Abstract
Raman imaging has the capability to provide unlabeled, spatially aware analysis of chemical components, with no a priori assumptions. Several lifestyle diseases such as nonalcoholic steatohepatitis (NASH) can appear in the liver as changes in the nature, abundance, and distribution of lipids, proteins, and other biomolecules and are detectable by Raman imaging. In order to identify which of these liver-associated changes occur as a direct result of the diet and which are secondary effects, we developed correlative imaging and analysis of diet and liver samples. Oleic acid was found to be a direct contributor to NASH liver composition, whereas protein and collagen distributions were found to be affected in a manner consistent with early fibrotic transformation, as a secondary consequence of the high-fat diet.
Keywords: Raman imaging, MCR−ALS, liver, NASH, lipids, collagen, fibrosis
Introduction
Nonalcoholic fatty liver disease and the more severe form of nonalcoholic steatohepatitis (NASH) are diseases where fat buildup in the liver can lead to cirrhosis and potentially fatal consequences.1,2 There is currently a lack of understanding especially in terms of the molecular mechanisms of the disease.3 While it is known that diet plays a significant role,2 complex metabolic pathways can make it challenging to trace direct links between diet and resulting liver pathology.4 Fatty acids can be present both in the diet and in the liver, with the types and ratios being particularly significant to NASH progression,5,6 and are emerging as therapeutic targets.7,8 Understanding how fatty acids and other components cause and/or correlate with NASH is important, but there are few methods to map different biochemical components across the liver. In particular, microscopic stains are limited in liver imaging, and, with a few exceptions,9 are not specific to individual fatty acids. However, unlabeled optical methods show promise.
Raman spectroscopy is an unlabeled method that can be used as an imaging mode which is particularly sensitive to different fatty acids and can make quantitative comparisons between them,10 while also being able to visualize other relevant cell/tissue components.11
Links between diet and liver function are usually studied in humans by a range of nonimaging analysis methods,12 compared with assays of biomarkers for liver function.13 Diet has been considered in the context of disease, by Raman imaging of metabolic processes,14 but has not to our knowledge been directly included in the same chemometric model as the target organism. Here, we aimed to develop a simple system of Raman imaging and analysis and use it to directly show links between the dietary intake and the molecular distributions in the liver without requiring staining, a priori information, or destruction of the samples.
To achieve this, Raman imaging uses a spectrometer to detect the optical spectrum scattered from molecules in a sample.15 Excited by laser light, the molecular composition of the sample can then be determined with high spatial resolution as the excitation beam probes each location. Biological tissue can be challenging to image due to autofluorescence but has been achieved in ex vivo liver tissue16 or simpler tissue models such as organoids.17 Using longer wavelengths in the near-infrared, the Raman scattering signal is reduced; however, the lower autofluorescence and reduced susceptibility to photodamage can actually allow for higher quality imaging and spectral analysis. This is especially useful with ex vivo samples where the sample composition may be both complex and unknown. While overall composition can be measured by homogenizing samples and measuring chemical components, using Raman imaging allows comparison of the chemical components as well as their distributions. To more directly link dietary components and the resulting spatial distributions in the liver, we therefore used Raman imaging with an excitation wavelength of 785 nm and measured regions in feed pellets as well as in 10 μm thick sections of liver, for both control (chow) and NASH diet. By first analyzing a joint data set of similar numbers of pellet and liver spectra, we obtained chemical components present in pellet or liver (or both) by multivariate curve resolution–alternating least-squares (MCR–ALS). The choice of MCR–ALS from the many different multivariate analysis approaches was made due to its ability to effectively isolate spectrally pure non-negative components that can be easily compared to reference spectra.18 Using the generated model, we then quantitatively determined the presence or absence of those components in the larger liver data set, along with their respective spatial distributions.
Methods
Sample Preparation
Chow (4.78% crude fat, 0.19% choline) and NASH (28.2% crude fat, 0.11% methionine) mouse food pellets were obtained from CLEA Japan and Oriental Yeast, respectively, sterilized with 30 kGy γ-ray. Pellets were cut in half immediately prior to Raman measurements, and the Raman spectra were recorded from the freshly opened surface of the pellets. Mice were maintained under specific pathogen-free conditions on a 12 h light–dark cycle with free access to food (chow or NASH) and water. We used C57BL/6JmsSlc male 6-week-old mice (Japan SLC). After 8 weeks, mice were sacrificed by cervical dislocation. Liver was extracted, embedded in O.C.T compound (Sakura Finetek) in a plastic mold, immediately frozen in liquid nitrogen and stored at −80 °C, and then later microtomed into 10 μm thick sections before imaging. These sections were placed on a quartz-bottomed cell culture dish (FPI, Japan) and then immersed in 2 mL of Dulbecco’s phosphate-buffered saline without calcium or magnesium (D-PBS(−)) (Nacalai Tesque, Japan) immediately prior to Raman measurement. All mice experiments were approved by the Animal Research Committee of the Research Institute for Microbial Diseases, Osaka University.
