Abstract
Aichi virus is the type species of a new genus, Kobuvirus, of the family Picornaviridae. In this study, we constructed a full-length cDNA clone of Aichi virus whose in vitro transcripts were infectious to Vero cells. During construction of the infectious cDNA clone, a novel sequence of 32 nucleotides was identified at the 5′ end of the genome. Computer-assisted prediction of the secondary structure of the 5′ end of the genome, including the novel sequence, suggested the formation of a stable stem-loop structure consisting of 42 nucleotides. The function of this stem-loop in virus replication was investigated using various site-directed mutants derived from the infectious cDNA clone. Our data indicated that correct folding of the stem-loop at the 5′ end of the positive strand, but not at the 3′ end of the negative strand, is critical for viral RNA replication. The primary sequence in the lower part of the stem was also suggested to be crucial for RNA replication. In contrast, nucleotide changes in the loop segment did not so severely reduce the efficiency of virus replication. A double mutant, in which both nucleotide stretches of the middle part of the stem were replaced by their complementary nucleotides, had efficient RNA replication and translation abilities but was unable to produce viruses. These results indicate that the stem-loop at the 5′ end of the Aichi virus genome is an element involved in both viral RNA replication and production of infectious virus particles.
Aichi virus was first isolated in 1989 from a stool specimen from a patient with oyster-associated nonbacterial gastroenteritis in Aichi, Japan (42). The complete genome sequence of this virus was determined, and the genome organization revealed that this virus is a member of the family Picornaviridae (45). However, the deduced amino acid sequences of Aichi virus proteins exhibited only 15 to 36% homology to those of other picornaviruses, suggesting that Aichi virus belongs to a distinct genus from the previously identified six genera of Picornaviridae (45). In 1999, this virus was classified into a new genus, Kobuvirus (18), whose name is derived from the characteristic morphology of the virus particles (kobu means bump in Japanese). Sequence analysis of 519-base reverse transcription-PCR (RT-PCR) products corresponding to the 3C-3D junction for 17 isolates of Aichi virus revealed that these isolates could be divided into two groups with an approximately 90% sequence homology (46).
Aichi virus has often been detected by enzyme-linked immunosorbent assay of stool specimens collected during oyster-associated gastroenteritis outbreaks in Japan. Between 1989 and 1991, 13 of 47 stool samples from adult patients in five of nine oyster-associated gastroenteritis outbreaks were positive for the Aichi virus antigen (44). Aichi virus has also been isolated from Pakistani children with gastroenteritis and from Japanese travelers with gastroenteritis from Southeast Asia (43). These findings suggest that this virus is widely distributed in Asia and that it is one of the causative agents of human gastroenteritis. A large-scale epidemiological survey is being performed to elucidate the impact of the virus worldwide.
In addition to the significance of Aichi virus as a possible human pathogen, a previous study demonstrated that this virus has some unique molecular features compared with other picornaviruses (45). Most picornaviruses have four capsid proteins, VP1 to -4 (34), and cleavage of a precursor protein VP0 into VP4 and VP2 occurs late in capsid assembly (14). In contrast, VP0 of Aichi virus is present in mature particles without being cleaved into VP4 and VP2 (45), as found in parechovirus (16, 36). The functions of the 2A and L proteins are diverse among picornaviruses. It is known that some of them exhibit proteolytic activity. Protein 2A of entero- and rhinoviruses is a trypsin-like protease (5, 39), and protein L of aphthovirus is a papain-like thiol protease (27, 31, 37). Aphtho- and cardiovirus 2A mediate the cleavage at its C terminus (7, 33, 35), and the autocatalytic motif NPGP is conserved at the cleavage site (7, 8). Aichi virus L and 2A have neither protease motifs nor the autocatalytic motif, and their functions remain unknown (45). Recently, it was reported that the 2A proteins of Aichi virus, as well as human parechoviruses and avian encephalomyelitis virus, have conserved motifs that are characteristic of a family of cellular proteins involved in the control of cell proliferation (15).
In this study, as the first step for studying the molecular basis of the replication and pathogenicity of Aichi virus, we attempted to construct a full-length cDNA clone whose in vitro transcripts are infectious to cultured cells. During construction of the full-length cDNA clone, we identified a novel sequence of 32 nucleotides at the 5′ end of the genome, and this finding enabled us to construct an infectious cDNA clone. The 5′ end of the poliovirus genome is shown to be an element that is involved in the initiation of positive-strand RNA synthesis. The first approximately 90 nucleotides (nt) of the poliovirus RNA fold into a cloverleaf-like (CL) structure (3, 30). The CL structure interacts with the viral protein 3CDpro (2, 3, 13) and either of another viral protein 3AB (13, 40, 41) or a cellular protein, poly(rC)-binding protein (PCBP) (2, 3, 10, 24), and this CL/3CDpro/PCBP or CL/3CDpro/3AB ribonucleoprotein (RNP) formation is essential to viral RNA replication. Unlike poliovirus, other enteroviruses, and rhinoviruses, the 5′-terminal sequences of the cardio-, aphtho-, hepato-, and parechovirus genomes fold into a stem-loop structure (6, 9, 11, 21, 28). It has been reported that the 5′-end 150 nt of the hepatitis A virus (HAV) genome containing three stem-loop structures and a polypyrimidine-rich sequence interact with the viral proteins, 3C, 3AB, and 3ABC (19, 20) and the cellular protein, PCBP2 (12). However, there has not been provided a direct evidence that the RNP formation at the 5′ end of the genome is involved in HAV RNA replication.
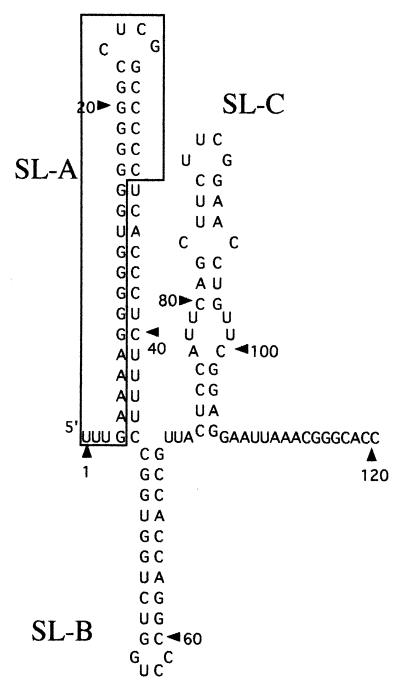
Here, using the infectious cDNA clone, we carried out a mutational analysis of the 5′ end of the Aichi virus genome. The computer-assisted prediction of the secondary structure of the 5′-terminal 120 nt, including the novel 32 nt of the viral genome, suggested the existence of three stem-loop structures (termed SL-A, SL-B, and SL-C). Both the secondary structure of the first stem-loop SL-A and the primary sequence of the bottom region of the stem of SL-A were found to be important for viral RNA replication. In addition, it was shown that SL-A plays an essential role in production of infectious virus particles.
MATERIALS AND METHODS
Cloning of the novel 5′-end sequence of the Aichi virus genome.
