Abstract
Targeted stable transduction of specific cells is a highly desirable goal for gene therapy applications. We report an efficient and broadly applicable approach for targeting retroviral vectors to specific cells. We find that the envelope of the alphavirus Sindbis virus can pseudotype human immunodeficiency virus type 1- and murine leukemia virus-based retroviral vectors. When modified to contain the Fc-binding domain of protein A, this envelope gives a significant enhancement in specificity in combination with antibodies specific for HLA and CD4 relative to that without antibody. Unlike previous targeting strategies for retroviral transduction, the virus titers are relatively high and stable and can be further increased by ultracentrifugation. This study provides proof of principle for a targeting strategy that would be generally useful for many gene therapy applications.
Efficient targeting of specific cells to achieve stable transduction has been attempted by various strategies. Inserting ligands or single-chain antibodies into the retroviral receptor binding envelope subunit has been the most common approach used to alter and/or restrict the host range of retroviral vectors (1, 5, 7, 8, 13, 14, 17, 24–26). Bridging virus vector and cell by antibodies or ligands is another approach (3, 20). In general, most strategies have suffered from inconsistent specificity and low viral titers as a result of modification of the retroviral envelope (1, 5, 9, 13, 17, 24–26). The modified envelope proteins appear to have specific binding activity but have low fusion activity (14, 28), resulting in inefficient entry into cells.
The alphavirus Sindbis virus encodes two transmembrane envelope proteins, E1 and E2. E2 is responsible for receptor binding; E1 is responsible for pH-dependent fusion. Unlike retroviruses, the Sindbis virus fusogenic E1 protein can fuse to cells independently of the receptor binding E2 protein (23). Recently, vectors based upon the Sindbis virus RNA genome were constructed whereby the Sindbis virus E2 envelope protein was modified by insertion of an Fc-binding portion (ZZ domain) (12) of protein A (6, 18). These Sindbis virus vectors would bind to and enter cells bearing specific cell surface antigens only in the presence of the appropriate monoclonal antibody (MAb). However, as a lytic RNA virus, Sindbis virus is not suitable for applications requiring stable transduction (6, 21). We tested the possibility that human immunodeficiency virus type 1 (HIV-1)-based vectors could potentially be pseudotyped with Sindbis virus envelope, thereby conferring the targeting properties of the modified Sindbis virus envelope to the HIV-1 vector.
MATERIALS AND METHODS
Plasmid construction.
The expression vector of Sindbis virus envelope protein, plntron SINDBIS, was made by cloning Sindbis virus envelope into pHCMV G (27), replacing the vesicular stomatitis virus G protein. The Sindbis virus envelope fragment was derived from the plasmid TOTO 2000 (kindly provided by Henry Huang). The envelope region of TOTO 2000 was derived from TOTO 1000 (19). The expression vector of the fusion protein, plntron ZZ SINDBIS, was derived from plntron SINDBIS and pEZZ 18 (Pharmacia Biotech). First, a BstEII site was introduced into plntron SINDBIS between the codons for amino acids 71 and 74 of the E2 glycoprotein as described before (18) by PCR-based mutagenesis, and the resulting construct was designated plntron Bst SINDBIS. A sequence corresponding to the region of protein A containing two synthetic immunoglobulin G (IgG) binding domains (ZZ domain) was amplified by PCR using pEZZ 18 as the template and cloned into the BstEII site of plntron Bst SINDBIS, and the resulting construct was designated plntron ZZ SINDBIS.
Cell lines.
HLtat CD4 cells were generated by cotransfection of CDM8CD4 (4) and pPUR (Clontech) into HLtat cells (NIH AIDS Research and Reference Reagent Program). Briefly, HLtat cells were plated the day prior to transfection at a density of 2 × 106/well in a six-well plate. Cells were cotransfected with 20 μg of CDM8CD4 and 1 μg of pPUR using Lipofectin (Gibco BRL, Grand Island, N.Y.) according to the manufacturer's instructions. Two days posttransfection, cells were trypsinized and cultured in 96-well plates at a density of 0.1 cells/well with Dulbecco modified Eagle medium (DMEM) supplemented with 10% calf serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 1 mg of puromycin/ml. Two weeks after the transfection, the puromycin-resistant clones were stained with anti-CD4–fluorescein isothiocyanate at 4°C for 30 min and washed twice with fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline plus 2% fetal calf serum). CD4 expression was analyzed by flow cytometry, and the clone with the highest expression of CD4 (clone 18) was designated HLtat CD4 cells. HLtat cells and HLtat CD4 cells were maintained in DMEM with 10% fetal calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml.
