ABSTRACT
Introduction:
Bloodstream infections (BSIs), encompassing both self-limiting bacteremia and potentially fatal septicaemia, make up the majority of healthcare-associated ailments worldwide. The organisms encountered are mostly multidrug-resistant (MDROs), leading to increased hospital stays. Our study aims to collect data about blood culture isolates from a medical college in eastern Uttar Pradesh, India.
Materials and Methods:
A retrospective analysis of blood culture isolates obtained at our laboratory for ten months from patients with clinical suspicion of sepsis or infection with the possibility of haematogenous spread was done. We only considered consecutive and patient-specific, non-duplicate isolates. Blood samples were initially incubated in BacT/ALERT® and then manually processed once they flagged positive.
Results:
A total of 1,033 blood samples were received, of which 217 (21%) showed the growth of a pathogenic organism. The positivity rate varied significantly across different age groups, locations, and departments (P value < 0.001). It was higher among in-patients, those with central venous access, and patients with diabetes mellitus (DM). Staphylococcus aureus [n = 105, 48.38%] was isolated most commonly, with a high prevalence of methicillin resistance (83%). Enterococcus demonstrated a high degree of resistance. MDROs accounted for 68% of the detected Gram-negatives.
Discussion:
This study comprehensively analyses blood culture results from a diverse group of patients and emphasizes the association between risk factors and positive blood cultures. Gram-positive and Gram-negative isolates demonstrated low sensitivity to common antibiotics, urging vigilant monitoring and specific therapy.
Conclusion:
Our study reveals important insights guiding clinical practices, antimicrobial stewardship, and infection control strategies.
Keywords: Bacteremia, Gram-positive bacteria, sepsis
Introduction
Bloodstream infections (BSIs) make up the majority of healthcare-associated ailments worldwide.[1] Around 11.3 million sepsis cases and 2.9 million fatalities occurred in India in 2017.[2] Additionally, 7.3% of Asians had community-onset BSIs (range, 2.0 to 48.4%), according to a recent meta-analysis.[3] The Surviving Sepsis Campaign recommends collecting blood cultures within the first hour of suspected sepsis. Knowing the bacterial sensitivity patterns is crucial for initiating empirical therapy.[4,5]
Distribution and sensitivity trends in bacteria fluctuate across geographical locations and various medical facilities.[6] Our study aims to analyse data about the prevalence and antibiotic susceptibility patterns of blood culture isolates.
Materials and Methods
Study area and period: This research aims to retrospectively analyse the blood culture isolates obtained at the Department of Microbiology, All India Institute of Medical Sciences (AIIMS), located in Raebareli, Uttar Pradesh. Our medical college is a teaching institution with a 600-bed capacity. It caters to the district’s and surrounding areas’ medical requirements, ensuring top-notch healthcare services. The study covers ten months, starting from May 2022 and ending in February 2023, and was approved by the institutional ethics committee. This study comprised blood culture specimens obtained from inpatients and outpatients (paediatric and adult) for bacteriological inquiry as a standard part of their patient care. A comprehensive inquiry into the records of all relevant cases was carried out using the laboratory register and the medical records department at the hospital.
Inclusion and exclusion criteria: The attending clinician requested blood cultures based on clinical symptoms that were deemed indicative of sepsis or infection with the possibility of haematogenous spread. The diagnosis of BSI required the presence of positive blood cultures in conjunction with systemic indications of infection.[7] To identify sepsis in newborns, medical professionals searched for distinct clinical manifestations such as fever (≥38.0°C), respiratory distress, convulsions, hypothermia (≤36.5°C), lethargy, poor feeding, vomiting, jaundice, purulent infections in the umbilical region.[8] Patients who were excluded from the analysis had autoimmune/chronic diseases, including tuberculosis and sarcoidosis, weakened immune systems, steroid use, heat stroke, or suspected viral and parasitic infections. We only considered consecutive and patient-specific non-duplicate isolates.