A standard Aichi virus strain, A846/88, was grown in Vero cells. The virion was purified by CsCl centrifugation as described previously (42). RNA was extracted by proteinase K treatment, followed by phenol-chloroform extraction and ethanol precipitation. Direct sequencing of the virion RNA was performed using primers, AV-86M and AV-93M (complementary to nt 67 to 86 and 75 to 93 of the previously published sequence in the DDBJ, EMBL, and GenBank databases [accession number AB010145], respectively) and reverse transcriptase (Seikagaku Kogyo, Japan), as described elsewhere (38). 5′-RACE was carried out using a 5′-RACE System for Rapid Amplification of cDNA Ends (Life Technologies, Inc.) according to the instructions of the manufacturer. Briefly, first-strand cDNA was synthesized at 50°C for 45 min using the GSP1 primer (complementary to nt 573 to 594 of the previously published sequence) and the virion RNA and then was tailed with dA or dT. Second-strand cDNA was synthesized using primer AP-dA (GGCCACGCGTCGACTAGTACTA17) or primer AP-dT (GGCCACGCGTCGACTAGTACT17). PCR was then performed with AP primer (GGCCACGCGTCGACTAGTACT) and GSP2 primer (complementary to nt 513 to 537 of the previously published sequence) using second-strand synthesis products as templates. The PCR products were ligated into pCRII-Topo (Invitrogen), and then clones were sequenced with an ABI PRIZM 310 genetic analyzer using a BigDye terminator cycle sequencing kit (Applied Biosystems) or a DYEnamic ET terminator cycle sequencing kit (Amersham Pharmacia Biotech).
Construction of full-length cDNA clones with a previously reported or the novel 5′-end sequence.
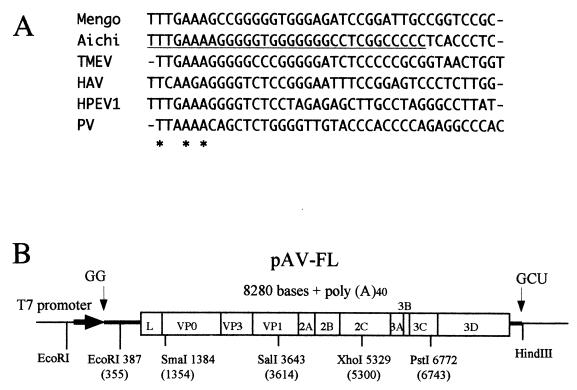
cDNA of the Aichi virus genome was synthesized by RT-PCR. To synthesize first-strand cDNA, 10 pmol of a primer and 50 ng of the genomic RNA were heated at 95°C for 2 min, chilled on ice, and then added to a reaction mixture (125 mM Tris-HCl [pH 8.3], 125 mM KCl, 25 mM magnesium acetate, 25 mM dithiothreitol, 250 μM of each deoxynucleoside triphosphate, and 10 U of avian myeloblastosis virus reverse transcriptase [Seikagaku Kogyo]), and then the mixture was incubated at 42°C for 50 min. A part of this RT reaction mixture was used for PCR. Five cDNA fragments corresponding to nt 1 to 1544, 1321 to 3803, 3524 to 5505, 5173 to 6784, and 6707 to 8251 (these nucleotide numbers refer to the sequence deposited previously in the DDBJ, EMBL, and GenBank databases) were amplified by PCR using primers sets T7AV1-1544M, 1321P-3803M, 3524P-5505M, 5173P-6784M, and 6707P-3′polyA, respectively. The sequences of the primers were as follows: T7AV1, 5′-AAGATATCTAATACGACTCACTATAGGtcaccctctttcccggtg-3′, plus-strand sequence with T7 promoter (underlined) and GG, followed by the previously reported 5′-end sequence (lowercase); 1544M, 5′-GCTTCCGCGTGATGGCCTTGGA-3′, minus-strand sequence, nt 1523 to 1544; 1321P, 5′-TGGTCCCGTCTCATGCACTCCG-3′, plus-strand sequence, nt 1321 to 1342; 3803M, 5′-GATCGTCGGGGTCCACATCGG-3′, minus-strand sequence, nt 3783 to 3803; 3524P, 5′-TACTTCGGATGGGAGGACTGGT-3′, plus-strand sequence, nt 3524 to 3545; 5505M, 5′-GCGGTGAAGTATTTAGATTGGGTTCC-3′, minus-strand sequence, nt 5480 to 5505; 5173P, 5′-CTTCGATGGGTACACGGGTCAA-3′, plus-strand sequence, nt 5173 to 5194; 6784M, 5′-TTTGAGGAAGAGCTGGGTGTCAAG-3′, minus-strand sequence, nt 6761 to 6784; 6707P, 5′-AAACAACCCGCTCCCCTCAAG-3′, plus-strand sequence, nt 6707 to 6727; and 3′polyA, 5′-GGAAGCTTT38GTAAGAACAGT-3′, minus-strand sequence, nt 8241 to 8251, with HindIII site (italic). The PCR products were ligated into the pCRII-Topo vector or pGEM-T vector (Promega), and then the obtained clones were sequenced. When nucleotide differences were found between the previously published sequence and the sequences of the clones, another RT-PCR was performed and the sequences of the obtained clones were confirmed. Clones with the sequence that is shared by most of the clones sequenced were used for the construction of a full-length cDNA clone. A SacI (present in the vector sequence)-SmaI fragment containing the T7 promoter and nt 1 to 1354, an SmaI-SalI fragment corresponding to nt 1354 to 3614, an SalI-XhoI fragment (nt 3614 to 5300), a XhoI-PstI fragment (nt 5300 to 6743) and a PstI-HindIII fragment containing nt 6743 to 8251, and a poly(A) tract were ligated into the SacI-HindIII sites of pUC118. The generated plasmid, pAV-1, has the Aichi virus cDNA sequence with the 5′-end sequence published previously. A full-length cDNA clone with the novel 5′-end sequence was constructed as follows. PCR was performed with the T7-5′F primer (TGTAATACGACTCACTATAGGtttgaaaagggggtggggg), which has the T7 promoter sequence (underlined), followed by two guanosine residues and the novel 5′-end sequence (lowercase), and the GSP2 primer using a cDNA clone obtained by 5′-RACE as a template. The amplified fragment was ligated into pCRII-Topo, and then the sequences of the generated clones were confirmed. A clone with the accurate sequence was digested with EcoRI, and the resulting ∼0.4-kb fragment containing the T7 promoter and 5′-end 387 nt was ligated into pAV-1 from which the T7 promoter and 5′-end 355 nt had been removed by digestion with EcoRI; the clone thus obtained is referred to as pAV-FL (see Fig. 1B). pAV-1 and pAV-FL have the same sequence with two exceptions that the latter has the newly identified 32 nt at the 5′ end and contains a C-T substitution at nt 12 of the previously published sequence. pAV-1 and pAV-FL have Aichi virus genome sequences of 8248 and 8280 nt, respectively.
FIG. 1.