Tissue Staining
As a qualitative morphology comparison, Oil Red O (for lipid imaging) and Sirius Red (for collagen) stains were done after equilibrating at room temperature on liver sections that were then sealed under a coverslip. Imaging was done using a ×2.5/0.06 NA A-Plan objective lens and a Canon EOS R detector. Between 12 and 26 images were collected for each liver slice and stitched together using Adobe Photoshop CS5. Stitching was only used for the bright-field imaging in this step and not for any Raman data or analysis.
Raman Imaging
Raman measurements were performed on unstained tissue sections using a Raman-11 laser microscope (Nanophoton, Japan) at 785 nm, grating with 600 l/mm, resulting in a spectral range of 728.9– 1806.6 cm–1, and collected onto a Pixis 400 CCD camera (Princeton, USA). Spectrometer calibration was performed using the 520 cm–1 peak from a silicon sample as well as measuring an ethanol (99.5%, Nacalai Tesque, Japan) spectrum and verifying the positions of the five main peaks in the fingerprint region. For pellet measurements, spectra were recorded using a ×20/0.45 NA magnification objective (Nikon, Japan), 3 s per spectrum and with a laser power of 105 mW at the sample plane. Spectra were collected in sets of nine (3 × 3 spectra with a spacing of 9 μm between spectra) from 16 different regions across the inner surface of the pellet. For chow pellets, due to the relatively high levels of background caused by autofluorescent materials in the sample, the Raman spectra were collected after first irradiating the sample collection spot with 785 nm excitation at 105 mW with four exposures of 3 s each, with a 10 s pause between each exposure to allow for any built-up heat to dissipate, resulting in a total of 12 s of exposure at each sampling point before the Raman measurement was taken. This autofluorescence reduction step was not required for the NASH pellet samples. For the tissue measurements, images were obtained from liver sections from two mice for each diet (i.e., two mice fed on chow diet and two mice fed on NASH diet). Positions were selected to cover two regions close to the edge of the liver as well as two regions deeper into the organ for both chow and NASH fed liver samples. Raman spectra were recorded using a ×40/0.8 NA water immersion lens (Nikon, Japan) for 5 s per spectrum and a laser power of 95 mW at the sample plane. For each tissue image, spectra were recorded at 2 μm spacing over an area of 160 μm × 160 μm square, resulting in a Raman image size of 80 × 80 pixels square. No damage was observed in any of the samples during Raman measurements.
Data Analysis
Raman spectra were preprocessed in MATLAB by baseline correction with PLS Toolbox (Eigenvector Research) using a fourth-order weighted least-squares algorithm and “filter and despike” cosmic ray removal (with parameters as follows, Algorithm: Despike, Window: 5, Mode: 2, Tolerance: 2, TRB Flag: Bottom, Threshold: 3). In order to avoid potential biases in the MCR model that could originate from using vastly different numbers of spectra from each sample type, we generated subsets of the Raman spectra contained in each of the tissue images (created by taking every 160th spectrum from each of the measured tissue sections, creating a 40 pixel image for each tissue section). To then generate the MCR model, a pseudoimage was constructed containing all spectral data from the two pellet samples and the two subsets of the tissue spectra. This produced a data set where the number of pixels from each sample type was roughly similar (144 pixels per food pellet sample type and 160 pixels per tissue sample type). A graphical representation of the data set used to create the MCR model is shown in Figure S1. By using both pellet and liver tissue in the model, we ensure that common spectral features can be determined and relative abundance can be quantified. Since we have both the relative abundance as well as the spatial distribution, both sets of information can be used as to whether a component is present and whether the distribution is random or aggregated, as well as whether it excludes or is colocalized with other components. MCR was performed using the “fasternnls” non-negativity constraints for both spectra and concentration. The number of components extracted was set to 9 as this was the minimum number of components to achieve a model fit of ≥95% (nine components = 95.07% cumulative fit). The Raman images taken from the liver tissue samples from mice fed on chow and NASH food pellets were then projected onto the MCR model in order to quantify the presence/absence of the individual MCR components in the liver tissue sections and assess their distribution, if present.
