Abstract
Plant viruses have movement protein (MP) gene(s) essential for cell-to-cell movement in hosts. Cucumber mosaic virus (CMV) requires its own coat protein (CP) in addition to the MP for intercellular movement. Our present results using variants of both CMV and a chimeric Brome mosaic virus with the CMV MP gene revealed that CMV MP truncated in its C-terminal 33 amino acids has the ability to mediate viral movement independently of CP. Coexpression of the intact and truncated CMV MPs extremely reduced movement of the chimeric viruses, suggesting that these heterogeneous CMV MPs function antagonistically. Sequential deletion analyses of the CMV MP revealed that the dispensability of CP occurred when the C-terminal deletion ranged between 31 and 36 amino acids and that shorter deletion impaired the ability of the MP to promote viral movement. This is the first report that a region of MP determines the requirement of CP in cell-to-cell movement of a plant virus.
Plant viruses encode proteins that control their movement from cell to cell. These proteins are called movement proteins (MPs) and interact with the normal symplastic connections between plant cells, the plasmodesmata, by modifying the plasmodesmal structure and function. Consequently, the highly regulated passage of small molecules through plasmodesmata is altered to allow the passage of large nucleoprotein complexes containing the viral genome (8, 28).
Some viruses, including Tobacco mosaic virus (TMV) and Red clover necrotic mosaic virus, do not require the viral coat protein (CP) for cell-to-cell movement. The MPs of these viruses have a nucleic acid binding activity (10, 37), and an MP-RNA nucleoprotein complex is thought to pass through the modified plasmodesmata to adjacent uninfected cells (8, 26). Other viruses do not move from cell to cell in the absence of viral CP. Cauliflower mosaic virus and Cowpea mosaic virus are known to move as virus-like particles through tubules that pass through plasmodesmata into neighboring cells. These tubules are composed of MP (40, 49). Nepo-, Tospo-, and Fabaviruses are also thought to move similarly with the tubule-mediated mechanism. There are viruses that are considered to move as a nucleoprotein complex different from virus particles despite their requirement of viral CP for cell-to-cell movement. Cucumber mosaic virus (CMV) is competent to induce tubules protruding from infected protoplasts like the viruses that move as virus-like particles (6). Nevertheless, no such tubules have been found in planta by electron microscopy (3), and mutants incapable of virion formation move successfully from cell to cell (22, 44, 46).
CMV is the type member of the genus Cucumovirus and is one of the most common plant viruses of substantial agricultural significance. CMV infects more than 1,000 species of plants, shrubs, and trees and both monocots and dicots (41). The genomic RNAs of CMV are designated as RNAs 1, 2, and 3, by diminishing size (39). All the RNAs have a cap structure at the 5′ terminus. The 3′ portion of all the RNAs is also highly conserved in virus-specific manner and can form a tRNA-like structure that can be aminoacylated with tyrosine. RNAs 1 and 2, encoding the 1a and 2a proteins, respectively, are involved in viral replication (18, 36). CMV RNA 2 encodes a second protein, 2b, which is translated from a subgenomic RNA, RNA 4A, and plays a role in systemic spread of the virus and virulence determination, possibly by suppressing a host RNA silencing mechanism (4, 14). RNA 3 encodes two proteins dispensable for viral replication in protoplasts. The 5′-proximal open reading frame (ORF) on RNA 3 is for the 3a protein, the MP of CMV (15). The 3′-proximal ORF is for the CP that is translated from a subgenomic RNA 4 (5).
We have investigated viral cell-to-cell movement mediated by the CMV MP using chimeric viruses derived from Brome mosaic virus (BMV), the type member of the genus Bromovirus. CMV and BMV are distantly related (42) and show many similarities, including the size and form of virus particles (24, 50), genome organizations (1, 41), and the requirement of the CP for cell-to-cell movement (7, 43). Our previous results indicate that the wild-type (wt) CMV MP requires its cognate CP to mediate cell-to-cell movement of the chimeric BMV genome (34), while the CMV MP with a deletion of its C-terminal 33 amino acids is able to mediate the movement of the chimeric BMV genome in the absence of the CMV CP (35). These suggest that the truncated CMV MP is different from the wt one in the requirement of viral CP. In this study, the difference in the CP requirement is characterized and the antagonism between the wt and truncated CMV MPs is tested. Based on the results, the role of CP in the CMV MP-mediated viral cell-to-cell movement is discussed.
MATERIALS AND METHODS
cDNA clones.
Plasmids pBTF1, pBTF2, and pBTF3W contain the full-length cDNAs of RNAs 1, 2, and 3, respectively, of the wt BMV (the KU2 strain) (30, 31, 32). The plasmids pT7B3CKY3 and pB3C3a247T contain the full-length cDNA of chimeric BMV RNA 3 with the genes of wt and the C-terminal 33 amino acid-truncated CMV MP, respectively (35). Plasmids pCY1-T7 and pCY2-T7 (generous gifts from S. Kuwata, Meiji University, and M. Suzuki, Yokohama National University) and plasmid pT7CKY3 contain the full-length cDNAs of CMV-Y RNAs 1, 2, and 3, respectively (35, 46).
There is a SalI site in the 5′-proximal end of the CP ORF in either BMV or CMV. Four bases (TCGA) within the SalI site were duplicated by T4 DNA polymerase treatment after digestion with the enzyme in order to make CP-defective variants of CMV RNA 3 and chimeric BMV RNA 3.
The RNA 3 variants of chimeric BMV and CMV with various lengths of C-terminal deletions of CMV MP were created by PCR-based in vitro mutagenesis (20). A translational stop codon, TGA, was introduced at the appropriate positions in the subcloned 124-bp region between the two HpaI sites in the CMV MP gene by using synthetic oligonucleotides. After we confirmed that each nucleotide substitution was successfully introduced without any undesired mutations, we used the 124-bp HpaI fragment to replace the corresponding region in pT7B3CKY3 or pT7CKY3. Variants with the 3-amino-acid deletion from the CMV MP C terminus were created by the mutagenesis against the 566-bp region between the NheI and SalI sites in pT7B3CKY3 and pT7CKY3, since the codon encoding the amino acid 277 is overlapped with the 3′-proximal HpaI site. The SalI site exists at the 5′-proximal end of the CP gene of either BMV or CMV as mentioned above. After we confirmed that the mutagenesis was successfully done, we used the HpaI-SalI fragments with the stop codon (385 bp from the chimeric BMV RNA 3 cDNA and 472 bp from the CMV RNA 3 cDNA) to replace the corresponding regions in pT7B3CKY3 and pT7CKY3.