Vector production.
SINDBIS- or ZZ SINDBIS-pseudotyped lentivirus vectors expressing the luciferase reporter gene were produced by calcium phosphate-mediated transfection of 293T cells as described previously (2). 293T cells (2 × 107) were transfected with 12.5 μg of pCMVdR8.2DVPR (2), 12.5 μg of the lentivirus reporter vector pHRCMVLuc (16), and 5 μg of the envelope protein expression vector (plntron SINDBIS or plntron ZZ SINDBIS). For generation of ZZ SINDBIS-pseudotyped lentivirus vectors expressing the enhanced green fluorescent protein (EGFP) reporter gene, 5 μg of plntron SINDBIS ZZ, 12.5 μg of pCMVdR8.2DVPR (2), and 12.5 μg of the lentivirus reporter vector pHRCMV EGFP (10) or pCS-Rh-MLV-E (10) were used. For the generation of ZZ SINDBIS-pseudotyped murine leukemia virus vector, 5 μg of plntron SINDBIS ZZ, 12.5 μg of pSVψ−env−MLV (10), and 12.5 μg of pSRαLEGFP (10) were used. 293T cells were cultured in DMEM with 10% ultra-low-IgG fetal bovine serum (Gibco BRL), 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Supernatants were collected on days 2 and 3 posttransfection and filtered through a 0.22-μm-pore-size filter; stocks were maintained at −70°C.
Vector concentration was achieved by ultracentrifugation at 40,000 × g for 90 min at 4°C. The pellet was resuspended in AIM-V medium (Gibco BRL). The EGFP transduction units of ZZ SINDBIS-pseudotyped HIV-1 vector and murine leukemia virus vector were titrated on 293T cells. 293T cells (2 × 105) were infected with 200 μl of HRCMVEGFP (ZZ SINDBIS) or 100 μl of SRαLEGFP (ZZ SINDBIS) (100× concentrated) with anti-human leukocyte antigen class I molecules A, B, and C (HLA ABC) (1 μg/ml) for 2 h at 37°C in 5% CO2. The virus was removed, and cells were cultured in DMEM with 10% calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Three days postinfection, the cells were analyzed for EGFP expression by flow cytometry.
MAbs.
Anti-HLA ABC was purchased from Sigma (St. Louis, Mo.). Anti-CD4 used for targeting was produced by the hybridoma OKT4 (ATCC CRL8002) and purified by protein A (Pierce, Rockford, Ill.). Conjugated anti-CD4 antibodies (phycoerythrin [PE] or fluorescein isothiocyanate) that recognize a CD4 epitope different from OKT4 were purchased from Becton Dickinson (San Jose, Calif.). For targeting, antibodies were selected from the IgG subclasses that are known to bind protein A.
Infection by luciferase vectors.
SINDBIS or ZZ SINDBIS-pseudotyped HIV-1 vectors HRCMVLuc (SINDBIS) and HRCMVLuc (ZZ SINDBIS) were incubated with anti-HLA ABC at various concentrations for 1 h on ice prior to the infection. 293T cells (2 × 105) or BHK cells (6 × 104) were infected with these vectors with or without MAb for 2 h at 37°C with 5% CO2. The vectors were subsequently removed and replaced with 1 ml of DMEM with 10% calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Four days postinfection, 293T cells (1 × 105) or BHK cells (5 × 105) were lysed in 200 μl of cell culture lysis reagent (Promega Corp., Madison, Wis.). The lysate was centrifuged to remove nuclei. The supernatant (20 μl) was analyzed for luciferase activity as described before (15).
Infection of mixed culture of HLtat cells and HLtat CD4 cells.
HRCMVEGFP (ZZ SINDBIS) was incubated with anti-CD4 (1 μg/ml) or anti-HLA (1 μg/ml) for 1 h on ice prior to infection. The mixed culture of HLtat cells and HLtat CD4 cells was infected with HRCMVEGFP (ZZ SINDBIS) with or without the MAb for 2 h at 37°C with 5% CO2. The virus was subsequently removed and replaced with 1 ml of fresh medium.
