a schematic representation of the enzymatic steps of flavonoid biosynthesis. The table below summarizes the results of the quantitative real time RT-PCR of mutant plants and the cotransfection analyses performed. “+” indicates weak, “++” medium, and “+++” strong gene expression or activation of reporter gene transcription, respectively; “−” indicates no effect on target gene expression or on activation of reporter gene transcription, “---” indicates strong reduction in target gene expression.
Recent studies of the protein-protein interaction specificities of the R2R3-MYB protein family revealed specific conserved amino acid signatures to be the structural basis for interaction of R2R3-MYB and R/B-like BHLH proteins (Grotewold et al., 2000; Hernandez et al., 2004; Zimmermann et al., 2004). None of the identified amino acid motifs was found to be present in the MYB12 or ZmP amino acid sequences. Moreover, yeast-two-hybrid experiments showed no interaction between MYB12 or ZmP and R/B-like BHLH transcription factors (Zimmermann et al., 2004). Also, the results of transfection assays using mutated versions of the CHS minimal promoter (Fig. 3B) support the view of MYB12 as a cofactor-independent activator, again similar to ZmP. Taken together, our results suggest that AtMYB12 is the functional homolog of P from maize. However, we cannot exclude that ZmP might have additional regulatory functions beyond controlling phlobaphene accumulation.
In contrast to MYB12 and ZmP, ZmC1 showed a different behavior. As described before, ZmC1 regulates together with a R/B-like BHLH partner (e.g. ZmSn) anthocyanin biosynthesis in maize kernels (Mol et al., 1996, 1998; Procissi et al., 1997). In addition to the previously reported activation of the AtCHS promoter by the combination of ZmC1/ZmSn (Hartmann et al., 2005), we show that also the promoters of CHI, F3H, and DFR are activated by ZmC1 plus ZmSn. ZmSn alone is not able to activate reporter expression in cotransfection experiments. The exception is the weak activation of the DFR promoter that might be caused by either Sn alone or by an endogenous ZmSn-interacting factor (potentially a R2R3-MYB) present in the protoplasts.
The FLS gene, which encodes the enzyme flavonol synthase that catalyzes the committing step toward flavonols, is not responsive to ZmC1 plus ZmSn but is, as mentioned above, fully responsive to MYB12. This result again supports the notion of differential control of flavonol and anthocyanin biosynthesis by pathway-specific transcription factors also in Arabidopsis. The principle mode of regulation of anthocyanin and tannin accumulation via the combinatorial action of R2R3-MYB and a BHLH-type transcription factor is a conserved feature in both species (Baudry et al., 2004 and refs. therein; Zimmermann et al., 2004), while phlobaphene synthesis in maize is controlled by ZmP and flavonol synthesis in Arabidopsis by the R2R3-MYB factor MYB12. Taken together, these results strengthen the view of a close correlation between structural and functional similarity of R2R3-MYB transcription factors across plant species, with modifications in the target gene set mainly determined by the sequence of the target gene promoter.
The myb12-1f allele is not functional in transient expression assays, which may indicate that an activation domain is located in the C-terminal part of MYB12. Sequence analysis of the C-terminal domain of MYB12 revealed no substantial sequence similarities to previously described activation domains. However, the modular structure of MYB12 is similar to that of other R2R3-MYB factors, with the MYB DNA binding domain positioned at the N terminus and the C-terminal part of the protein harboring the transactivation domain. This includes AtMYB75 for which the transactivation domain was mapped to the 58 C-terminal residues of the protein (Zimmermann et al., 2004).