Results and Discussion
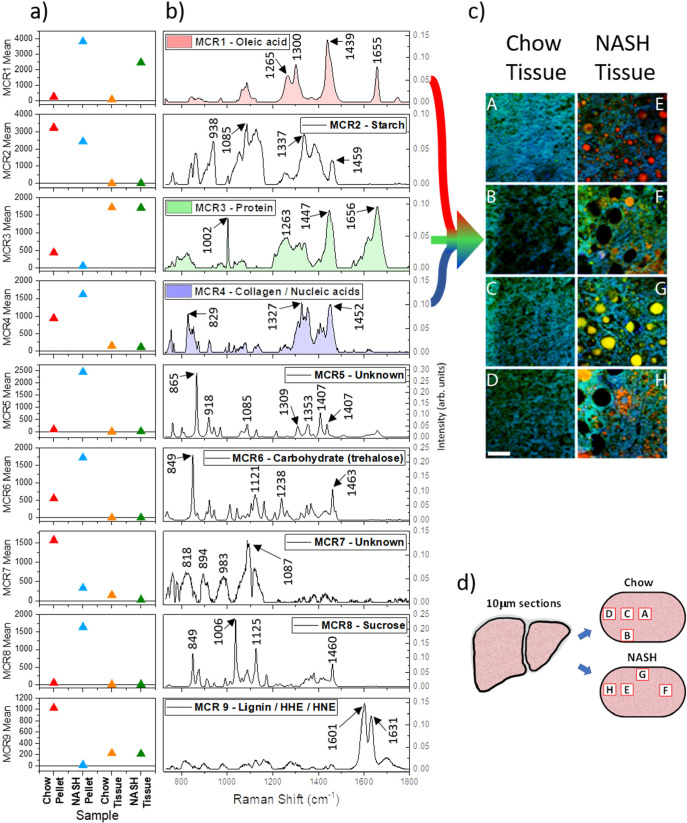
The mean scores for the nine MCR components are shown in Figure 1a and are derived from the mean intensities score for all pixels from each sample: chow pellet, NASH pellet, and resultant liver sections for each diet type. The corresponding spectra for each MCR component are shown in Figure 1b. Each spectrum is derived from the combined diet/tissue data set, with no a priori information used. Several spectra match closely to reference spectra available in the literature and/or spectra measured from purified samples in our laboratory. A summary of assignments and supporting spectral references is shown in Table 1. For reference, a selection of spectra averaged from different regions (i.e., high lipid vs low lipid) are also shown in the Supporting Information (Figure S2).
Figure 1.
(a) Mean scores for the nine chemical components generated by MCR–ALS analysis. The colored triangles indicate the contribution of each component for chow pellet (red triangles), NASH pellet (blue triangles), and corresponding chow tissue (orange triangles) and NASH tissue (green triangles). (b) MCR component spectral profiles for all nine emergent components. Component 1 (red undershading), component 3 (green undershading), and component 4 (blue undershading) have been used as color channels to create the RGB image shown in (c). (c) False color RGB image of the distribution of component 1 (red), component 3 (green), and component 4 (blue) in the chow (panels A–D) and NASH (panels E–H) tissue sections. The scale bar represents 40 μm. (d) Schematic showing the approximate relative positions within liver sections that correspond to locations of Raman images presented in panel (c).
Table 1. Assignments Linking MCR Derived Components to Molecular Contenta.
| MCR component | chemical assignment |
|---|---|
| 1 | oleic acid |
| 2 | starch |
| 3 | protein |
| 4 | collagen/nucleic acids |
| 5 | unknown |
| 6 | carbohydrate/trehalose |
| 7 | unknown |
| 8 | sucrose |
| 9 | lignin/HHE/HNE |
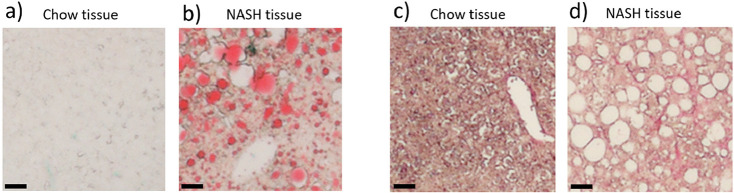
The spectral profile of MCR component 1 is very similar to that of the Raman spectrum of oleic acid, suggesting that the NASH diet is relatively rich in oleic acid10 and that these high oleic acid levels in the feed are absorbed by the body and eventually incorporated into cells. Although the chow pellet and tissue do not show strong intensities for component 1, it should be noted that there is some intensity present throughout the chow pellet and tissue, reflecting the fact that oleic acid is the major dietary monounsaturated fatty acid and the most abundant fatty acid in the human body,29 suggesting we would also expect to see some levels of oleic acid in healthy mouse tissues. For comparison, Figures 2a and b show example regions of liver tissue from chow and NASH fed mice that have been stained for lipids by Oil Red O. Although this stain is not specific to oleic acid, the distributions of Oil Red O staining and MCR component 1 in the Raman data (see Figure 1c, red channel and Figure S5) are similar, i.e., negligible in the chow fed liver, while notable as droplets throughout the NASH liver. In addition, the Oil Red O staining in the NASH liver can be observed as both small droplets within the tissue and larger droplets forming in the holes of the liver tissue, which is consistent with our observations for MCR component 1.