The chimeric BMV RNA 3 derivatives in which the CMV MP genes are in tandem were created as follows. First, the BMV CP gene in the NsiI/BlnI-introduced pBTF3W (34) was replaced with the CMV MP gene from pCMP-NX (35) as described previously (33) to create pB3(CPtoCMP). The BglII/EcoRI fragments of pT7B3CKY3 and pB3C3a247T were replaced with the corresponding fragment containing the CMV MP gene from pB3(CPtoCMP). The resulting plasmids were designated pB3(Cmp/Cmp) and pB3(CmpDC33/Cmp), respectively. On the other hand, the 124-bp HpaI fragment in pB3(CPtoCMP) was replaced with that from pB3C3a247T to create pB3(CPtoCMPDC33). The BglII/EcoRI fragments of pT7B3CKY3 and pB3C3a247T were replaced with the corresponding fragment containing the CMV MP gene with mutation from pB3(CPtoCMPDC33). The resulting plasmids were designated pB3(Cmp/CmpDC33) and pB3(CmpDC33/CmpDC33), respectively.
Inoculation of plants and protoplasts.
Chenopodium quinoa plants and protoplasts were inoculated with in vitro transcripts synthesized from the plasmids by T7 RNA polymerase after linearization with appropriate restriction endonuclease as previously described (34). In all cases, each inoculum contained an RNA 3 variant of CMV or chimeric BMV, together with its cognate RNAs 1 and 2. Inoculation onto Nicotiana benthamiana plants was done as described elsewhere (16).
Analysis of RNA and protein.
Northern, tissue-printing, and press blot hybridizations were done as described previously (35). Probes used to detect BMV RNAs (21) and CMV RNAs (35) were as described previously. The 32P radioactive signals on the membrane were quantified with a digital radioactive imaging analyzer (Fujix BAS 2000; Fuji Photo Film).
Electrophoresis and immunodetection of proteins extracted from protoplasts and plant tissues were done as described previously (34, 35). Polyclonal rabbit antiserum raised against BMV, CMV (35), or the CMV MP (a generous gift from P. Palukaitis and I. B. Kaplan) was used as the first antibody. The second antibody used was alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G.
RESULTS
The CMV MP with a deletion of the C-terminal 33 amino acids do not require CP to mediate viral cell-to-cell movement.
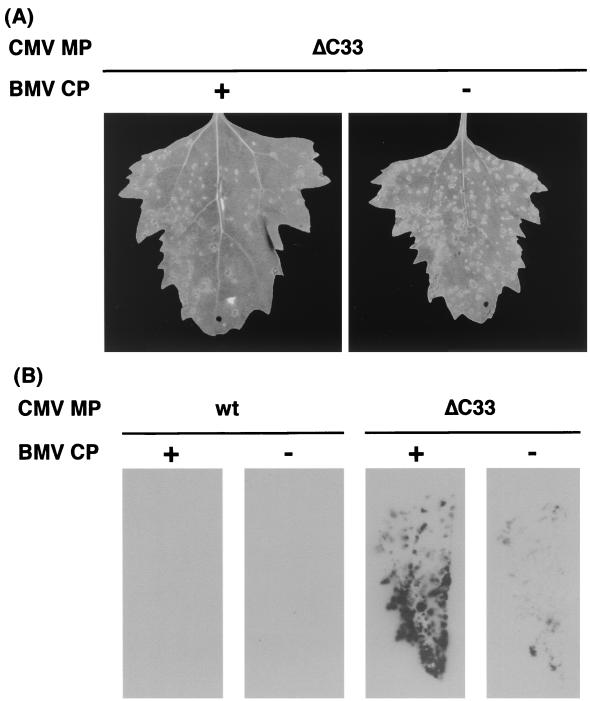
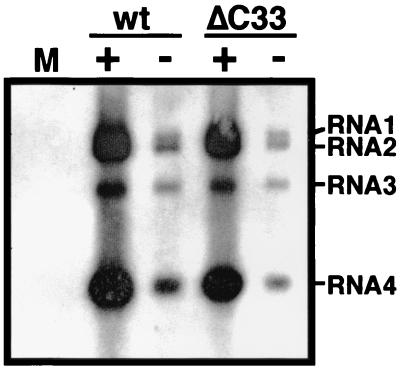
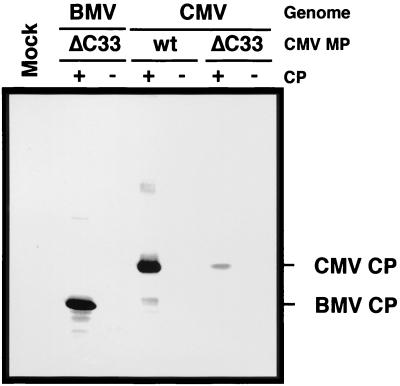
Our previous results demonstrated that the wt CMV MP requires its cognate CP to mediate viral cell-to-cell movement (34). However, a chimeric BMV containing the gene of CMV MP from which the C-terminal 33 amino acids are deleted (ΔC33-CMV MP) can move from cell to cell, although this chimeric virus expresses the BMV CP instead of the CMV CP (35). Also, a CMV variant with the ΔC33-CMV MP gene can move from cell to cell. Therefore, it was considered that viral cell-to-cell movement was mediated by the ΔC33-CMV MP independently of or cooperatively with either BMV CP or CMV CP. We first tested whether the BMV CP was required for the movement mediated by the truncated CMV MP. The CP-defective variant of chimeric BMV with the ΔC33-CMV MP gene induced chlorotic lesions in inoculated C. quinoa leaves. The lesions were indistinguishable from lesions induced by the CP-intact chimeric BMV with the ΔC33-CMV MP gene (Fig. 1A). Distribution of viral RNA in these leaves was analyzed by the press blot method. Although the size and number of visible lesions were comparable between the two chimeric viruses with the ΔC33-CMV MP gene (Fig. 1A and data not shown), the CP-defective virus accumulated viral RNA to lower level than the CP-intact virus (Fig. 1B). This is probably due to the absence of RNA protection by the BMV CP since the absence of CP resulted in approximately 10-fold decrease of viral RNA accumulation in protoplasts (Fig. 2). Immunoblot analysis with anti-BMV antiserum confirmed that the intact BMV CP did not accumulate either in the protoplasts or in the leaves inoculated with the CP-defective virus (Fig. 3; data not shown). The above results indicate that the ΔC33-CMV MP functions in cell-to-cell movement of the chimeric virus independently of the BMV CP. On the other hand, a CP-defective variant, as well as a CP-intact variant, of the chimeric BMV with the wt-CMV MP gene did not induce lesions in the inoculated C. quinoa leaves (data not shown). No viral RNA accumulation was detected in these leaves (Fig. 1B). However, viral RNA accumulated in protoplasts infected by either CP-intact or -defective variant to the level comparable to the corresponding variants of the chimeric BMV with the ΔC33-CMV MP gene (Fig. 2). Thus, it was confirmed that the wt-CMV MP alone does not function in cell-to-cell movement of the chimeric BMV genome.