The mixture of HLtat cells and HLtat CD4 cells was detached by 2 mM EDTA–phosphate-buffered saline 3 days postinfection. The cells (2 × 105) were stained with 50 μl of anti-CD4–PE (5 μg/ml) for 30 min on ice. The cells were then washed with FACS buffer and analyzed by flow cytometry.
Infection of PBMC.
Preparation of peripheral blood mononuclear cells (PBMC) and stimulation of them by anti-CD3 MAb and anti-CD28 MAb were done as previously described (10). In brief, PBMC were activated by anti-CD3 and anti-CD28 antibodies for 2 days. After activation by the antibodies, PBMC were cultured for 1 day in growth medium (RPMI 1640 supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 1 mg of interleukin 2/ml) in the absence of the antibodies.
CS-Rh-MLV-E (ZZ SINDBIS) was incubated with anti-CD4 (10 μg/ml) or anti-HLA (10 μg/ml) for 1 h on ice prior to the infection. CD3- and CD28-activated PBMC (105) were infected with the vector with or without MAb for 2 h at 37°C with 5% CO2. The vector was subsequently removed and replaced with 1 ml of growth medium. Three days postinfection, the PBMC (2 × 105) were stained with 50 μl of anti-CD4–PE (5 μg/ml) for 30 min on ice. The cells were then washed with FACS buffer and analyzed by flow cytometry.
Immunoblot assay.
HIV vector (HRCMVEGFP) with no envelope, pseudotyped with Sindbis virus envelope, or pseudotyped with ZZ SINDBIS was concentrated by ultracentrifugation and resuspended in electrophoresis loading buffer (20% glycerol, 10% 2-mercaptoethanol, 4% sodium dodecyl sulfate [SDS] 125 mM Tris-HCl [pH 6.8], 0.02% bromophenol blue) and boiled for 5 min. The amount of viral sample was normalized to the amount of HIV p24 (90 ng of p24/sample). The samples were subjected to electrophoresis through an SDS–10% polyacrylamide gel as described previously (15). Immunoblot analysis was performed with anti-Sindbis virus ascitic fluid (ATCC VR-1248) and horseradish peroxidase-conjugated anti-mouse antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.). The protein bands were visualized by enhanced chemiluminescence (Amersham, Piscataway, N.J.).
Flow cytometry.
Flow cytometry was performed with a FACScan flow cytometer (Becton Dickinson). The data were analyzed with the Cell Quest program (Becton Dickinson).
RESULTS AND DISCUSSION
We first tested whether the wild-type Sindbis virus envelope would pseudotype with HIV-1. The Sindbis virus envelope proteins were expressed in a DNA-based vector using the cytomegalovirus immediate early promoter (Fig. 1). We produced HIV-1 vectors bearing luciferase reporter genes encoding wild-type Sindbis virus envelope [HRCMVLuc (SINDBIS)] via cotransfection of 293T cells. The resulting virus was used to infect human 293T and hamster BHK cells (Fig. 2). These results demonstrate that the wild-type Sindbis virus envelope forms pseudotypes with HIV-1 efficiently.
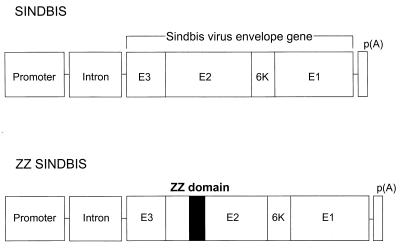
FIG. 1.
Schematic representation of Sindbis virus envelope (SINDBIS) and chimeric envelope between SINDBIS and the IgG binding domain of protein A (ZZ SINDBIS). Sindbis virus envelope proteins (E3, E2, 6K, and E1) were cloned into an expression vector derived from pHCMV G (27). The envelope protein is first synthesized as a polypeptide and subsequently cleaved by cellular proteases to generate the E3, E2, 6K, and E1 proteins. E1 and E2 are incorporated into virions as envelope proteins. E3 and 6K are leader sequences for E2 and E1, respectively (22). ZZ SINDBIS is a modified Sindbis virus envelope in which the IgG binding domain of protein A (ZZ) was inserted in the E2 region (see Material and Methods). ZZ domain was inserted between amino acids 71 and 74 of E2. The promoter is the cytomegalovirus immediate early promoter, and the intron is intron II of the rabbit β-globin gene. p(A), polyadenylation signal.