Figure 2.
Liver tissue sections from chow and NASH fed mice stained with Oil Red O or Sirius Red: (a) Oil Red O—chow, (b) Oil Red O—NASH, (c) Sirius Red—chow, and (d) Sirius Red—NASH. Scale bar represents 40 μm. An overview of a whole liver section for each tissue type for both stains is shown in Figure S2.
The spectral profile of component 2 suggests the majority of this component is associated with the presence of starch,19 and the mean values for each of the samples support this assignment as both the diet pellets show high mean values with a relatively even distribution throughout the pellets, whereas the tissue sections have average values very close to zero.
Component 3 exhibits a combination of a sharp band close to 1000 cm–1, relatively broad bands at 1447 and 1658 cm–1, and a broad band envelope between ∼1200 and 1350 cm–1, making this typical of proteins.20 Therefore, component 3 reflects the bulk of the protein distribution within the samples, where the chow pellet contains slightly more protein than the NASH pellet and where the protein contents in the tissues are similar in abundance to each other (although with changes in distribution induced by the presence of holes in the NASH tissue).
Component 4 is found predominantly in the two pellet samples but is also present at lower levels in the two tissue samples. The molecular components that are contributing to component 4 are not clear. The spectral profile does share some similarities with collagen in the 1200–1500 cm–1 region,30 but the corresponding Amide I band expected around 1650 cm–1 is not present.20,30 Additionally, nucleic acids are known to provide Raman signals at approximately 800–850 cm–1 from the phosphate backbone, while bands originating from the bases can be observed in the 1300–1500 cm–1 region.21,22 However, nucleic acid bands from bases, particularly adenine and guanine, would also be expected between 1500 and 1700 cm–1, and the phosphate backbone in both RNA and DNA gives rise to a relatively strong band around the 1100 cm–1 region,21,22 all of which are missing in the spectral profile of component 4. Figure 2b shows Sirius Red staining for the chow and NASH liver tissue, showing the distribution of collagen within these samples. The pattern of holes is clearly evident in the NASH liver tissue. The appearance of fibril-like structures in the NASH tissue is consistent with patterns visible in the Raman data, with MCR component 4 (which we assigned to collagen and nucleic acids in Figure 1c, blue channel and Figure S5) showing similar fibril-like structures.
Component 5 is mainly present in the two diet pellets, with low amounts observed in the chow pellet and high levels in the NASH pellet. These high values are not evenly distributed within the NASH pellet and are instead located in a few of the regions measured (Figure S4). This spectrum is currently unassigned.
The spectral profile of component 6 shows notable similarity to that of trehalose,23 with possible additional contributions from pectin (particularly the very intense band in the 848 cm–1 region24), suggesting component 6 reflects some of the carbohydrate content of the diet pellets, particularly disaccharides and soluble fiber. This component is found throughout both the chow and NASH pellets, with particularly high levels found in two measurement regions for the NASH pellet (Figure S4b), but is not strongly observed in either tissue type (figure S5).
The region up to 1000 cm–1 in the spectral profile of component 7 is affected by distortions introduced into the spectra by baseline correction of spectra with very high baseline distortions from autofluorescent molecules present in the measured positions. This primarily affected one measurement region within the chow pellet (Figure S4). However, the pair of bands between 1000 and 1050 cm–1 is observed in the raw spectra from multiple regions within the diet pellets. Unfortunately, due to the limited spectral information it is unclear what molecules are contributing to this component.
The spectral profile of component 8 is a close match to that of sucrose in its crystalline form.25 The highest levels of this component are present in the NASH pellet, and it is found predominantly within one measurement area, although all areas measured in the NASH tissue do show some contrast for component 8 (Figure S4b), while the chow pellet contains significantly less. Therefore, similarly to component 6, component 8 reflects carbohydrate content, this time in the form of sucrose. As it is not observed in either of the tissue samples (Figure S5), if sucrose is present in these samples, it is at levels too low to be detected by Raman/MCR.
