Abstract
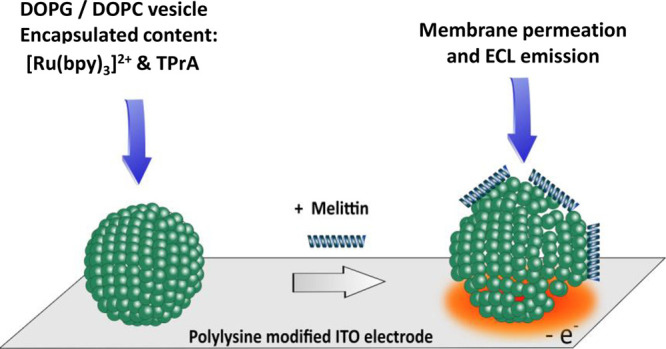
The permeabilization of liposomes by melittin, an antimicrobial peptide (AMP), has been studied by an electrochemiluminescence (ECL) imaging strategy. The methodology consisted first of encapsulating ECL reagents in sealed giant asymmetrical liposomes (100 μm in diameter) made of DOPG/DOPC phospholipids (i.e., 1,2-dioleoyl-sn-glycerol-3-phospho-(1′-rac-glycerol) sodium salt/1,2-dioleolyl-sn-glycero-3-phosphocholine). Then liposomes were placed on an indium tin oxide electrode coated with poly-l-lysine to avoid any membrane poration/permeabilization through polarization of the electrode surface. Finally, the addition of melittin (from 10 μM to 100 nM in concentration) enabled the permeabilization of the lipid membrane followed by the liposome content release and subsequent light generation through the ECL reagents oxidation processes. Interestingly, at a melittin concentration of 10 μM, two successive leakages occurring on the same liposome could be imaged. Combination of ECL and photoluminescence imaging allowed comprehensive monitoring of the permeabilization and content release of a single liposome. This ECL imaging approach opens interesting perspectives to characterize the instant release of vesicle content upon permeabilization by AMPs or other membrane-active species.
Keywords: Electrochemiluminescence, Liposome, ECL imaging, Permeabilization, Membrane active peptide, Melittin
Introduction
In recent years, antimicrobial peptides (AMPs) have gained significant attention due to their clinical potential as a new class of antibiotics to fight antimicrobial resistance.1−5 These peptides, which are involved in the innate immune systems of living organisms, play important roles in defense against bacterial, viral, and fungal infections, as well as adaptive immunity during the development of cancers and autoimmune diseases. Unlike most conventional antibiotics, AMPs frequently destabilize biological membranes causing lysis of cells and therefore have potential as new therapeutic agents. Within this framework, numerous biophysical techniques (including fluorescence, circular dichroism, solid-state NMR spectroscopy) have been developed to study membrane permeabilization by AMPs.6 Among the numerous available model systems (e.g., large/giant unilamellar vesicles (LUVs/GUVs), planar supported bilayers) to investigate membrane permeabilizing peptides, GUVs are particularly appropriate because their large size allows (i) their manipulation and (ii) their visualization by optical microscopy techniques.7−11 They can therefore be used in single vesicle experiments to monitor morphological changes.
So far, the GUV suspension method is considered as the more traditional approach to study membrane-permeabilizing peptides with micrometric-sized vesicles. In this method, GUVs are first suspended in a solution containing a water-soluble membrane-impermeant fluorescent probe in the presence of a peptide of interest.12−16 Then, fluorescence inside and outside the vesicles is measured and analyzed by confocal microscopy throughout the permeabilization process. Alternatively, the water-soluble fluorescent probe may be encapsulated inside GUVs. In this recently developed method, the fluorescent probe leakage is determined by the loss of fluorescence from inside the vesicle. The structure and fluorescence variations of the GUVs are continuously observed throughout the process by phase contrast and confocal fluorescence microscopy.17−24 Very recently, the GUV leakage has been investigated using an original method called photovoltage transients, which consists in monitoring the time-evolution of the charge/discharge pulses of photoinduced electrons.25 This approach was also combined with fluorescence microscopy observations to image the leakage events at single GUV.
Compared to fluorescence microscopy, electrochemiluminescence (ECL) appears particularly relevant to image liposome permeabilization by peptides such as AMPs. ECL is a light-emitting process that has become a powerful tool in analytical chemistry notably for immunoassays and clinical diagnosis.26−30 In ECL, the light emission is initiated by an electron transfer reaction occurring at an electrode surface providing thus a very sensitive optical detection method because no excitation light source is required.31,32 Therefore, the photonic background remains extremely low, which requires simple instrumentation that can be coupled to the microscopy and allows a very sensitive detection.33,34 In addition, electrochemical stimulation allows control of the duration of the ECL emitting region, while optical readout makes the method extremely sensitive and allows localization of the event of interest. These remarkable characteristics have led to the development of ECL methods in analytical chemistry and more particularly in bioanalysis.35−39 Mechanistically, the combination of a ruthenium polypyridine luminophore, [Ru(bpy)3]2+, and tripropylamine (TPrA) as a sacrificial co-reactant is a model ECL system with high efficiency.40−42 Given the optical readout of ECL, a major development of this technique is currently focused on single object imaging.43 On the one hand, single entities such as nanowires,44 beads for immunoassays,42,45,46 attoliter droplets collisions,47 graphene oxide nanoparticles,48 and Janus metallic nanoparticles49 could be visualized by ECL. More recently, ECL has been successfully implemented to image not only membrane proteins,50 but also whole plasma membranes of single cells,40,51 and finally single mitochondria.41 On the other hand, transient events can be imaged by ECL such as hydrogen peroxide efflux, which can also be observed by ECL at the single cell level.52,53
As a result, we have recently developed an original strategy based on ECL to image in real-time the dynamic permeabilization of GUVs which was triggered by the polarization of the electrode.54 Based on a similar methodology, the work described herein deals with the ECL imaging of GUVs permeabilized by an archetype of antimicrobial peptides, i.e., melittin.
Melittin, the main venom component of the European Honeybee, is a cationic linear peptide-amide composed of 26 amino acid residues. Melittin is often used as a model system to investigate the structure, folding, and function of peptides and proteins in membranes.55 It also has been studied preclinically as an anticancer agent for its broad lytic effects in multiple tumor types.56,57 Despite numerous investigations, the membrane permeabilization mechanisms triggered by melittin are still under debate58−61 and may be multiple. Under specific conditions, it has been suggested that melittin may assemble into stable transmembrane helical bundles to form pores.62 However, in synthetic bilayers melittin generally adopts an equilibrium orientation that is parallel to the membrane surface.63,64 This leads to a transient permeabilization of membranes with a burst of leakage occurring rather rapidly after the peptide addition (<1 min). Interestingly, after this initial burst, the leakage generally slows down or stops.58,59,65,66 Actually, it is very difficult and even impossible to rationalize mechanisms involved during membrane permeabilization by AMPs because processes are dependent on the contribution of many experimental conditions including the lipid composition, peptide concentration, temperature, ionic strength, pH, etc.. GUVs have been largely used to investigate the mechanistic processes involved in their permeabilization when in contact with an AMP such as magainin 2, melittin, LL37, or lactoferricin B.67−70,23 These studies show that the permeabilization followed by the leakage of a fluorescent probe (initially present inside the GUV) starts stochastically at different times for different vesicles. Nevertheless, once it begins, complete leakage rapidly occurs. If a permeabilization event can be schematized upon successive AMP binding, membrane permeation and content release, this behavior suggests that the permeabilization event (as opposed to peptide binding or vesicle leakage) is the rate-determining step in the process.18,19,70,71 Importantly, and compared to LUVs, the stochastic leakage phenomenon is specifically detected with GUVs because they can be observed individually.