FIG. 1.
The ability of wt- and ΔC33-CMV MPs to mediate cell-to-cell movement of chimeric BMV with or without expression of BMV CP in C. quinoa leaves. (A) Symptom induced by the chimeric BMV variants with the gene of ΔC33-CMV MP in the presence (+) or the absence (−) of BMV CP expression at 7 days p.i. (B) Press blotting analysis of C. quinoa leaves inoculated with chimeric BMV variants at 7 days p.i. Viral RNA was detected by using a probe specific for the conserved 3′ sequence of BMV RNAs. wt and ΔC33 denote the chimeric BMV variants harboring the corresponding MP gene. + and − denote the variants that expressed and did not express BMV CP, respectively.
FIG. 2.
Northern hybridization analysis of C. quinoa protoplasts inoculated with the chimeric BMV variants using a probe specific for the conserved 3′ sequence of BMV RNAs. Total RNA was extracted at 24 hours p.i. and analyzed. The positions of the chimeric BMV genomic and subgenomic RNAs are indicated on the right. M, mock. For other symbols, refer to the legend of Fig. 1.
FIG. 3.
Immunodetection of the CP of BMV and of CMV in infected C. quinoa protoplasts at 24 hours p.i. BMV and CMV denote the genomic background of RNA3 from which CMV MP was expressed. The positions of the CPs of CMV and BMV are indicated on the right. For other symbols, refer to the legend of Fig. 1.
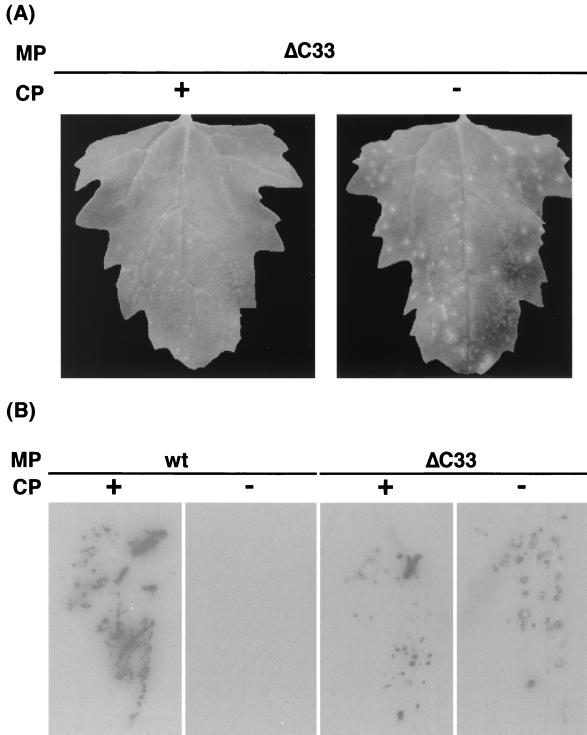
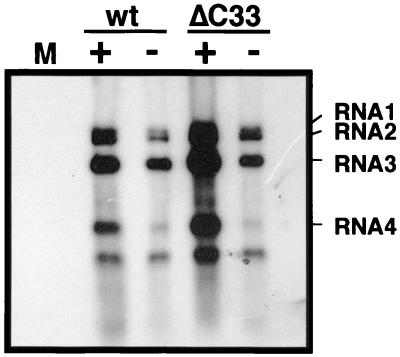
Next, to investigate whether the ΔC33-CMV MP requires CMV CP in CMV movement, a frameshift mutation was introduced into the CP gene of a CMV variant harboring the ΔC33-CMV MP gene that is able to move from cell to cell (35). The CP-defective CMV variant with the ΔC33-CMV MP gene induced necrotic lesions on the inoculated leaves (Fig. 4A). The size of lesions induced by the CP-defective variant was slightly larger than that by the CP-intact variant; the former was approximately 0.5 to 1.3 mm and the latter was 0.3 to 0.7 mm in diameter at 7 days postinoculation (p.i.). Viral RNA in these leaves was detected by press-blot analysis (Fig. 4B). On the other hand, when the same frameshift mutation was introduced into the CP gene of wt-CMV, the CP-defective CMV did not move from cell to cell (Fig. 4B) as reported previously (7, 46). Immunoblot analysis with anti-CMV antiserum confirmed that no intact CMV CP accumulated in the protoplasts inoculated with these CP-defective variants (Fig. 3). The results demonstrated that CMV CP was dispensable for cell-to-cell movement of the CMV variant with ΔC33-CMV MP gene. The CP-defective variant accumulated viral RNA in infected C. quinoa protoplasts, although the accumulation level was lower than from the variant with the intact CP gene (Fig. 5). Further, the CP-intact or the CP-defective variant of CMV with the ΔC33-CMV MP gene was tested for the ability to systemically infect a host by analyzing viral RNA distribution in N. benthamiana plants with the tissue-printing hybridization technique. The CP-intact variant systemically infected the plants while the infection by the CP-defective variant was limited to the inoculated leaves (data not shown), indicating that the ΔC33-CMV MP is able to mediate viral systemic movement, while the CP is necessary for the systemic but not for the cell-to-cell movement.
FIG. 4.
The ability of wt- and ΔC33-CMV MPs to mediate cell-to-cell movement of CMV with or without expression of CP in C. quinoa leaves. (A) Symptom induced by the CMV variants with the ΔC33-MP gene in the presence (+) or the absence (−) of CP expression at 7 days p.i. (B) Press blotting analysis of C. quinoa leaves inoculated with CMV variants at 7 days p.i. Viral RNA was detected by using a probe specific for the conserved 3′ sequence of CMV RNAs. For other symbols, refer to the legend of Fig. 1.
FIG. 5.
Northern hybridization analysis of C. quinoa protoplasts inoculated with the CMV variants using a probe specific for the conserved 3′ sequence of CMV RNAs. Total RNA was extracted at 24 hours p.i. and analyzed. Positions of CMV genomic and subgenomic RNAs are indicated on the right. The bands with no indication are likely RNA 4A. M, mock. For other symbols, refer to the legend of Fig. 1.