The spectral profile of component 9 is a close match to that of corn husk.31 Further investigation of literature in the area would indicate that the two main bands observed originate from lignin26 and, more specifically, the ferulic acid contained in lignin.27 Comparison of the two pellet samples shows that the chow pellet shows high mean values for component 9, whereas the NASH pellet values are close to zero, indicating that the chow pellet contains notable amounts of corn husk and the NASH pellet does not. If this component only originated from lignin, it would not be expected to be present in either of the tissue samples. However, it should be noted that as the spectral profile of component 9 is dominated by the two bands between 1550 and 1650 cm–1, if a molecule present in the tissue samples also has a Raman spectrum dominated by two bands in this region, this may also be seen as contrast in component 9. Two candidates for this are 4-hydroyxhexenal (HHE) and 4-hydroxynonenal (HNE), both of which have Raman spectra dominated by intense bands in this region.28 Both of these molecules formed during lipid peroxidation, occurring as a consequence of oxidative stress.32 Although component 9 appears to be present at similar levels within the two tissue samples, the distribution is notably different (Figure S5). Within the chow tissue, component 9 is relatively homogeneously distributed and present at similar levels throughout the tissue. In the NASH tissue, however, component 9 has a much more droplet like distribution, with high intensity within these droplets. These regions correspond closely to areas in the NASH tissue that are known to be lipid-rich (i.e., areas with high scores for component 1).
Overall, the largest difference seen in the tissue is the presence of MCR component 1 (oleic acid), prevalent in the NASH samples. This can be traced directly back to the diet and presents as lipid droplets of different morphologies, with a range of sizes between 2 and 30 μm. Tissue from the NASH diet also reveals significant holes, not shown in the chow (i.e., healthy) diet. MCR component 3 (protein) and component 4 (collagen/nucleic acids) are both involved in the surrounding tissue, with some, but not complete, spatial overlap between the two channels (green and blue in Figure 1c). This morphology, specifically the visible collagen/protein networks surrounding the holes in the tissue (Figure 1c and Figure 2d), is consistent with early stage fibrosis, which involves a complex series of changes in both morphology and related cellular processes.33
Several components appear present only in the diet with no corresponding presence in the liver. Some of these are as expected; carbohydrates (MCR 6) and simple sugars (MCR 8) were not expected in the liver and did not appear in significant quantities. However, these may be observable in biochemical imaging of hepatic tissue of other diseases, for example, abnormally high glucose levels.34 Starch (MCR 2) was also expected to appear in both diets but not significantly in the liver samples, which is confirmed in the results (Figure 1).
Two out of the nine spectral components are currently unidentified. These are component 5 and 7, present in the NASH and chow diet, respectively. Neither of these were abundant in either tissue type. Finally, component 9 has a spectral profile that appears to share features between lignin, likely from corn husk in the feed, and bands related to lipid oxidation. Although this component appears in both chow and NASH tissue samples, it is highly aggregated in the NASH tissue, colocalized with other lipid features, and may indeed originate from lipid-peroxidation products within the lipid-rich regions of the tissue.
The observations of high levels of oleic acid (MCR 1) in the diet correspondingly with high levels of oleic acid in the liver are consistent with previous studies using gas chromatography35 and manifest as lipid droplets of a variety of sizes. Evidence of lipid peroxidation appears (MCR 9) and is similar in terms of mean intensity across chow and NASH liver but with markedly different spatial distribution (Figure S5) and not colocalized with oleic acid. Biochemically, these components should also be independent since HHE and HNE are breakdown products from n-3 and n-6, respectively,32 whereas oleic acid is an n-9 fatty acid. Proteins (MCR 3) are visible in the chow diet but have a four times stronger intensity in both types of liver. The chow and NASH tissue have comparable levels of protein; however, the spatial distribution in the NASH tissue is notably altered, consistent with early fibrosis.
Conclusions
The results show the effectiveness of NIR Raman imaging and MCR–ALS with correlative analysis of diet and liver, detecting components specific to individual diets as well as those shared across different diets and highlighting the resulting liver composition and morphology. We should note that only components present in sufficiently high abundance will be detected. A more subtle limitation is that only components that have different concentrations in different samples can be discriminated as distinct components. Nevertheless, many components are evident, both spectrally and spatially. The components generated with MCR–ALS can be directly compared across samples within the same data set that was used to generate the model. This allows us to directly evaluate components that persist in the liver from the diet and those that do not. Here, oleic acid could be considered as persisting in the liver with comparable concentration to the diet. Sugars (sucrose and trehalose) while present in both diets are not observed in the liver. Secondary effects such as changes in the spatial distribution of molecules in the liver can also be observed with this approach, with collagen morphology undergoing significant change from the more homogeneous distribution in chow fed liver samples to the fibril-like structures that surround holes visible in the NASH diet liver samples.