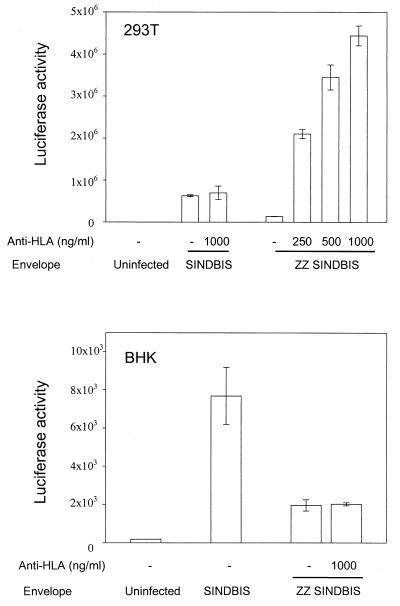
FIG. 2.
Anti-HLA MAb mediates gene transduction by ZZ SINDBIS pseudotyped HIV-1 vector. 293T cells (HLA antigen positive) and BHK cells (HLA antigen negative) were infected with HRCMVLuc (SINDBIS) and HRCMVLuc (ZZ SINDBIS). The p24 levels of HRCMVLuc (SINDBIS) and HRCMVLuc (ZZ SINDBIS) were 541 and 1,843 ng/ml, respectively. 293T cells (2 × 105) and BHK cells (6 × 104) were infected with 200 μl of each vector for 2 h with or without anti-HLA (1 μg/ml) as described in the text. Four days after infection, the cells were lysed by 200 μl of 1× luciferase lysis buffer (Promega), and 10 μl of each lysate was subjected to a luciferase assay. The luciferase activity of uninfected 293T cells was 1.8 × 102 relative light units. The results are the averages of three independent infections performed in parallel.
We modified the Sindbis envelope in an analogous fashion to that previously described (18) by inserting the IgG binding region of protein A (ZZ domain) into the E2 envelope protein at the receptor binding domain (Fig. 1). In the absence of antibody, the HIV-1 vector (HRCMVLuc) bearing the modified Sindbis virus envelope (ZZ SINDBIS) showed substantially lower levels of luciferase activity than wild-type Sindbis virus envelope, although still higher than background levels. In the presence of antibodies directed against HLA ABC, the levels of infection increased up to over a 30-fold enhancement over those obtained without antibody and to levels comparable to those obtained with wild-type Sindbis envelope (Fig. 2). As a control, no augmentation in infectivity was observed with BHK rodent cells, which do not bear HLA antigens.
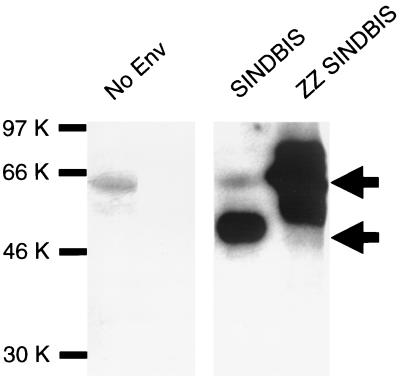
We investigated incorporation of Sindbis virus envelope and ZZ SINDBIS into the HIV-1 vector. Pseudotyped virions were subjected to Western blotting and probed using a mouse anti-Sindbis virus antibody that detects E2 protein (Fig. 3). This polyclonal antibody was generated by immunization of mice with Sindbis virus (ATCC VR-1248). HRCMVEGFP (SINDBIS) showed a 50- to 55-kDa band corresponding to the E2 protein, and HRCMVEGFP (ZZ SINDBIS) showed a major band around 65 kDa, which is the estimated molecular mass of the chimeric protein (Fig. 3). Consistent with a previous report (18), the E1 protein is not detected using this anti-Sindbis virus antibody.
FIG. 3.
