Abstract
The Arabidopsis (Arabidopsis thaliana) genome encodes a family of 51 proteins that are homologous to known serine carboxypeptidases. Based on their sequences, these serine carboxypeptidase-like (SCPL) proteins can be divided into several major clades. The first group consists of 21 proteins which, despite the function implied by their annotation, includes two that have been shown to function as acyltransferases in plant secondary metabolism: sinapoylglucose:malate sinapoyltransferase and sinapoylglucose:choline sinapoyltransferase. A second group comprises 25 SCPL proteins whose biochemical functions have not been clearly defined. Genes encoding representatives from both of these clades can be found in many plants, but have not yet been identified in other phyla. In contrast, the remaining SCPL proteins include five members that are similar to serine carboxypeptidases from a variety of organisms, including fungi and animals. Reverse transcription PCR results suggest that some SCPL genes are expressed in a highly tissue-specific fashion, whereas others are transcribed in a wide range of tissue types. Taken together, these data suggest that the Arabidopsis SCPL gene family encodes a diverse group of enzymes whose functions are likely to extend beyond protein degradation and processing to include activities such as the production of secondary metabolites.
Serine carboxypeptidases (SCPs) are members of the α/β hydrolase family of proteins, which make use of a Ser-Asp-His catalytic triad to cleave the carboxyterminal peptide bonds of their protein or peptide substrates (Hayashi et al., 1973, 1975; Bech and Breddam, 1989; Liao and Remington, 1990; Liao et al., 1992; Ollis et al., 1992). Proteins that contain this catalytic triad and are otherwise homologous to SCPs have been found in a variety of organisms (Doi et al., 1980; Kim and Hayashi, 1983; Breddam, 1986; Baulcombe et al., 1987; Galjart et al., 1988; Bradley, 1992; Degan et al., 1994; Endrizzi et al., 1994; Wajant et al., 1994; Jones et al., 1996; Li and Steffens, 2000). Many of these serine carboxypeptidase-like (SCPL) proteins have been annotated as peptidases based only on this shared sequence similarity; however, studies have shown that some of them function not as peptidases, but as acyltransferases and lyases (Wajant et al., 1994; Lehfeldt et al., 2000; Li and Steffens, 2000; Shirley et al., 2001). For example, several SCPL proteins have been shown to catalyze the production of plant secondary metabolites involved in herbivory defense and UV protection (Wajant et al., 1994; Lehfeldt et al., 2000; Li and Steffens, 2000), including the hydroxynitrile lyase from Sorghum bicolor (SbHNL) necessary for cyanogenesis, and the acyltransferases in wild tomato (Lycopersicon pennellii) that catalyze the formation 2,3,4-isobutyryl-Glc (Wajant et al., 1994; Lehfeldt et al., 2000). Two enzymes in Arabidopsis (Arabidopsis thaliana) responsible for sinapate ester biosynthesis are also SCPL proteins. Sinapoylglucose:malate sinapoyltransferase (SMT) catalyzes the formation of sinapoylmalate, a UV-protectant in seedlings (Landry et al., 1995; Lehfeldt et al., 2000) and sinapoylglucose:choline sinapoyltransferase (SCT) is required for the formation of sinapoylcholine in Arabidopsis seeds (Shirley et al., 2001).
Plants accumulate a broad array of secondary compounds and appear to have recruited many classes of proteins to serve in their synthesis and regulation. In the course of evolutionary history, the genes that encode these proteins have diversified to coordinate these metabolic pathways unique to plants. Examples that have been particularly well studied include the Cyt P450-dependent monooxygenases, MYB transcription factors, terpene synthases, and UDP-glucosyltransferases (Meyer et al., 1996; Mizutani et al., 1998; Jackson et al., 2001; Li et al., 2001b; Lim et al., 2001; Stracke et al., 2001; Aubourg et al., 2002; Jackson et al., 2002; Naur et al., 2003). SCPL proteins appear to represent a similar case of recruitment and diversification in plant lineages that has received limited attention to date.
In this paper, we present a bioinformatics-based examination of the Arabidopsis SCPL genes and their encoded proteins. Inspection of the 51 SCPL genes demonstrated that many of them required reannotation. A phylogenetic analysis of the corresponding corrected protein sequences suggested that Arabidopsis SCPL proteins fall into several distinct classes. Extending this analysis to include SCPL proteins from other organisms reveals a division between plant-specific SCPL proteins and those SCPL proteins common to a broader spectrum of organisms. Together with an expression profile for the entire SCPL gene family generated using reverse transcription (RT)-PCR, these analyses may help to assign roles to members of the SCPL gene family that are as yet of unknown function.
RESULTS
The Arabidopsis Genome Encodes 51 SCPL Proteins
Annotation of the Arabidopsis genome has identified 53 putative SCPL genes. One of these (At2g22960) was previously identified as a pseudogene because the ATPase-like sequences upstream of the first exon suggest that this SCPL gene lacks a promoter (Lehfeldt et al., 2000). The predicted translation products of another two (At3g56540 and At5g22960) terminate shortly after the putative catalytic Ser, and thus lack the active site Asp and His residues. A search of the downstream regions of these genes revealed no additional exons, suggesting that they are pseudogenes. As a result, they were excluded from further consideration, bringing the apparent total of Arabidopsis SCPL genes to 50. Finally, inspection of the Arabidopsis genome identified a gene annotated as a hydroxynitrile lyase-like protein (At4g15100), which encodes an SCPL polypeptide that is 74% identical to the protein encoded by At1g11080. Combined, these results suggest that there are a total of 51 functional SCPL proteins in Arabidopsis.
The Majority of SCPL Gene Annotation Errors Involve Splice Site Predictions
Based on a comparison of each RIKEN Arabidopsis full-length (RAFL) cDNA with the corresponding genomic sequence in GenBank, we found that eight SCPL genes are incorrectly annotated. The majority of the errors are located at splice junctions, resulting in incorrect exon lengths, the exclusion of entire exons, and the inclusion of portions of introns in the predicted mRNAs (Table I). Eight likely annotation errors in genes for which no cDNAs are currently available were identified as well, based on comparisons between the genes in question and other, closely related SCPL genes. For example, the predicted splice sites joining the penultimate and final exon of At1g73290 are in a slightly different position and alternate phase from the highly conserved exon splice sites of closely related Arabidopsis SCPL genes. Since inspection of the sequence of At1g73290 revealed alternate 5′ and 3′ intron splice sites in the same phase and position as the sites in these related genes, its annotation was revised. Similarly, the predicted protein sequence for At3g12240 contains an aberrant carboxyterminal region. Examination of the corresponding genomic sequence revealed that the 3′ end of the penultimate exon of the gene was not identified correctly during annotation, resulting in the inferred translation continuing through the final intron until a stop codon was reached. Translation of the 3′ untranslated region (UTR) revealed the presence of an additional exon virtually identical to the final exon of a closely related and physically linked SCPL protein (At3g12230); thus, the annotation of At3g12240 was updated accordingly. Conversely, exon 13 of gene At1g33540 appeared to be truncated when compared to those of related genes, and translation of the 5′ end of the subsequent intron revealed a sequence similar to those found in other closely related SCPL genes. These findings suggested that the 3′ end of the exon had been incorrectly predicted, and thus, a portion of the correct inferred sequence was missing from the database. A similar exon/intron boundary prediction error resulted in the extension of the 3′ end of the first exon of SCPL gene At2g24000 (Table I). Finally, the inferred translation products of At2g24010, At2g35770, and At4g15100 appeared to be truncated at the N termini, and selection of an upstream in-frame start codon yielded an amino acid sequence more closely in agreement with other SCPL proteins. All subsequent analyses described here were performed using these 16 corrected sequences.
Table I.