(A) Alignment of the 5′-terminal 40 nt of the genomes of Aichi virus, mengovirus (accession number L22089), Theiler's murine encephalomyelitis virus (TMEV, X56019), HAV (M14707), HPEV1 (L02971), and poliovirus (PV; J02281). These sequences were aligned using the CLUSTALW program. The newly identified 32 nt of the Aichi virus genome is underlined. Asterisks represent identical nucleotides in all viruses. (B) Schematic diagram of the full-length cDNA clone of Aichi virus, pAV-FL. Thick lines and an open box show the untranslated regions and the coding region, respectively. The thin line represents the vector sequence. The Aichi virus cDNA is under the control of the T7 promoter. Nonviral nucleotides predicted to be present at the 5′ and 3′ ends of the transcripts are shown. Restriction enzyme sites used for construction of the clone are indicated below the line and open box. Nucleotide numbers in parentheses refer to the previously published sequence.
In vitro transcription.
The full-length cDNA clone and its derivatives were linearized by digestion with HindIII, and RNA transcripts were synthesized with T7 RNA polymerase using a MEGAscript Kit (Ambion, Inc.). After the reaction mixture had been treated with DNase I, RNA was extracted with phenol-chloroform and then precipitated with 2-propanol. The integrity of the synthesized RNAs was confirmed by agarose gel electrophoresis. These transcripts would have two extra guanosine residues preceding the respective 5′-end sequences and three extra nucleotides, GCU, which are derived from the HindIII site, at the end of the poly(A) tail (40 A's) (see Fig. 1B).
Examination of infectivities of AV-FL RNA and the virion RNA and one-step growth analysis.
To examine the infectivities of AV-FL RNA and the virion RNA, Vero cell monolayers in 3.5-cm dishes were transfected with serial dilutions of the RNAs using Lipofectin reagent (Life Technologies, Inc.) according to the manufacturer's recommendations. After incubation for 6 h at 37°C, the cells were washed and then overlaid with Eagle minimum essential medium containing 1% agarose and 5% fetal calf serum (FCS). After incubation for 48 h, the cells were stained with neutral red and then, after an additional incubation for 24 h, the plaques were counted. The experiments were repeated three times.
For one-step growth assay, Vero cell monolayers were infected with viruses at a multiplicity of infection of 5, washed at 1 h postinfection, and then incubated at 37°C for 3, 6, 9, 12, and 24 h. Viruses were released by three successive cycles of freezing and thawing, and virus titers in the cultures were determined by plaque assay.
Site-directed mutagenesis of a stem-loop structure at the most 5′ end of the Aichi virus genome.
An EcoRI fragment of pAV-FL, containing the T7 promoter sequence and the 5′-end 384 nt of the genome, was subcloned into the EcoRI site of pUC118. Site-directed mutagenesis was carried out by inverse PCR using this subclone, a high-fidelity DNA polymerase (KOD–Plus [Toyobo, Inc.]), and appropriate oligonucleotide primers. Primer pairs used for construction of mutants and sequences of the primers were as follows: mut1, primers A (5′-CGGCCCCCTCACCCTCTTTTCCGGTGGTCT; plus sense) and B (5′-AGcgggggCCACCCCCTTTTCAAACCTATAGTGA; minus sense); mut2, primers C (5′-CGcgggggTCACCCTCTTTTCCGGTGGTCT; plus sense) and D (5′-AGGCCCCCCCACCCCCTTTTCAAACCTA; minus sense); mut3, primers C and B; mut4, primers A and E (5′-AGGCCCCCagtgggaCTTTTCAAACCTATAGTGAGTCGT; minus sense); mut5, primers F (5′-CGGCCCCCggtggggCTTTTCCGGTGGTCTGGTCCCGGA; plus sense) and D; mut6, primers F and E; mut7, primers A and G (5′-AGGCCCCCCCACCCCgaaaagAAACCTATAGTGAGTCGT; minus sense); mut8, primers H (5′-CGGCCCCCTCACCCTgaaaagCGGTGGTCTGGTCCCGGA; plus sense) and D; mut9, primers H and G; mut10, primers I (5′-CGGCCCCCTCAtCtTCTTTTCCGGTGGTCTGGTCCCGGA; plus sense) and D; mut11, primers J (5′-CGGCCCCCTCAaCaTCTTTTCCGGTGGTCTGGTCCCGGA; plus sense) and D; mut12, primers K (5′-aaGCCCCCTCACCCTCTTTTCCGGTGGTCT; plus sense) and L (5′-ttGCCCCCCCACCCCCTTTTCAAACCTA; minus sense), mut13: primers M (5′-tttccggagtccctcttggaCGGTGGTCTGGTCCCGGACCA; plus sense) and N (5′-ttcccggagacccctcttgaaCCTATAGTGAGTCGTATTACAATTCAAGG; minus sense); and mut14, primers O (5′-CGGCtCtCTCACCCTCTTTTCCGGTGGTCT; plus sense) and P (5′-AGaCCCCCCCACCCCCTTTTCAAACCTATA; minus sense). In these sequences, mutated nucleotides are indicated by lowercase letters. PCR products were self-ligated, and the derived clones were subjected to sequencing to confirm the presence of expected mutations and the absence of unexpected mutations. EcoRI fragments of those clones were ligated into pAV-FL from which the EcoRI fragment had been removed, generating various full-length cDNA clones carrying the site-directed mutations in SL-A, the first stem-loop structure at the 5′ end (see Fig. 4).
FIG. 4.
Predicted secondary structure of the 5′-end 120 nt of the Aichi virus genome. The newly identified 32 nt of the Aichi virus genome is boxed. Three stem-loops were termed SL-A, SL-B, and SL-C.
To investigate the potency of mutant RNAs to produce viable viruses, Vero cell monolayers in a 35-mm dish were transfected with 1 μg of RNA using Lipofectin reagent. At 6 h after transfection, cells were washed and then cultured in 2 ml of Eagle minimum essential medium containing 5% FCS at 37°C. After 72 h, cells were disrupted by three freeze-thaw cycles, and the virus titers in these cultures were determined by plaque assay.
Dot blot hybridization.
In vitro transcripts of 20 μg were electroporated into 107 Vero cells with a 0.4-cm cuvette at a setting of 980 V and 25 μF using a Gene Pulser (Bio-Rad), and then the cells were cultured in four or five 35-mm dishes. At several time points after electroporation, a total RNA was extracted from cells using Trizol Reagent (Life Technologies, Inc.). Each total RNA sample (3 μg) was denatured and then blotted onto a nylon membrane (Hybond-N+; Amersham-Pharmacia Biotech). A BamHI fragment of pAV-FL corresponding to nt 4790 to 5253 was subcloned into the same site of pGEM-3Z (Promega), and digoxigenin (DIG)-labeled minus-sense RNA was synthesized from the plasmid with T7 RNA polymerase in the presence of DIG-UTP using a DIG RNA labeling mix (Roche Molecular Biochemicals). The membrane was incubated with the DIG-labeled RNA in hybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% N-lauroylsarcosine, 0 .02% sodium dodecyl sulfate [SDS], 2% blocking reagent) at 75°C overnight. After hybridization and being washed with 2× SSC containing 0.1% SDS for 15 min twice at room temperature and 0.1× SSC containing 0.1% SDS for 15 min twice at 68°C, the membrane was incubated with anti-DIG alkaline phosphatase-conjugated antibody (Roche Molecular Biochemicals), and then chemiluminescent detection was performed using CDP-Star (Amersham Pharmacia Biotech).