myb12-ko Seedlings Show Reduced Flavonol Content
Seedlings of both the myb12-ko and the MYB12-OX line show an abnormal flavonol content. Flavonol accumulation is strongly reduced in myb12-ko seedlings (on average about 2-fold for quercetins and 3.5-fold for kaempferols; Fig. 6, A and B), which express a nonfunctional MYB12 protein variant (myb12-1f), and strongly increased in seedlings and plants when MYB12 is overexpressed (approximately 4-fold for quercetins and 3-fold for kaempferols; Fig. 6, A and B). Complementation of the myb12-ko line with a 4.5-kb genomic fragment containing the MYB12 wild-type allele restores wild-type flavonol levels, confirming that the myb12-1f mutation is responsible for the observed phenotype (Fig. 6D). However, even without the presence of functional MYB12 protein, the synthesis of flavonols was not eliminated entirely. This fits the results of the quantitative RT-PCR analyses in which some residual expression of the flavonoid biosynthesis genes was always detected in the seedlings analyzed. Clearly, the flavonoid biosynthesis genes are also the targets of other transcription factors, including other R2R3-MYB proteins. MYB12 belongs to subgroup 7 of the R2R3-MYB family, and other members of this subgroup are MYB11 and MYB111 (Stracke et al., 2001). Results from microarray experiments indeed show a simultaneous expression of the three factors in 10-d-old seedlings (AtGenExpress, http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenex.htm). The details of this anticipated redundancy and the exact contributions of MYB11 and MYB111 to the control of flavonol accumulation in seedlings remain to be elucidated. Preliminary experiments indicate a complex interaction of these related genes in various parts of Arabidopsis plants.
While most of the biosynthetic enzymes of the flavonoid pathway are encoded by single copy genes, there are five additional genes closely related to FLS present in Arabidopsis, two of which are seemingly not expressed (Winkel-Shirley, 2001). Up to now, it has not been proven that these related genes encode enzymes with FLS activity. Also, the interpretation of the fact that homozygous plants carrying the fls-1∷En allele still contain flavonols (Wisman et al., 1998) is complicated by the fairly high reversion rate of the transposon allele that causes mosaic plants that are able to express some functional FLS in the revertant sectors. Further studies are required to identify the reason for differential effects of MYB12 on the accumulation of kaempferol versus quercetin derivatives. In addition to differentially regulated FLS-like genes that may encode FLS activity, the action of the hydroxylase F3′H on kaempferols and the stability of the glycosylated end products also have to be considered.
The myb12-ko plants also display reduced flavonol content in other parts of the plant, and AtGenExpress as well as our own data show that MYB12 expression is not restricted to seedlings. These data suggest a role for MYB12 in the regulation of flavonol biosynthesis also in organs of adult plants, but with a less dominant activity compared to that observed in seedlings.
MYB12 Exerts Its Regulatory Function upon Flavonol Biosynthesis by Transcriptional Activation of CHS and FLS
The in planta effects of MYB12 on the level of gene expression were investigated using quantitative RT-PCR. The primary MYB12 targets were CHS and FLS. This is demonstrated by the clear reduction of the mRNA levels of both genes in myb12-ko seedlings (summarized in Fig. 8). CHS and FLS represent important branching points within flavonoid biosynthesis. CHS catalyzes the committing step toward flavonoids, the formation of 4,2′,4′,6′-tetrahydroxychalcone, and FLS the committing step toward flavonols by using dihydroflavonols as substrate (Fig. 1). Dihydroflavonols are also the substrate for DFR, which catalyzes the formation of leucoanthocyanidins. The leucoanthocyanidins are the first intermediates of the (pro-) anthocyanidin-specific branch of flavonoid biosynthesis. MYB12 directs the flux of flavonoid intermediates specifically toward the production of flavonols by activating FLS but not DFR.
Upon overexpression of MYB12 in plants, additional genes, namely CHI and F3H, fall under MYB12 control. This target gene selectivity could potentially be a result of the strong 35S promoter that may cause an accumulation of high levels of MYB12, which may activate promoters that are not controlled at lower factor levels. Such effects have been reported previously for c-Myb (Andersson et al., 1999) and AtMYB4 (Jin et al., 2000). Alternatively, CHI and F3H that are activated in MYB12-OX but not significantly reduced in the myb12-ko plants might be also switched on in the seedlings by other factors, so that the removal of MYB12 activity does not reduce their expression levels. The tight coregulation of CHS, CHI, F3H, and FLS that has been observed before (Hartmann et al., 2005), and the attractive model of MYB12 as a master switch for the accumulation of flavonols, argues in favor of redundant activation of CHI and F3H in seedlings. That MYB12 has the potential to direct the accumulation of flavonols is shown by the biochemical phenotype of the MYB12-OX plants.