Antagonism between the ΔC33- and wt-CMV MPs.
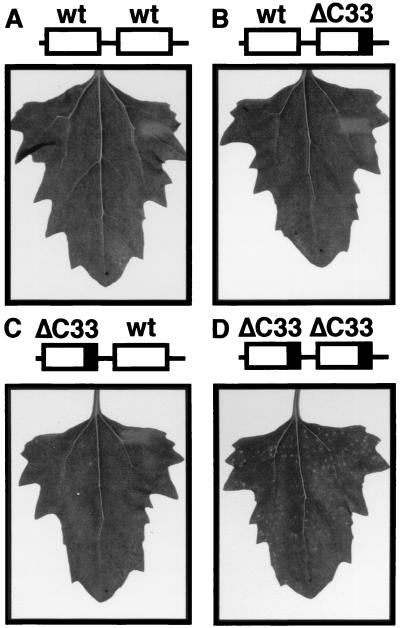
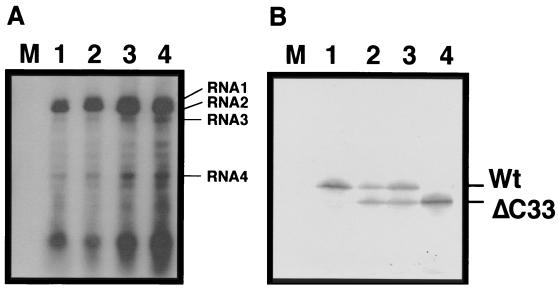
To test whether the ΔC33-CMV MP is antagonistic to the wt-CMV MP, chimeric BMV RNA3 variants with two MP genes, each in the position of the MP and CP ORFs, were constructed so that the variants could express both wt and ΔC33-CMV MPs from either the MP or the CP ORF of the viral genome (Fig. 6B and C). Similar variants having two copies of the wt or the ΔC33-CMV MP gene were also constructed as controls (Fig. 6A and D). The resulting four variants were tested for their ability to move from cell to cell in C. quinoa leaves. If the antagonism occurred between the two MPs, the movement of the chimeric viruses with the heterogeneous CMV MP genes should be suppressed. We repeated these experiments three times with four inoculated leaves in every experiment, and highly reproducible results were obtained. One of the chimeric BMV variants in which the ΔC33-CMV MP and wt- CMV MP genes lay at the position of MP and CP genes, respectively, induced two to six lesions in every inoculated leaf (Fig. 6C). The other chimeric BMV variant in which the two CMV MP genes lay at the reverse position induced no symptoms (Fig. 6B). On the other hand, a chimeric BMV variant with two ΔC33-CMV MP genes induced more than 50 lesions in every inoculated leaf, while another chimeric BMV variant with two wt-CMV MP genes induced no symptoms (Fig. 6A and D). Since a visible local lesion requires infection of many cells by invading virus (43), the symptomatic results are most likely to reflect the ability of the variants to move from cell to cell, although we failed to detect viral RNA accumulation in these infected leaves by press blot hybridization (data not shown). Thus, these results suggest that viral movement is strongly suppressed by coexpression of wt- and ΔC33-CMV MPs. Similar level of replication of the variants and the translation of MPs from the replaced ORFs were confirmed by protoplast infection (Fig. 7). Therefore, we concluded that the antagonism occurred between the wt- and ΔC33-CMV MPs.
FIG. 6.
Symptoms on C. quinoa leaves inoculated with the chimeric BMV variants with two CMV MP genes in RNA 3. Photographs were taken at 7 days p.i. The combinations of the CMV MPs in the inoculated chimeric RNA 3's are wt-wt (A), wt-ΔC33 (B), ΔC33-wt (C), and ΔC33-ΔC33 (D), respectively. Schematic diagrams of the inocula are shown above the photographs. Lines, noncoding regions derived from BMV RNA 3. Open boxes, wt-CMV MP gene; boxes with closed area, ΔC33-CMV MP gene.
FIG. 7.
Protoplast infection of the chimeric BMV variants with two CMV MP genes in RNA 3. (A) Northern hybridization analysis at 24 hours p.i. using a probe specific for the conserved 3′ sequence of BMV RNAs. Positions of the chimeric BMV genomic and subgenomic RNAs are indicated on the right. Since these variants have no CP gene, strong signals near the bottom in each lane are considered as degraded viral RNA. (B) Immunodetection of the wt- and ΔC33-CMV MP at 24 h p.i. Positions of those proteins are indicated on the right. The combinations of CMV MP genes in the chimeric RNA 3s are wt-wt (lane 1), wt-ΔC33 (lane 2), ΔC33-wt (lane 3), and ΔC33-ΔC33 (lane 4). For easy visualization of these inocula, see the schematic diagrams in Fig. 6. M, mock. For symbols, refer to the legend of Fig. 1.
The translation products from RNA 4 accumulated to a higher level than those from RNA 3 in protoplasts infected with the variants having the heterogeneous set of CMV MP genes (Fig. 7B, lanes 2 and 3). The difference in the ability to induce lesions between the variants having the heterogeneous set of CMV MP genes might be due to the timing lag of protein expression. To test this possibility, the accumulation kinetics of the MPs was examined in infected protoplasts every hour. MPs translated from RNA 3 or RNA 4 were detectable at 8 h p.i. and later (data not shown). We, thus, failed to detect difference in the timing of translation due to the genetic position of the two MP genes.
Mapping of the domain involved in the CP requirement for viral cell-to-cell movement.
The C-terminal 33-amino-acid region of CMV MP was predicted to be divided into three parts based on the analysis by the method of Chou and Fasman (9) (data not shown). From the prediction, a stop codon was introduced into the CMV MP gene in both CMV RNA 3 and the chimeric BMV RNA 3 with the CMV MP gene to create RNA 3 derivatives that express truncated CMV MP with deletions of 10, 19, and 25 amino acids from its C terminus. Northern hybridization analysis of infected protoplasts revealed that these variants accumulated progeny RNAs in C. quinoa protoplasts (data not shown). However, when inoculated onto C. quinoa plants, neither CMV nor the chimeric BMV variants induced local lesions (Table 1). No accumulation of viral RNA was detected in these leaves by press blotting hybridization analyses (Table 1). The results did not change even when the concentration of these inocula increased up to threefold (data not shown). The failure of the CMV variants, as well as of the chimeric BMV variants, to move from cell to cell suggests that the C-terminal deletions of 10, 19, and 25 amino acids abolish the ability of the CMV MP to mediate viral cell-to-cell movement, although the ΔC33-CMV MP promotes the movement.
TABLE 1.