Herein, we show that GUV permeabilization by melittin can be evidenced by an ECL imaging approach (Scheme 1).
Scheme 1. Strategy Adopted to Image by ECL the Membrane Permeation of Giant Vesicles by Melittin.
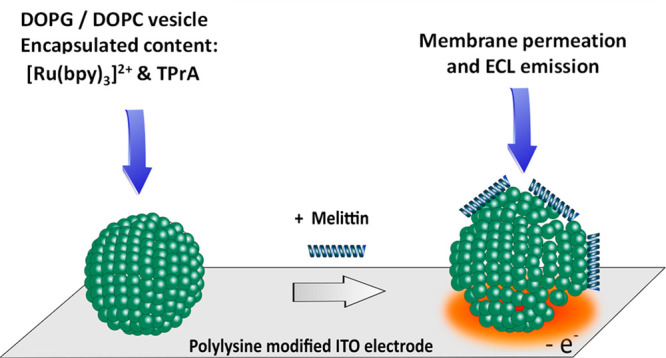
To achieve this, the ECL reagents (i.e., [Ru(bpy)3]2+ and TPrA) were first encapsulated in GUVs. The giant vesicles were deposited on an electrode surface previously coated with poly-l-lysine to avoid any membrane permeabilization induced by the surface polarization. Under these conditions, the addition of melittin triggered membrane permeabilization that led to the content leakage and subsequent ECL emission upon oxidation of ECL reagents at the electrode surface. It is important to note that the choice of lipids was dictated by their ability (i) to keep intact (i.e., impermeable) liposomes as long as they were not in contact with melittin and (ii) to improve the interaction with the positively charged AMP since the main driving forces for membrane binding are electrostatic attraction and hydrophobic interactions.72 Accordingly, asymmetrical liposomes composed of negatively charged DOPG (outer leaflet) and zwitterionic DOPC phospholipids (inner leaflet) were prepared. The status and location of liposomes (intact or permeabilized) was assessed by epifluorescence imaging. In a complementary way, ECL provided images of the efflux of matter after liposome permeabilization.
Results and Discussion
Stability of Giant Liposomes Lying on a Polarized ITO Electrode Coated with Poly-l-lysine
The preparation of asymmetrical giant liposomes (100 μm in diameter) encapsulating both the luminophore ([Ru(bpy)3]2+ at 250 μM) and the co-reactant (TPrA at 100 mM) for ECL has been recently described.54 Briefly, the preparation consists in two steps. First, aqueous droplets containing the intravesicular solution and surrounded by DOPC lipids to form the inner leaflet of giant liposomes were prepared with a microfluidic device. Second, the freshly prepared droplets were passed through a water/oil interface containing DOPG phospholipids to form the outer leaflet. Note that intra- and extra-liposomal solutions contain sucrose and glucose, respectively. Indeed, the different densities between sucrose and glucose is useful to both help droplets to cross the water/oil interface, and to allow liposome sedimentation on the electrode surface. Note that the lipids were chosen so that the liposomes remain impermeable in the absence of AMPs while remaining relevant in the exploration of membrane permeabilization by AMPs.73,74 On the one hand, and as recently demonstrated,54 the use of a zwitterionic phospholipid from the phosphocholine series, such as DOPC, on the internal leaflet is required to form stable liposomes. On the other hand, taking into consideration that AMPs are globally positively charged and that the main driving forces for peptide binding are electrostatic attraction and hydrophobic interaction,72 the use of the negatively charged DOPG on the outer-leaflet appeared perfectly adapted.
In a recent work, it was shown that DOPG/DOPC (outer/inner-leaflet) asymmetrical liposomes undergo permeabilization when they are directly in contact with an ITO material surface under polarization (+1.2 V vs. Ag/AgCl).54 Since the aim of this work is the characterization of the membrane permeabilization by melittin, it was crucial to prevent any electrode effect on the membrane stability. This was successfully achieved by covering the ITO electrode surface with poly-l-lysine. Under these conditions, and as shown in Figure 1, liposomes were found stable for at least 30 min.
Figure 1.
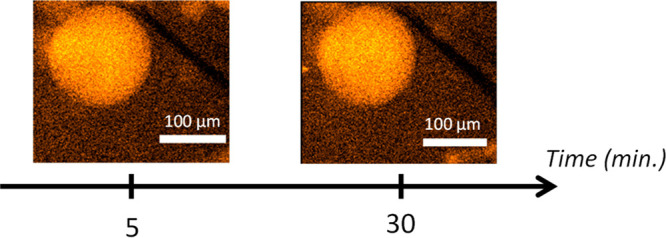
Photoluminescent (PL) images of a single giant asymmetrical liposome made of DOPG and DOPC on its outer and inner leaflets, respectively. It contains 250 μM [Ru(bpy)3]2+ luminophore as well as 100 mM TPrA and lies on a poly-l-lysine-coated ITO electrode surface polarized at +1.2 V vs Ag/AgCl. Images recorded at t = 5 min and t = 30 min of polarization. The PL microscopy images reveal the localization of the [Ru(bpy)3]2+ complex inside the liposome with an illumination at λex = 455 nm.
Imaging the Permeabilization of Liposomes by Melittin
After ensuring the stability of liposomes at a polarized poly-l-lysine-coated ITO electrode, their membrane integrity has been tested in the presence of melittin, a positively charged antimicrobial peptide known to also cause permeabilization in prokaryotic and eukaryotic cells or artificial vesicles. Accordingly, melittin was introduced in the electrochemical cell and let to interact with the negatively charged outer leaflets of liposomes while keeping the solution osmolality constant. The experiments were first carried out at a melittin concentration of 10 μM (final concentration in the electrochemical cell). Under these conditions, membrane permeabilization processes were observed both by photoluminescence (PL) and ECL after a few tens of seconds following the addition of peptides. Two sets of ECL and PL images are shown in Figure 2, before and during the membrane permeabilization of a representative liposome (see the liposome located in the dotted white square zone).
Figure 2.