Obviously, the next question is “What regulates the regulator?” So far, the RT-PCR experiments that show very similar expression levels of MYB12 transcripts in wild-type and myb12-ko seedlings (Fig. 7B) exclude an auto regulatory action of MYB12 on its own promoter. Also, our results do not support the view of MYB12 as a developmental regulator of flavonoid biosynthesis. MYB12 exerts mainly quantitative effects on target gene expression; no clear-cut effects on the timing of gene expression have been observed. The observation that MYB12 directly acts on the promoters of the flavonoid biosynthesis genes places MYB12 at the downstream end of the signaling chain that causes flavonol-specific gene activation. Now, we need to address the question whether MYB12 is involved in any of the many stress signaling pathways (Li et al., 1993; Christie et al., 1994; Dixon and Paiva, 1995; Ryan et al., 2001, 2002) that result in flavonol accumulation. The myb12-ko plants described and characterized here will be instrumental in these experiments.
CONCLUSION
Taken together, we have identified MYB12 as a flavonol-specific activator of flavonoid biosynthesis in developing seedlings. The interaction of MYB12 with a specific cis-acting promoter element, the MRE, provides the functional basis for transcriptional activation of a set of MYB12-responsive genes. We also show that MYB12 function does not depend on BHLH coactivators. An extended functional similarity between MYB12 and its maize homolog ZmP was observed, again indicating that structurally similar MYB transcription factors exhibit similar functions across different species. The biochemical data show a direct correlation between the expression level of functional MYB12 and the amount of flavonols in young seedlings. Changes in MYB12 activity do not effect anthocyanin formation, an observation that is in concordance with DFR not being responsive to MYB12. Finally, our results could be applied to the targeted manipulation of flavonoid metabolism toward the increased production of flavonols in plants. The expression of a single transgene, namely the MYB12 open reading frame (ORF) under a tissue- or even cell-specific promoter, should allow us to engineer plants with modified flavonol content in the addressed cells or tissue.
MATERIALS AND METHODS
Standard Molecular Biology Techniques
Standard molecular biology techniques were applied according to Sambrook et al. (1989). All DNA sequencing was performed by the MPI for Plant Breeding Research DNA core facility (ADIS) using BigDye terminator chemistry on Applied Biosystems 377 and 3700 sequencers (Applied Biosystems, Weiterstadt, Germany). Oligonucleotides were purchased from MWG (Ebersberg, Germany), metabion (Martinsried, Germany), or Invitrogen (Paisley, United Kingdom).
Cell Culture and Plant Material
The Arabidopsis (Arabidopsis thaliana) cell line At7 was described previously (Trezzini et al., 1993; Hartmann et al., 1998). Seedlings for quantitative real time RT-PCR, HPLC analysis, and photometric anthocyanin analysis were grown in petri dishes on Whatman-paper soaked with Murashige and Skoog medium lacking Suc under sterile conditions. After sowing, the plates were kept for 5 d in the dark at 4°C and then transferred to a phytochamber (22°C, continuous white light illumination at a fluency rate of 100 μmol m−2 s−1).
Isolation of the myb12-1f Mutant
The isolation of insertion mutants from the En/Spm mutagenized AMAZE2 population (Wisman et al., 1998) was described previously (Meissner et al., 1999). HPLC analysis of the progeny of the homozygous myb12∷En-1 line that did display wild-type-like PCR fragments led to the recovery of the homozygous myb12-1f line, which showed reduced levels of flavonols in the absence of an En-1 element at the MYB12 locus. This line was backcrossed five times to wild type (accession Col-0) to give rise to an En-free homozygous myb12-1f mutant. The myb12-1f allele was checked by sequencing. PCR analysis confirmed the absence of intact En-elements in the genome of myb12-1f plants.
Generation of MYB12-OX Transgenic Plants
The construct used for plant transformation was based on the binary vector pGPTV-TATA, a derivative of pGPTV (Becker et al., 1992) containing the TATA-GUS cassette of pBT10 (Sprenger-Haussels and Weisshaar, 2000). Plants were transformed by infiltration (Bechtold et al., 1993) using Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986). For overexpression, the 35S-MYB12 cassette of p35S-MYB12 (see below) was amplified with the primers HK122 (5′-TTCATTAAAGCTTCGGATCCATCGATG-3′; introduction of a HinDIII restriction site) and HK123 (5′-CATGATGAGCTCGTTGACAGAAGCCAAGCG-3′; introduction of a SacI restriction site). The 35S-MYB12 cassette was introduced into pGPTV-TATA by replacing the TATA-GUS sequences and transformed into Arabidopsis accession Col-0. Transformants were selected on media containing kanamycin. For further experiments, homozygous transformants were used.