Summary of annotation errors found in Arabidopsis SCPL genes and their encoded proteins
Dash indicates RIKEN cDNA sequences not available.
| SCPL Protein | Gene Accession No. | RIKEN cDNA Accession Nos. | Description of Annotation Error(s) |
|---|---|---|---|
| 1 | At5g36180 | AY093044 | Additional exon (14) predicted incorrectly |
| AY128925 | 22 bp at 3′ end of final exon (13) not predicted | ||
| 5 | At1g73290 | – | Incorrect splice phase between exons 12 and 13 (final exon) |
| 8 | At2g22990 | AF361601 | Incorrect splice phase between exons 12 and 13 (final exon) |
| AY035052 | |||
| AY051060 | |||
| AY143880 | |||
| 15 | At3g12240 | – | Additional 114 bp at 3′ end of exon 13 predicted incorrectly |
| Entire final exon (14) not predicted | |||
| 18 | At1g33540 | – | 54 bp at 3′ end of final exon (11) not predicted |
| 19 | At5g09640 | AY063833 | 156 bp at 5′ end of exon 9 not predicted |
| AY091309 | 5′ and 3′ ends, and reading frame of exon 11 predicted incorrectly | ||
| 5′ and 3′ ends, and reading frame of exon 13 predicted incorrectly | |||
| Final exon (14) not predicted | |||
| 20 | At4g12910 | AY136365 | 15 bp at 5′ end of exon 1 not predicted |
| BT000181 | Exon 6 skipped0 | ||
| 22 | At2g24000 | – | Additional 30 bp at 3′ end of exon 1 predicted incorrectly |
| 23 | At2g24010 | – | Start codon predicted incorrectly |
| Signal peptide not predicted | |||
| 28 | At2g35770 | – | Start codon predicted incorrectly |
| Signal peptide not predicted | |||
| 29 | At4g30810 | AY050958 | Exon 5 skipped |
| AY133774 | 6 bp at 5′ end of exon 6 not predicted | ||
| 28 bp at 3′ end of exon 6 not predicted | |||
| 17 bp at 5′ end of exon 7 not predicted | |||
| 41 bp at 3′ end of exon 7 not predicted | |||
| Additional 14 bp at 5′ end of exon 8 predicted incorrectly | |||
| 30 | At4g15100 | – | Start codon predicted incorrectly |
| Signal peptide predicted incorrectly | |||
| 32 | At1g61130 | – | Additional 39 bp at 3′ end of exon 5 predicted incorrectly |
| 43 | At2g12480 | BT004306 | Additional 78 bp at 5′ end of exon 7 predicted incorrectly |
| 45 | At1g28110 | AY059854 | 15 bp at 3′ end of exon 6 not predicted |
| BT008763 | |||
| 46 | At2g33530 | AY074317 | 21 bp at 3′ end of exon 6 not predicted |
Arabidopsis SCPL Proteins Exhibit Conserved Sequence Motifs
In addition to the Ser-Asp-His catalytic triad, 15 amino acid residues are conserved in carboxypeptidase Y (CPY) and all Arabidopsis SCPL proteins. Of these, three align with residues that have been shown to be involved in substrate binding in CPY from yeast (Saccharomyces cerevisiae; Endrizzi et al., 1994; Table II; Supplemental Fig. 1), and an additional three CPY active site residues are conserved in all but a few SCPL proteins. In CPY, all of these residues either bind the terminal carboxylate group of the enzyme's substrate or comprise the binding pockets for the substrate's terminal and penultimate amino acid residues, referred to as the S1′ and S1 sites, respectively. Interestingly, only one of the seven residues that contribute to the S1 site in CPY is conserved in the Arabidopsis SCPL proteins compared to four of twelve S1′ site residues. A further five conserved amino acids correspond to residues that are thought to orient S1 or S1′ site residues in CPY such that they make appropriate substrate contacts (Endrizzi et al., 1994). There are no amino acids that can be identified as putative substrate binding residues within the region that aligns with CPY between L178 and W312, despite the fact that this region contributes nine residues to the S1 and S1′ sites of CPY. Overall, this region has low amino acid identity among all Arabidopsis SCPL proteins and corresponds to the primarily α-helical region between amino acids 180 and 317 of CPY that has been identified as an ancient insertion in the α/β hydrolase fold family (Endrizzi et al., 1994). Six Cys residues appear in conserved positions in almost all of the sequences and align near the Cys that form disulfide bridges in CPY. Results from SignalP (Bendtsen et al., 2004) indicate that all of the 51 SCPL proteins contain signal peptides that are likely to target them to the endoplasmic reticulum (Supplemental Fig. 1), possibly for posttranslational modification and subsequent transport to the vacuole or secretion from the cell. Consistent with this observation, many potential glycosylation sites that fit the consensus sequence NX(S/T) are found within the SCPL protein sequences (Marshall, 1972). Further, an N-terminal sequence found in eight of the SCPL proteins aligns with the N-terminal propeptide in CPY (Endrizzi et al., 1994). Finally, over one-half of the SCPL proteins, including SCT, contain a region of low sequence identity between the fourth and fifth conserved Cys residues (columns 400–460, Supplemental Fig. 1), which aligns within the α-helical region between amino acids 180 and 317 of CPY mentioned previously. When these Arabidopsis SCPL proteins are aligned with carboxypeptidase II from wheat (Triticum aestivum; CPDW-II; GenBank accession no. A29639), a well-studied enzyme thought to be involved in the mobilization of protein reserves in seeds (Breddam et al., 1987), this variable region corresponds to a sequence that is proteolytically removed from the mature wheat protein (Breddam et al., 1987; Liao and Remington, 1990), suggesting that there may be few constraints on amino acid changes that occur within this region.
Table II.
Conservation of amino acids in Arabidopsis SCPL proteins that align with residues making substrate contacts in the active site of CPY from yeast
In CPY, these residues are either involved in catalysis, binding of the C-terminal carboxylate of the substrate, or comprise the binding sites for R-groups of the C-terminal or penultimate amino acid residues of the substrate (S1′ and S1, respectively).
| Residuea | Conserved | Columnb | Function in CPY |
|---|---|---|---|
| N51 | Only in Clades IB and II | 168 | Carboxylate binding |
| G52 | Yes | 169 | Carboxylate binding |
| G53 | Yes | 170 | S1′ |
| C56 | Yes, except A in SCPL protein 51 | 173 | S1′ |
| S57 | Yes, except T in SCPL protein 18 | 174 | S1′ |
| T60 | No | 177 | S1′ |
| F64 | No | 182 | S1′ |
| E65 | Yes, except A or Q in 3 SCPL proteins | 183 | S1′ |
| E145 | No, N,D, or Q in 22 SCPL proteins | 275 | Carboxylate binding |
| S146 | Yes | 276 | Catalytic |
| Y147 | Yes | 277 | S1 |
| L178 | No | 322 | S1 |
| Y185 | No | 329 | S1 |
| Y188 | No | 332 | S1 |
| N254 | No | 402 | S1′ |
| Y256 | No | 447 | S1′ |
| Y269 | No | 460 | S1′ |
| L272 | No | 463 | S1′ |
| S297 | No | 492 | S1′ |
| W312 | No | 507 | S1 |
| D338 | Yes | 533 | Catalytic |
| I340 | No | 535 | S1 |
| C341 | No | 536 | S1 |
| H397 | Yes | 594 | Catalytic |
| M398 | No | 595 | S1′ |
Designated by amino acid and position in CPY.
Refers to column position in Supplemental Figure 2.