Although in this study we looked specifically at the links between diet and liver, the same approach can be applied to other organs from which tissue sections can be prepared. Additionally, the correlative imaging approach is well-suited to targeted or diagnostic studies such as how chemical components or environmental contaminants such as microplastics or pesticides present in the diet can transfer to organs. Interestingly, the nonsupervised algorithms employed here can identify emergent or unexpected changes in tissue if the spectral strength is sufficient for detection.
Acknowledgments
This work was funded by the Japan Society for the Promotion of Science (JSPS) through the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program); the JSPS World Premier International Research Center Initiative Funding Program; and the Uehara Memorial Foundation. S.A. acknowledges funding from JSPS KAKENHI Grant-in-Aid for Scientific Research S (20H05693). The authors thank C. Smith for helpful discussion.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/cbmi.4c00027.
Graphical representation of the data and workflow to create the MCR model based on pellet and tissue data; average Raman spectra obtained from chow and NASH fed liver samples; Oil Red O and Sirius Red staining of chow and NASH fed liver tissue sections; scores plots for each of the components from the MCR–ALS model generated from the pellet and liver tissue combined images; scores plots for each of the components from the full chow and NASH liver tissue samples projected onto the MCR–ALS model (DOCX)
Author Contributions
A.J.H., T.S., S.A., and N.I.S. conceived of and designed the study. T.S. and S.A. provided and prepared feed and mouse liver samples for imaging, with feed samples sourced from CLEA Japan and Oriental Yeast. T.S. performed tissue staining. A.J.H. and N.I.S. carried out the measurements, analyzed, and interpreted the data. A.J.H. and N.I.S. wrote the manuscript. All authors contributed to revising and editing the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Sheka A. C.; Adeyi O.; Thompson J.; Hameed B.; Crawford P. A.; Ikramuddin S. Nonalcoholic Steatohepatitis: A Review. JAMA 2020, 323 (12), 1175–1183. 10.1001/jama.2020.2298. [DOI] [PubMed] [Google Scholar]
- Kwanten W. J. Diet and Non-Alcoholic Fatty Liver Disease, a Short Narrative Review. Acta Gastroenterol Belg 2023, 86 (2), 306–310. 10.51821/86.2.11547. [DOI] [PubMed] [Google Scholar]
- Wei S.; Wang L.; Evans P. C.; Xu S. NAFLD and NASH: Etiology, Targets and Emerging Therapies. Drug Discov Today 2024, 29 (3), 103910. 10.1016/j.drudis.2024.103910. [DOI] [PubMed] [Google Scholar]
- Wei W.; Wong C. C.; Jia Z.; Liu W.; Liu C.; Ji F.; Pan Y.; Wang F.; Wang G.; Zhao L.; Chu E. S. H.; Zhang X.; Sung J. J. Y.; Yu J. Parabacteroides Distasonis Uses Dietary Inulin to Suppress NASH via Its Metabolite Pentadecanoic Acid. Nat. Microbiol 2023, 8 (8), 1534–1548. 10.1038/s41564-023-01418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K.; Mizukoshi E.; Sunagozaka H.; Arai K.; Yamashita T.; Takeshita Y.; Misu H.; Takamura T.; Kitamura S.; Zen Y.; Nakanuma Y.; Honda M.; Kaneko S. Characteristics of Hepatic Fatty Acid Compositions in Patients with Nonalcoholic Steatohepatitis. Liver Int. 2015, 35 (2), 582–590. 10.1111/liv.12685. [DOI] [PubMed] [Google Scholar]
- Gentile C. L.; Pagliassotti M. J. The Role of Fatty Acids in the Development and Progression of Nonalcoholic Fatty Liver Disease. J. Nutr Biochem 2008, 19 (9), 567–576. 10.1016/j.jnutbio.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma I.; Vatner D. F. Fatty Acid Esterification as a NASH Therapeutic Target. Cell Mol. Gastroenterol Hepatol 2024, 17 (2), 311–312. 10.1016/j.jcmgh.2023.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser D. A.; Wang X.; Lund J.; Nikolić N.; Iruarrizaga-Lejarreta M.; Skjaeret T.; Alonso C.; Kastelein J. J. P.; Rustan A. C.; Kim Y. O.; Schuppan D. A Structurally Engineered Fatty Acid, Icosabutate, Suppresses Liver Inflammation and Fibrosis in NASH. J. Hepatol 2022, 76 (4), 800–811. 10.1016/j.jhep.2021.12.004. [DOI] [PubMed] [Google Scholar]