Generation of a MYB12-COMP Line
A 4.5-kb genomic fragment containing the MYB12 gene was amplified using the primers FM189 (GTGTGTGGTTGGTAAGCTTTAAATTAGCTAGTG; introduction of a HinDIII restriction site) and FM 190 (TTCCGAAAGAGCTCCATATAATAGATGCTTC; introduction of a SacI restriction site). The fragment was subcloned into the pCR 2.1. TOPO vector (Invitrogen) and checked for the correct sequence. After restriction digestion with EcoRI and SacI, the genomic fragment was cloned into a derivative of pPAM (GenBank accession no. AY027531). myb12-ko plants were transformed by infiltration (Bechtold et al., 1993) using A. tumefaciens strain GV3101 (Koncz and Schell, 1986). The presence of the MYB12 wild-type sequence in the selected transgenic line was confirmed by PCR and subsequent sequencing. Extracts from T2 seedlings of the complementation line, designated MYB12-COMP, were used for HPLC analyses.
RNA Sample Preparation and cDNA Synthesis for Quantitative Real Time RT-PCR
Total RNA was isolated using the RNeasy Mini kit from Qiagen (Hilden, Germany). RNA was treated with DNaseI (Qiagen) according to the manufacturer's specifications and precipitated with LiCl. RNA concentrations were determined using the RiboGreen RNA quantitation kit (Molecular Probes, Leiden, The Netherlands) following the manufacturer's specifications. cDNA synthesis was carried out following a protocol from Invitrogen with some minor modifications. A typical reaction mix (20 μL) consisted of 7 μL total RNA (maximum 5 μg), 0.5 μL of 0.5 μg/μL oligo(dT)(12–18), 0.5 μL random hexamers (50 ng/μL), 4 μL SuperScript II 5× reaction buffer, 4 μL MgCl2 (25 mm), 1 μL dNTPs (10 mm each), 2 μL dithiothreitol (0.1 mm), and 1 μL SuperScript II reverse transcriptase (50 U/μL)]. The reaction mix was incubated at room temperature for 10 min and subsequently for 30 to 45 min at 42°C, followed by an incubation at 70°C for 10 min, after which the samples were cooled on ice or kept at −20°C for later use.
Quantitative Real Time RT-PCR
Quantitative real time RT-PCR using double labeled fluorogenic hybridization probes (TaqMan probes, see below) was performed utilizing an ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The reaction volume of PCR samples was 25 μL. In all cases, Platinum Quantitative PCR Supermix-UDG (Invitrogen) was used for amplification following the supplier's instructions. Quantification of 18S rRNA for normalization of cDNA concentrations was performed using the TaqMan Ribosomal RNA Control Reagent (Applied Biosystems) according to manufacturer's instructions. Reaction mixes included the following components: 12.5 μL Platinum Quantitative PCR Supermix-UDG, 1.25 μL TaqMan Ribosomal Control Reagent, 0.5 μL ROX (carboxy-X-rhodamine) passive reference dye (Invitrogen), 5.75 μL water, and 5 μL cDNA template. Cycling conditions were: 2 min 50°C, 2 min 95°C followed by 30 cycles of denaturation for 15 s at 95°C and annealing/extension for 1 min at 60°C. In total, the threshold cycle numbers for the 18S rRNA target [Ct(18S)] were determined six times in two independent PCR runs per cDNA sample, one performed before the quantitation of the target genes and one performed afterward to check for variations in sample quality. The average Ct(18S) value including the sd Ct(18S) was calculated. The Ct values for the different target genes were determined using the PCR profile described above, but 40 instead of 30 two-step amplification cycles were carried out. The PCR reaction mixes comprised Platinum Quantitative PCR Supermix-UDG (12.5 μL), forward primer (variable, final concentration 0.9 μm), reverse primer (variable, final concentration 0.9 μm), TaqMan probe (variable, final concentration 0.2 μm), ROX passive reference dye (0.5 μL), water, and cDNA template (5 μL) to give a final volume of 25 μL. Target gene Ct values were determined as triplicates in two biological replicates, respectively. The Ct mean value including the sd Ct(target) was calculated. Subsequently ΔCt values [ΔCt = Ct(18S) − Ct(target)] were calculated to specify the relative expression of the individual target genes compared to the 18S rRNA standard. The error of ΔCt (ΔΔCt) was calculated according to Gauss' error propagation law. The relative expression of the target genes to the 18S standard are given by the equation: R(target/18S) = 2ΔCt. The error ΔR was calculated by the same method as ΔΔCt before. R(target/18S) values for the individual targets were normalized to the respective R(target/18S) value of the calibrator cDNA (from 2-d-old wild-type seedlings). Again, the errors of the resulting relative expression values were calculated by application of Gauss' error propagation law. The procedure was performed twice, once for every biological replicate. Finally, the average relative expression of the target genes from the two independent biological replicates was calculated. The errors given are the sds of the mean values.