Detection of Sindbis virus envelope and ZZ SINDBIS on virions. Ultracentrifuged and 100-fold-concentrated samples of HIV vector (HRCMVEGFP) pseudotyped by SINDBIS, ZZ SINDBIS, or no envelope (Env) were lysed and subjected to SDS-polyacrylamide gel electrophoresis and Western blotting as described in Materials and Methods. The amount of virus for each sample was normalized by the amount of HIV p24 antigen (90 ng/sample). Viral proteins were detected with anti-Sindbis virus mouse immune ascitic fluid (18). The bands around 60 kDa seen in the no-envelope and SINDBIS samples are nonspecific bands. The upper and lower arrows show bands corresponding to the chimeric protein and the E2 protein, respectively.
Of practical importance for gene therapy applications, the titers of HIV-1 vector bearing ZZ SINDBIS [HRCMVLuc (ZZ SINDBIS)] were stable over at least two freeze-thaw cycles in which the vector was frozen at −70°C for 2 to 4 h and thawed in a 37°C water bath. Virus recovery was 103% and 115% of titers of untreated virus after one and two freeze- thaw cycles, respectively. These viruses could also be concentrated at least 100-fold by ultracentrifugation without loss of infectivity: the titer of 2 μl of 100×-concentrated virus was 2.2 × 107 EGFP transduction units/ml, or 96% of the titer of unconcentrated virus (2.3 × 105 EGFP transduction units/ml).
Targeting efficiencies were similar when HRCMVEGFP (ZZ SINDBIS) was concentrated after incubation of antibody with virus (30 min at 25°C) and when it was concentrated before addition of antibody (data not shown). As expected, no targeting was observed when wild-type Sindbis virus envelope was concentrated in the presence of antibody. This result also demonstrates stable binding of antibody to the HRCMVEGFP (ZZ SINDBIS) virions.
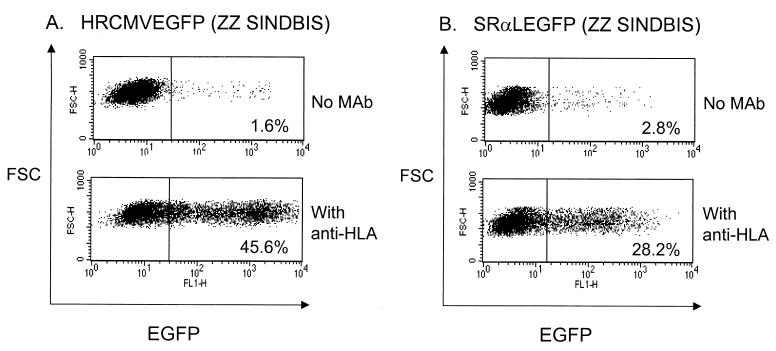
More quantitative data on efficiencies of transduction were derived by the use of an HIV-1 vector bearing the EGFP gene as a reporter gene that allows detection and enumeration by flow cytometry. Virus stocks in the absence of concentration were about 1 × 105 to 3 × 105 EGFP+ units/ml on 293T cells [HRCMVEGFP (ZZ SINDBIS), in the presence of anti-HLA antibody and as determined by flow cytometry]. Consistent with our initial results utilizing the luciferase gene as a reporter gene, the EGFP-based HIV-1 vector specifically infected human cells that bear HLA molecules in the presence of anti-HLA ABC antibodies (Fig. 4A). Since the efficiency of pseudotyping depends upon the retroviral species, we also tested the ability of the Sindbis virus envelope to pseudotype with murine retroviruses. We utilized the murine retroviral vector SRαLEGFP to assess targeting to HLA. We found that SRαLEGFP (ZZ SINDBIS) transduced human cells efficiently only in the presence of anti-HLA (Fig. 4B). With both HIV-1 and murine vectors, addition of the reverse transcriptase inhibitors zidovudine and nevirapine blocked the infection (data not shown), substantiating the idea that expression of EGFP is a result of a normal retroviral infection process and not due to reported “pseudotransduction” of EGFP protein derived from producer cells (11).
FIG. 4.
HIV-1 vector (A) and MLV vector (B) can be pseudotyped with ZZ SINDBIS. 293T cells (2 × 105) were infected with 200 μl of HRCMVEGFP (ZZ SINDBIS) or 100 μl of SRαLEGFP (ZZ SINDBIS) (100× concentrated) with or without anti-HLA (1 μg/ml). Three days after infection, EGFP expression was analyzed by flow cytometry. The titers of HRCMVEGFP (ZZ SINDBIS) and SRαLEGFP (ZZ SINDBIS) (100× concentrated) with 1 μg of anti-HLA are 4.5 × 105 and 2.8 × 105 EGFP transduction units/ml, respectively.