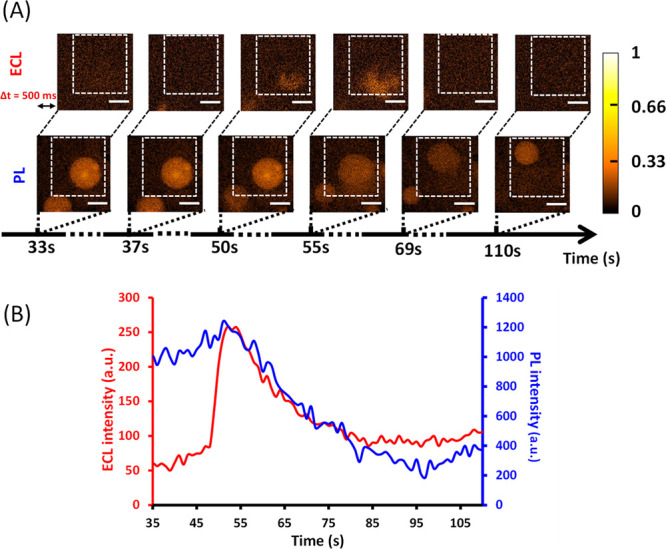
(A) Parallel time-lapse by ECL (top) and PL (below) showing the permeation by 10 μM melittin of a DOPG/DOPC liposome (the one isolated in the dotted white square). The liposome lies on a polarized ITO electrode (+1.2 V vs Ag/AgCl) coated with poly-l-lysine. It initially contains both 250 μM [Ru(bpy)3]2+ and 100 mM TPrA. Note that ECL and PL images have a short difference in initial acquisition time (Δt = 500 ms) to avoid overlap between the two types of visual information. During ECL and PL imaging, the focus was on the surface of the ITO electrode. Scale bar: 100 μm. (B) Evolution of PL (blue curve) and ECL (red curve) signals obtained as a function of time for a single liposome [shown in (A)] permeabilized by melittin. PL and ECL intensities were measured from a typical ROI outlined by the dashed white square shown in (A). Melittin was added at t = 0 s.
Before t = 50 s, the single liposome appears intact (see PL images) confirming the absence of detectable leakage of its content near the electrode (see the corresponding ECL images for t < 50 s). For 50 < t < 69 s, an ECL signal is detected at the bottom of the image suggesting a liposomal leakage of ECL reagents next to the electrode surface. This is confirmed by the PL images that clearly show a strong concomitant fluorescence decay followed by a displacement of the liposome in the direction opposite to the leakage point (i.e., to the top of the image) most likely due to the release of the content outside the liposome. Then, the liposome seems to stop leaking in agreement with the absence of ECL signal (see the PL and ECL images at t = 110 s). Compared to the PL images taken before the leakage, the image at 110 s clearly shows that the size of the liposome decreases in accordance with a loss of its content. At this point, it is important to note that in most cases, a complete leakage is observed (two videos are included in the Supporting Information (SI) showing typical behaviors both in ECL and in PL). Therefore, the behavior observed in Figure 2 represents a minority of events, thus confirming the stochastic character of the melittin-promoted permeation of the vesicle. However, it shows the possibility to monitor the transient nature of the permeabilization process in some cases. These qualitative observations, both in ECL and PL, clearly indicate a membrane permeabilization by the added melittin in agreement with the appearance of an ECL signal resulting from the liposomal content release and oxidation. It shows that the poly-l-lysine layer deposited on the ITO electrode is thin enough or/and nonperfectly uniform to allow electron transfer between the released luminophore/co-reactant and the electrode. In addition, considering the stability of DOPC/DOPG liposomes at the polarized ITO electrode surface coated with poly-l-lysine, this result confirms that the membrane permeabilization is generated by the introduction of melittin in the solution. To our knowledge, this is the first report of the ECL imaging of a permeabilization event by an antimicrobial peptide at a phospholipidic membrane.
Liposome permeabilization can also be quantified from PL and ECL intensities in specific regions of interest (ROIs) like the one outlined in Figure 2A. The combination of the two signals allows complete monitoring of the permeabilization and release of the content of a single liposome. As shown in Figure 2B, ECL appears as soon as PL decreases. Compared to the brutal opening of liposomes observed at a naked and polarized ITO electrode,54 the PL signal observed here decreases for several tens of seconds (around 30 s) in agreement with times observed in other studies75 indicating a rather slow leakage and explaining why the amperometric current could not be monitored in this case. Moreover, it is important to underline that the simultaneous ECL increase and PL decrease confirm that both responses are correlated and come from the same liposome. Also, the signal duration and shape are similar for ECL and PL in agreement with the fact that both emissions depend on the luminophore concentration and diffusion. In addition, the TPrA concentration (i.e., 100 mM) is large enough to not be considered as a limiting step in the ECL reactions and light emission.
Although the leakage observed in Figure 2 is incomplete within this time window, the newly formed liposome leaked again approximately 2 min later. The two successive leakages occurring on the same liposome are gathered in Figure S1 of the SI. Compared to the first leakage, and as shown on the ECL images, the second one appears on the opposite face of the liposome (top of the image), but the liposome is not displaced in this case. The second leakage is, this time, complete in accordance with the disappearance of the liposome (see the PL image at t = 250 s). Taking the entire image as the ROI, the second ECL signal (red curve) is less intense than the first one, showing that most of the liposome content has been lost during the first leak. Taking the same ROI for PL images (i.e., the entire image) the second leakage is less intense due to a lower signal/noise ratio.
Following the experiments performed at a melittin concentration of 10 μM, the sensitivity of our original approach has been tested by decreasing the peptide concentration. Accordingly, a series of experiments have been performed at 1 μM and 100 nM in melittin (Figures 3 and 4).
Figure 3.
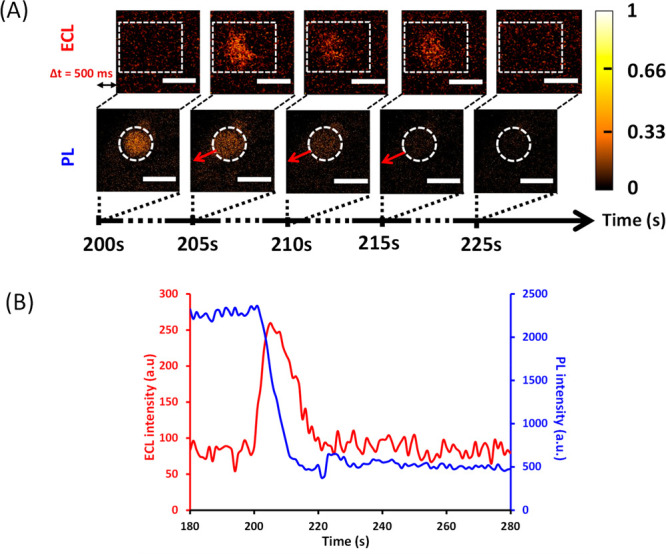
(A) Parallel time-lapse imaging by ECL (top) and PL (below) showing the permeation by 1 μM melittin of a DOPG/DOPC liposome (the one isolated in the dotted white circle). The liposome lies on a polarized ITO electrode (+1.2 V vs Ag/AgCl) coated with poly-l-lysine. It initially contains both 250 μM [Ru(bpy)3]2+ and 100 mM TPrA. Note that ECL and PL images have a short difference in initial acquisition time (Δt = 500 ms) to avoid overlap between the two types of visual information. During ECL and PL imaging, the focus was on the surface of the ITO electrode. Scale bar: 100 μm. (B) Evolution of PL (blue curve) and ECL (red curve) signals obtained as a function of time for a single liposome [shown in (A)] permeabilized by melittin. PL and ECL intensities were measured from a typical ROI outlined by the dashed white zones shown in (A). Melittin was added at t = 0 s.