Ability of wt and truncated CMV MPs to promote viral movement in inoculated leaves of C. quinoa
| No. of deleted amino acids from the CMV MP C terminus | CMV
|
Chimeric BMV
|
||
|---|---|---|---|---|
| Symptom inductiona | Distribution of viral RNAb | Symptom inductiona | Distribution of viral RNAb | |
| 0 (wt) | +++ | + | − | − |
| 3 | − | − | − | − |
| 5 | − | − | − | − |
| 7 | − | − | − | − |
| 10 | − | − | − | − |
| 19 | − | − | − | − |
| 25 | − | − | − | − |
| 27 | − | − | − | − |
| 29 | − | − | − | − |
| 31 | ± | − | − | ± |
| 32 | ± | − | − | ± |
| 33 | ++ | + | ++++ | + |
| 35 | + | + | +++ | + |
| 36 | + | + | + | + |
| 37 | − | − | − | − |
| 39 | − | − | − | − |
| 43 | − | − | − | − |
Symptom induction was estimated from the results of three independent experiments and scored as follows: ++++, >50; +++, 21 to 50; ++, 11 to 20; and +, 1 to 10 (lesions per leaf induced on average). −, no lesion appeared; ±, A few lesions appeared in one or two experiments.
RNA distribution was estimated from press blotting hybridization of three independent experiments. + and − indicate detection and no detection of viral RNA, respectively. ±, viral RNA was successfully detected but in an unusual fashion.
To delimit the CMV MP regions involved in virus movement, more CMV variants with various sizes of the C-terminal deletion in the CMV MP were created (Table 1). These variants all replicated similarly in C. quinoa protoplasts (data not shown). The ability of the variants to move from cell to cell and to induce local lesions is summarized in Table 1. In short, CMV variants moved from cell to cell if the C-terminal deletion in CMV MP was from 33 to 36 amino acids, although the degrees of movement were distinctive. When 31 or 32 amino acids were deleted, a few local lesions were occasionally induced in the inoculated leaves, although viral RNA was not detected by press blotting analysis. Longer and shorter C-terminal deletions abolished viral movement. It is noteworthy that the function of CMV MP to mediate viral cell-to-cell movement was abolished when only three amino acids were deleted from its C terminus. A movement-incapable CMV variant with the C-terminal deletion of 43 amino acids corresponds to a CMV mutant that infects tobacco plants systemically (23). To test this, a CMV variant was created by the manner identical to that for Kaplan's mutant. This variant, however, did not move from cell to cell and did not induce any symptom in C. quinoa plants (data not shown).
The ability of the CMV MP with the various sizes of C-terminal deletion to mediate cell-to-cell movement was also tested in chimeric BMV. As with CMV variants, the chimeric BMV variants with the gene of truncated CMV MP moved from cell to cell and induced local lesions in the inoculated leaves when the C-terminal deletion was from 33 to 36 amino acids, although the degrees of movement were distinctive (Table 1). In contrast to the case of CMV, however, the chimeric BMV with a deletion of either 31 or 32 amino acids in the CMV MP C terminus induced no symptom, but press blotting analyses of the inoculated leaves showed accumulation and unusual distribution of viral RNA. The shape of RNA signals reproducibly looked like a trail of a comet (data not shown). The difference in the shape of signals should reflect the difference in the localization of virus RNA. Further studies are required to determine the distribution of these variants. The chimeric BMV variants competent for cell-to-cell movement had the CP gene of BMV but not of CMV, suggesting that the CMV MP with C-terminal deletion ranging from 31 to 36 amino acids can promote viral cell-to-cell movement independently of viral CP. The independence from CP of viral movement mediated by the CMV MP with those C-terminal deletions was further confirmed by the ability of the CP-defective CMV variants to move from cell to cell and to induce lesions in the inoculated leaves (Table 2). Although viral RNA was not detected in the leaves inoculated with the CP-defective CMV variants having a deletion of 31 or 32 amino acids in the C terminus of the MP, local lesions with pinpoint size were induced on the inoculated leaves (data not shown).
TABLE 2.
Ability of truncated CMV MP to mediate viral movement in C. quinoa in the absence of CMV CP
| No. of deleted amino acids | Local lesion induction
|
RNA distributionc | |
|---|---|---|---|
| No.a | Avg diamb (mm) ± SD | ||
| 36 | +++ | 0.95 ± 0.15 | + |
| 35 | ++ | 0.60 ± 0.10 | + |
| 34 | ++++ | 1.45 ± 0.85 | + |
| 33 | +++ | 2.00 ± 1.00 | + |
| 32 | + | 0.45 ± 0.10 | − |
| 31 | ++ | 0.55 ± 0.15 | − |
The number of local lesions was estimated from the results of three independent experiments and was scored as follows: ++++, >50; +++, 21 to 50; ++, 11 to 20; and +, 1 to 10 (lesions per leaf induced on average).
Average diameter of local lesions with the standard deviation from the results of three independent experiments.
RNA distribution was estimated from press blotting hybridization of three independent experiments; + and − indicate detection and no detection of viral RNA, respectively.
DISCUSSION
This study demonstrates that the CMV MP truncated in its C terminus can promote viral cell-to-cell movement independently of CP. The CP-independent viral movement occurs when the deletion length was within 31 to 36 amino acids. To our knowledge, this is the first report to show a region of MP that determines the requirement of CP in cell-to-cell movement of a plant virus.
The functions of CMV MP have been compared with those of TMV MP, the best-characterized virus MP. The two MPs share the abilities to (i) traffic through plasmodesmata (19, 48), (ii) increase plasmodesmal size exclusion limit (15, 48), (iii) bind single-stranded nucleic acids in vitro (10, 27), and (iv) complement their respective movement-deficient mutants (13, 23). In contrast to these similarities, CMV MP cannot promote viral cell-to-cell movement without its cognate CP, although TMV MP can. Replacement of both MP and CP genes is required to make a movement-competent chimeric virus with wt-CMV MP gene (34). In contrast, movement-competent chimeric viruses can be made by replacing the MP gene with that of TMV in dianthovirus and hordeivirus (17, 45). In addition, CMV MP expressed in transgenic plants cannot promote a movement-defective variant of TMV (12). These indicate that the wt-CMV MP lacks one or more functions in the absence of the CMV CP compared with the TMV MP.