Determination of RT-PCR Efficiencies
For all primer and probe systems, standard curves were generated using serial dilutions of DNA known to contain the respective target gene sequences. Ct values were measured as described above and plotted against the natural logarithm of DNA concentration. Regression analysis provided a linear function from which the PCR efficiency could be calculated using the term E = e−1/m − 1, where E is the PCR efficiency, e Euler's number, and m the slope of the regression function.
Primer and Probes for Quantitative Real Time RT-PCR
All hybridization probes listed below were purchased from Eurogentec (Seraing, Belgium). Probes were labeled with the 5′- dye FAM (6-carboxyfluorescein) and the 3′-dye TAMRA (6-carboxytetramethylrhodamine; quencher). Only HPLC purified primers were used. Target gene: MYB12 (At2g47460), forward primer 5′-AACCAAGGGAATCTCGACTGTCT-3′, reverse primer 5′-CCCAATCGATAAACTCATCCGT-3′, probe 5′-ACGACCACCAAGTTAACGATGCGTCG-3′; target gene: CHS (At5g13930), forward primer 5′-CGCATCACCAACAGTGAACAC-3′, reverse primer 5′-TCCTCCGTCAGATGCATGTG-3′, probe 5′-CGACTTGTCGCACATGCGCTTGAA-3′; target gene: CHI (At3g55120), forward primer 5′-CCGGTTCATCGATCCTCTTC-3′, reverse primer 5′-ATCCCGGTTTCAGGGATACTATC-3′, probe 5′-CCTACCGGCTCTCTTACGGTTGCGTT-3′; target gene: F3H (At3g51240), forward primer 5′-CAGATCGTTGAGGCTTGTGAGA-3′, reverse primer 5′-GACGAGTCATATCCGCCACTAAGT-3′, probe 5′-TCCAAGTGGTCGATCACGGCGTC-3′; target gene: FLS (At5g08640), forward primer 5′-CCGTCGTCGATCTAAGCGAT-3′, reverse primer 5′-CGTCGGAATCCCGTGGT-3′, probe 5′-ACCGCGCGCCTCACGCTT-3′; target gene: F3′H (At5g07990), forward primer 5′-GCTCTCGCCGGAGTATTCAA-3′, reverse primer 5′-CCAGCGACGCCTTGTAAATC-3′, probe 5′-TCGGAGACTTCGTGCCGTCCACTTGA-3′; target gene: DFR (At5g42800), forward primer 5′-CTCCTATCACTCGGAACGAGGCGCATTAC-3′, reverse primer 5′-TGGTCGGTCCATTCATCACA-3′, probe 5′-AGCGTTGCATAAGTCGTCCAA-3′.
Preparation of Methanolic Extracts for HPLC Analysis
A total of 100 to 200 mg of seedlings were placed in a 1.5-mL reaction tube, and 300 μL of 80% methanol and about 12 zirconium beads (diameter 1 mm; Roth, Karlsruhe, Germany) were added. Samples were homogenized at maximum speed for at least 1 min using a Mini BeadBeater-8 (BioSpec Products, Bartlesville, OK). Homogenized samples were centrifuged for a minimum of 15 min at 14,000 rpm in a standard table centrifuge at 4°C. The supernatants were vacuum dried in a SpeedVac (Thermo Savant, Holbrook, NY). Afterward, the dried pellets were dissolved in 100 μL of 80% methanol containing 0.02% (w/v) naringenin (Roth)/100 mg starting material. Naringenin was used as an internal standard for the relative quantification of flavonoid compounds. Pilot experiments showed that no endogenous naringenin was detectable in the seedlings analyzed at any time point (data not shown), making it a suitable calibrator for relative quantification.