Specimen collection and processing: By the hospital’s established protocol for sample collection, the blood samples were obtained after adhering to aseptic precautions. Notably, before the blood collection, the skin was thoroughly cleansed with a 2% chlorhexidine solution, as stipulated by the protocol. This practice aims to maintain the sample’s integrity and minimize the risk of contamination. Optimal sites for blood sampling included the antecubital and median cubital fossae. If the patient had central venous access, blood was collected from one of the lumens after thoroughly cleaning the port hub with 2% chlorhexidine. For each sample, 8-10 ml and 2–4 ml of blood were obtained from adult and paediatric patients, respectively. The blood samples were promptly added to BacT/ALERT® aerobic blood culture bottles, adult (BacT/ALERT® FA plus and BacT/ALERT® FN plus) or paediatric (BacT/ALERT® PF plus), as required, made by BioMérieux, France followed by immediate transport to the Microbiology laboratory wherein these were incubated in a BacT/ALERT® 3D Microbial Detection System an automated blood culture analyser (BioMérieux, France). The samples were processed in compliance with the manufacturer’s recommended standard criteria.
The BacT/ALERT-positive broths were immediately inoculated onto blood agar and MacConkey agar (Hi-Media, Mumbai). These inoculated agar plates were incubated at 37°C for 18 to 24 hours. The culture was deemed contaminated when diphtheroids, Bacillus spp., Micrococcus spp., and viridans streptococci were detected. The bacterial growth on the agar plates in the case of pathogens was determined and characterized based on the colony morphology, Gram staining, and traditional biochemical testing procedures, all conducted using established laboratory techniques.[9,10] Antibiotic susceptibility testing was carried out using the Kirby-Bauer disk diffusion method, employing antibiotic discs (Hi-Media) and interpreting the results based on the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI).[11] In this study, multidrug resistance (MDR) is defined as resistance to three or more classes of tested antibiotics.[12] The Staphylococcal isolates underwent additional testing for methicillin resistance as per CLSI.[11]
Quality Control and Data Analysis: The strains, namely Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Pseudomonas aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 25923, were employed to ensure quality control in biochemical assays and assessment of antibiotic sensitivity. The percentage of susceptible isolates for each antibiotic was calculated by dividing the number of susceptible isolates by the total number of isolates. The data was compiled into a single chart using Microsoft Excel and analysed.
Results
During the study period, a total of 1,033 blood samples were collected. Among the patients, 54.70% were male, and about one-third belonged to the paediatric age group. Out of all the blood samples, 78.61% were obtained from patients admitted in the in-patient department (IPD) and 21.39% from patients of the out-patient department (OPD). The General Medicine department contributed to over one-third of the overall samples. Departments like Dermatology, Urology, Obstetrics and Gynaecology, and Ophthalmology had sent less than ten samples in the 10-month study period and were clubbed together in the analysis. About 15.39% of the patients had central venous access, and a quarter of them had a history of diabetes mellitus (DM). Patients exhibited a diverse array of signs and symptoms, so those involving the same system were combined in the data presentation. The most common presentation of the patients was fever (46%) followed by respiratory signs and symptoms (12.58%). Table 1 shows the demographic distribution of the patients.
Table 1.
Distribution of the patients on the basis of demographic and clinical profile (n = 1033)
| Number of patients | Parameter | Percentage (%) |
|---|---|---|
| Gender | ||
| Male | 565 | 54.70% |
| Female | 468 | 45.30% |
| Age groups (years) | ||
| 0-10 | 186 | 18.01% |
| 11-20 | 161 | 15.59% |
| 21-30 | 140 | 13.55% |
| 31-40 | 95 | 9.20% |
| 41-50 | 105 | 10.16% |
| 51-60 | 128 | 12.39% |
| 61-70 | 143 | 13.84% |
| >70 | 75 | 7.26% |
| Location | ||
| OPD | 221 | 21.39% |
| IPD | 812 | 78.61% |
| Departments | ||
| Gen Medicine | 386 | 37.37% |
| Trauma & Emer. | 263 | 25.46% |
| Paediatrics | 240 | 23.23% |
| Paediatric Surgery | 46 | 4.45% |
| Neurosurgery | 38 | 3.68% |
| Gen Surgery | 22 | 2.13% |
| Others | 38 | 3.68% |
| Central venous access | ||
| Present | 159 | 15.39% |
| Absent | 874 | 84.61% |
| History of Diabetes Mellitus | ||
| Present | 259 | 25.07% |
| Absent | 774 | 74.93% |
| Clinical Presentation | ||
| Fever | 476 | 46.08% |
| Respiratory symptoms (shortness of breath, tachypnoea, cough) | 130 | 12.58% |
| Meningeal signs (headache, vomiting) | 118 | 11.42% |
| Urinary symptoms (Burning micturition, frequency) | 88 | 8.52% |
| Gastro-intestinal symptoms (pain in abdomen, diarrhoea) | 87 | 8.42% |
| Cellulitis | 49 | 4.74% |
| Sepsis | 48 | 4.65% |
| Others | 37 | 3.58% |
Of the 1,033 samples, 217 (21%) showed growth of a pathogenic organism, while 26 (2.5%) showed growth of contaminants. The remaining 790 samples (76.5%) did not show any growth, as per Figure 1. The 26 samples which showed contamination were excluded from further analysis.