Figure 4.
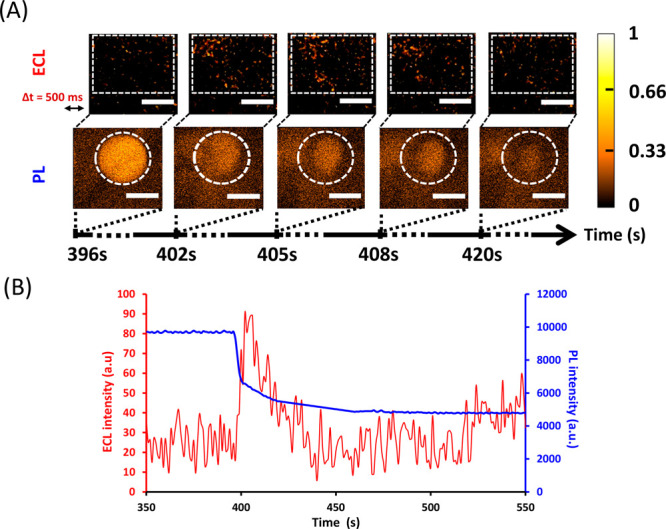
(A) Parallel time-lapse imaging by ECL (top) and PL (below) showing the permeation by 100 nM melittin of a DOPG/DOPC liposome (the one isolated in the dotted white circle). The liposome lies on a polarized ITO electrode (+1.2 V vs Ag/AgCl) coated with poly-l-lysine. It initially contains both 250 μM [Ru(bpy)3]2+ and 100 mM TPrA. Note that ECL and PL images have a short difference in initial acquisition time (Δt = 500 ms) to avoid overlap between the two types of visual information. During ECL and PL imaging, the focus was on the surface of the ITO electrode. Scale bar: 100 μm. (B) Evolution of PL (blue curve) and ECL (red curve) signals obtained as a function of time for a single liposome [shown in (A)] permeabilized by melittin. PL and ECL intensities were measured from a ROI outlined by the dashed white zones shown in (A). Melittin was added at t = 0 s.
At a melittin concentration of 1 μM, it is still possible to observe by ECL the location of the leak, whereas it is almost impossible by PL (compare ECL and PL images in Figure 3A). Based on ECL images, red arrows have been added on PL images to show where the leakage comes from on the liposome of interest. This confirms the higher sensitivity of ECL compared to PL (thanks to a better signal/noise ratio) that allows the location of events. Nevertheless, vesicle permeabilization can be quantified not only by ECL, but also by PL provided an adequate ROI is selected (see Figure 3). As observed at a melittin concentration of 10 μM, the concomitant PL decrease and ECL increase confirm that both signals originate from the same liposome. The duration of the ECL signal falls in the same time range as previously observed and a complete leakage occurs.
At a melittin concentration of 100 nM, permeation can still be detected by both ECL and PL, but the ECL signal becomes too diffuse to precisely localize which part of the liposome is leaking (see Figures 4A and 3A for comparison). In this case, the ROI size has to be increased to extract the leakage event from the background signal (Figure 4B). Nevertheless, the duration of the ECL signal is still in the same time window as those previously observed at higher melittin concentrations. Besides, ECL and PL signals remain concomitant.
Globally, whatever the melittin concentration, similar behaviors were observed both in PL and ECL. Nevertheless, and as a general rule, the lower the peptide concentration, the lower the ECL intensity and the longer the delay between peptide addition and permeabilization are.
Conclusion
The ECL imaging of a liposome permeabilization event triggered by an antimicrobial peptide has been achieved. The combination of ECL and photoluminescence imaging allowed a comprehensive monitoring of the permeabilization and content release of a single liposome. Furthermore, the ECL approach has proven to be very sensitive since it allows the localization of permeabilization events. Like fluorescence microscopy, the ECL approach highlights the stochastic nature of AMP-promoted membrane permeabilization.
More generally, this original ECL imaging strategy may be considered as a new and complementary biophysical technique35 (additionally to fluorescence, circular dichroism, solid-state NMR spectroscopy)76,77 to investigate any cell or vesicle permeabilization or poration events, by membrane-active peptides or proteins, a central issue in the general topic of cell transmembrane trafficking.
Experimental Section
Lipids
1,2-Dioleoyl-sn-glycerol-3-phospho-(1′-rac-glycerol) sodium salt (DOPG, 10 mg/mL in chloroform) and 1,2-dioleolyl-sn-glycero-3-phosphocholine (DOPC, 10 mg/mL in chloroform) were purchased from Avanti Polar Lipids.
Chemicals
Poly-l-lysine (0.01% solution), tris(2,2′-bipyridyl)dichlororuthenium(II) hexahydrate, mineral oil, trin-propylamine (TPrA), d-(+)-glucose, sucrose, and phosphate buffer saline (PBS) tablets (10 mM phosphate, 137 mM sodium chloride; 2.7 mM potassium chloride; pH 7.4) were purchased from Aldrich. Fmoc-protected amino acids were purchased from Iris Biotech. Rink Amide AM resin, N,N′-diisopropylcarbodiimide (DIC), 1-hydroxybenzotriazole hydrate (HOBt), triisopropylsilane (TIS), and α-cyano-4-hydroxycinnamic acid (CHCA) were obtained from Merck. Acetonitrile (HPLC grade), dichloromethane, diethyl ether, dimethylformamide (DMF, peptide synthesis grade), piperidine (peptide synthesis grade), and trifluoroacetic acid (TFA) were purchased from Carlo Erba. All chemicals were used without further modification.
Instrumentation
ECL experiments were performed using a μAUTOLAB Type III and an inverted Zeiss Observer.Z1 microscope placed in a Faraday cage. The detection and collection of luminescent signals were performed with an EMCCD camera (C9100-13, Hamamatsu, Japan) connected to the microscope.Photoluminescence (PL) excitation was performed with a diode (THORLABS M455L3, λex = 455 nm). The [Ru(bpy)3]2+ luminophore (λex = 450 nm; λem= 610 nm) was illuminated with a Zeiss filter set 74HE. The melittin peptide was assembled on a CS3365 CSBio automated peptide synthesizer. Peptide purification was performed on a WATERS preparative HPLC system equipped with a 2489 UV/vis detector and a 2535 quaternary gradient module. Analytical HPLC was performed on an Agilent 1220 LC System. MALDI-TOF MS peptide characterization was done at the MS3U platform (Sorbonne Université) on a Bruker Autoflex Speed.
Reactant Aqueous Solutions
Highly purified water (resistivity = 18 MΩ.cm; Milli-Q system; Millipore, Billerica, MA, USA) was used to prepare extra- and intravesicular solutions. The extravesicular solution was composed of PBS in which glucose (0.7 M) was dissolved. The intravesicular solution was made of PBS/sucrose (0.7 M)/[Ru(bpy)3]2+ (250 μM)/TPrA (100 mM). Importantly, the pH value was adjusted at a value of 7.4 by addition of phosphoric acid. If necessary, osmolality was adjusted by adding small amounts of PBS solution to ensure isotonic conditions.