As shown in this study, CMV MP comes to promote cell-to-cell movement of CMV and chimeric BMV independently of viral CP by deletion of the C terminus. This suggests that the truncated CMV MP is comparable to TMV MP in the independence of CP in movement function. However, the C-terminal region of the MP is conserved among all strains of CMV whose sequences have been published to date in the GeneBank, EMBL, and DDBJ databases. This suggests that the C-terminal region has a biological significance necessary for the CMV life cycle. Indeed, a CMV variant with ΔC33-CMV MP gene does not move as efficiently as wt-CMV (35).
On the other hand, we have reported that a chimeric BMV with the ΔC33-CMV MP gene moves efficiently to induce chlorotic lesions comparably to wt BMV in their size and number (34) and that another chimeric BMV with both the CMV MP and CP genes moves less efficiently than wt BMV (35). In addition, when the BMV MP and CP genes were replaced with the ΔC33-CMV MP and CMV CP genes, respectively, the resulting chimeric BMV moved more efficiently than the chimeric BMV with wt-CMV MP and CMV CP (unpublished results). These results indicate that ΔC33-CMV MP can promote more cell-to-cell movement of the chimeric BMV genome than CMV movement equipment composed of wt-CMV MP and CP. Therefore, the two kinds of CMV equipment for cell-to-cell movement different in regard to the involvement of the CP show different specialties with respect to the genomes of CMV and chimeric BMV.
It is known that a nonfunctional MP expressed in transgenic plants confers resistance to multiple viruses by inhibiting their cell-to-cell movement (2, 11, 25, 29). The inhibitory effect on virus movement has been considered due to antagonism between competent and incompetent MPs. The chimeric BMV variants containing both wt- and ΔC33-CMV MP genes were remarkably inferior in cell-to-cell movement to the variant containing two ΔC33-CMV MP genes. This suggests antagonism between wt- and ΔC33-CMV MPs. Therefore, it is most likely that these MPs share common functions, although the wt-CMV MP is dominant negative in the absence of the CMV CP.
Sequential deletion analysis of the CMV MP C terminus revealed that the CMV MPs with truncation ranging from 31 to 36 amino acids are able to promote viral movement independently of viral CP. The wt-CMV MP is also able to promote viral movement in the presence of the CMV CP. However, the C-terminal deletion of fewer than 31 amino acids abolishes the ability of the CMV MP to mediate viral movement even in the presence of the CMV CP. These results indicate that the CP requirement in cell-to-cell movement of CMV is conferred by the C terminus of the MP and, further, that the participation of the CP in cell-to-cell movement is regulated in a highly complicated manner. A similar complex relationship between the size of C-terminal deletion and the competence of the MP to promote viral movement has been observed in the MP of Cowpea chlorotic mottle virus, a member of the genus Bromovirus (38). The existence of CP affects the movement, although this virus does not require the CP for cell-to-cell movement. The complicated results from the sequential deletion analysis may suggest that the C terminus of MP of these viruses affects the conformation of the protein.
Deletion of the C-terminal three amino acids in CMV MP is sufficient to impair viral movement. This suggests that the full-length C terminus is required to mediate viral cell-to-cell movement in a movement mechanism supported with CMV CP. Similarly, the MP of Alfalfa mosaic virus (AlMV), a member of the genus Alfamovirus belonging to the family Bromoviridae, is rendered nonfunctional by a deletion of the C-terminal three amino acids (47). AlMV is considered to move as virus-like particles via tubular structure, and a similar C-terminal deletion interfered with tubule formation by the MP (51). While the CMV MP also has an ability to induce tubules on the surface of infected protoplasts, a mutation impairing the ability to assemble tubules does not affect the systemic spread of CMV in tobacco and N. benthamiana (6). Therefore, although it is uncertain whether the C-terminal deletion of the CMV MP impairs the ability for the tubule formation, it is not likely that the impaired ability to assemble tubules affects the local spread mediated by the CMV MP.
Our results reveal that the C-terminal deletion more than 37 amino acids impairs the ability of the CMV MP to promote viral movement as well as local lesion induction. Kaplan et al. (23) have reported a CMV mutant with the C-terminal deletion of 43 amino acids in the MP that infects tobacco systemically. The mutation was maintained in the progeny virus in the systemically infected leaves. To the contrary, our corresponding variant of CMV constructed identically with the previous report of Kaplan et al. (23) did not move from cell to cell. The reason for the different results is not clear, although the CMV strains used in these studies are not identical. Since a variant of chimeric BMV with the gene of the CMV MP truncated in its C-terminal 43 amino acids is also unable to move from cell to cell, 1-amino-acid difference in the sequence of the truncated CMV MP might cause the different results. Alternatively, there might be a second-site mutation in the movement-competent CMV mutant.
Coexistence of the CMV CP with the CMV MP can mediate the cell-to-cell movement of heterogeneous as well as homogeneous viral genomes (34). As discussed above, the wt-CMV MP alone is considered nonfunctional. However, it is suggested that the MP itself and/or the putative MP-viral RNA complex are altered by the existence of the CP, although the mechanism remains unclear. A likely possibility is that the CMV CP interacts with the wt-CMV MP. The interaction might be mediated by the C terminus of the wt-CMV MP. However, our preliminary study on the interaction between these proteins in vitro failed to detect the binding of CP to the C terminus of the CMV MP (unpublished data). Further experiments are needed to clarify how the CP is involved in CMV movement.
Since a positive correlation has been seen between viral movement and the induction of local lesions in C. quinoa leaves, the lesions appearing on the leaves should reflect viral cell-to-cell movement and amplification (references 34, 35, and 43 and the present study). The local lesions also appear on the leaves inoculated with either CMV or chimeric BMV variants that did not express viral CP (Fig. 1, 4, and 7). This indicates that the CPs of BMV and CMV are dispensable for local lesion induction in C. quinoa. Similar observations have been reported in necrotic reaction in C. amalanticolor, in which viral movement from initially infected epidermal cells, and not the simultaneous expression of the MP and the CP of CMV in the cells, is required for the induction of cell death (6).
ACKNOWLEDGMENTS
We are grateful to Sigeru Kuwata (Meiji University) and Masashi Suzuki (Yokohama National University) for the infectious clones of CMV-Y and to Peter Palukaitis (Scottish Crop Research Institute) and Igor Boris Kaplan (Cornell University) for antiserum for CMV MP.