HPLC Analysis of Methanolic Extracts
Samples were analyzed using a 522 HPLC system equipped with a DAD detector 540 (Bio-Tek Kontron, Winooski, VT). Reversed phase chromatography was carried out using a Macherey & Nagel (Dueren, Germany) Nucleosil 120-5 C18 250/2 column. HPLC parameters were as follows: column temperature 30°C; solvent A = 0.1% trifluoroacetic acid in water, solvent B = 98% acetonitrile with 0.1% trifluoroacetic acid; solvent gradient, 0 min = 0% B, 3 min = 6% B, 12 min = 18% B, 25 min = 25% B, 35 min = 100% B, 40 min = 100% B. The flow rate was 0.35 mL/min. Peaks were classified as corresponding to kaempferol or quercetin derivatives by UV spectral analysis. The precise chemical nature of the detected substances, i.e. the glycosylation pattern of the flavonol aglyca, has not been elucidated. The areas of the flavonol peaks were normalized to the peak area of the internal standard naringenin resulting in relative flavonol amounts. Error bars indicate the sd of the average of relative flavonol amounts determined as triplicates in two independent biological replicates.
Photometric Determination of Anthocyanins
Extraction of anthocyanins of 6-d-old Arabidopsis seedlings was performed following the protocols of Rabino and Mancinelli (1986) and Kubasek et al. (1992) with minor modifications. One milliliter of acidic methanol (1% HCl, w/v) was added to 300 mg of fresh plant material. Samples were incubated for 18 h at room temperature under moderate shaking. Plant material was sedimented by centrifugation (14,000 rpm, room temperature, 1 min) and 400 μL of the supernatant was added to 600 μL of acidic methanol. Absorption of the extracts at 530- and 657-nm wavelength was determined photometrically (UVIKON 922, Bio-Tek Kontron). Quantification of anthocyanins was performed using the following equation: QAnthocyanins = (A530 − 0.25 × A657) × M−1, where QAnthocyanins is the amount of anthocyanins, A530 and A657 is the absorption at the indicated wavelengths and M is the weight of the plant material used for extraction [g]. All samples were measured as triplicates in two independent biological replicates. Error bars indicate the sd of the average of the anthocyanin content.
Isolation of Protoplasts and Cotransfection Experiments
Protoplast isolation, transfection experiments, and illumination conditions for transient expression were described previously (Hartmann et al., 1998, 2005) with the exception that for the determination of luciferase activity a Mini-Lum luminometer (Bioscan, Washington DC) was used, and that GUS activity was measured using a FluoroCount microtiter plate reader (Packard Instrument, Meriden, CT). Differently from the previously described procedure aliquots of the GUS reaction mixes were transferred to 200 μL of 0.2 m Na2CO3 placed in a 96-well microtiter plate. Also, for every measurement of GUS activity, a 4-methylumbelliferone standard curve was prepared to link the obtained relative fluorescence unit values to 4-methylumbelliferone amounts. Cotransfected protoplasts were kept 8 or 20 h in the dark prior to harvest.