Phospholipid Solutions
The giant liposomes consisted of DOPG (outer leaflet) and DOPC (inner leaflet). To form them (see below), phospholipids in oil suspensions had to be prepared as follows: first, DOPG or DOPC (121 μL; 10 mg/mL in CHCl3) was added in a glass vial (2 mL) to evaporate chloroform under vacuum (2 h). Then, mineral oil (2 mL) was added to dried phospholipids and the corresponding suspension was sonicated (1 h).
Preparation of [Ru(bpy)3]2+/TPrA Encapsulated Giant Liposomes
Giant liposomes were prepared in two steps. First, aqueous droplets containing the intravesicular solution and surrounded by DOPC lipids to form the inner leaflet of giant liposomes were prepared with a microfluidic device. Second, the freshly prepared droplets were passed through a water/oil interface containing DOPG phospholipids to form the outer leaflet. More details of this preparation can be found in ref (54).
Imaging of GUVs
The observation of giant liposomes lying on the poly-l-lysine-coated ITO electrode was performed by fluorescence microscopy (Zeiss Observer.Z1), using N-Achroplan 10× (aperture 0.25) and LD Plan-Neofluar 20× (numerical aperture 0.6) objectives. A mercury lamp (HXP 120C) was used for fluorescence excitation while selecting the appropriate filter to highlight the ruthenium complex (Zeiss filter set 74 HE). A flashing diode (THORLABS M455L3, λex = 455 nm) was used to collect PL images during ECL experiments.
ITO Microelectrodes
Optical glass slides (22 mm × 22 mm × 0.13 mm) with 150 nm thick ITO films (90% In2O3/10% SnO2, ACM, Villiers Saint Frédéric, France) were used to prepare microelectrodes with a low electrical resistance (≤20 Ω per square) and high transparency. The ITO electrode surface (∼800 μm in diameter) was delimited by a PDMS well in which a micropipette tip (1 mL) was inserted vertically to the glass surface. Details of the device fabrication can be found in ref (54).
Combination of ECL/Photoluminescence (PL) Experiments
The combination of a potentiostat (μAUTOLAB Type III) and an e-corder 401 system (eDAQ Pty Ltd., Australia, associated with the eDAQ Chart software) was used to perform ECL and photoluminescence experiments. ECL signals generated by the permeabilization of liposome, were detected by using an EMCCD camera (Hamamatsu, Japan) connected to an inverted Zeiss LSM 710 microscope placed in a Faraday cage. The ITO working electrode was set at a potential value of +1.2 V vs Ag/AgCl (used as the reference electrode); the counter electrode was a 1 cm length platinum wire. By adjusting the gain, sensitivity, and exposure time, the quality of the luminescent images was optimized. HCImage Live and ImageJ software were used to record and process images, respectively. The use of a flashing diode (λex = 455 nm) was necessary to combine ECL and PL investigations during permeabilization of liposomes on the electrode surface. This revealed the ruthenium complex inside the liposomes and correlated the ECL signal with the correct liposome. The appropriate time-sequences for the combined detection of ECL and PL signals is given in Figure S2 of the SI.
Modification of ITO Electrodes with Poly-l-lysine
Before each experiment, the micrometer-sized PDMS well with the ITO electrode at the bottom was filled with the poly-l-lysine solution (0.01%; 20 μL) and let in contact with the electrode surface for 1 h. To limit solvent evaporation, the device was placed inside a closed Petri dish. The well was finally rinsed with PBS then glucose solutions.
Melittin Synthesis
Melittin (H-GIGAVLKVLTTGLPALISWIKRKRQQ-NH2) was synthesized by standard automated Fmoc solid phase peptide synthesis on Rink-amide-AM resin (resin loading 0.26 mmol/g, 0.05 mmol scale). Couplings were carried out at room temperature for 2 h using 10-fold excess Fmoc-protected amino acid activated by DIC/HOBt in DMF. Final peptide deprotection and cleavage from the resin was performed by treatment for 3 h with a cocktail containing TFA and as scavengers TIS and H2O, in the ratio 95:2.5:2.5 (v/v). The cleavage mixture was then filtered to remove the resin, the TFA was sparged under a stream of argon, the peptide precipitated in cold diethyl ether (−20 °C, 2 h) and freeze-dried. Melittin was purified by semipreparative HPLC on a RP-C18 column (ACE C18–300, 250 × 10 mm) using a linear gradient of 35 to 60% B in A over 30 min with a flow rate of 6 mL/min (A = 0.1% TFA in H2O and B = 0.1% TFA in CH3CN). Detection was performed at 220 nm. The purity of the collected fractions was checked by analytical HPLC on a RP-C18 column (ACE C18-300, 100 × 4.6 mm) using a linear gradient of 40 to 65% B in A over 10 min (flow rate of 1 mL/min). Fractions with purity > 95% were pooled and freeze-dried, yielding 37.7 mg of peptide (isolated yield = 21.4%). The peptide was characterized by MALDI-TOF MS in the ion positive reflector mode using the CHCA matrix (10 mg/mL in CH3CN/H2O/TFA, 50:50:0.1): measured m/z = 2845.4 [M + H]+ (first isotope); calculated m/z = 2845.7.
Acknowledgments
This work was supported in parts by CNRS UMR 8640, Ecole Normale Supérieure, PSL University and Sorbonne Université. F.B.T. thanks the doctoral school ED388 “Chimie Physique et de Chimie Analytique de Paris Centre” for a PhD grant. We thank Gilles Clodic (MS3U platform, Sorbonne Université) for MALDI-TOF mass spectrometry analysis. N.S. acknowledges the financial support from Agence Nationale de la Recherche (ELISE - ANR-21-CE42).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/cbmi.3c00003.
The authors declare no competing financial interest.