Figure 1.
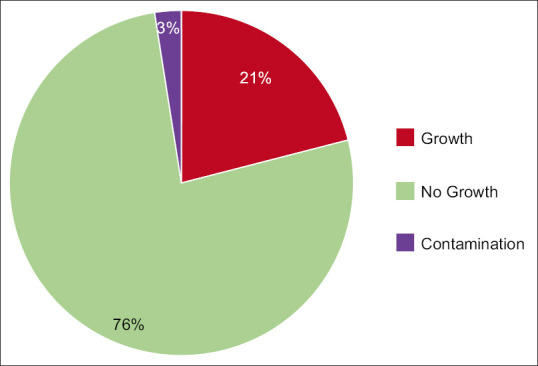
Growth in blood culture
Table 2 presents the distribution of positive blood cultures based on demographic parameters. The positivity rate was similar in both genders but varied significantly across different age groups, locations, and departments (P value < 0.001). It was notably higher among IPD, patients with central venous access, and those with a history of DM (Odds ratio >1).
Table 2.
Distribution of positive blood culture across various demographic parameters
| Growth | No Growth | Total | |
|---|---|---|---|
| Gender | |||
| Male | 119 (21.6%) | 432 (78.4%) | 551 (100%) |
| Female | 98 (21.5%) | 358 (78.5%) | 456 (100%) |
| P=0.97 | |||
| Age groups (years) | |||
| 0-10 | 28 (15.38%) | 154 (84.62%) | 182 (100%) |
| 11-20 | 18 (11.32%) | 141 (88.68%) | 159 (100%) |
| 21-30 | 25 (18.52%) | 110 (81.48%) | 135 (100%) |
| 31-40 | 26 (27.96%) | 67 (72.04%) | 93 (100%) |
| 41-50 | 33 (31.73%) | 71 (68.27%) | 104 (100%) |
| 51-60 | 35 (28.23%) | 89 (71.77%) | 124 (100%) |
| 61-70 | 30 (21.28%) | 111 (78.72%) | 141 (100%) |
| >70 | 22 (31.88%) | 47 (68.12%) | 69 (100%) |
| P≤0.001 | |||
| Location | |||
| OPD | 23 (10.5%) | 196 (89.5%) | 219 (100%) |
| IPD | 194 (24.62%) | 594 (75.38%) | 788 (100%) |
| P≤0.001; Odds ratio: 2.78 | |||
| Departments | |||
| Gen Medicine | 57 (15.2%) | 318 (84.8%) | 375 (100%) |
| Trauma & Emer. | 105 (41.5%) | 148 (58.5%) | 253 (100%) |
| Paediatrics | 29 (12.29%) | 207 (87.71%) | 236 (100%) |
| Paediatric Surgery | 9 (20%) | 36 (80%) | 45 (100%) |
| Neurosurgery | 6 (15.79%) | 32 (84.21%) | 38 (100%) |
| Gen Surgery | 4 (18.18%) | 18 (81.82%) | 22 (100%) |
| Others | 7 (18.42%) | 31 (81.58%) | 38 (100%) |
| P≤0.001 | |||
| Central venous access | |||
| Present | 39 (25.66%) | 113 (74.34%) | 152 (100%) |
| Absent | 178 (20.82%) | 677 (79.18%) | 855 (100%) |
| P=0.18; Odds ratio: 1.31 | |||
| History of Diabetes Mellitus | |||
| Present | 71 (28.17%) | 181 (71.83%) | 252 (100%) |
| Absent | 146 (19.34%) | 609 (80.66%) | 755 (100%) |
| P=0.003; Odds ratio: 1.63 | |||
| Clinical Presentation | |||
| Fever | 360 (77.25%) | 106 (22.75%) | 466 (100%) |
| Respiratory symptoms (shortness of breath, tachypnoea, cough) | 99 (77.34%) | 29 (22.65%) | 128 (100%) |
| Meningeal signs (headache, vomiting) | 89 (79.46%) | 23 (20.54%) | 112 (100%) |