Most Arabidopsis SCPL Proteins Can Be Grouped into Two Major Clades
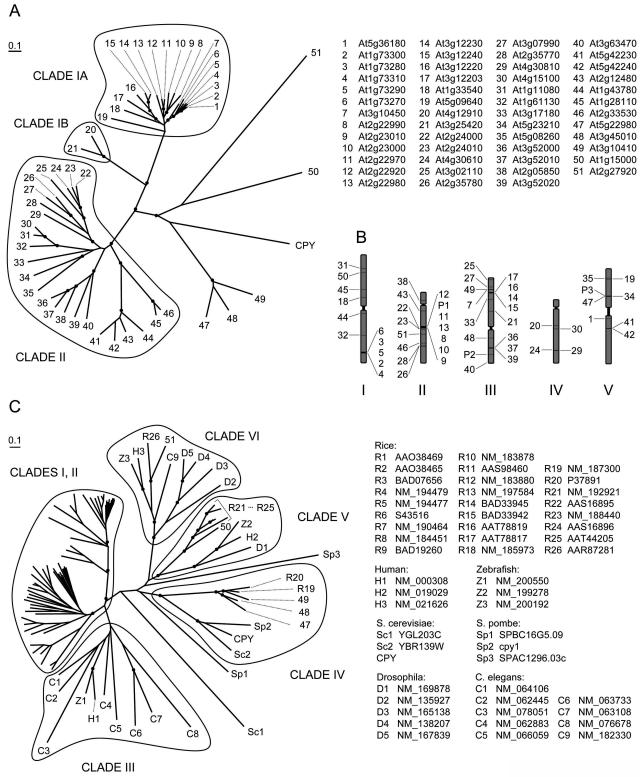
Phylogenetic analysis of the 51 Arabidopsis SCPL proteins showed that the majority of sequences cluster into two major clades, although there are five additional members that are less closely related (Fig. 1A). For the sake of convenience and brevity in subsequent discussion, these proteins have been numbered counterclockwise as they are presented in Figure 1, beginning from the top right of Clade I. Clade I can be further subdivided into Clades IA (SCPL proteins 1 through 19) and IB (SCPL proteins 20 and 21) based upon the degree of amino acid identity shared between these two subgroups. The SCPL proteins of Clade IA include the known acyltransferases SMT (SCPL protein 8) and SCT (SCPL protein 19; Fig. 1A; Lehfeldt et al., 2000; Shirley et al., 2001).
Figure 1.
Unrooted trees of SCPs and the SCPL protein family. A, Phylogenetic tree of the 51 Arabidopsis SCPL proteins and CPY from yeast. SCPL proteins are arbitrarily numbered proceeding counterclockwise. The GenBank gene accession numbers for each corresponding SCPL protein are shown on the right. B, Chromosomal positions of the Arabidopsis SCPL genes (pseudogenes are designated as P1–3). C, Phylogenetic tree of Arabidopsis, rice, fungal, and animal SCPL proteins. The GenBank identification numbers are provided on the right. Rice SCPL proteins 1 to 18 (not labeled) fall into Clades I and II. Tree nodes with bootstrap values above 90% are indicated in both A and C with black dots.
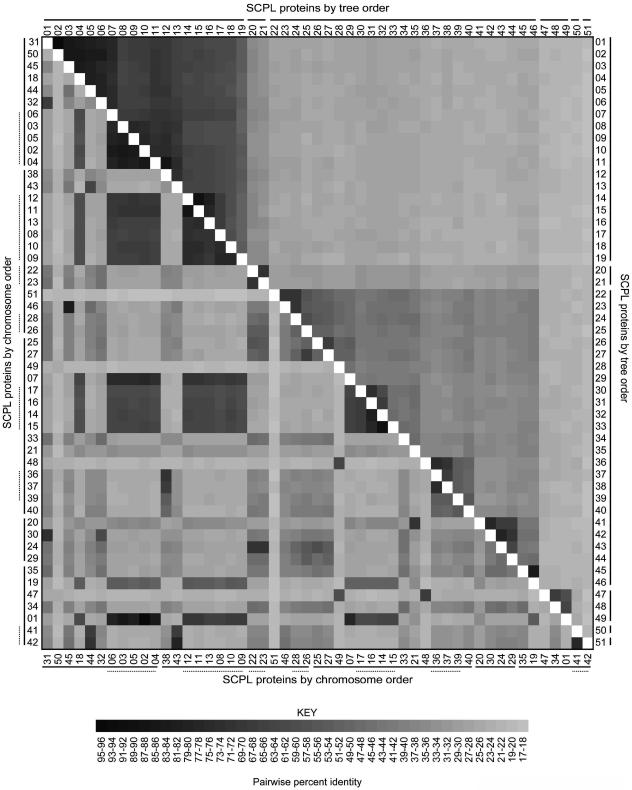
Several subclades of Clade IA are associated with clusters of SCPL genes that are tandemly arranged in the genome. For example, SCPL proteins 2 to 6 are between 79% and 86% identical, and their corresponding genes are located together at the bottom of chromosome I (Fig. 1B). Interestingly, SCPL gene 1 is located on chromosome 5, but its encoded protein is 96% identical to that of SCPL gene 2 on chromosome 1 (Fig. 2). The mRNA sequences of these genes are 97% identical, nine of their 12 introns are greater than 74% identical (e.g. intron 10 of SCPL gene 1 is 97% identical to intron 10 of SCPL gene 2), and their 5′ and 3′ UTRs are highly similar. Furthermore, this identity extends through the region downstream of the two genes to include a portion of the gene that follows SCPL gene 2 (At1g72290) duplicated in the region downstream of SCPL gene 1. Taken together, these data suggest that SCPL gene 1 represents a duplication of SCPL gene 2 onto chromosome 5. Another SCPL gene cluster includes genes 8 through 13 on chromosome II. This group includes SNG1 (SCPL gene 8), which encodes SMT (Fig. 1A). Given that proteins encoded by the genes flanking SNG1 are over 70% identical to SMT (Fig. 2), it is tempting to speculate that these enzymes may share some functional characteristics.
Figure 2.
Pairwise identity matrix for the Arabidopsis SCPL proteins. Percent identity values are shown for each pairwise alignment of the 51 Arabidopsis SCPL proteins coded on the gray scale shown at bottom. In the top right half of the matrix, the proteins are arranged by their numbered tree positions given in Figure 1. Bars next to the numbers on the top horizontal and right axes indicate clade divisions as designated in Figure 1. In the bottom left half of the matrix, the proteins are arranged in order of their chromosomal positions, from the top of chromosome 1 to the bottom of chromosome 5. The solid bars next to the SCPL protein numbers on the left and lower horizontal axes designate each of the five chromosomes, and the dashed bars designate groups of clustered SCPL genes.
The pairwise percent identity between any two Clade IA SCPL proteins ranges from 51% to 96%, averaging 66% overall (Fig. 2). The exon/intron splicing patterns of the Clade IA SCPL genes are also alike, with the phase, number, and location of splice sites being identical for each SCPL gene with the exception of only two sites (Supplemental Fig. 1). Clade II SCPL proteins are more varied in sequence, with any two proteins being 41% identical on average (Fig. 2). In contrast to the 261 residues uniquely conserved among 75% of Clade IA members, only 56 amino acids meet the same criteria for Clade II proteins. The exon/intron splicing patterns of Clade II genes also show more variability, with fewer splice sites of the same phase and location conserved among them (Supplemental Fig. 1).
SCPL proteins 20 and 21 (Clade IB) are between 32% and 38% identical to Clade IA SCPL proteins and 20% to 32% identical to Clade II members (Fig. 2), but among those shared residues are a high percentage of the residues conserved only among the Clade IA proteins (Supplemental Fig. 1). Further, all but two of the consensus intron/exon splice sites in the Clade IA genes are conserved in position and phase in SCPL genes 20 and 21. These observations suggest that they are more closely related to SCPL proteins 1 to 19 than the others, leading us to designate them as Clade IB (Fig. 1A).