This work was supported in part by a Grant-in-Aid (12052201) for Scientific Research on Priority Area (A) and a Grant in-Aid (09NP1501) for Creative Basic Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and a Grant-in-Aid (JSPS-RFTF96L00603) from the Research for the Future program of the Japan Society for the Promotion of Science. H.N. was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
REFERENCES
- 1.Ahlquist P. Bromoviruses. In: Granoff A, Webster R G, editors. Encyclopedia of virology. 2nd ed. Vol. 1. San Diego, Calif: Academic Press, Inc.; 1999. pp. 198–204. [Google Scholar]
- 2.Beck D L, Van Dolleweerd C J, Lough T J, Balmori E, Voot D M, Andersen M T, O'Brien I E, Forster R L. Disruption of virus movement confers broad-spectrum resistance against systemic infection by plant viruses with a triple gene block. Proc Natl Acad Sci USA. 1994;91:10310–10314. doi: 10.1073/pnas.91.22.10310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackman L M, Boevink P, Santa Cruz S, Palukaitis P, Oparka K J. The movement protein of cucumber mosaic virus traffics into sieve elements in minor veins of Nicotiana clevelandii. Plant Cell. 1998;10:525–537. doi: 10.1105/tpc.10.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brigneti G, Voinnet O, Li W X, Ji L H, Ding S W, Baulcombe D C. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;16:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Boccard F, Baulcombe D C. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology. 1993;193:563–578. doi: 10.1006/viro.1993.1165. [DOI] [PubMed] [Google Scholar]
- 6.Canto T, Palukaitis P. Are tubules generated by the 3a protein necessary for cucumber mosaic virus movement? Mol Plant-Microbe Interact. 1999;12:985–993. [Google Scholar]
- 7.Canto T, Prior D A M, Hellwald K H, Oparka K J, Palukaitis P. Characterization of cucumber mosaic virus. IV. Movement protein and coat protein are both essential for cell-to-cell movement of CMV. Virology. 1997;237:237–248. doi: 10.1006/viro.1997.8804. [DOI] [PubMed] [Google Scholar]
- 8.Carrington J C, Kasschau K D, Mahajan A K, Schaad M C. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell. 1996;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou P Y, Fasman G. Prediction of protein conformation. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 10.Citovsky V, Knorr D, Schuster G, Zambryski P. The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell. 1990;60:637–647. doi: 10.1016/0092-8674(90)90667-4. [DOI] [PubMed] [Google Scholar]
- 11.Cooper B, Lapidot M, Heick J A, Dodds J A, Beachy R N. A defective movement protein of TMV in transgenic plants confers resistance to multiple viruses whereas the functional analog increases susceptibility. Virology. 1995;206:307–313. doi: 10.1016/s0042-6822(95)80046-8. [DOI] [PubMed] [Google Scholar]
- 12.Cooper B, Schmitz I, Rao A L N, Beachy R N, Dodds J A. Cell-to-cell transport of movement-defective cucumber mosaic and tobacco mosaic viruses in transgenic plants expressing heterologous movement protein gene. Virology. 1996;216:208–213. doi: 10.1006/viro.1996.0048. [DOI] [PubMed] [Google Scholar]
- 13.Deom C M, Oliver M J, Beachy R N. The 30-kilodalton gene product of tobacco mosaic virus potentiates virus movement. Science. 1987;237:389–394. doi: 10.1126/science.237.4813.389. [DOI] [PubMed] [Google Scholar]
- 14.Ding S, Li W, Symons R H. A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 1995;14:5762–5772. doi: 10.1002/j.1460-2075.1995.tb00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding B, Li Q, Nguyen L, Palukaitis P, Lucas W J. Cucumber mosaic virus (CMV) 3a protein potentiates cell-to-cell trafficking of CMV RNA in tobacco plants. Virology. 1995;207:345–353. doi: 10.1006/viro.1995.1093. [DOI] [PubMed] [Google Scholar]
- 16.Fujita Y, Mise K, Okuno T, Ahlquist P, Furusawa I. A single codon change in a conserved motif of a bromovirus movement protein gene confers compatibility with a new host. Virology. 1996;223:283–291. doi: 10.1006/viro.1996.0480. [DOI] [PubMed] [Google Scholar]
- 17.Giesman-Cookmeyer D, Silver S, Vaewhongs A A, Lommel S A, Deom C M. Tobamovirus and dianthovirus movement proteins are functionally homologous. Virology. 1995;213:38–45. doi: 10.1006/viro.1995.1544. [DOI] [PubMed] [Google Scholar]
- 18.Hayes R J, Buck K W. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell. 1990;63:363–368. doi: 10.1016/0092-8674(90)90169-f. [DOI] [PubMed] [Google Scholar]
- 19.Itaya A, Hickman H, Bao Y, Nelson R, Ding B. Cell-to-cell trafficking of cucumber mosaic virus movement protein: green fluorescent protein fusion produced by biolistic gene bombardment in tobacco. Plant J. 1997;12:1223–1230. [Google Scholar]
- 20.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 21.Kaido M, Mori M, Mise K, Okuno T, Furusawa I. Inhibition of brome mosaic virus (BMV) amplification in protoplasts from transgenic tobacco plants expressing replicable BMV RNAs. J Gen Virol. 1995;76:2827–2833. doi: 10.1099/0022-1317-76-11-2827. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan I B, Zhang L, Palukaitis P. Characterization of cucumber mosaic virus. V. Cell-to-cell movement requires capsid protein but not virions. Virology. 1998;246:221–231. doi: 10.1006/viro.1998.9192. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan I B, Shintaku M H, Li Q, Zhang L, Marsh L E, Palukaitis P. Complementation of virus movement in transgenic tobacco expressing the cucumber mosaic virus 3a gene. Virology. 1995;209:188–199. doi: 10.1006/viro.1995.1242. [DOI] [PubMed] [Google Scholar]
- 24.Krol M A, Olson N H, Tate J, Johnson J E, Baker T S, Ahlquist P. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc Natl Acad Sci USA. 1999;96:13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapidot M, Gafny R, Ding B, Wolf S, Lucas W, Beachy R N. A dysfunctional movement protein of tobacco mosaic virus that partially modifies the plasmodesmata and limits virus spread in transgenic plants. Plant J. 1993;4:959–970. [Google Scholar]