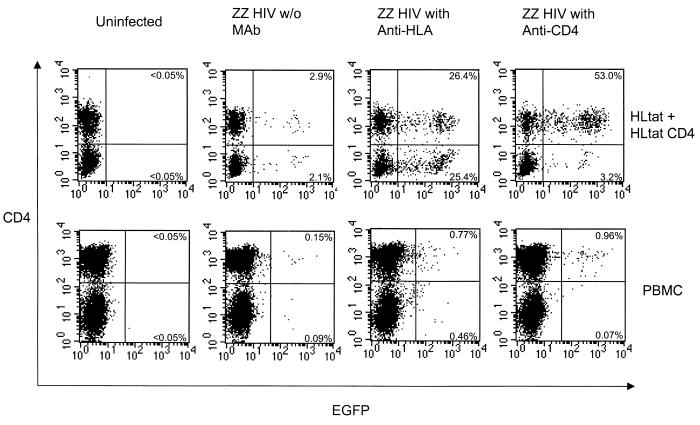
We demonstrated targeting in heterogeneous cell populations. A mixed culture of HLtat cells and HLtat CD4 cells was infected with HRCMVEGFP (ZZ SINDBIS) either alone or with anti-CD4 antibody (Fig. 5). Virus with anti-CD4 antibody resulted in preferential infection of HLtat CD4 cells. As a control, virus treated with antibody directed against HLA ABC present on both HLtat cells and HLtat CD4 cells allowed efficient transduction but did not confer preferential transduction of either population.
FIG. 5.
MAbs mediate gene transduction of a heterogeneous cell population by ZZ SINDBIS-pseudotyped HIV vector (ZZ HIV). A mixture of HLtat cells (1.5 × 104) and HLtat CD4 cells (1.5 × 104) was seeded in the same well a day before infection. They were infected with HRCMVEGFP (ZZ SINDBIS) (200 μl, 5 × 104 EGFP transduction units) either alone, with anti-HLA (1 μg/ml), or with anti-CD4 (1 μg/ml) for 2 h. Three days after infection, cells (5 × 105) were stained with anti-CD4–PE. The cells were then analyzed for CD4 and EGFP expression by flow cytometry. CD3- and 28-activated PBMC (1 × 105) were infected with CS-Rh-MLV-E (ZZ SINDBIS) (200 μl, 2 × 105 EGFP transduction units) either alone, with anti-HLA (10 μg/ml), or with anti-CD4 (10 μg/ml). Three days after infection, cells (5 × 105) were stained with anti-CD4–PE and analyzed by flow cytometry. The percentage of EGFP-positive cells in the CD4 positive population was calculated as the number of events in the upper right quadrant divided by the total number of events in the right and left upper quadrants. For the CD4-negative population, the percentage of EGFP-positive cells was calculated by dividing the number of events in the lower right quadrant by the number of events in the right and left lower quadrants.
Human primary PBMC consist of both CD4+ and CD4− subpopulations. We determined whether we could specifically target the CD4-positive population of lymphocytes by using an anti-CD4 MAb (Fig. 5). The HIV vector (CS-Rh-MLVE) pseudotyped by ZZ SINDBIS pretreated with anti-CD4 antibody resulted in preferential infection of the CD4-positive subpopulation. As a control, antibody directed against HLA ABC present on both CD4+ and CD4− subpopulations did not confer preferential transduction of either CD4+ or CD4− subpopulations of cells.
The approach described here for specific targeting of cells by retroviral vector transduction overcomes many of the limitations of previous targeting strategies. The preparation of targeting vector is not limited by the introduction of modifications into native retroviral envelopes that usually result in substantial decreases in infectivity. This strategy is applicable to both lentivirus and murine retroviral vectors. The Sindbis virus pseudotyped virions can be produced at relatively high titers and are also highly stable to ultracentrifugation and freeze-thaw cycles, thus facilitating the enhancement of titers and storage. Finally, the approach described here should in theory be generally applicable to any cell surface molecule for which there are specific reagents that bind. MAbs were used here; however, other applications could involve ligands for specific receptors, peptides, or nucleic acid reagents selected for specific binding properties. Depending on the particular application, the Sindbis virus E2 protein could be modified to directly encode the ligand (21) and/or include other affinity reagents, such as streptavidin. Such reagents would be more amenable to in vivo applications where the presence of plasma antibodies would complicate use of the chimeric ZZ domain.