Western blotting.
Vero cells were electroporated with AV-FL and mut6 RNAs as described above. After 3 and 9 h, cells were scraped, washed with ice-cold phosphate-buffered saline (PBS), and lysed with cell lysis buffer (20 mM Tris-HCl [pH 7.4], 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate). The lysates were heated in SDS sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 2.5% 2-mercaptoethanol, 0.001% bromophenol blue, 10% glycerol) at 97°C for 4 min, and proteins were electrophoresed on an SDS–10% polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane using a semidry electroblotting apparatus (Trans-Blot SD; Bio-Rad). The membrane was blocked in PBS-T (PBS containing 0.1% Tween 20) containing 5% skim milk for 2 h at room temperature and then incubated in PBS-T containing guinea pig antiserum against Aichi virus particles for 1 h. After being washed with PBS-T, the membrane was incubated with secondary antibody (horseradish peroxidase-conjugated anti-guinea pig immunoglobulin G) for 1 h. The membrane was washed with PBS-T and detection was performed using chemiluminescence reagents (Roche Molecular Biochemicals).
Nucleotide sequence accession number.
The complete Aichi virus cDNA sequence in pAV-FL has been submitted to the DDBJ, EMBL, and GenBank databases under accession no. AB040749.
RESULTS
Identification of a novel sequence at the 5′ end of the genome.
The Aichi virus genome was previously reported to be 8,248 nt long (45). We first constructed a full-length cDNA clone with the 5′-end sequence published previously (pAV-1). However, transcripts derived from this clone did not exhibit any infectivity toward Vero cells. To clarify the reason for this failure, we analyzed the 5′ end of the genome. We first performed direct sequencing of the virion RNA, and the result showed that approximately 30 nt are present upstream of the previously reported 5′ terminus of the genome (data not shown), although we failed to obtain accurate sequence data, probably due to the stable secondary structure formation at the 5′ end of the RNA. Next, 5′-RACE was carried out. To identify the most 5′-end nucleotide of the genome, the homopolymeric tailing reaction of the first-strand cDNA was performed with dA and dT. Although most of 24 dA-tailed clones had the previously reported 5′-end or truncated sequences, 5 clones had a TnGAAAA⋯ sequence at the 5′ end. To determine the number of T residue at the 5′ end, dT-tailed 12 clones were sequenced. As a result, two clones had a TTTGAAAA⋯ sequence at the 5′ end, and no clone with one, two, or more than three T residues at the 5′ end was found. Based on the results, we determined the authentic 5′-end sequence to be TTTGAAAA⋯. Finally, a GC-rich sequence of 32 nt was newly identified at the 5′ end of the genome (Fig. 1A). The number of the newly identified nucleotides was almost consistent with that predicted by direct sequencing of the virion RNA. In the 5′ end of the genomes of cardio- and parechoviruses, the (T)TTGAAA sequence is conserved, which is followed by the sequence containing four or five G residues (Fig. 1A), and the Aichi virus genome had also this feature.
Comparison of the nucleotide and deduced amino acid sequences of the full-length cDNA clone with the previously published sequences.
Full-length cDNA clone with the novel 5′ end was constructed and termed pAV-FL (Fig. 1B). Compared with the nucleotide sequence published previously in the DDBJ, EMBL, and GenBank databases (accession no. AB010145), pAV-FL had 22 nt differences throughout the genome besides the 5′-end 32 nt. These differences included ones that cause alterations in the amino acid sequence. The cDNA clone lacked three nucleotides, i.e., the A residues at nt 1333 and 1337 and a T residue at nt 1420 of the previously published sequence. These deletions led to frameshifts and, as a result, the deduced amino acid sequence of pAV-FL had an altered amino acid sequence of 28 residues compared with the previously published sequence (Fig. 2A). We performed three independent RT-PCR analyses to amplify the cDNA fragment corresponding to nt 1 to 1544 and found the deletion of the 3 nt in all 12 clones sequenced. In addition, we confirmed that the cDNA clone used for sequence determination in the previous study (45) also lacked these three nucleotides. This 28-amino-acid sequence is predicted to correspond to amino acids 38 to 65 of VP0. Using the Maximum Matching program in the software Genetyx-Mac 10.1 (Software Development Co., Ltd.), the VP0 sequence of the Aichi virus cDNA clone was individually aligned with those of poliovirus type 1 (PV1; accession number J02281), human rhinovirus 2 (HRV2; X02316), encephalomyocarditis virus (EMCV; M81861), foot-and-mouth disease virus (FMDV) type OK1 (X00871), HAV (M14707), and human parechovirus 1 (HPEV1; L02971), the amino acid identities being calculated. The Aichi virus VP0 sequence was most similar to that of FMDV (25.0% identity), followed by EMCV (24.1%), HRV2 (22.3%), PV1 (22.1%), HAV (20.3%), and HPEV1 (19.5%). In these alignments, 10 and 8 of the 28 amino acids identified in this study were identical to those of the EMCV and HRV2 sequences, respectively (Fig. 2B).
FIG. 2.
(A) Nucleotide and deduced amino acid sequences of the region in which deletion of three nucleotides was found. Nucleotide numbers in parentheses refer to the previously published sequence. The positions of the deleted nucleotides are indicated by open triangles. The 28 amino acids and 2 nt that differ from those published previously are boxed. (B) Alignment of the amino acid sequence of Aichi virus VP0 with those of EMCV and HRV2. The amino acid numbering starts with the first amino acid of VP0. Only the N-terminal parts of the aligned sequences are shown. The altered 28 amino acids of Aichi virus sequence are underlined, with asterisks representing identical residues.
The other nucleotide differences were present at nt 44, 143, 173, 184, 798, 955, 1268, 1394, 1424, 3000, 3084, 3259, 4259, 4796, 5002, 5052, 6836, 7302, and 7467 of pAV-FL. We sequenced the cDNA clones used for sequence determination in the previous study (45) and confirmed that all of these nucleotides except for nt 44, 173, 1268, 1394, and 1424 were different between pAV-FL and the cDNA clones previously prepared. Therefore, these differences seem to result from mutations that occurred during the several passages of the virus preparation in this laboratory. The differences at nt 955 (in the L-coding region), 1268 (VP0), 3259 (VP1), 4259 (2B), 4796 (2C), 5002 (2C), and 6836 (3D) led to amino acid changes, and nt 1394 and 1424 were located in the region coding for the altered 28 amino acids.
Infectivity of transcripts from pAV-FL toward Vero cells and growth kinetics of AV-FL virus.