| Urinary symptoms (Burning micturition, frequency) | 68 (79.07%) | 18 (20.93%) | 86 (100%) |
| Gastro-intestinal symptoms (pain in abdomen, diarrhoea) | 70 (82.35%) | 15 (17.65%) | 85 (100%) |
| Cellulitis | 38 (79.17%) | 10 (20.83%) | 48 (100%) |
| Sepsis | 38 (79.17%) | 10 (20.83%) | 48 (100%) |
| Others | 28 (82.35%) | 6 (17.65%) | 34 (100%) |
| P=0.97 |
The results of the blood cultures taken from patients revealed that the frequency of Gram-positive bacteria (76.9%) isolated was over three times that of Gram-negative bacteria (23.1%). Staphylococcus aureus [n = 105, 48.38%] was the most commonly isolated organism, followed by Coagulase-Negative Staphylococcus (CoNS) [n = 55, 25.34%], accounting for almost three-quarters of all isolates. The most commonly isolated Gram-negative organisms were Escherichia coli and Acinetobacter baumannii complex, each with 15 (6.91%) isolates. Figure 2 illustrates the distribution of different organisms isolated from blood cultures.
Figure 2.
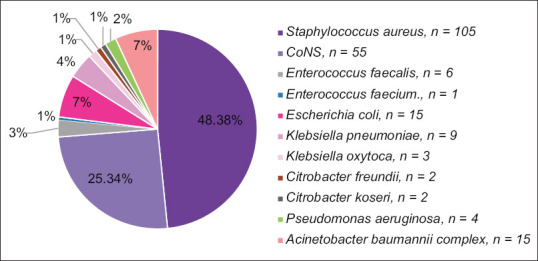
Distribution of various organisms isolated from blood cultures
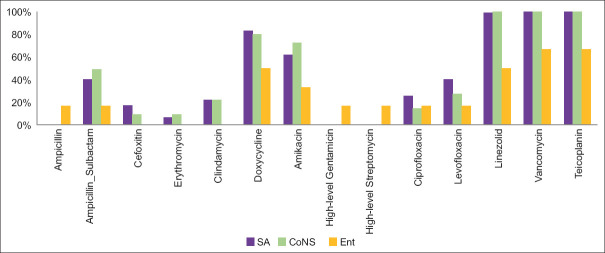
Our research found a high prevalence of methicillin resistance among Staphylococcus aureus (83%) and Coagulase-Negative Staphylococci (81%). The sensitivity of all Staphylococcus isolates to Penicillin, Ampicillin-Sulbactam, Quinolones, Clindamycin, and Erythromycin was less than 50%; however, it was 100% for Vancomycin and Teicoplanin, and only one Staphylococcus aureus isolate was resistant to Linezolid. Enterococcus demonstrated a higher degree of resistance to Vancomycin and Teicoplanin (33%), and Linezolid (50%). Its sensitivity to all the other antibiotics tested was less than 50%, except for Doxycycline (50%). [Figure 3]
Figure 3.
Sensitivity pattern of Gram-positive bacteria
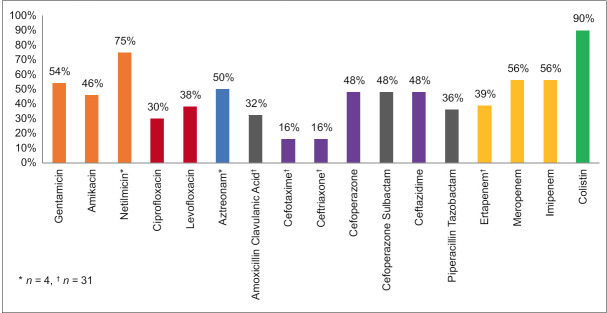
Due to the limited number of Gram-negative isolates, their sensitivity pattern was calculated as a group. Some drugs were not tested against all isolates because they are not recommended for those organisms. Netilmicin and Aztreonam were only tested against Pseudomonas aeruginosa (n = 4). Similarly, Cefotaxime, Ceftriaxone, Ertapenem, and Amoxicillin-clavulanic acid were not tested against Pseudomonas aeruginosa and Acinetobacter baumannii complex (n = 31). Please refer to Figure 4 for further details.