Arabidopsis SCPL Proteins Have Homologues in Other Plants
To determine whether the clade structure observed for Arabidopsis SCPL proteins reflected the diversity and relationships between similar proteins in other plants, we included other plant SCPL proteins in a separate sequence analysis. In this analysis, the sequences of the isobutyryl-Glc acyltransferase from wild tomato and three similar enzymes from wild potato (GenBank accession nos. AF248647, AF006078, AF006079, and AF006080) were grouped into Clade IA. The pairwise identities between these individual proteins and most Clade I proteins range from 38% to 46%. Similarly, a wound/jasmonate-inducible SCPL protein from Lycopersicon esculentum (accession no. AF242849) is highly similar to SCPL proteins 20 and 21 from Arabidopsis with pairwise identity values of 62% and 66%, respectively (Moura et al., 2001).
Several relatively well-studied SCPs and SCPL proteins from other plant species fell into Clade II when included in the phylogenetic analysis of the Arabidopsis SCPL proteins. CPDW-II (Breddam et al., 1987) is 58% and 60% identical with respect to SCPL proteins 26 and 27. The hydroxynitrile lyase from Sorghum bicolor (accession no. AJ421152; Wajant et al., 1994) is also 45% identical to proteins 26 and 27. Further, SCPL protein 40 is 50% identical to an SCP from barley grain (CP-MII; accession no. AAB31589) and 57% identical to a gibberellin-induced SCP from pea (CP-A; accession no. AJ251969; Degan et al., 1994; Jones et al., 1996). Finally, among 26 nonredundant SCPL genes annotated in the rice (Oryza sativa) genome, 12 of the encoded proteins group among the Clade II Arabidopsis SCPL proteins and six cluster into Clade I (Fig. 1C).
Animal and Fungal SCPL Proteins Help to Define Four Additional SCPL Clades
Extending the analysis of SCPL proteins to sequences from organisms outside the plant kingdom revealed that homologs of SCPL proteins 47 to 51 are found in diverse eukaryotic lineages (Fig. 1C) and cluster into an additional four clades. Arabidopsis SCPL proteins 47 to 49 are most similar to CPY and an SCPL protein from Schizosaccharomyces pombe (YBR139W) in Clade IV (Fig. 1B; Endrizzi et al., 1994). SCPL protein 50 is encoded by an intronless gene and lacks many of the conserved Cys residues found in the majority of other Arabidopsis SCPL proteins (Supplemental Fig. 1). It clusters into Clade V with five SCPL proteins from rice, four of which are encoded by genes with no introns, and the other by a gene with a single intron. Also grouped into Clade V is an SCPL protein from Drosophila (lacking introns) and SCPs from human and zebrafish (Fig. 1C). As with Clade V, Clade VI includes SCPL proteins from both plants and animals including Arabidopsis, human, Drosophila, zebrafish, and Caenorhabditis elegans. In fact, Arabidopsis SCPL protein 51 and rice SCPL protein 26 (accession no. AAR87281) are each 41% identical to the human retinoid-inducible SCP (Chen et al., 2001) but only 21% to 25% identical to any other plant SCPL proteins outside of Clade VI. Clade III stands apart from the other clades in that it contains exclusively animal SCPL proteins, including the human SCP lysosomal protective protein also known as cathepsin A (Galjart et al., 1988; Fig. 1C). Finally, it is noteworthy that a query of the 240 sequenced genomes of prokaryotes available through BLASTP using the amino acid sequence of SMT, retinoid-inducible Ser carboxypeptidase, and CPY yielded only a single protein (NP_902682) with an E value lower than 1e-7. Since even this protein did not appear to include the catalytic Ser, Asp, and His residues, as determined by alignment, it would appear that Ser carboxypeptidase-like proteins are not commonly found in prokaryotes.
Arabidopsis SCPL Genes Exhibit Tissue-Specific Expression Patterns
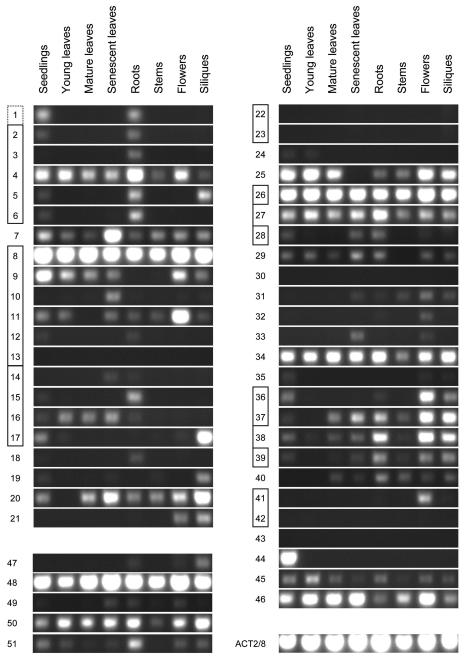
To provide a foundation for determining the function of the Arabidopsis SCPL proteins, RT-PCR analyses were carried out using RNA extractions from eight different tissue types. These experiments revealed a diversity of expression patterns for Arabidopsis SCPL genes, even among members of the five SCPL gene clusters. For example, each member of the cluster on chromosome 1 (SCPL genes 2–6) is expressed in roots and seedlings, but SCPL gene 5 is also expressed in siliques, and SCPL gene 4 is expressed in all tissues surveyed (Fig. 3). The shared expression pattern in seedlings and roots exhibited by SCPL gene 1 on chromosome 5 is consistent with its recent duplication from SCPL gene 2 on chromosome 1.
Figure 3.
RT-PCR analysis of Arabidopsis SCPL gene expression. RNA was extracted from the tissues indicated and used for RT-PCR to evaluate SCPL gene expression. The Arabidopsis SCPL genes are identified according to the numbering scheme used in Figure 1. Genes that are tandemly arranged in the genome are boxed, with the dotted box surrounding gene 1 reflecting its close sequence identity with SCPL gene 2. Results for SCPL genes 1 to 21 and 22 to 46 are grouped together because they represent Clades I and II, respectively. Results for the ACT2/8 positive control are given on bottom left.
The members of each of the other two Clade I gene clusters are also differentially expressed (Fig. 3). Consistent with previous findings (Lehfeldt et al., 2000), SCPL gene 8 (SNG1) is expressed in all tissue types. On the other hand, expression of SCPL gene 9 is below detectable limits in roots and stems, SCPL gene 10 shows positive results only for senescent leaves, and the PCR results for SCPL gene 11 suggest that it is highly expressed in leaves. The SCPL gene cluster on chromosome 3 likewise includes a gene expressed primarily in roots (SCPL gene 15), one expressed in all leaf stages examined (SCPL gene 16), and one expressed primarily in siliques (SCPL gene 17). The Clade II gene cluster on chromosome 3 containing SCPL genes 36, 37, and 39 exhibits the most uniformity of expression, with each gene expressed in roots, seedlings, flowers, and siliques; however, 36 is more limited in its expression than the other two.
Thirteen SCPL genes appear to be expressed chiefly in single tissue types: two in senescent leaves (genes 7 and 10), four in roots (genes 3, 6, 15, and 18), two in siliques (genes 17 and 19), four in flowers (genes 11, 32, 36, and 41), and one in seedlings (gene 41; Fig. 3). Twenty-three of the 51 SCPL genes appear to be expressed in both roots and seedlings, an observation that may reflect the fact that the intact seedlings used for mRNA preparation included root tissue. In contrast, SCPL gene 41 is one of three genes expressed in seedlings but not in roots, the other two being 9 and 17. The data for these genes suggest that the actual site of their expression is either the hypocotyls or cotyledons.