The development of specific targeting reagents for stable gene transfer raises a number of possible applications which were not previously available. For example, current hematopoietic stem cell gene transfer experiments utilize amphotropic and vesicular stomatitis virus G-pseudotyped murine or lentiviral vectors to infect hematopoietic stem cells. These studies require technically demanding and expensive procedures to purify CD34+ cells and transduce these cells ex vivo. By targeting CD34, hematopoietic stem cells can in principle be transduced ex vivo without extensive purification. One of the primary advantages of lentiviral vectors is to allow direct in situ transduction of cells in vivo (16). In combination with the appropriate choice of targeting reagents, lentiviral vectors potentially provide a means for transduction of specific cells in vivo. One can envision a number of applications, including targeting of specific cells within tissues to correct metabolic defects and targeting of specific pathologic cells in infectious diseases or cancer. Future studies will further enhance the capabilities of this model system.
ACKNOWLEDGMENTS
We thank Dong Sung An, Saskia Boisot, and Scott Kitchen for technical support, Takao Masuda for advice, and Liz Duarte and Rosie Taweesup for manuscript preparation. We thank Henry Huang and Akira Nakanishi for providing reagents.
This work was supported by NIH grants AI399975-01 and AI36555 and the Japanese Foundation for AIDS Prevention. S.K.-P.K. is a Research Fellow of the National Cancer Institute of Canada supported with funds provided by the Terry Fox Run.
REFERENCES
- 1.Ager S, Nilson B H, Morling F J, Peng K W, Cosset F L, Russell S J. Retroviral display of antibody fragments; interdomain spacing strongly influences vector infectivity. Hum Gene Ther. 1996;7:2157–2164. doi: 10.1089/hum.1996.7.17-2157. [DOI] [PubMed] [Google Scholar]
- 2.An D S, Morizono K, Li Q X, Mao S H, Lu S, Chen I S. An inducible human immunodeficiency virus type 1 (HIV-1) vector which effectively suppresses HIV-1 replication. J Virol. 1999;73:7671–7677. doi: 10.1128/jvi.73.9.7671-7677.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerger A L, Snitkovsky S, Young J A. Retroviral vectors preloaded with a viral receptor-ligand bridge protein are targeted to specific cell types. Proc Natl Acad Sci USA. 1999;96:9867–9872. doi: 10.1073/pnas.96.17.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camerini D, Seed B. A CD4 domain important for HIV-mediated syncytium formation lies outside the virus binding site. Cell. 1990;60:747–754. doi: 10.1016/0092-8674(90)90089-w. [DOI] [PubMed] [Google Scholar]
- 5.Han X, Kasahara N, Kan Y W. Ligand-directed retroviral targeting of human breast cancer cells. Proc Natl Acad Sci USA. 1995;92:9747–9751. doi: 10.1073/pnas.92.21.9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iijima Y, Ohno K, Ikeda H, Sawai K, Levin B, Meruelo D. Cell-specific targeting of a thymidine kinase/ganciclovir gene therapy system using a recombinant Sindbis virus vector. Int J Cancer. 1999;80:110–118. doi: 10.1002/(sici)1097-0215(19990105)80:1<110::aid-ijc21>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 7.Jiang A, Chu T H, Nocken F, Cichutek K, Dornburg R. Cell-type-specific gene transfer into human cells with retroviral vectors that display single-chain antibodies. J Virol. 1998;72:10148–10156. doi: 10.1128/jvi.72.12.10148-10156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang A, Dornburg R. In vivo cell type-specific gene delivery with retroviral vectors that display single chain antibodies. Gene Ther. 1999;6:1982–1987. doi: 10.1038/sj.gt.3301043. [DOI] [PubMed] [Google Scholar]