Effector and Reporter Constructs for Transient Transfection Experiments
All effector and reporter constructs used, except p35S-MYB12, p35S-myb12-1f, pF3′H, and pDFR, have been described before (Hartmann et al., 1998, 2005). The MYB12-ORF and the myb12-1f-ORF were amplified using the primers FM1 (5′-ATAGGTACCTCCGGAACCATGGGAAGAGCGCCATGTTGC-3′; introduction of a NcoI restriction site) and FM2 (5′-TATGGATCCACCGGTTGACAGAAGCCAAGCGACCAAAGC-3′; introduction of a PinAI restriction site) and inserted into the p35S-ZmSn-effector construct replacing the ZmSn-ORF. A 1.3-kb promoter fragment of the F3′H gene for the pF3′H-uidA reporter construct was amplified using the primers FM179 (5′-CTCTTGCTAAGCTTGACTGTATAATAACCTC-3′; introduction of a HinDIII restriction site) and FM184 (5′-GAGAAATAGAGTTGCCATGGTGTTGG-3′; introduction of a NcoI restriction site). This promoter cassette was introduced into the multiple cloning site of the recipient vector pBT10-GUS (Sprenger-Haussels and Weisshaar, 2000). Similarly, a 1.1-kb promoter fragment of the DFR gene was amplified using the primers FM31 (5′-GGTTGAAGAAGAAGAAGGAAAGCTTTGAAG-3′; introduction of a HinDIII restriction site) and FM32 (5′-CTGACTAACCATGGTTGTGGTTATATG-3′; introduction of a NcoI restriction site). Because of an internal HinDIII restriction site within the PCR product, a 520-bp fragment of the promoter was cloned into the pBT10 vector.
Acknowledgments
We thank Ute Tartler for excellent technical assistance. We are grateful to Chiara Tonelli for providing the cDNAs encoding ZmC1 and ZmSn, and Erich Grotewold for providing the cDNA encoding ZmP. We also thank Ralf Stracke for many inspiring discussions during the writing of this paper and the anonymous reviewers of the first version of the manuscript for constructive criticisms. We are grateful to Klaus Hahlbrock for continuous support throughout the project.
This work was supported by the Max-Planck-Society, and in part by EC-BIOTECH (grant no. BIO4–CT95–0129 to B.W.) and by the Deutsche Forschungsgemeinschaft.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.058032.
References
- Andersson KB, Berge T, Matre V, Gabrielsen OS (1999) Sequence selectivity of c-Myb in vivo. Resolution of a DNA target specificity paradox. J Biol Chem 274: 21986–21994 [DOI] [PubMed] [Google Scholar]
- Armstrong GA, Weisshaar B, Hahlbrock K (1992) Homodimeric and heterodimeric leucine zipper proteins and nuclear factors from parsley recognize diverse promoter elements with ACGT cores. Plant Cell 4: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194–1199 [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR (1998) A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci USA 95: 5655–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra S, Athma P, Peterson T (1996) Alleles of the maize P gene with distinct tissue specificities encode Myb-homologous proteins with C-terminal replacements. Plant Cell 8: 1149–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Alfenito MR, Walbot V (1994) Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194: 541–549 [Google Scholar]
- Cone KC, Cocciolone SM, Burr FA, Burr B (1993) Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Lacomme C, Morel J-B, Roby D (1999) A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J 20: 57–66 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch JJ, Oppenheimer DG, Marks MD (1994) Characterization of a weak allele of the GL1 gene of Arabidopsis thaliana. Plant Mol Biol 24: 203–207 [DOI] [PubMed] [Google Scholar]
- Feldbrügge M, Sprenger M, Hahlbrock K, Weisshaar B (1997) PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. Plant J 11: 1079–1093 [DOI] [PubMed] [Google Scholar]
- Gonda TJ, Gough NM, Dunn AR, de Blaquiere J (1985) Nucleotide sequence of cDNA clones of the murine myb proto-oncogene. EMBO J 4: 2003–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL (2000) Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci USA 97: 13579–13584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55: 481–504 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B (2005) Differential combinatorial interactions of ACE-, MRE- and RRE-binding transcription factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57: 155–171 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Valentine WJ, Christie JM, Hays J, Jenkins GI, Weisshaar B (1998) Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol Biol 36: 741–754 [DOI] [PubMed] [Google Scholar]
- Hernandez JM, Heine GF, Irani NG, Feller A, Kim MG, Matulnik T, Chandler VL, Grotewold E (2004) Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J Biol Chem 279: 48205–48213 [DOI] [PubMed] [Google Scholar]
- Higginson T, Li SF, Parish RW (2003) AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J 35: 177–192 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K (1996) A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 13371–13376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Clegg MT, Jiang T (2004) Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol 134: 575–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Gu X, Peterson T (2004) Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol 5: R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB -gene family. Plant Mol Biol 41: 577–585 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kranz H, Scholz K, Weisshaar B (2000) c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J 21: 231–235 [DOI] [PubMed] [Google Scholar]
- Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM (1992) Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4: 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 24: 473–483 [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (2001) Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128: 1539–1546 [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee T-M, Raba R, Amundson RG, Last RL (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J (1997) MYB transcription factors in plants. Trends Genet 13: 67–73 [DOI] [PubMed] [Google Scholar]
- Meissner RC, Jin H, Cominelli E, Denekamp M, Fuertes A, Greco R, Kranz HD, Penfield S, Petroni K, Urzainqui A, et al (1999) Function search in a large transcription factor gene family in Arabidopsis: assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell 11: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217 [Google Scholar]
- Mol J, Jenkins GI, Schäfer E, Weiss D (1996) Signal perception, transduction, and gene expression involved in anthocyanin biosynthesis. Crit Rev Plant Sci 15: 525–557 [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N, Takahashi M, Matsui M, Ishii S, Date T, Sasamoto S, Ishizaki R (1988) Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res 16: 11075–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, McCollum D, Hirani B, Haese GJD, Zhang X, Burke JD, Turner K, Gould KL (1994) The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J 13: 471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietta PG (2000) Flavonoids as antioxidants. J Nat Prod 67: 1035–1042 [DOI] [PubMed] [Google Scholar]
- Procissi A, Dolfini S, Ronchi A, Tonelli C (1997) Light-dependent spatial and temporal expression of pigment regulatory genes in developing maize seeds. Plant Cell 9: 1547–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE (1993) Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5: 1497–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JNM, Koes R (1998) Analysis of bHLH and MYB domain proteins: species specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J 13: 475–488 [DOI] [PubMed] [Google Scholar]
- Rabino I, Mancinelli AL (1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz P, Braun E, Wolfe A, Bowen B, Grotewold E (1999) Maize R2R3 Myb genes: Sequence analysis reveals amplification in the higher plants. Genetics 153: 427–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Ryan KG, Swinney EE, Markham KR, Winefield C (2002) Flavonoid gene expression and UV photoprotection in transgenic and mutant petunia leaves. Phytochemistry 59: 23–32 [DOI] [PubMed] [Google Scholar]
- Ryan KG, Swinney EE, Winefield C, Markham KR (2001) Flavonoids and UV photoprotection in Arabidopsis mutants. Z Naturforsch C 56: 745–754 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schwinn K, Davies K, Alm V, Mackay V, Martin C (2001) Regulation of anthocyanin biosynthesis in antirrhinum. Acta Hortic 560: 201–206 [Google Scholar]
- Shikazono N, Tanaka A, Yokota Y, Watanabe H, Tano S (1998) Nucleotide sequence of the GLABROUS1 gene of Arabidopsis thaliana ecotype Columbia. DNA Seq 9: 177–181 [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Boone TC, Murdock DC, Keith DE, Press MF, Larson RA, Souza LM (1986) Studies of the human c-myb gene and its product in human acute leukemias. Science 233: 347–351 [DOI] [PubMed] [Google Scholar]
- Sprenger-Haussels M, Weisshaar B (2000) Transactivation properties of parsley proline rich bZIP transcription factors. Plant J 22: 1–8 [DOI] [PubMed] [Google Scholar]
- Stober-Grässer U, Brydolf B, Bin X, Grässer F, Firtel RA, Lipsick JS (1992) The Myb DNA-binding domain is highly conserved in Dictyostelium discoideum. Oncogene 7: 589–596 [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Styles ED, Ceska O (1977) The genetic control of flavonoid synthesis in maize. Can J Genet Cytol 19: 289–302 [Google Scholar]
- Tice-Baldwin K, Fink GR, Arndt KT (1989) BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science 246: 931–935 [DOI] [PubMed] [Google Scholar]
- Trezzini GF, Horrichs A, Somssich IE (1993) Isolation of putative defense-related genes from Arabidopsis thaliana and expression in fungal elicitor-treated cells. Plant Mol Biol 21: 385–389 [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar B, Jenkins GI (1998) Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol 1: 251–257 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman E, Cardon GH, Fransz P, Saedler H (1998) The behaviour of the autonomous maize transposable element En/Spm in Arabidopsis thaliana allows efficient mutagenesis. Plant Mol Biol 37: 989–999 [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40: 22–34 [DOI] [PubMed] [Google Scholar]