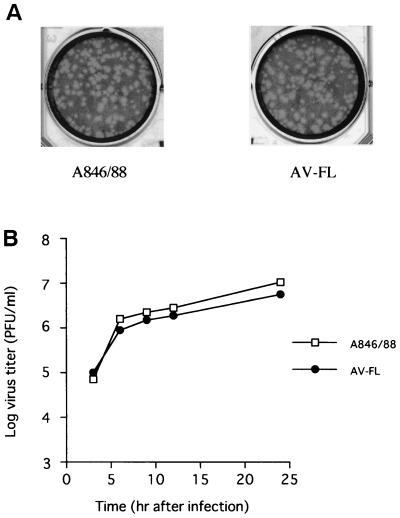
To investigate whether transcripts derived from pAV-FL are infectious toward Vero cells, the transcripts and the virion RNA were employed for transfection using Lipofectin reagent, and their plaque formation efficiency was examined. The infectivity of transcripts derived from pAV-FL averaged 5.3 × 104 PFU/μg of RNA. This infectivity was about 2% of that of the virion RNA, which produced 2.6 × 106 PFU/μg. When viruses recovered from cells transfected with the virion RNA or transcripts derived from pAV-FL were inoculated into cells, they generated similar-sized plaques (Fig. 3A). To further compare growth characteristics of the parent virus (A846/88) and the virus derived from the transcripts (AV-FL virus), growth kinetics of these viruses were examined. As shown in Fig. 3B, AV-FL virus had the similar growth property to the parent virus.
FIG. 3.
(A) Plaques formed with Aichi virus A846/88 and AV-FL virus. Cells were stained with neutral red 48 h after infection and incubated for another 24 h. (B) One-step growth curves of A846/88 and AV-FL. Vero cells were infected with viruses at a multiplicity of infection of 5. At several time points, cells were harvested and the titers of the viruses were determined by plaque assay.
Site-directed mutagenesis of the stem-loop structure at the most 5′ end of the Aichi virus genome.
The 5′-untranslated region of picornavirus genomes contains two genetic elements (32). The short 5′-terminal element is involved in RNA replication (2, 3), and the longer element is termed the internal ribosome entry site (IRES), which directs cap-independent translation (17, 25). For the 5′-terminal element, two types of secondary structure are known: one is a CL structure found in the entero- and rhinovirus genomes (3, 30), and the other is the stem-loop structure found in the cardio-, aphtho-, hepato-, and parechovirus genomes (6, 9, 11, 21, 28). The secondary structure of the 5′-terminal 120 nt, including the novel 32 nt of the Aichi virus genome, was predicted using the MFOLD program (22). The analysis suggested that the 5′ end of the genome folds into three stem-loop structures (Fig. 4). The first stem-loop structure (termed SL-A) consisting of 42 nt was similar in size to that of HAV.
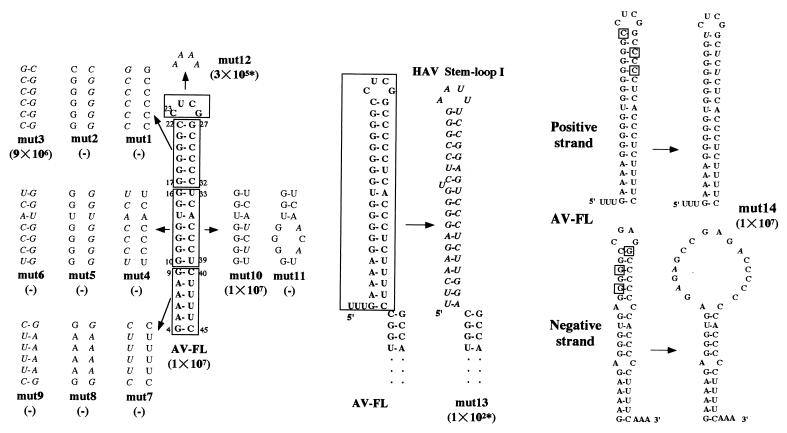
To examine the functional importance of SL-A at the most 5′ end of the Aichi virus genome on virus replication, site-directed mutational analysis was carried out (Fig. 5). First, single or double mutations to disrupt or restore the predicted structure of SL-A were introduced into the upper (mut1, mut2, and mut3), middle (mut4, mut5, and mut6) and lower (mut7, mut8, and mut9) parts of the stem segment by replacing 6 or 7 nt in each part with their complementary nucleotides. In addition, mut10 and mut11 were constructed by changing C residues at nt 36 and 38 to U and A residues, respectively. The stem structure of mut10 is predicted to be preserved, while that of mut11 would be disrupted. mut12 was constructed to assess the effect of the primary sequence of the loop segment on virus replication. Furthermore, SL-A was replaced with the HAV stem-loop I (6), which is similar to SL-A in size and shape, generating mut13.
FIG. 5.
Mutational analysis of SL-A of Aichi virus. The regions or nucleotides of wild-type SL-A, into which mutations were introduced, are boxed, and mutated nucleotides in mutants are shown in italic. These mutant RNAs were transfected into Vero cells using Lipofectin reagent. At 72 h after transfection, cells were harvested and virus titers were determined by plaque assay. The titers (in PFU/milliliter) are shown in parentheses. The asterisk indicates a small-plaque phenotype.
To test the effect of mutations on the translation efficiency, in vitro transcription-translation reactions of mutant cDNA clones were carried out in rabbit reticulocyte lysate using a TNT T7 quick-coupled transcription-translation system (Promega). No significant difference in the translation efficiency among AV-FL and its mutants was observed (data not shown).
The ability of these mutant RNAs to produce viable viruses was examined. RNAs were transfected into Vero cells using Lipofectin reagent, and virus titers in the cell cultures at 72 h after transfection were determined by plaque assay (Fig. 5). All the mutants in which the stem structure was disrupted (mut1, mut2, mut4, mut5, mut7, mut8, and mut11) lacked the ability to produce viruses. A double mutant (mut3), in which the structure in the upper part is restored, was capable of producing viruses with almost the same efficiency as AV-FL. mut10 containing the mutations to preserve the base-pairing in the middle part also produced viable viruses with high efficiency. These results suggest that the maintenance of the secondary structure is primarily essential for virus replication. The double mutations in the middle and lower parts of the stem eliminated their infectivity (mut6 and mut9), although the mutants maintained the stem structure. This suggests that the primary sequence of the middle and lower parts of the stem-loop structure is also crucial for virus replication. The virus yield of mut12, in which 4 nt in the loop segment were changed, was approximately 30-fold lower than that of AV-FL, and its plaque size was smaller. We confirmed the maintenance of the mutation in the mut12 virus by direct sequencing of a product derived from RT-PCR using a total RNA extracted from mut12 virus-infected cells. An Aichi virus-HAV chimera, in which SL-A was exchanged with the HAV stem-loop I (mut13), exhibited very low infectivity and a small-plaque phenotype. The difference of the primary sequence between the stem-loop structures of the two viruses may lead to the low efficiency of virus replication.
A stem-loop structure was predicted not only at the 5′ end of the positive strand of the Aichi virus RNA but also at the 3′ end of its negative strand (Fig. 5). It is reported that the CL structure of poliovirus is functionally required in the positive strand but not in the negative strand (3). To determine whether the secondary structure formed in the negative strand of the Aichi virus RNA is required for virus replication, we constructed mut14 harboring so-called asymmetric mutations. By changing G-C pairs of the positive strand to G-U pairs the stem segment formed at the 5′ end of the positive strand is maintained, but that formed at the 3′ end of the negative strand is disrupted in this mutant (Fig. 5). mut14 RNA produced viruses as efficiently as AV-FL RNA. Nucleotide changes in mut10 are also asymmetric mutations. SL-A of mut10 is maintained (Fig. 5), but the middle part of the stem segment of the stem-loop structure at the 3′ end of the negative strand is disrupted (data not shown). As described above, mut10 produced viruses efficiently. These results suggest that the stem-loop structure at the 5′ end of the positive strand, not at the 3′ end of the negative strand, is critical for virus replication.