- Dapper C.; Liu J.; Klemba M. Leveraging a Fluorescent Fatty Acid Probe to Discover Cell-Permeable Inhibitors of Plasmodium Falciparum Glycerolipid Biosynthesis. Microbiol Spectr 2022, 10 (6), e0245622 10.1128/spectrum.02456-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T.; Hobro A. J.; Pavillon N.; Umakoshi T.; Verma P.; Smith N. Label-Free Raman Mapping of Saturated and Unsaturated Fatty Acid Uptake, Storage, and Return toward Baseline Levels in Macrophages. Analyst 2021, 146 (4), 1268–1280. 10.1039/D0AN02077J. [DOI] [PubMed] [Google Scholar]
- Krafft C.; Diderhoshan M. A.; Recknagel P.; Miljkovic M.; Bauer M.; Popp J. Crisp and Soft Multivariate Methods Visualize Individual Cell Nuclei in Raman Images of Liver Tissue Sections. Vib. Spectrosc. 2011, 55 (1), 90–100. 10.1016/j.vibspec.2010.09.003. [DOI] [Google Scholar]
- Mondal D. D.; Chakraborty U.; Bera M.; Ghosh S.; Kar D. An Overview of Nutritional Profiling in Foods: Bioanalytical Techniques and Useful Protocols. Front Nutr 2023, 10, 1124409. 10.3389/fnut.2023.1124409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George E. S.; Reddy A.; Nicoll A. J.; Ryan M. C.; Itsiopoulos C.; Abbott G.; Johnson N. A.; Sood S.; Roberts S. K.; Tierney A. C. Impact of a Mediterranean Diet on Hepatic and Metabolic Outcomes in Non-Alcoholic Fatty Liver Disease: The MEDINA Randomised Controlled Trial. Liver International 2022, 42 (6), 1308–1322. 10.1111/liv.15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beton-Mysur K.; Kopec M.; Brozek-Pluska B. Raman Imaging—A Valuable Tool for Tracking Fatty Acid Metabolism—Normal and Cancer Human Colon Single-Cell Study. Int. J. Mol. Sci. 2024, 25 (8), 4508. 10.3390/ijms25084508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicerone M. T.; Camp C. H. Histological Coherent Raman Imaging: A Prognostic Review. Analyst 2018, 143 (1), 33–59. 10.1039/C7AN01266G. [DOI] [PubMed] [Google Scholar]
- Huang L.; Sun H.; Sun L.; Shi K.; Chen Y.; Ren X.; Ge Y.; Jiang D.; Liu X.; Knoll W.; Zhang Q.; Wang Y. Rapid, Label-Free Histopathological Diagnosis of Liver Cancer Based on Raman Spectroscopy and Deep Learning. Nat. Commun. 2023, 14 (1), 48. 10.1038/s41467-022-35696-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLone V.; Aizenshtadt A.; Goertz J.; Skottvoll F. S.; Mota M. B.; You J.; Zhao X.; Berg H. E.; Stokowiec J.; Yu M.; Schwendeman A.; Scholz H.; Wilson S. R.; Krauss S.; Stevens M. M. Quantitative Chemometric Phenotyping of Three-Dimensional Liver Organoids by Raman Spectral Imaging. Cell Reports Methods 2023, 3 (4), 100440. 10.1016/j.crmeth.2023.100440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos V.; Benítez L.; Marro M.; Loza-Alvarez P.; Piña B.; Tauler R.; de Juan A. Relevant Aspects of Unmixing/Resolution Analysis for the Interpretation of Biological Vibrational Hyperspectral Images. TrAC Trends in Analytical Chemistry 2017, 94, 130–140. 10.1016/j.trac.2017.07.004. [DOI] [Google Scholar]
- Vuiblet V.; Nguyen T. T.; Wynckel A.; Fere M.; Van-Gulick L.; Untereiner V.; Birembaut P.; Rieu P.; Piot O. Contribution of Raman Spectroscopy in Nephrology: A Candidate Technique to Detect Hydroxyethyl Starch of Third Generation in Osmotic Renal Lesions. Analyst 2015, 140 (21), 7382–7390. 10.1039/C5AN00821B. [DOI] [PubMed] [Google Scholar]
- Kuhar N.; Sil S.; Verma T.; Umapathy S. Challenges in Application of Raman Spectroscopy to Biology and Materials. RSC Adv. 2018, 8 (46), 25888–25908. 10.1039/C8RA04491K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobro A. J.; Standley D. M.; Ahmad S.; Smith N. I. Deconstructing RNA: Optical Measurement of Composition and Structure. Phys. Chem. Chem. Phys. 2013, 15 (31), 13199–13208. 10.1039/c3cp52406j. [DOI] [PubMed] [Google Scholar]