Expression in all but one or two tissue types appears to be a characteristic of several SCPL genes. For instance, SCPL gene 20 is expressed in all tissues except young leaves, and 25 is not expressed in senescent leaves. In contrast to the consistently positive PCR results for the majority of SCPL genes, several of them yielded only negative PCR results: 13, 22, 23, 30, 42, and 43 (Fig. 3). Although we cannot exclude the possibility that some of these reactions did not yield product due to technical issues associated with primer design, it is likely that some or all of these genes are expressed only at low levels or in a highly tissue-specific manner, or only in response to abiotic or biotic stimuli that were not studied here. Finally, our expression data were in very good agreement with both Massively Parallel Signature Sequencing (MPSS) expression data (http://mpss.udel.edu/at/java.html; Brenner et al., 2000) and microarray data (http://www.weigelworld.org/resources/microarray/AtGenExpress/) made available to the Arabidopsis research community.
DISCUSSION
Many Arabidopsis SCPL Proteins Are Misannotated
Although the advent of completely sequenced and annotated genomes has become an indispensable resource for the scientific community, independent verification of gene annotation is required to ensure database reliability. Within the 51 annotated genes analyzed in this study, most of the predicted splice sites appear to have been identified correctly. Given that the likelihood of a splice site annotation error within a gene is directly dependent upon the number of introns it contains, even a low error rate can still be problematic when studying genes with multiple introns. For example, misannotations were found in almost one-third of the SCPL genes examined, with many of the sequences containing more than one error. Moreover, many of the errors are substantial, resulting in large deletions, insertions, and truncations. Further, we identified several annotated SCPL genes that are almost certainly pseudogenes and an additional SCPL gene not annotated as such. These data demonstrate that careful reexamination of sequence annotation is necessary prior to detailed molecular and phylogenetic analyses, and that studies completed in the absence of this type of analysis are likely to contain a substantial number of errors. Errors of this type can be found in a previous analysis of the Arabidopsis SCPL protein family, in addition to three protein sequence duplications: SMT, SCT, and At1g73310 (Milkowski and Strack, 2004).
Arabidopsis SCPL Proteins Have Structural Features in Common
Some of the common sequence characteristics of the Arabidopsis SCPL proteins may provide preliminary insights into structural features required for activity and/or subcellular localization. For example, the presence of the Ser-Asp-His triad in every SCPL protein suggests that some or all of them may employ a similar catalytic mechanism. Indeed, inhibition studies conducted with SMT and SCT indicate that the Ser residue is required for the catalytic activity of both enzymes (Lehfeldt et al., 2000; Shirley et al., 2001), and the kinetic analysis of SCT indicates that a fundamental alteration of the catalytic mechanism is not required to bring about transacylation activity (Shirley and Chapple, 2003). Thus, it may be that the majority of SCPL enzymes, including both acyltransferases and hydrolases, utilize the Ser-Asp-His triad in catalysis, although our alignment suggests that with regard to other residues involved in substrate binding, these enzymes have diverged substantially over evolutionary time.
Two observations suggest that the six Cys residues present in almost all of the Arabidopsis SCPL proteins may be important in maintaining the tertiary structure of the functional enzymes. First, the six residues align with Cys in CPY that are known to form three disulfide bonds that are likely to contribute to the stability of the protein. Second, two of these residues are situated on either side of a region that is highly variable among many SCPL family members. This variable region aligns with a sequence in CPDW-II that is similarly bordered by disulfide bond-forming Cys and is proteolytically excised from the mature enzyme (Breddam et al., 1987; Liao and Remington, 1990). Since this posttranslational modification results in the production of a heterodimeric protein molecule, the disulfides that cross-link the monomers are probably important to prevent their dissociation.
The identification of signal peptides via SignalP in 49 of the 51 SCPL proteins is consistent with experimental work showing that CPY and SMT are transported to the vacuole (Marshall, 1972; Stevens et al., 1982; Sharma and Strack, 1985; Valls et al., 1987; Hause et al., 2002). The SignalP results are also supported by the fact that the signal peptide sequence removed from the mature form of SMT (Lehfeldt et al., 2000) is accurately predicted by the algorithm. An additional N-terminal feature found in eight of the SCPL proteins is the presence of a variable-length peptide sequence that aligns with the propeptide of CPY (Ramos et al., 1994). These peptides could serve a chaperone-like role similar to that of the CPY propeptide (Winther and Sorensen, 1991). Consistent with the presence of the N-terminal signal peptides in SCPL proteins are the many putative glycosylation sites found in most of their sequences. These data suggest that many SCPL proteins may be posttranslationally modified as they are processed through the endoplasmic reticulum and Golgi. Finally, and as mentioned above, the inferred translations of many SCPL proteins contain a variable region known to be removed in CPDW-II (Breddam et al., 1987; Liao and Remington., 1990). Protein gel blots of SCT expressed in planta and in yeast suggest that this region is also removed during the maturation of this SCPL acyltransferase (Shirley and Chapple, 2003). Taken together, these data suggest a pattern of subcellular targeting and posttranslational processing that may be required to produce functional enzymes.
SCPL Gene Expression Profiles May Help to Reveal the Function of Their Encoded Proteins
RT-PCR reveals that members of the SCPL gene family are expressed in all major tissue types examined: seedlings, roots, stems, leaves, flowers, and siliques. Thus, it seems likely that SCPL genes and their encoded proteins have diversified in Arabidopsis to function differently both in terms of their activities and their tissue-specific expression. Alternatively, some groups of SCPL proteins may have maintained the same metabolic roles, with different members serving the same function in different tissues. In any case, the diversification of SCPL gene expression patterns appears to have continued until fairly recently, as indicated by the distinct regulation exhibited by SCPL genes found in four of the six chromosomal clusters.
Our RT-PCR results may also serve as a guide for identifying the functions of Arabidopsis SCPL proteins. These results will be particularly important in experiments aimed at identifying metabolic pathways blocked in T-DNA insertional mutants. For example, in both the sng1 and sng2 mutants, the activated Glc ester substrate for the SCPL protein in question accumulates in a tissue-specific manner. If phenotypes of this kind can be expected to occur in other SCPL acyltransferase-deficient mutants, SCPL gene expression patterns can be used to direct metabolic profiling experiments, and the identity of the Glc ester accumulated will help to identify the pathway in which the enzyme is involved.
Finally, some reactions that might be catalyzed by SCPL proteins are known to occur in specific tissues. As a result, our RT-PCR gene expression survey may help to identify candidate genes for these enzymes. For example, sinapoylcholinesterase activity is found only in germinating seedlings (Strack et al., 1980) where it functions to hydrolyze sinapoylcholine, liberating choline and sinapate for the biosynthesis of membrane lipids and UV-protective sinapoylmalate, respectively (Strack, 1981). Among the 51 SCPL genes of Arabidopsis, the expression of only one (SCPL 44, At1g43780) is seedling specific, making it an excellent candidate for the SCE (sinapoylcholinesterase) gene.
SCPL Proteins in Clades I and II Appear to Represent Plant-Specific Proteins
Our analyses of Arabidopsis SCPL proteins and related sequences from other organisms suggest that these proteins can be divided into six clades. Of these, Clades I and II appear to be plant specific, based upon the presence of highly similar proteins identified in an assortment of plants and the absence of closely related coding sequences in the genomes of insects, yeast, and animals that were examined. Consistent with this hypothesis, a preliminary phylogenetic analysis of three annotated SCPL proteins from the initial draft of the Chlamydomonas reinhardtii genome (http://genome.jgi-psf.org/chlre2/chlre2.home.html) suggested that they fall into Clades I, II, and VI. These findings suggest that at some point in the evolutionary history of the plant lineage, an ancestral SCPL protein may have acquired an enzymatic activity that added to the catalytic repertoire of plants. This seems like a particularly likely explanation for Clade IA proteins given that SMT, SCT, and the SCPL isobutyryl acyltransferases are grouped within this clade. Indeed, considering that SMT and SCT are both acyltransferases but are among the most distantly related of Clade IA proteins, it seems likely that some or all of the as-yet-uncharacterized Clade IA proteins are also acyltransferases.