RNA replication ability of mutants.
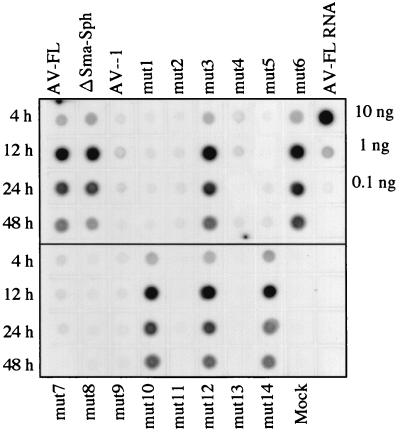
We next investigated whether the inability or low efficiency of the mutants to produce viruses is due to a defect in RNA replication. Mutant RNAs were transfected into Vero cells by electroporation and dot blot hybridization of total RNAs extracted from cells at 4, 12, 24, and 48 h after transfection was carried out using a DIG-labeled RNA complementary to nt 4790 to 5253 as a probe. As a positive control for detecting viral RNA replication, we constructed a mutant which contains an in-frame deletion spanning from nt 1387 to 1848 within the VP0 coding region (ΔSma-Sph). This mutant was unable to produce viable viruses. AV-1 that lacks the 5′-end 32 nt was also included in the assay. The signals of AV-FL, ΔSma-Sph and mutants capable of producing viruses efficiently (mut3, mut10, mut12, and mut14) was readily detected, while RNA replication in AV-1 and mut1, mut2, mut4, mut5, mut7, mut8, mut9, mut11, and mut13, which exhibited low or no infectivity, was not observed (Fig. 6). Since ΔSma-Sph RNA was detected clearly and definitely, the sensitivity in the present assay is sufficient to detect RNA replication in transfected cells. Thus, these results indicate that the inability or low efficiency of mut1, mut2, mut4, mut5, mut7, mut8, mut9, mut11, and mut13 to produce viruses is due to a defect in RNA replication.
FIG. 6.
RNA replication of AV-FL and its mutants. Total RNAs were extracted from Vero cells transfected with AV-FL and mutant RNAs at the indicated time points after transfection, and plus-strand viral RNA accumulation was examined by dot blot hybridization. As controls, 10, 1, and 0.1 ng of AV-FL RNA were dotted. The decrease of signal intensity observed at 24 and 48 h after transfection in AV-FL, ΔSma-Sph, mut3, mut6, mut10, mut12, and mut14 is thought to be due to cell death.
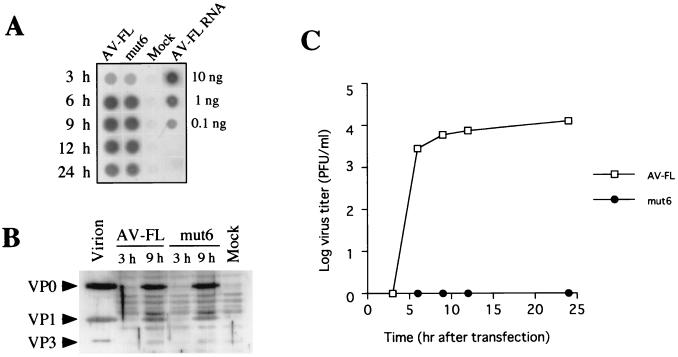
Unexpectedly, efficient RNA replication of mut6, which exhibited no infectivity (Fig. 5), was observed (Fig. 6). A defect of mut6 in the production of viable viruses was further investigated. AV-FL and mut6 RNAs were transfected into Vero cells by electroporation, and plus-strand viral RNA accumulation, synthesis of capsid proteins, and virus titers in transfected cells were examined. Dot blot hybridization using the extracted total RNAs showed that RNA replication in mut6 was as efficient as AV-FL (Fig. 7A). To examine the synthesis of capsid proteins in cells transfected with AV-FL and mut6, Western blotting using antiserum against purified virus particles was carried out. At 9 h after electroporation, synthesis of VP0 and VP1 was clearly detected in cells transfected with mut6 as well as in AV-FL (Fig. 7B). In contrast, the plaque assay showed that mut6 did not generate viruses even at 24 h after electroporation, while AV-FL RNA produced viruses at 6 h after electroporation (Fig. 7C). We repeated this experiment by using transcripts derived from two distinct mut6 clones that were constructed independently. The results were the same. This would rule out the possibility of the presence of unexpected mutations in other region of the genome. Thus, these results showed that the compensatory mutation of the both nucleotide stretches in the middle part of the stem did not affect the RNA replication and translation efficiencies but abolished production of viable viruses.
FIG. 7.
RNA replication, protein synthesis, and virus production in Vero cells electroporated with AV-FL and mut6. (A) Dot blot analysis of plus-sense viral RNA accumulation in cells. Total RNAs were extracted from cells transfected with AV-FL and mut6 at the indicated time points after electroporation. The total RNA samples were dotted and probed with DIG-labeled minus-sense viral RNA. (B) Detection of capsid proteins by Western blotting. Cells were lysed at 3 and 9 h after electroporation with AV-FL and mut6. Each lysate was subjected to SDS–10% polyacrylamide gel electrophoresis, and capsid proteins were detected by Western blotting using antiserum raised against purified virus particles. As a control, purified virion was analyzed. Positions of VP0, VP1, and VP3 are indicated on the left. (C) Virus yields in cells electroporated with AV-FL and mut6. At the indicated time points after electroporation, cells were harvested and the virus titer was examined by plaque assay.
DISCUSSION
In this study, we constructed a full-length cDNA clone of Aichi virus, pAV-FL, and in vitro transcripts from this clone was shown to produce infectious viruses. The efficiency of plaque formation in cells transfected with transcripts derived from pAV-FL was about 50-fold lower than that in cells transfected with the virion RNA. However, since the plaque morphology and growth kinetics of AV-FL virus and the parent virus were similar to each other (Fig. 3), we considered that we successfully cloned an Aichi virus full-length cDNA. The extra nucleotides GG at the 5′ end of the transcripts may affect the first cycle of virus replication.
During construction of pAV-FL, we identified a novel sequence of 32 nt at the 5′ end of the genome (Fig. 1A). The computer-aided prediction of the 5′-end 120 nt of the genome containing the newly identified 32 nt suggested that the sequence folds into three stem-loops (Fig. 4). The 5′ end of picornavirus genomes has been reported to maintain two types of secondary structure: one is a CL structure found in the entero- and rhinovirus genomes (3, 30), and the other is the stem-loop structure found in the cardio-, aphtho-, hepato-, and parechovirus genomes (6, 9, 11, 21, 28). The CL structure at the 5′ end of the poliovirus genome is known to be necessary for viral RNA replication (3). On the other hand, the functional importance of the stem-loop found in the cardio-, aphtho-, hepato-, and parechovirus genomes in virus replication has not been sufficiently investigated. In the present study using various site-directed mutants, we examined the importance of SL-A, the most 5′-end stem-loop, in virus replication.