Figure 4.
Sensitivity pattern of Gram-negative bacteria
| Penicillin | Ampicillin | Cefoxitin | Erythromycin | Clindamycin | Doxycycline | Amikacin | |
|---|---|---|---|---|---|---|---|
| SA | 12% | NA | 17.14% | 6.66% | 21.90% | 82.86% | 61.90% |
| CoNS | 5.45% | NA | 9.09% | 9.09% | 21.81% | 80% | 72.72% |
| Ent | 16.67% | 16.67% | NA | NA | NA | 50% | 33% |
|
| |||||||
| High-level Gentamicin | High-level Streptomycin | Ciprofloxacin | Levofloxacin | Vancomycin | Teicoplanin | Linezolid | |
|
| |||||||
| SA | NA | NA | 25.71% | 40% | 100% | 100% | 99% |
| CoNS | NA | NA | 14.54% | 27.27% | 100% | 100% | 100% |
| Ent | 16.67% | 16.67% | 16.67% | 16.67% | 66.67% | 66.67% | 50% |
SA: Staphylococcus aureus; CoNS: Coagulase-Negative Staphylococcus; Ent: Enterococcus faecalis. NA: Not Applicable
The high prevalence of multidrug-resistant organisms (MDROs) among Gram-negative bacteria found in blood isolates has reached an alarming rate of up to 68%. This poses a severe threat to public health as the situation is further complicated because all Klebsiella pneumoniae, Klebsiella oxytoca, and Citrobacter freundii isolates are MDROs. Moreover, over half of the Escherichia coli and Acinetobacter baumannii complex isolates are MDROs, as shown in Figure 5.
Figure 5.
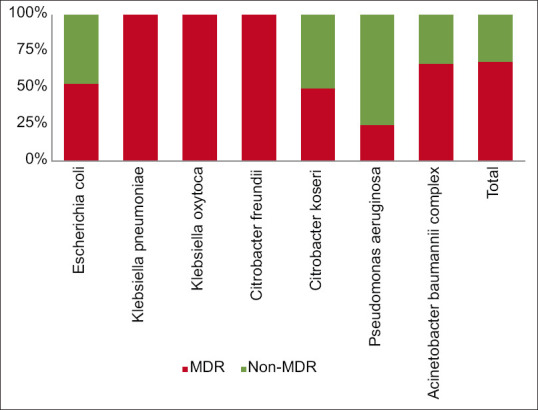
Percentage of MDROs among various Gram-negative isolates
Discussion
Monitoring the blood-borne organisms in a specific area is essential to identify infectious pathogens and understand their sensitivity patterns. By doing so, we can appropriately use antibiotics and curtail the spread of antibiotic resistance.[13,14] The current study comprehensively analyses blood culture results from a diverse group of patients. It reveals the prevalence of BSIs, the distribution of causative organisms, and antibiotic susceptibility patterns.
In our study, out of a total of 1033 blood culture samples, 217 (21%) came positive, similar to a study in Nepal by Pokhrel et al.[15] (20.5%). However, other studies had slightly higher (Ejaz et al.:28.26%) or lower (Kajumbula et al.: 14%).[16,17] The American Society of Microbiology (ASM) suggests an adequate positive blood culture rate of 6–12%.[18] A study in five Belgian tertiary care hospitals showed a positive blood culture rate of 9.8–12.9%.[19]
The observed variability in positive blood culture results can be attributed to several factors, including the quantity or volume of blood culture samples collected for analysis.[20] Only cultures showing monomicrobial growth were included in our analysis.