Our grouping of SCPL proteins 20 and 21 with proteins 1 to 19 to form Clade I is based primarily on sequence characteristics not reflected by pairwise percent identities. Whereas there are only slightly higher shared identities between SCPL proteins 20, 21, and the proteins in Clade IA versus the members of Clades II to VI (Fig. 2), the patterns of amino acid and splice site conservation sets 20 and 21 apart more clearly from Clades II to VI. The similarity of SCPL proteins 20 and 21 to the wound/jasmonate-inducible Ser carboxypeptidase from tomato suggest that they may be important to defense responses in Arabidopsis. As in tomatoes, jasmonate acts as a signaling molecule in Arabidopsis in the defense against insects, pathogens, and abiotic stress (McConn et al., 1997; Thomma et al., 1999; Overmyer et al., 2000; Rao et al., 2000). SCPL proteins 20 and 21 might therefore play a role similar to their tomato homolog, with the diverse expression of SCPL gene 20 making its encoded protein a particularly good candidate for involvement in plant defense.
Aside from the wound-inducible SCP from tomato, all of the other known plant SCPs included in our analysis fall into Clade II. This suggests that Arabidopsis SCPL proteins 22 to 46 act as genuine carboxypeptidases, although some members may be esterases or lyases (Degan et al., 1994; Wajant et al., 1994; Jones et al., 1996; Moura et al., 2001). Although its true substrate and the nature of its catalytic activity remain unknown, the finding that SCPL protein 24 is involved in brassinosteroid signaling suggest that Clade II members may play important roles in plant growth and development (Li et al., 2001a). Whatever the exact function of the Clade II SCPL proteins may be, the fact that they do not appear to be encoded within the genomes of animals and fungi suggests that these proteins have become specialized to perform functions that are likely to be unique to plants.
Candidate Reactions for SCPL Acyltransferases Can Be Found throughout Plant Metabolism
Our understanding of the reactions catalyzed by plant SCPL proteins is limited at best; however, some reactions likely to be catalyzed by SCPL proteins can be identified in the biochemical literature. For example, it has been shown that the most abundant anthocyanin in wild carrot (Daucus carota) is a sinapoylated cyanidin glycoside (Harborne et al., 1983; Glässgen et al., 1992). The enzyme that catalyzes the sinapoylation of this anthocyanin uses sinapoyl-Glc as the activated sinapate donor (Glässgen and Seitz, 1992; Nakayama et al., 2003; Suzuki et al., 2004). As in Daucus, the major anthocyanin in Arabidopsis is acylated with a sinapoyl moiety (Bloor and Abrahams, 2002), which may be added by a sinapoyl-Glc-utilizing SCPL protein. Further, since isolated Daucus vacuoles actively accumulate the sinapoylated anthocyanin, but not the nonacylated form, the sinapate moiety may function as a vacuolar uptake or retention tag (Hopp and Seitz, 1987). Alternatively, the sinapate moiety may be required for, or be the site of, glutathione derivatization by a glutathione S-transferase analogous to that encoded by the maize Bronze2 gene (Marrs et al., 1995). Since anthocyanins are accumulated in Arabidopsis at high levels in senescing leaves, SCPL genes 7 and 10 are good candidates for genes encoding this putative anthocyanin sinapoyltransferase because they both belong to Clade 1A and are expressed at high levels in the appropriate tissue.
Another potential role for Clade IA SCPL proteins is in the acylation of glucosinolates. Glucosinolates are amino acid-derived sulfated thioglucosides that are characteristic of the members of the Brassicaceae, including Arabidopsis (Wittstock and Halkier, 2002). These compounds are substrates for myrosinases, which convert the various glucosinolates into an array of toxic and volatile substances that are important in plant-insect interactions (Rask et al., 2000). The enzymes that catalyze the core reactions of glucosinolate biosynthesis have been identified (Wittstock and Halkier, 2002); however, a number of glucosinolates are further modified by additions of other substituents that include sinapoyl and benzoyl groups (Linscheid et al., 1980; Reichelt et al., 2002; Brown et al., 2003). This acylation may have an impact on glucosinolate turnover and plant-herbivore interactions because they are not readily degradable targets for myrosinases (Taipalensuu et al., 1997). The presence of benzoylated glucosinolates in Arabidopsis has already been demonstrated, and sinapoylated glucosinolates have been detected in Arabidopsis seed extracts (Graser et al., 2001; Reichelt et al., 2002). Sinapoyl-Glc and benzoyl-Glc are potential donor molecules in the acylation reaction, making Clade IA SCPL enzymes good candidates for the catalysts involved.
Finally, Glc ester transesterification reactions are found in a number of other pathways leading to plant secondary metabolites, including chlorogenic acid in sweet potato and gallotannins in oak (Gross, 1983; Villegas and Kojima, 1986). Similarly, indole-3-acetic acid (IAA)-Glc synthesized by indole-3-acetic acid glucosyltransferase serves as an activated form of IAA, which is used in a transesterification reaction to form IAA-inositol (Michalczuk and Bandurski, 1982; Jackson et al., 2001, 2002; Ljung et al., 2002). This latter reaction is analogous to sinapoylmalate biosynthesis and preliminary indications suggest that it may be catalyzed by an SCPL enzyme (Kowalczyk et al., 2003).
SCPL Proteins in Clades III to VI May Function in Pathways Conserved among Animals, Plants, and Fungi
All of the SCPL proteins and known SCPs outside of the plant kingdom fall into Clades III to VI. The similarity of Arabidopsis SCPL proteins 47 to 51 to fungal and animal SCPs suggests that these proteins may perform similar or identical functions in these divergent organisms and may thus be true orthologs. In fact, the analysis of Arabidopsis SCPL proteins within the context of rice, animal, and fungal SCPL proteins suggests that Clade III is specific to animals, Clade IV is specific to only plants and fungi, and Clades V and VI represent proteins that serve a function conserved among all eukaryotic organisms. We have not included two SCPL proteins from S. pombe and one from yeast in Clade IV because they are only distantly related to the members of this clade, and the relevant branch points are not supported by high bootstrap values. Further analysis of fungal SCPL proteins may reveal that these proteins define additional SCPL clades that are not characteristic of plants and animals.
CONCLUSION
Although the biological role of the majority of Arabidopsis SCPL proteins remains to be determined, our research and the research of others suggests that SCPL proteins are likely to function in a broad range of biochemical pathways including those involved in secondary metabolite biosynthesis. SCPL proteins may therefore be important to normal plant growth and development, as well as to the synthesis of compounds that protect plants against pathogens and UV light and for resistance to natural and man-made xenobiotics. Regardless of the actual enzymatic function, RT-PCR results indicate that few Arabidopsis SCPL genes are expressed predominantly in tissues undergoing extensive protein breakdown such as cotyledons and senescent leaves, suggesting that previous assumptions about the roles of these proteins being confined solely to protein turnover may be in error.
MATERIALS AND METHODS
Search of On-Line Sequence Databases
To identify all SCPL and SCP sequences in the Arabidopsis (Arabidopsis thaliana) genome and in other fully or partially sequenced genomes, nonredundant genomic, cDNA, and protein sequences with the keywords “Ser carboxypeptidase” in their definition fields were obtained from GenBank (Benson et al., 2000). To identify any SCPL genes that had not been annotated as such, we used a subset of Arabidopsis SCPL amino acid sequences to again query the genome databases using PSIBlast (Altschul et al., 1997). Once identified, sequences were downloaded directly into a Vector NTI database (Informax/Invitrogen, Carlsbad, CA) using the on-line retrieval tool. GenBank identification numbers for all sequences included in our analyses are provided in Figure 1, with the exception of CPY (GenBank accession no. 576359).