Our data indicated that SL-A is an element involved in viral RNA replication. This means that the two different types of structure at the 5′ end of picornavirus genomes, a stem-loop and a cloverleaf structure, have a common function. As reported in the CL structure of poliovirus (3), proper folding of the secondary structural element at the 5′ end of the positive strand of Aichi virus RNA, but not that at the 3′ end of the negative strand, was found to be required for its function. In addition, the primary sequence of the bottom region of SL-A was functionally significant. It is also reported in the poliovirus CL that a compensatory mutation by replacing the both nucleotide stretches of “Stem A,” which consist of nt 2 to 8 and 82 to 88 in PV1, with their complementary nucleotides is lethal (3). SL-A could not be functionally substituted by the HAV stem-loop I, which is similar to SL-A in size and shape (mut13). This result would be explained by the sequence difference of the bottom region between the Aichi virus and the HAV stem-loops. The effect of an unpaired uridine residue in the middle part of the HAV stem-loop I (Fig. 5) on the infectivity of mut13 remains to be tested.
On the other hand, mutation of the nucleotides in the loop segment of SL-A had only moderate effect on virus replication. Although mut12 had a small-plaque phenotype, it produced viruses with high efficiency (Fig. 5). In the dot blot hybridization analysis, no significant difference in the RNA replication efficiency was observed between AV-FL and mut12 (Fig. 6). This is in contrast to the finding that the nucleotide sequences of loop segments of the poliovirus CL structure have significant roles in RNA replication. It is reported that mutations of the sequences of the loop segments in stem-loops B and D of the CL structure, such as nucleotide changes or insertions, abolish interaction with PCBP and 3CD, respectively, and affect RNA replication severely (2, 3, 24, 26). If some factors involved in viral RNA replication interact with the 5′ end of the Aichi virus genome, the primary sequence of the loop segment of SL-A may not be a determinant responsible for the recognition by the factors. The 5′ end of the HAV genome folds into three stem-loop structures. A precursor polypeptide 3ABC of HAV interacts with the second stem-loop, and a more stable RNP complex is formed with the sequence containing the three stem-loop structures (19). Therefore, although SL-A of Aichi virus is an important structural element for RNA replication, it is possible that a longer sequence containing the three stem-loop structures is required for interaction with viral or cellular factors essential for viral RNA replication. Further studies to investigate what viral or cellular proteins interact with the 5′ end of the Aichi virus genome containing the three stem-loop structures will be needed in order to understand how the 5′ end of the Aichi virus genome functions as a replication signal.
This study also demonstrated that SL-A plays an essential role in the production of viable viruses at some stage other than viral RNA replication during virus infection. In mut6, which has a double mutation in the middle part of SL-A, RNA replication and protein synthesis occurred efficiently (Fig. 6 and 7A and B), but the mutant was unable to produce viable viruses (Fig. 5 and 7C). This result indicates that the primary sequence of the middle part of the stem is not crucial for viral RNA replication but for production of infectious viruses, if SL-A is folded properly. Considering that mut10, in which the downstream nucleotide stretch (nt 33 to 39) of the middle part of the stem was mutated, showed high infectivity (Fig. 5), nt 10 to 16 may be more important for the production of viable viruses than nt 33 to 39. It is now unknown whether the secondary structure of SL-A as well as the primary sequence of the middle part of the stem has an important role in the production of infectious viruses because disruption of base pairing of the stem abolished RNA replication.
A possible explanation for a defect of mut6 in the production of viable viruses is that mut6 RNA is not encapsidated. During poliovirus infection, since only newly synthesized positive strands are packaged, it is thought that RNA replication and packaging are directly coupled (23). Specific interactions between capsid proteins and proteins of the viral RNA replication complex occur, and newly synthesized viral RNAs that emerges from the replication complex are encapsidated through interaction between capsid proteins and the viral RNA (23). According to this model, interaction between capsid proteins and proteins of the RNA replication complex would occur in the case of mut6. The sequence of the middle part of SL-A may be involved in the next step, which is responsible for the initiation of encapsidation of RNA. One possible role of this RNA sequence of SL-A in encapsidation would be to interact with capsid proteins. Since the poliovirus RNA in which the capsid-coding region was replaced with a foreign gene was encapsidated by capsid proteins that were provided in trans by a helper virus (4, 29), the capsid-coding region is thought not to be involved in the specific encapsidation process. The IRES region of poliovirus (nt 109 to 742) can be substituted by the EMCV IRES without a significant defect in encapsidation, suggesting that the poliovirus IRES also does not contain a signal essential for encapsidation of the poliovirus RNA (1). However, RNA sequences specifically recognized by the capsid proteins have not yet been identified in picornaviruses. The result obtained in this study suggests the possibility that the 5′ end of picornavirus genomes plays an important role in encapsidation. Alternatively, mut6 RNA can be encapsidated, but the resultant virus particles may not be infectious due to a defect in certain early step of the infection cycle, e.g., an uncoating step. Further studies are required to clarify a role of SL-A in production of infectious viruses.
ACKNOWLEDGMENTS
This work was supported in part by a grant for the Human Science Research Foundation of Japan and a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture, Tokyo, Japan.