- Benevides J. M.; Overman S. A.; Thomas G. J. Jr Raman, Polarized Raman and Ultraviolet Resonance Raman Spectroscopy of Nucleic Acids and Their Complexes. J. Raman Spectrosc. 2005, 36 (4), 279–299. 10.1002/jrs.1324. [DOI] [Google Scholar]
- De Gussem K.; Vandenabeele P.; Verbeken A.; Moens L. Raman Spectroscopic Study of Lactarius Spores (Russulales, Fungi). Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2005, 61 (13), 2896–2908. 10.1016/j.saa.2004.10.038. [DOI] [PubMed] [Google Scholar]
- Szymańska-Chargot M.; Chylińska M.; Pieczywek P. M.; Rösch P.; Schmitt M.; Popp J.; Zdunek A. Raman Imaging of Changes in the Polysaccharides Distribution in the Cell Wall during Apple Fruit Development and Senescence. Planta 2016, 243 (4), 935–945. 10.1007/s00425-015-2456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Liao Y.; Meng Y.; Jiao X.; Huang W.; Liu T. C. The Early, Rapid, and Non-Destructive Detection of Citrus Huanglongbing (HLB) Based on Microscopic Confocal Raman. Food Anal. Methods 2019, 12 (11), 2500–2508. 10.1007/s12161-019-01598-1. [DOI] [Google Scholar]
- Agarwal U. P.; McSweeny J. D.; Ralph S. A. FT-Raman Investigation of Milled-Wood Lignins: Softwood, Hardwood, and Chemically Modified Black Spruce Lignins. Journal of Wood Chemistry and Technology 2011, 31 (4), 324–344. 10.1080/02773813.2011.562338. [DOI] [Google Scholar]
- Prats Mateu B.; Hauser M. T.; Heredia A.; Gierlinger N. Waterproofing in Arabidopsis: Following Phenolics and Lipids In Situ by Confocal Raman Microscopy. Front. Chem. 2016, 4, 26973831 10.3389/fchem.2016.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie J. R.; Pawlak A. M.; Boulton M. E.; Zhang J.; Monnier V. M.; McGarvey J. J.; Stitt A. W. Multiplex Analysis of Age-Related Protein and Lipid Modifications in Human Bruch’s Membrane. FASEB J. 2010, 24 (12), 4816–4824. 10.1096/fj.10.166090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez S.; Bermudez B.; Montserrat-de la Paz S.; Jaramillo S.; Varela L. M.; Ortega-Gomez A.; Abia R.; Muriana F. J. G. Membrane Composition and Dynamics: A Target of Bioactive Virgin Olive Oil Constituents. Biochim. Biophys. Acta 2014, 1838 (6), 1638–1656. 10.1016/j.bbamem.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Frushour B. G.; Koenig J. L. Raman Scattering of Collagen, Gelatin, and Elastin. Biopolymers 1975, 14 (2), 379–391. 10.1002/bip.1975.360140211. [DOI] [PubMed] [Google Scholar]
- Jin Y.; Tian H.; Gao Z.; Yang G.; Dong D. Oil Content Analysis of Corn Seeds Using a Hand-Held Raman Spectrometer and Spectral Peak Decomposition Algorithm. Front. in Plant Sci. 2023, 14, 1174747. 10.3389/fpls.2023.1174747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikado A.; Morino K.; Nishio Y.; Nakagawa F.; Mukose A.; Sono Y.; Yoshioka N.; Kondo K.; Sekine O.; Yoshizaki T.; Ugi S.; Uzu T.; Kawai H.; Makino T.; Okamura T.; Yamamoto M.; Kashiwagi A.; Maegawa H. 4-Hydroxy Hexenal Derived from Docosahexaenoic Acid Protects Endothelial Cells via Nrf2 Activation. PLoS One 2013, 8 (7), e69415 10.1371/journal.pone.0069415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppan D.; Surabattula R.; Wang X. Y. Determinants of Fibrosis Progression and Regression in NASH. J. Hepatol 2018, 68 (2), 238–250. 10.1016/j.jhep.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Sharabi K.; Tavares C. D. J.; Rines A. K.; Puigserver P. Molecular Pathophysiology of Hepatic Glucose Production. Mol. Aspects Med. 2015, 46, 21–33. 10.1016/j.mam.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappini F.; Coilly A.; Kadar H.; Gual P.; Tran A.; Desterke C.; Samuel D.; Duclos-Vallée J.-C.; Touboul D.; Bertrand-Michel J.; Brunelle A.; Guettier C.; Le Naour F. Metabolism Dysregulation Induces a Specific Lipid Signature of Nonalcoholic Steatohepatitis in Patients. Sci. Rep 2017, 7 (1), 46658. 10.1038/srep46658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.