- 26.Lazarowitz S G, Beachy R N. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell. 1999;11:535–548. doi: 10.1105/tpc.11.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Palukaitis P. Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic cucumovirus and tobacco mosaic tobamovirus. Virology. 1996;216:71–79. doi: 10.1006/viro.1996.0035. [DOI] [PubMed] [Google Scholar]
- 28.Lucas W J, Gilbertson R L. Plasmodesmata in relation to viral movement within leaf tissues. Annu Rev Phytopathol. 1994;32:387–411. [Google Scholar]
- 29.Malyshenko S I, Kondakova O A, Nazarova J V, Kaplan I B, Taliansky M E, Atabekov J G. Reduction of tobacco mosaic virus accumulation in transgenic plants producing non-functional viral transport proteins. J Gen Virol. 1993;74:1149–1156. doi: 10.1099/0022-1317-74-6-1149. [DOI] [PubMed] [Google Scholar]
- 30.Mise K, Tsuge S, Nagao K, Okuno T, Furusawa I. Nucleotide sequence responsible for the synthesis of a truncated coat protein of brome mosaic virus strain ATCC66. J Gen Virol. 1992;73:2543–2551. doi: 10.1099/0022-1317-73-10-2543. [DOI] [PubMed] [Google Scholar]
- 31.Mise K, Mori M, Nakayashiki H, Koyama T, Okuno T, Furusawa I. Nucleotide sequence of a set of cDNA clones derived from the brome mosaic virus ATCC66 strain and comparison with the Russian strain genome. Ann Phytopathol Soc Jpn. 1994;60:454–462. [Google Scholar]
- 32.Mori M, Mise K, Kobayashi K, Okuno T, Furusawa I. Infectivity of plasmids containing brome mosaic virus cDNA linked to the cauliflower mosaic virus 35S RNA promoter. J Gen Virol. 1991;72:243–246. doi: 10.1099/0022-1317-72-2-243. [DOI] [PubMed] [Google Scholar]
- 33.Mori M, Zhang G, Kaido M, Okuno T, Furusawa I. Efficient production of human gamma interferon in tobacco protoplasts by genetically engineered brome mosaic virus RNAs. J Gen Virol. 1993;74:1255–1260. doi: 10.1099/0022-1317-74-7-1255. [DOI] [PubMed] [Google Scholar]
- 34.Nagano H, Mise K, Okuno T, Furusawa I. The cognate coat protein is required for cell-to-cell movement of a chimeric brome mosaic virus mediated by the cucumber mosaic virus movement protein. Virology. 1999;265:226–234. doi: 10.1006/viro.1999.0065. [DOI] [PubMed] [Google Scholar]
- 35.Nagano H, Okuno T, Mise K, Furusawa I. Deletion of the C-terminal 33 amino acids of cucumber mosaic virus movement protein enables a chimeric brome mosaic virus to move from cell to cell. J Virol. 1997;71:2270–2276. doi: 10.1128/jvi.71.3.2270-2276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitta N, Takanami Y, Kuwata S, Kubo S. Inoculation with RNAs 1 and 2 of cucumber mosaic virus induces viral RNA replicase activity in tobacco mesophyll protoplasts. J Gen Virol. 1988;69:2695–2700. [Google Scholar]
- 37.Osman T A, Hayes R J, Buck K W. Cooperative binding of the red clover necrotic mosaic virus movement protein to single-stranded nucleic acids. J Gen Virol. 1992;73:223–227. doi: 10.1099/0022-1317-73-2-223. [DOI] [PubMed] [Google Scholar]
- 38.Osman F, Schmitz I, Rao A L N. Effect of C-terminal deletions in the movement protein of cowpea chlorotic mottle virus on cell-to-cell and long-distance movement. J Gen Virol. 1999;80:1357–1365. doi: 10.1099/0022-1317-80-6-1357. [DOI] [PubMed] [Google Scholar]
- 39.Palukaitis P, Roossinck M J, Dietzgen R G, Francki R I B. Cucumber mosaic virus. Adv Virus Res. 1992;41:281–348. doi: 10.1016/s0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- 40.Perbal M C, Thomas C L, Maule A J. Cauliflower mosaic virus gene I product (P1) forms tubular structures which extend from the surface of infected protoplasts. Virology. 1993;195:281–285. doi: 10.1006/viro.1993.1375. [DOI] [PubMed] [Google Scholar]
- 41.Roossinck M. Cucumoviruses. In: Granoff A, Webster R G, editors. Encyclopedia of virology. 2nd ed. Vol. 1. San Diego, Calif: Academic Press, Inc.; 1999. pp. 315–320. [Google Scholar]
- 42.Rybicki E P. Bromoviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Sixth Report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 450–457. [Google Scholar]
- 43.Schmitz I, Rao A L N. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement-defective RNA3 variants of brome mosaic virus. Virology. 1996;226:281–293. doi: 10.1006/viro.1996.0656. [DOI] [PubMed] [Google Scholar]
- 44.Schmitz I, Rao A L N. Deletions in the conserved amino-terminal basic arm of cucumber mosaic virus coat protein disrupt virion assembly but do not abolish infectivity and cell-to-cell movement. Virology. 1998;248:323–331. doi: 10.1006/viro.1998.9257. [DOI] [PubMed] [Google Scholar]
- 45.Solovyev A G, Zelenina D A, Savenkov E I, Grdzelishvili V Z, Morozov S Y, Lesemann D E, Maiss E, Casper R, Atabekov J G. Movement of a barley stripe mosaic virus chimera with a tobacco mosaic virus movement protein. Virology. 1996;217:435–441. doi: 10.1006/viro.1996.0137. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki M, Kuwata S, Kataoka J, Masuta C, Nitta N, Takanami Y. Functional analysis of deletion mutants of cucumber mosaic virus RNA3 using an in vitro transcription system. Virology. 1991;183:106–113. doi: 10.1016/0042-6822(91)90123-s. [DOI] [PubMed] [Google Scholar]
- 47.van der Vossen E A, Notenboom T, Bol J F. Characterization of sequences controlling the synthesis of alfalfa mosaic virus subgenomic RNA in vivo. Virology. 1995;212:663–672. doi: 10.1006/viro.1995.1524. [DOI] [PubMed] [Google Scholar]
- 48.Waigmann E, Lucas W J, Citovsky V, Zambryski P. Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc Natl Acad Sci USA. 1994;91:1433–1437. doi: 10.1073/pnas.91.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wellink J, van Lent J W, Verver J, Sijen T, Goldbach R W, van Kammen A. The cowpea mosaic virus M RNA-encoded 48-kilodalton protein is responsible for induction of tubular structures in protoplasts. J Virol. 1993;67:3660–3664. doi: 10.1128/jvi.67.6.3660-3664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wikoff W R, Tsai C J, Wang G, Baker T S, Johnson J E. The structure of cucumber mosaic virus: cryoelectron microscopy, X-ray crystallography, and sequence analysis. Virology. 1997;232:91–97. doi: 10.1006/viro.1997.8543. [DOI] [PubMed] [Google Scholar]
- 51.Zeng H, Wang G, Zhang L. Alfalfa mosaic virus movement protein induces tubules in plant protoplasts. Mol Plant-Microbe Interact. 1997;10:1010–1014. [Google Scholar]