Reannotation of SCPL Genes
Two different approaches were taken to evaluate the annotation of the Arabidopsis SCPL genes. For each of the 20 SCPL genes with one or more available RAFL cDNAs, the predicted open reading frame was aligned with the RAFL cDNA sequence to determine whether differences existed between the two. In cases where these sequences were not identical, the genomic sequence was then examined to determine how this discrepancy may have arisen, such as by the erroneous addition, deletion, extension, or truncation of exons. In these cases, the RAFL cDNA was adopted as the revised SCPL coding sequence. To evaluate the annotation of SCPL genes for which RAFL cDNA sequences were not available, their inferred amino acid sequences were aligned together with those of the RAFL cDNAs described above to identify likely sites of misannotation.
Phylogenetic and Sequence Analyses
All alignments were performed using the default parameters in AlignX, a part of the Vector NTI program suite 9.0.0, which uses ClustalW as a base alignment algorithm (Thompson et al., 1994). Alignments were exported to the PHYLIP software package (Felsenstein, 1985) and used to generate unrooted neighbor joining trees and bootstrap values, also using the default parameters. AlignX was also used to generate pairwise identity values for SCPL protein sequences. SignalP (http://www.cbs.dtu.dk/services/SignalP; Bendtsen et al., 2004) was used with the default parameters to identify signal peptides. Tree graphics were generated using the TreeView program (Page, 1996).
RT-PCR
RNA extractions were performed as described previously (Lehfeldt et al., 2000). RT reactions were carried out using Promega ImProm-II (Promega, Madison, WI) reverse transcriptase for each tissue type following the protocol provided by the manufacturer. Four-fold dilutions of cDNA were used for each PCR reaction. Oligonucleotides were chosen to minimize the likelihood of nonspecific annealing by designing the forward primer within regions that were most divergent between any subset of closely related SCPL proteins (Supplemental Table I). Each reverse primer was designed against the similarly diverse 3′ UTR of each gene. BLASTN (Altschul et al., 1990) was then used to search the Arabidopsis genome with each primer pair to ensure a low risk of nonspecific PCR amplification. For all but one gene, the region to be amplified included at least one intron in the genomic sequence, such that amplicons from contaminating genomic DNA could be distinguished from those derived from cDNA. Each PCR reaction was run with an annealing temperature at the lower primer Tm for each primer pair. A primer pair common to actin genes 2 and 8 (An et al., 1996) was used for positive control PCR reactions.
Supplementary Material
Acknowledgments
The authors are grateful to J. Lohmann, M. Schmid, and D. Weigel, MPI for Developmental Biology, Tuebingen, Germany, for release of their microarray dataset prior to publication.
This work was supported by the National Science Foundation (grant to C.C.) and by Purdue University (graduate fellowships to C.M.F.). This is journal paper number 2005–17562 from the Purdue University Agricultural Experiment Station.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.057950.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An YQ, McDowell JM, Huang SR, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTP) gene family in Arabidopsis thaliana. Mol Genet Genomics 267: 730–745 [DOI] [PubMed] [Google Scholar]
- Baulcombe DC, Barker RF, Jarvis MG (1987) A gibberellin responsive wheat gene has homology to yeast carboxypeptidase Y. J Biol Chem 262: 13726–13735 [PubMed] [Google Scholar]
- Bech LM, Breddam K (1989) Inactivation of carboxypeptidase Y by mutational removal of the putative essential histidyl residue. Carlsberg Res Commun 54: 165–171 [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL (2000) Genbank. Nucleic Acids Res 28: 15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloor SJ, Abrahams S (2002) The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59: 343–346 [DOI] [PubMed] [Google Scholar]
- Bradley D (1992) Isolation and characterization of a gene encoding a carboxypeptidase Y-like protein from Arabidopsis thaliana. Plant Physiol 98: 1526–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breddam K (1986) Serine carboxypeptidases: a review. Carlsberg Res Commun 51: 83–128 [Google Scholar]
- Breddam K, Sørensen SB, Svendsen IB (1987) Primary structure and enzymatic properties of carboxypeptidase-II from wheat bran. Carlsberg Res Commun 52: 297–311 [Google Scholar]
- Brenner S, Williams SR, Vermaas EH, Storck T, Moon K, McCollum C, Mao J, Luo S, Kirchner JJ, Eletr S, et al (2000) In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs. Proc Natl Acad Sci USA 97: 1665–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62: 471–481 [DOI] [PubMed] [Google Scholar]
- Chen J, Streb JW, Maltby KM, Kitchen CM, Miano JM (2001) Cloning of a novel retinoid-inducible serine carboxypeptidase from vascular smooth muscle cells. J Biol Chem 276: 34175–34181 [DOI] [PubMed] [Google Scholar]
- Degan FD, Rocher A, Cameron-Mills V, von Wettstein D (1994) The expression of serine carboxypeptidases during maturation and germination of barley grain. Proc Natl Acad Sci USA 91: 8209–8213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi E, Komori N, Matoba T, Morita Y (1980) Purification and some properties of a carboxypeptidase in rice bran. Agric Biol Chem 44: 85–92 [Google Scholar]
- Endrizzi JA, Breddam K, Remington SJ (1994) 2.8-Å structure of yeast serine carboxypeptidase. Biochemistry 33: 11106–11120 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Galjart NJ, Gillemans N, Harris A, van der Horst GT, Verheijen FW, Galjaard H, d'Azzo A (1988) Expression of cDNA encoding the human “protective protein” associated with lysosomal beta-galactosidase and neuraminidase: homology to yeast proteases. Cell 54: 755–764 [DOI] [PubMed] [Google Scholar]
- Glässgen WE, Seitz HU (1992) Acylation of anthocyanins with hydroxycinnamic acids via 1-O-acylglucosides by protein preparations from cell cultures of Daucus carota L. Planta 186: 582–585 [DOI] [PubMed] [Google Scholar]
- Glässgen WE, Wray V, Strack D, Metzger JW, Seitz HU (1992) Anthocyanins from cell suspension cultures of Daucus carota. Phytochemistry 31: 1593–1601 [DOI] [PubMed] [Google Scholar]
- Graser G, Oldham NJ, Brown PD, Temp U, Gershenzon J (2001) The biosynthesis of benzoic acid glucosinolate esters in Arabidopsis thaliana. Phytochemistry 57: 23–32 [DOI] [PubMed] [Google Scholar]
- Gross GG (1983) Synthesis of mono-, di- and trigalloyl-β-D-glucose by β-glucogallin-dependent galloyltransferases from oak leaves. Z Naturforsch 38: 519–523 [Google Scholar]
- Harborne JB, Mayer AM, Bar-Nun N (1983) Identification of the major anthocyanin of carrot cells in tissue culture as cyanidin 3-(sinapoylxylosylglucosylgalactoside). Z Naturforsch 38: 1055–1056 [Google Scholar]
- Hause B, Meyer K, Viitanen PV, Chapple C, Strack D (2002) Immunolocalization of 1-O-sinapoylglucose:malate sinapoyltransferase in Arabidopsis thaliana. Planta 215: 26–32 [DOI] [PubMed] [Google Scholar]
- Hayashi R, Bai Y, Hata T (1975) Evidence for an essential histidine in carboxypeptidase Y. Reaction with the chloromethyl ketone derivative of benzyloxycarbonyl-L-phenylalanine. J Biol Chem 250: 5221–5226 [PubMed] [Google Scholar]
- Hayashi R, Moore S, Stein WH (1973) Serine at the active center of yeast carboxypeptidase. J Biol Chem 248: 8366–8369 [PubMed] [Google Scholar]