REFERENCES
- 1.Alexander L, Lu H H, Wimmer E. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosome entry site elements: genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci USA. 1994;91:1406–1410. doi: 10.1073/pnas.91.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 4.Ansardi D C, Moldoveane Z, Porter D C, Walker D E, Conry R M, LoBuglio A F, McPherson S, Morrow C D. Characterization of poliovirus replicons encoding carcinoembryonic antigen. Cancer Res. 1994;54:6359–6364. [PubMed] [Google Scholar]
- 5.Bazan J F, Fletterick R J. Viral cysteine proteases are homologous to the trypsin-like serine proteases: structural and functional implications. Proc Natl Acad Sci USA. 1988;85:7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown E A, Day S P, Jansen R W, Lemon S M. The 5′ nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J Virol. 1991;65:5828–5638. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke B E, Sanger D V. Processing and assembly of foot-and-mouth disease virus proteins using subgenomic RNA. J Gen Virol. 1988;69:2313–2325. doi: 10.1099/0022-1317-69-9-2313. [DOI] [PubMed] [Google Scholar]
- 8.Dougherty W G, Semler B L. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duke G M, Hoffman M A, Palmenberg A C. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 11.Ghazi F, Hughes P J, Hyypia T, Stanway G. Molecular analysis of human parechovirus type 2 (formerly echovirus 23) J Gen Virol. 1998;79:2641–2650. doi: 10.1099/0022-1317-79-11-2641. [DOI] [PubMed] [Google Scholar]
- 12.Graff J, Cha J, Blyn L B, Ehrenfeld E. Interaction of poly(rC) binding protein 2 with the 5′ noncoding region of hepatitis A virus RNA and its effects on translation. J Virol. 1998;72:9668–9675. doi: 10.1128/jvi.72.12.9668-9675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome: identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 14.Hellen C U T, Wimmer E. Maturation of poliovirus capsid proteins. Virology. 1992;187:391–397. doi: 10.1016/0042-6822(92)90440-z. [DOI] [PubMed] [Google Scholar]
- 15.Hughes P J, Stanway G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J Gen Virol. 2000;81:201–207. doi: 10.1099/0022-1317-81-1-201. [DOI] [PubMed] [Google Scholar]
- 16.Hyypia T, Horsnell C, Maaronen M, Khan M, Kalkkinen N, Auvinen P, Kinnunen L, Stanway G. A distinct picornavirus group identified by sequence analysis. Proc Natl Acad Sci USA. 1992;89:8847–8851. doi: 10.1073/pnas.89.18.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang S K, Krausslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King A M Q, Brown F, Christian P, Hovi T, Hyypia T, Knowles N J, Lemon S M, Minor P D, Palmenberg A C, Skern T, Stanway G. Picornaviridae. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carsten E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. New York, N.Y: Academic Press, Inc.; 1999. p. 996. [Google Scholar]
- 19.Kusov Y Y, Morace G, Probst C, Gauss-Muller V. Interaction of hepatitis A virus (HAV) precursor proteins 3AB and 3ABC with the 5′ and 3′ termini of the HAV RNA. Virus Res. 1997;51:151–157. doi: 10.1016/s0168-1702(97)00089-0. [DOI] [PubMed] [Google Scholar]
- 20.Kusov Y Y, Gauss-Muller V. In vitro RNA binding of the hepatitis A virus proteinase 3C (HAV 3Cpro) to secondary structure elements within the 5′ terminus of the HAV genome. RNA. 1997;3:291–302. [PMC free article] [PubMed] [Google Scholar]
- 21.Le S-Y, Chen J-H, Sonenberg N, Maizel J V., Jr Conserved tertiary structural elements in the 5′ nontranslated region of cardiovirus, aphthovirus and hepatitis A virus RNAs. Nucleic Acids Res. 1993;21:2445–2451. doi: 10.1093/nar/21.10.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 23.Nugent C I, Johnson K L, Sarnow P, Kirkegaard K. Functional coupling between replication and packaging of poliovirus replicon RNA. J Virol. 1999;73:427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsley T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 25.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 26.Percy N, Barclay W S, Sullivan M, Almond J F. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J Virol. 1992;66:5040–5046. doi: 10.1128/jvi.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piccone M E, Zellner M, Kumosinski T F, Mason P W, Grubman M J. Identification of the active-site residues of the L proteinase of foot-and-mouth disease virus. J Virol. 1995;69:4950–4956. doi: 10.1128/jvi.69.8.4950-4956.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilipenko E V, Blinov V M, Agol V I. Gross rearrangements within the 5′-untranslated region of the picornaviral genomes. Nucleic Acids Res. 1990;18:3371–3375. doi: 10.1093/nar/18.11.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porter D C, Ansardi D C, Morrow C D. Encapsidation of poliovirus replicons encoding the complete human immunodeficiency virus type 1 gag gene by using a complementation system which provides the P1 capsid protein in trans. J Virol. 1995;69:1548–1555. doi: 10.1128/jvi.69.3.1548-1555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivera V M, Welsh J D, Maizel V. Comparative sequence analysis of the 5′ noncoding region of the enteroviruses and rhinoviruses. Virology. 1988;165:42–50. doi: 10.1016/0042-6822(88)90656-3. [DOI] [PubMed] [Google Scholar]
- 31.Roberts P J, Belsham G J. Identification of critical amino acids within the foot-and-mouth disease virus leader protein, a cysteine protease. Virology. 1995;213:140–146. doi: 10.1006/viro.1995.1554. [DOI] [PubMed] [Google Scholar]
- 32.Rohll J B, Percy N, Ley R, Evans D J, Almond J W, Barclay W S. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J Virol. 1994;68:4384–4391. doi: 10.1128/jvi.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roos R P, Kong W, Semler B L. Polyprotein processing of Theiler's murine encephalomyelitis virus. J Virol. 1989;63:5344–5353. doi: 10.1128/jvi.63.12.5344-5353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Field's virology. New York, N.Y: Raven Press, Ltd.; 1996. pp. 609–654. [Google Scholar]
- 35.Ryan M D, Belsham G J, King A M Q. Specificity of substrate-enzyme interactions in foot-and-mouth disease virus polyprotein processing. Virology. 1989;173:35–45. doi: 10.1016/0042-6822(89)90219-5. [DOI] [PubMed] [Google Scholar]
- 36.Stanway G, Kalkkinen N, Roivainen M, Ghazi F, Khan M, Smyth M, Meurman O, Hyypia T. Molecular and biological characteristics of echovirus 22, a representative of a new picornavirus group. J Virol. 1994;68:8232–8238. doi: 10.1128/jvi.68.12.8232-8238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strebel K, Beck E. A second protease of foot-and-mouth disease virus. J Virol. 1986;58:893–899. doi: 10.1128/jvi.58.3.893-899.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi K, Kojima K, Urasawa S. Nondefective rotavirus mutants with an NSP1 gene which has a deletion of 500 nucleotides, including a cysteine-rich zinc finger motif-encoding region (nucleotides 156–248), or which has a nonsense codon at nucleotides 153–155. J Virol. 1996;70:4125–4130. doi: 10.1128/jvi.70.6.4125-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toyoda H, Nicklin M J H, Murray M G, Anderson C W, Dunn J J, Studier F W, Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45:761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 40.Xiang W, Cuconati A, Paul A V, Cao X, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA. 1995;1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang W, Harris K S, Alexander L, Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. 1991;164:954–957. doi: 10.1093/infdis/164.5.954. [DOI] [PubMed] [Google Scholar]
- 43.Yamashita T, Sakae K, Kobayashi S, Ishihara Y, Miyake T, Mubina A, Isomura S. Isolation of cytopathic small round virus (Aichi virus) from Pakistani children and Japanese travelers from Southeast Asia. Microbiol Immunol. 1995;39:433–435. doi: 10.1111/j.1348-0421.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 44.Yamashita T, Sakae K, Kobayashi S, Isomura S, Utagawa E. Prevalence of newly isolated, cytopathic small round virus (Aichi strain) in Japan. J Clin Microbiol. 1993;31:2938–2943. doi: 10.1128/jcm.31.11.2938-2943.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita T, Sakae K, Tsuzuki H, Suzuki Y, Ishikawa N, Takeda N, Miyamura T, Yamazaki S. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J Virol. 1998;72:8408–8412. doi: 10.1128/jvi.72.10.8408-8412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamashita T, Sugiyama M, Suzuki H, Sakae K, Suzuki Y, Miyazaki Y. Application of a reverse transcription-PCR for identification and differentiation of Aichi virus, a new member of the picornavirus family associated with gastroenteritis in humans. J Clin Microbiol. 2000;38:2955–2961. doi: 10.1128/jcm.38.8.2955-2961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]