Supplementary Material
References
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Mookherjee N.; Anderson M. A.; Haagsman H. P.; Davidson D. J. Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Disc. 2020, 19, 311–332. 10.1038/s41573-019-0058-8. [DOI] [PubMed] [Google Scholar]
- Magana M.; Pushpanathan M.; Santos A. L.; Leanse L.; Fernandez M.; Ioannidis A.; Giulianotti M. A.; Apidianakis Y.; Bradfute S.; Ferguson A. L.; Cherkasov A.; Seleem M. N.; Pinilla C.; de la Fuente-Nunez C.; Lazaridis T.; Dai T.; Houghten R. A.; Hancock R. E. W.; Tegos G. P. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. 10.1016/S1473-3099(20)30327-3. [DOI] [PubMed] [Google Scholar]
- Gan B. H.; Gaynord J.; Rowe S. M.; Deingruber T.; Spring D. R. The multifaceted nature of antimicrobial peptides: current synthetic chemistry approaches and future directions. Chem. Soc. Rev. 2021, 50, 7820–7880. 10.1039/D0CS00729C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. Y.; Yan Z. B.; Meng Y. M.; Hong X. Y.; Shao G.; Ma J. J.; Cheng X. R.; Liu J.; Kang J.; Fu C. Y. Antimicrobial peptides: mechanism of action, activity and clinical potential. Military Med. Res. 2021, 8, 48. 10.1186/s40779-021-00343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K. A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria?. Nat. Rev. Microbiol. 2005, 3, 238–250. 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Toyota T.; Zhang Y. Identifying and Manipulating Giant Vesicles: Review of Recent Approaches. Micromachines 2022, 13, 644. 10.3390/mi13050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menger F. M.; Keiper J. S. Chemistry and physics of plant vesicles as biomembrane models. Curr. Opin. Chem. Biol. 1998, 2, 726–732. 10.1016/S1367-5931(98)80110-5. [DOI] [PubMed] [Google Scholar]
- Döbereiner H.-G. Properties of giant vesicles. Curr. Opin. Colloid Interface Sci. 2000, 5, 256–263. 10.1016/S1359-0294(00)00064-9. [DOI] [Google Scholar]
- Messina P.; Lemaître F.; Huet F.; Ngo K. A.; Vivier V.; Labbé E.; Buriez O.; Amatore C. Monitoring and Quantifying the Passive Transport of Molecules Through Patch-Clamp Suspended Real and Model Cell Membranes. Angew. Chem., Int. Ed. 2014, 53, 3192–3196. 10.1002/anie.201308990. [DOI] [PubMed] [Google Scholar]
- Perez Jimenez A. I.; Challier L.; Aït-Yahiatène E.; Delacotte J.; Labbé E.; Buriez O. Selective Electrochemical Bleaching of the Outer Leaflet of Fluorescently Labeled Giant Liposomes. Chem.—Eur. J. 2017, 23, 6781–6787. 10.1002/chem.201605786. [DOI] [PubMed] [Google Scholar]
- Ambroggio E. E.; Separovic F.; Bowie J. H.; Fidelio G. D.; Bagatolli L. A. Direct Visualization of Membrane Leakage Induced by the Antibiotic Peptides: Maculatin, Citropin, and Aurein. Biophys. J. 2005, 89, 1874–1881. 10.1529/biophysj.105.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A.; Lai G. H.; Schmidt N. W.; Sun V. Z.; Rodriguez A. R.; Tong R.; Tang L.; Cheng J.; Deming T. J.; Kamei D. T.; et al. Translocation of HIV TAT Peptide and Analogues Induced by Multiplexed Membrane and Cytoskeletal Interactions. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 16883–16888. 10.1073/pnas.1108795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaten S. A.; Ablan F. D. O.; Spaller B. L.; Trieu J. M.; Almeida P. F. Translocation of Cationic Amphipathic Peptides across the Membranes of Pure Phospholipid Giant Vesicles. J. Am. Chem. Soc. 2013, 135, 16517–16525. 10.1021/ja407451c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön P.; García-Sáez A. J.; Malovrh P.; Bacia K.; Anderluh G.; Schwille P. Equinatoxin II Permeabilizing Activity Depends on the Presence of Sphingomyelin and Lipid Phase Coexistence. Biophys. J. 2008, 95, 691–698. 10.1529/biophysj.108.129981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apellániz B.; Nieva J. L.; Schwille P.; García-Sáez A. J. Allor-None versus Graded: Single-Vesicle Analysis Reveals Lipid Composition Effects on Membrane Permeabilization. Biophys. J. 2010, 99, 3619–3628. 10.1016/j.bpj.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam J. M.; Kobayashi T.; Yamazaki M. The Single-Giant Unilamellar Vesicle Method Reveals Lysenin-Induced Pore Formation in Lipid Membranes Containing Sphingomyelin. Biochemistry 2012, 51, 5160–5172. 10.1021/bi300448g. [DOI] [PubMed] [Google Scholar]
- Islam M. Z.; Alam J. M.; Tamba Y.; Karal M. A. S.; Yamazaki M. The Single GUV Method for Revealing the Functions of Antimicrobial, Pore-Forming Toxin, and Cell-Penetrating Peptides or Proteins. Phys. Chem. Chem. Phys. 2014, 16, 15752–15767. 10.1039/C4CP00717D. [DOI] [PubMed] [Google Scholar]
- Tamba Y.; Yamazaki M. Single Giant Unilamellar Vesicle Method Reveals Effect of Antimicrobial Peptide Magainin 2 on Membrane Permeability. Biochemistry 2005, 44, 15823–15833. 10.1021/bi051684w. [DOI] [PubMed] [Google Scholar]
- Karal M. A. S.; Alam J. M.; Takahashi T.; Levadny V.; Yamazaki M. Stretch-Activated Pore of the Antimicrobial Peptide, Magainin 2. Langmuir 2015, 31, 3391–3401. 10.1021/la503318z. [DOI] [PubMed] [Google Scholar]
- Moghal M. M. R.; Islam M. Z.; Sharmin S.; Levadnyy V.; Moniruzzaman M.; Yamazaki M. Continuous Detection of Entry of Cell-Penetrating Peptide Transportan 10 into Single Vesicles. Chem. Phys. Lipids 2018, 212, 120–129. 10.1016/j.chemphyslip.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Islam M. Z.; Sharmin S.; Levadnyy V.; Alam Shibly S. U.; Yamazaki M. Effects of Mechanical Properties of Lipid Bilayers on the Entry of Cell-Penetrating Peptides into Single Vesicles. Langmuir 2017, 33, 2433–2443. 10.1021/acs.langmuir.6b03111. [DOI] [PubMed] [Google Scholar]
- Lee M. T.; Sun T. L.; Hung W. C.; Huang H. W. Process of inducing pores in membranes by melittin. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 14243–14248. 10.1073/pnas.1307010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Xiao S.; Li J.; Yuan B.; Yang K.; Ma Y. Molecular details on the intermediate states of melittin action on a cell membrane. BBA – Biomembranes 2018, 1860, 2234–2241. 10.1016/j.bbamem.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Sun S.; Xia Y.; Liu J.; Dou Y.; Yang K.; Yuan B.; Kang Z. Real-time monitoring the interfacial dynamic processes at model cell membranes: Taking cell penetrating peptide TAT as an example. J. Colloid Interface Sci. 2022, 609, 707–717. 10.1016/j.jcis.2021.11.076. [DOI] [PubMed] [Google Scholar]