- Hopp W, Seitz HU (1987) The uptake of acylated anthocyanin into isolated vacuoles from a cell suspension culture of Daucus carota. Planta 170: 74–85 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, Bowles DJ (2002) Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid, phenotypic characterization of transgenic lines. Plant J 32: 573–583 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Lim E-K, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ (2001) Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Jones CG, Lycett GW, Tucker GA (1996) Protease inhibitor studies and cloning of a serine carboxypeptidase cDNA from germinating seeds of pea (Pisum sativum L.). Eur J Biochem 235: 574–578 [DOI] [PubMed] [Google Scholar]
- Kim Y, Hayashi R (1983) Properties of a serine carboxypeptidase in cauliflower. Agric Biol Chem 47: 2655–2667 [Google Scholar]
- Kowalczyk S, Jakubowska A, Zielinska E, Bandurski RS (2003) Bifunctional indole-3-acetyl transferase catalyses synthesis and hydrolysis of indole-3-acetyl-myo-inositol in immature endosperm of Zea mays. Physiol Plant 119: 165–174 [Google Scholar]
- Landry LG, Chapple CCS, Last R (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12: 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AX, Steffens JC (2000) An acyltransferase catalyzing the formation of diacylglucose is a serine carboxypeptidase-like protein. Proc Natl Acad Sci USA 97: 6902–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC (2001. a) BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 5916–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim E-K, Bowles DJ (2001. b) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Liao D, Breddam K, Sweet RM, Bullock T, Remington SJ (1992) Refined atomic model of wheat serine carboxypeptidase II at 2.2-Å resolution. Biochemistry 31: 9796–9812 [DOI] [PubMed] [Google Scholar]
- Liao D, Remington SJ (1990) Structure of wheat serine carboxypeptidase II at 3.5-Å resolution. A new class of serine proteinase. J Biol Chem 265: 6528–6531 [DOI] [PubMed] [Google Scholar]
- Lim E-K, Li Y, Parr A, Jackson R, Ashford DA, Bowles DJ (2001) Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J Biol Chem 276: 4344–4349 [DOI] [PubMed] [Google Scholar]
- Linscheid M, Wendisch D, Strack D (1980) The structures of sinapic acid esters and their metabolism in cotyledons of Raphanus sativus. Z Naturforsch 35: 907–914 [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G (2002) Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol 50: 309–332 [DOI] [PubMed] [Google Scholar]
- Marrs KA, Alfenito MR, Lloyd AM, Walbot V (1995) A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375: 397–400 [DOI] [PubMed] [Google Scholar]
- Marshall RD (1972) Glycoproteins. Annu Rev Biochem 41: 673–702 [DOI] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J (1997) Jasmonate is essential for insect defense. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Cusumano JC, Somerville C, Chapple CCS (1996) Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA 93: 6869–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalczuk L, Bandurski RS (1982) Enzymic synthesis of 1-O-indol-3-ylacetyl-β-D-glucose and indol-3-ylacetyl-myo-inositol. Biochem J 207: 273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkowski C, Strack D (2004) Serine carboxypeptidase-like acyltransferases. Phytochemistry 65: 517–524 [DOI] [PubMed] [Google Scholar]
- Mizutani M, Ward E, Ohta D (1998) Cytochrome P450 superfamily in Arabidopsis thaliana: isolation of cDNAs, differential expression, and RFLP mapping of multiple cytochromes P450. Plant Mol Biol 37: 39–52 [DOI] [PubMed] [Google Scholar]
- Moura DS, Bergey DR, Ryan CA (2001) Characterization and localization of a wound-inducible type I serine-carboxypeptidase from leaves of tomato plants (Lycopersicon esculentum Mill.). Planta 212: 222–230 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Suzuki H, Nishino T (2003) Anthocyanin acyltransferases: specificities, mechanism, phylogenetics, and applications. J Mol Catal 23b: 117–132 [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, Halkier BA (2003) CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol 133: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al (1992) The α/β hydrolase fold. Protein Eng 5: 197–211 [DOI] [PubMed] [Google Scholar]
- Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Kangasjarvi J (2000) Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signalling pathways in regulating superoxide-dependent cell death. Plant Cell 12: 1849–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM (1996) An application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Ramos C, Winther JR, Kiellandbrand MC (1994) Requirement of the propeptide for in-vivo formation of active yeast carboxypeptidase Y. J Biol Chem 269: 7006–7012 [PubMed] [Google Scholar]
- Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR (2000) Jasmonic acid signalling modulates ozone-induced hypersensitive cell death. Plant Cell 12: 1633–164611006337 [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J (2000) Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol 42: 93–113 [PubMed] [Google Scholar]
- Reichelt M, Brown PD, Schneider B, Oldham NJ, Stauber E, Tokuhisa J, Kliebenstein DJ, Mitchell-Olds T, Gershenzon J (2002) Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 59: 663–671 [DOI] [PubMed] [Google Scholar]
- Sharma V, Strack D (1985) Vacuolar localization of 1-sinapoylglucose:L-malate sinapoyltransferase in protoplasts from cotyledons of Raphanus sativus. Planta 163: 563–568 [DOI] [PubMed] [Google Scholar]
- Shirley AM, Chapple C (2003) Biochemical characterization of sinapoylglucose:choline sinapoyltransferase, a serine carboxypeptidase-like protein that functions as an acyltransferase in plant secondary metabolism. J Biol Chem 278: 19870–19877 [DOI] [PubMed] [Google Scholar]
- Shirley AM, McMichael CM, Chapple C (2001) The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose:choline sinapoyltransferase. Plant J 28: 83–94 [DOI] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R (1982) Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell 30: 439–448 [DOI] [PubMed] [Google Scholar]
- Strack D (1981) Sinapine as a supply of choline for the biosynthesis of phosphatidylcholine in Raphanus sativus. Z Naturforsch 36: 215–221 [Google Scholar]
- Strack D, Nurmann G, Sachs G (1980) Sinapine esterase. I. Characterization of sinapine esterase from cotyledons of Raphanus sativus. Z Naturforsch 35: 963–966 [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-Myb gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Sawada S, Watanabe K, Nagae S, Yamaguchi MA, Nakayama T, Nishino T (2004) Identification and characterization of a novel anthocyanin malonyltransferase from scarlet sage (Salvia splendens) flowers: an enzyme that is phylogenetically separated from other anthocyanin acyltransferases. Plant J 38: 994–1003 [DOI] [PubMed] [Google Scholar]
- Taipalensuu J, Andreasson E, Eriksson S, Rask L (1997) Regulation of the wound-induced myrosinase-associated protein transcript in Brassica napus plants. Eur J Biochem 247: 963–971 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Tierens KFMJ, Broekaert WF (1999) Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls LA, Hunter CP, Rothman JH, Stevens TH (1987) Protein sorting in yeast: the localization determinant of yeast vacuolar carboxypeptidase Y resides in the propeptide. Cell 48: 887–897 [DOI] [PubMed] [Google Scholar]
- Villegas RJA, Kojima M (1986) Purification and characterization of hydroxycinnamoyl D-glucose quinate hydroxycinnamoyl transferase in the root of sweet potato, Ipomoea batatas Lam. J Biol Chem 261: 8729–8733 [PubMed] [Google Scholar]
- Wajant H, Mundry K, Pfizenmaier K (1994) Molecular cloning of hydroxynitrile lyase from Sorghum bicolor (L.). Homologies to serine carboxypeptidases. Plant Mol Biol 26: 735–746 [DOI] [PubMed] [Google Scholar]
- Winther JR, Sorensen P (1991) Propeptide of carboxypeptidase Y provides a chaperone-like function as well as inhibition of the enzymatic activity. Proc Natl Acad Sci USA 88: 9330–9334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7: 263–270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.