- 9.Kasahara N, Dozy A M, Kan Y W. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science. 1994;266:1373–1376. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 10.Kung S, An D S, Chen I S Y. A murine leukemia virus (MuLV) long terminal repeat derived from rhesus macaques in the context of a lentivirus vector and MuLV Gag sequence results in high-level gene expression in human T lymphocytes. J Virol. 2000;74:3668–3681. doi: 10.1128/jvi.74.8.3668-3681.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M L, Winther B L, Kay M A. Pseudotransduction of hepatocytes by using concentrated pseudotyped vesicular stomatitis virus G glycoprotein (VSV-G)-Moloney murine leukemia virus-derived retrovirus vectors: comparison of VSV-G and amphotropic vectors for hepatic gene transfer. J Virol. 1996;70:2497–2502. doi: 10.1128/jvi.70.4.2497-2502.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowenadler B, Jansson B, Paleus S, Holmgren E, Nilsson B, Moks T, Palm G, Josephson S, Philipson L, Uhlen M. A gene fusion system for generating antibodies against short peptides. Gene. 1987;58:87–97. doi: 10.1016/0378-1119(87)90032-1. [DOI] [PubMed] [Google Scholar]
- 13.Marin M, Noel D, Valsesia-Wittman S, Brockly F, Etienne-Julan M, Russell S, Cosset F L, Piechaczyk M. Targeted infection of human cells via major histocompatibility complex class I molecules by Moloney murine leukemia virus-derived viruses displaying single-chain antibody fragment-envelope fusion proteins. J Virol. 1996;70:2957–2962. doi: 10.1128/jvi.70.5.2957-2962.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin F, Neil S, Kupsch J, Maurice M, Cosset F, Collins M. Retrovirus targeting by tropism restriction to melanoma cells. J Virol. 1999;73:6923–6929. doi: 10.1128/jvi.73.8.6923-6929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda T, Planelles V, Krogstad P, Chen I S. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 17.Nilson B H, Morling F J, Cosset F L, Russell S J. Targeting of retroviral vectors through protease-substrate interactions. Gene Ther. 1996;3:280–286. [PubMed] [Google Scholar]
- 18.Ohno K, Sawai K, Iijima Y, Levin B, Meruelo D. Cell-specific targeting of Sindbis virus vectors displaying IgG-binding domains of protein A. Nat Biotechnol. 1997;15:763–767. doi: 10.1038/nbt0897-763. [DOI] [PubMed] [Google Scholar]
- 19.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux P, Jeanteur P, Piechaczyk M. A versatile and potentially general approach to the targeting of specific cell types by retroviruses: application to the infection of human cells by means of major histocompatibility complex class I and class II antigens by mouse ecotropic murine leukemia virus-derived viruses. Proc Natl Acad Sci USA. 1989;86:9079–9083. doi: 10.1073/pnas.86.23.9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawai K, Meruelo D. Cell-specific transfection of choriocarcinoma cells by using Sindbis virus hCG expressing chimeric vector. Biochem Biophys Res Commun. 1998;248:315–323. doi: 10.1006/bbrc.1998.8922. [DOI] [PubMed] [Google Scholar]
- 22.Schlesinger S, Schlesinger M J. Togaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 523–539. [Google Scholar]
- 23.Smit J M, Bittman R, Wilschut J. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J Virol. 1999;73:8476–8484. doi: 10.1128/jvi.73.10.8476-8484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somia N V, Zoppe M, Verma I M. Generation of targeted retroviral vectors by using single-chain variable fragment: an approach to in vivo gene delivery. Proc Natl Acad Sci USA. 1995;92:7570–7574. doi: 10.1073/pnas.92.16.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valsesia-Wittmann S, Drynda A, Deleage G, Aumailley M, Heard J M, Danos O, Verdier G, Cosset F L. Modifications in the binding domain of avian retrovirus envelope protein to redirect the host range of retroviral vectors. J Virol. 1994;68:4609–4619. doi: 10.1128/jvi.68.7.4609-4619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valsesia-Wittmann S, Morling F J, Nilson B H, Takeuchi Y, Russell S J, Cosset F L. Improvement of retroviral retargeting by using amino acid spacers between an additional binding domain and the N terminus of Moloney murine leukemia virus SU. J Virol. 1996;70:2059–2064. doi: 10.1128/jvi.70.3.2059-2064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yee J K, Friedmann T, Burns J C. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43(Part A):99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Zhu L, Lee S, Li L, Chang E, Soong N W, Douer D, Anderson W F. Identification of the block in targeted retroviral-mediated gene transfer. Proc Natl Acad Sci USA. 1999;96:4005–4010. doi: 10.1073/pnas.96.7.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]