- Ma X.; Gao W.; Du F.; Yuan F.; Yu J.; Guan Y.; Sojic N.; Xu G. Rational design of electrochemiluminescent devices. Acc. Chem. Res. 2021, 54, 2936–2945. 10.1021/acs.accounts.1c00230. [DOI] [PubMed] [Google Scholar]
- Fiorani A.; Han D.; Jiang D.; Fang D.; Paolucci F.; Sojic N.; Valenti G. Spatially resolved electrochemiluminescence through a chemical lens. Chem. Sci. 2020, 11, 10496–10500. 10.1039/D0SC04210B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanut A.; Palomba F.; Rossi Scota M.; Rebeccani S.; Marcaccio M.; Genovese D.; Rampazzo E.; Valenti G.; Paolucci F.; Prodi L. Dye-Doped Silica Nanoparticles for Enhanced ECL-Based Immunoassay Analytical Performance. Angew. Chem., Int. Ed. 2020, 59, 21858–21863. 10.1002/anie.202009544. [DOI] [PubMed] [Google Scholar]
- Du F.; Chen Y.; Meng C.; Lou B.; Zhang W.; Xu G. Recent advances in electrochemiluminescence immunoassay based on multiple-signal strategy. Curr. Opin. Electrochem. 2021, 28, 100725. 10.1016/j.coelec.2021.100725. [DOI] [Google Scholar]
- Dutta P.; Han D.; Goudeau B.; Jiang D.; Fang D.; Sojic N. Reactivity mapping of luminescence in space: insights into heterogeneous electrochemiluminescence bioassays. Biosens. Bioelectron. 2020, 165, 112372. 10.1016/j.bios.2020.112372. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Qi W.; Xu G. Recent advances in electrochemiluminescence. Chem. Soc. Rev. 2015, 44 (10), 3117–3142. 10.1039/C5CS00086F. [DOI] [PubMed] [Google Scholar]
- Ma C.; Cao Y.; Gou X.; Zhu J.-J. Recent Progress in Electrochemiluminescence Sensing and Imaging. Anal. Chem. 2020, 92, 431–454. 10.1021/acs.analchem.9b04947. [DOI] [PubMed] [Google Scholar]
- Bard A. J.Electrogenerated Chemiluminescence; M. Dekker: New York, 2004. [Google Scholar]
- Dong J.; Lu Y.; Xu Y.; Chen F.; Yang J.; Chen Y.; Feng J. Direct imaging of single-molecule electrochemical reactions in solution. Nature. 2021, 596, 244–249. 10.1038/s41586-021-03715-9. [DOI] [PubMed] [Google Scholar]
- Abdussalam A.; Xu G. Recent advances in electrochemiluminescence luminophores. Anal. Bioanal. Chem. 2022, 414, 131–146. 10.1007/s00216-021-03329-0. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Arbault S.; Sojic N.; Jiang D. Electrochemiluminescence Imaging for Bioanalysis. Annual Rev. Anal. Chem. 2019, 12, 275–295. 10.1146/annurev-anchem-061318-115226. [DOI] [PubMed] [Google Scholar]
- Qi H.; Zhang C. Electrogenerated Chemiluminescence Biosensing. Anal. Chem. 2020, 92, 524–534. 10.1021/acs.analchem.9b03425. [DOI] [PubMed] [Google Scholar]
- Blackburn G F; Shah H P; Kenten J H; Leland J; Kamin R A; Link J; Peterman J; Powell M J; Shah A; Talley D B Electrochemiluminescence Detection for Development of Immunoassays and DNA Probe Assays for Clinical Diagnostics. Clin. Chem. 1991, 37, 1534–1539. 10.1093/clinchem/37.9.1534. [DOI] [PubMed] [Google Scholar]
- Sardesai N.; Pan S.; Rusling J. Electrochemiluminescent immunosensor for detection of protein cancer biomarkers using carbon nanotube forests and [Ru-(bpy)(3)](2+)-doped silica nanoparticles. Chem. Commun. 2009, 4968–4970. 10.1039/b909220j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voci S.; Goudeau B.; Valenti G.; Lesch A.; Jovic M.; Rapino S.; Paolucci F.; Arbault S.; Sojic N. Surface-Confined Electrochemiluminescence Microscopy of Cell Membranes. J. Am. Chem. Soc. 2018, 140, 14753–14760. 10.1021/jacs.8b08080. [DOI] [PubMed] [Google Scholar]
- Ma Y.; Colin C.; Descamps J.; Arbault S.; Sojic N. Shadow Electrochemiluminescence Microscopy of Single Mitochondria. Angew. Chem., Int. Ed. 2021, 60, 18742–18749. 10.1002/anie.202105867. [DOI] [PubMed] [Google Scholar]
- Deiss F.; LaFratta C. N.; Symer M.; Blicharz T. M.; Sojic N.; Walt D. R. Multiplexed Sandwich Immunoassays Using Electrochemiluminescence Imaging Resolved at the Single Bead Level. J. Am. Chem. Soc. 2009, 131, 6088–6089. 10.1021/ja901876z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.; Chen H.-Y.; Xu J.-J. Electrogenerated chemiluminescence detection of single entities. Chem. Sci. 2021, 12, 5720–5736. 10.1039/D0SC07085H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. J.; Marchuk K.; Willets K. A. Imaging Electrogenerated Chemiluminescence at Single Gold Nanowire Electrodes. Nano Lett. 2015, 15, 6110–6115. 10.1021/acs.nanolett.5b02383. [DOI] [PubMed] [Google Scholar]
- Sentic M.; Milutinovic M.; Kanoufi F.; Manojlovic D.; Arbault S.; Sojic N. Mapping electrogenerated chemiluminescence reactivity in space: mechanistic insight into model systems used in immunoassays. Chem. Sci. 2014, 5, 2568–2572. 10.1039/C4SC00312H. [DOI] [Google Scholar]
- Valenti G.; Zangheri M.; Sansaloni S. E.; Mirasoli M.; Penicaud A.; Roda A.; Paolucci F. Transparent Carbon Nanotube Network for Efficient Electrochemiluminescence Devices. Chem.—Eur. J. 2015, 21, 12640–12645. 10.1002/chem.201501342. [DOI] [PubMed] [Google Scholar]
- Dick J. E.; Renault C.; Kim B.-K.; Bard A. J. Simultaneous Detection of Single Attoliter Droplet Collisions by Electrochemical and Electrogenerated Chemiluminescent Responses. Angew. Chem., Int. Ed. 2014, 53, 11859–11862. 10.1002/anie.201407937. [DOI] [PubMed] [Google Scholar]
- Fan F. -R. F.; Park S.; Zhu Y.; Ruoff R. S.; Bard A. J. Electrogenerated Chemiluminescence of Partially Oxidized Highly Oriented Pyrolytic Graphite Surfaces and of Graphene Oxide Nanoparticles. J. Am. Chem. Soc. 2009, 131, 937–939. 10.1021/ja8086246. [DOI] [PubMed] [Google Scholar]
- Zhu M.-J.; Pan J.-B.; Wu Z.-Q.; Gao X.-Y.; Zhao W.; Xia X.-H.; Xu J.-J.; Chen H.-Y. Electrogenerated Chemiluminescence Imaging of Electrocatalysis at a Single Au-Pt Janus Nanoparticle. Angew. Chem., Int. Ed. 2018, 57, 4010–4014. 10.1002/anie.201800706. [DOI] [PubMed] [Google Scholar]
- Liu Y. J.; Zhang H. D.; Li B. X.; Liu J. W.; Jiang D. C.; Liu B. H.; Sojic N. Single Biomolecule Imaging by Electrochemiluminescence. J. Am. Chem. Soc. 2021, 143, 17910–17914. 10.1021/jacs.1c06673. [DOI] [PubMed] [Google Scholar]
- Valenti G.; Scarabino S.; Goudeau B.; Lesch A.; Jovic M.; Villani E.; Sentic M.; Rapino S.; Arbault S.; Paolucci F.; Sojic N. Single Cell Electrochemiluminescence Imaging: From the Proof-of-Concept to Disposable Device-Based Analysis. J. Am. Chem. Soc. 2017, 139, 16830–16837. 10.1021/jacs.7b09260. [DOI] [PubMed] [Google Scholar]
- Xu J.; Huang P.; Qin Y.; Jiang D.; Chen H.-Y. Analysis of Intracellular Glucose at Single Cells Using Electrochemiluminescence Imaging. Anal. Chem. 2016, 88, 4609–4612. 10.1021/acs.analchem.6b01073. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Ma G.; Chen Y.; Fang D.; Jiang D.; Chen H.-Y. Electrochemiluminescence Imaging for Parallel Single-Cell Analysis of Active Membrane Cholesterol. Anal. Chem. 2015, 87, 8138–8143. 10.1021/acs.analchem.5b00542. [DOI] [PubMed] [Google Scholar]
- Ben Trad F.; Wieczny V.; Delacotte J.; Morel M.; Guille-Collignon M.; Arbault S.; Lemaître F.; Sojic N.; Labbé E.; Buriez O. Dynamic Electrochemiluminescence Imaging of Single Giant Liposome Opening at Polarized Electrodes. Anal. Chem. 2022, 94, 1686–1696. 10.1021/acs.analchem.1c04238. [DOI] [PubMed] [Google Scholar]
- Guha S.; Ferrie R. P.; Ghimire J.; Ventura C. R.; Wu E.; Sun L.; Kim S. Y.; Wiedman G. R.; Hristova K.; Wimley W. C. Applications and evolution of melittin, the quintessential membrane active peptide. Biochem. Pharmacol. 2021, 193, 114769. 10.1016/j.bcp.2021.114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Wan C.; Cheng J.; Huang H.; Lovell J. F.; Jin H. Delivery Strategies for Melittin-Based Cancer Therapy. ACS Appl. Mater. Interfaces. 2021, 13, 17158–17173. 10.1021/acsami.1c03640. [DOI] [PubMed] [Google Scholar]
- Wang A.; Zheng Y.; Zhu W.; Yang L.; Yang Y.; Peng J. Melittin-Based Nano-Delivery Systems for Cancer Therapy. Biomolecules. 2022, 12, 118. 10.3390/biom12010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley W. C.; Hristova K. The mechanism of membrane permeabilization by peptides: still an enigma. Aust. J. Chem. 2020, 73, 96–103. 10.1071/CH19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley W. C. How does melittin permeabilize membranes?. Biophys. J. 2018, 114, 251–253. 10.1016/j.bpj.2017.11.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Choi H.; Weisshaar J. C. Melittin-induced permeabilization, re-sealing, and re-permeabilization of E. coli membranes. Biophys. J. 2018, 114, 368–379. 10.1016/j.bpj.2017.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benachir T.; Lafleur M. Study of vesicle leakage induced by melittin. Biochim. Biophys. Acta 1995, 1235, 452–460. 10.1016/0005-2736(95)80035-E. [DOI] [PubMed] [Google Scholar]
- Yang L.; Harroun T. A.; Weiss T. M.; Ding L.; Huang H. W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001, 81, 1475–1485. 10.1016/S0006-3495(01)75802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S.; Tamm L. K. Orientation of melittin in phospholipid bilayers: a polarized attenuated total reflection infrared study. Biophys. J. 1991, 60, 922–930. 10.1016/S0006-3495(91)82126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristova K.; Dempsey C. E.; White S. H. Structure, location, and lipid perturbations of melittin at the membrane interface. Biophys. J. 2001, 80, 801–811. 10.1016/S0006-3495(01)76059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedman G.; Herman K.; Searson P.; Wimley W. C.; Hristova K. The electrical response of bilayers to the bee venom toxin melittin: evidence for transient bilayer permeabilization. Biochim. Biophys. Acta 2013, 1828, 1357–1364. 10.1016/j.bbamem.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladokhin A. S.; Wimley W. C.; White S. H. Leakage of membrane vesicle contents: determination of mechanism using fluorescence requenchin. Biophys. J. 1995, 69, 1964–1971. 10.1016/S0006-3495(95)80066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. W. Action of Antimicrobial Peptides: Two-State Model. Biochemistry 2000, 39, 8347–8352. 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- Lee C. C.; Sun Y.; Qian S.; Huang H. W. Transmembrane Pores Formed by Human Antimicrobial Peptide LL-37. Biophys. J. 2011, 100, 1688–1696. 10.1016/j.bpj.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-T.; Hung W.-C.; Chen F.-Y.; Huang H. W. Mechanism and Kinetics of Pore Formation in Membranes by Water-Soluble Amphipathic Peptides. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 5087–5092. 10.1073/pnas.0710625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M.; Alam J. M.; Dohra H.; Yamazaki M. Antimicrobial Peptide Lactoferricin B-Induced Rapid Leakage of Internal Contents from Single Giant Unilamellar Vesicles. Biochemistry 2015, 54, 5802–5814. 10.1021/acs.biochem.5b00594. [DOI] [PubMed] [Google Scholar]
- Tamba Y.; Ohba S.; Kubota M.; Yoshioka H.; Yoshioka H.; Yamazaki M. Single GUV Method Reveals Interaction of Tea Catechin (2)-Epigallocatechin Gallate with Lipid Membranes. Biophys. J. 2007, 92, 3178–3194. 10.1529/biophysj.106.097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K. Membrane Permeabilization Mechanisms. Adv. Exp. Med. Biol. 2019, 1117, 9–16. 10.1007/978-981-13-3588-4_2. [DOI] [PubMed] [Google Scholar]
- Ladokhin A. S.; White S. H. ’Detergent-Like’ Permeabilization of Anionic Lipid Vesicles by Melittin. Biochim. Biophys. Acta 2001, 1514 (2), 253–260. 10.1016/S0005-2736(01)00382-0. [DOI] [PubMed] [Google Scholar]
- Paterson D. J.; Tassieri M.; Reboud J.; Wilson R.; Cooper J. M. Lipid Topology and Electrostatic Interactions Underpin Lytic Activity of Linear Cationic Antimicrobial Peptides in Membranes. Proc. Natl. Acad. Sci. U S A 2017, 114 (40), E8324–E8332. 10.1073/pnas.1704489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S.; Ghimire J.; Wu E.; Wimley W. C. Mechanistic Landscape of Membrane-Permeabilizing Peptides. Chem. Rev. 2019, 119, 6040–6085. 10.1021/acs.chemrev.8b00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger B.; Salnikov E. S. The membrane interactions of antimicrobial peptides revealed by solid-state NMR spectroscopy. Chem. Phys. Lipids. 2012, 165, 282–301. 10.1016/j.chemphyslip.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Marquette A.; Bechinger B. Biophysical Investigations Elucidating the Mechanisms of Action of Antimicrobial Peptides and Their Synergism. Biomolecules 2018, 8, 18. 10.3390/biom8020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