The demographic distribution of the study population reflects a balanced representation of both genders, with a slight male predominance. The preponderance of paediatric patients in the study, constituting about one-third of the total, was a unique observation, allowing us to analyse the patterns of BSIs in this group. It is essential to highlight that negative blood cultures do not necessarily mean no infection is present. Physicians sending one set of blood cultures, the presence of anaerobes, fungi, or viruses could account for the negative cultures in sepsis patients.[21] The male-to-female ratio in our study was 1.2:1, but blood culture positivity rates were similar in both genders, around 20%. Similar observations have been documented by Ombelet et al. and Banik et al.[22,23]
Approximately, 80% of our samples were from patients admitted to the IPD. The positivity rate among these samples was more than twice as high as those received from the OPD, and this difference was statistically significant. It is worth noting that our findings contradict those of Tarai et al.[24] This increased rate could be explained by the vulnerable inpatients contracting hospital-acquired sepsis, which needs to be explored in further studies. Although the General Medicine department had the highest number of patients in the sample pool (n = 386, 37.37%), the Trauma and Emergency department had the highest percentage of positive cases (48.8%) among all the departments. This was because, in our centre, patients with septicaemia are initially admitted to the Trauma and Emergency department, where they undergo initial investigations, are stabilized, and then are transferred to other departments, based on their age and underlying conditions, like General Medicine (positivity rate 26.1%), and Paediatrics (13.7%).
The presence of central venous access and a history of DM significantly increase the risk of positive blood culture (Odds ratio >1), highlighting their association with BSIs. In our study, 25.66% of patients with central venous access (n = 152) developed positive blood culture, which was similar to the findings of Kaur et al. (21.73%)[25] but relatively high compared to the findings of Lona-Reyes et al.[26] (7.35%, 15/204). The relation of bacteremia with DM is well studied. Though a study by Stoeckle et al. reported only 25.8 episodes of bacteremia/1000 admissions among diabetic patients, their relative frequency of BSI was much higher than non-diabetic patients (Odds ratio = 4.4).[27] Ghonim et al.[28] reported a high incidence of bacteremia among diabetic patients (66.67%), but their sample size was small (n = 60). Our study observed a blood culture positivity rate of 28.17% among diabetic patients with an Odds ratio of 1.63.
The majority of the patients presented with only fever (46%), followed by respiratory symptoms like cough and shortness of breath (12.58%); however, positivity rate of blood culture did not differ significantly among the varied clinical presentations of patients, suggesting any infection can spread to blood, and there is no fixed clinical presentation for BSIs. Primary physicians should also be sceptical about patients presenting with trivial symptoms like fever or cough as they may progress to BSIs.
The contamination rate observed (2.5%) in our study is below the desired level proposed by Hall. The contaminants primarily consist of isolates from the genera Bacillus spp., Corynebacterium spp., and Micrococcus spp., as well as polymicrobial growth.[29] The overall positivity rate of blood cultures was 21%, with Gram-positive bacteria being the most common (76.9%). This discovery aligns with a comparable investigation in a tertiary hospital in Ghana, where Gram-positive bacteria outnumbered the Gram-negative. Staphylococcus aureus was the most frequently isolated organism (n = 105). The prevalence of Gram-positive cocci has increased as the leading cause of BSIs since the early 1980s with the advent of modern medical treatment.[21] Coagulase-negative Staphylococci (S. epidermidis and S. haemolyticus) were the second most common Gram-positive cocci to be isolated. It is expected to find coagulase-negative staphylococci in blood cultures, which can be due to the normal bacterial flora present on the skin.[1] This typically occurs when the skin is not adequately disinfected. Our research discovered an increased isolation of coagulase-negative staphylococci in blood cultures, likely caused by the same contamination.
It is important to note that Staphylococcus isolates had low sensitivity to commonly prescribed antibiotics such as Penicillin, Ampicillin-Sulbactam, Quinolones, Clindamycin, and Erythromycin, heightening the need to tailor antibiotic therapy based on local resistance patterns. These bacteria exhibited good sensitivity to Doxycycline (>80%). Among the Staphylococcus aureus isolates, 83.65% were MRSA, a cause for great concern. This percentage is higher than documented in other studies.[1,30] This observation suggests that infection control practices need to be more stringent and robust in our institute. Herein, we want to highlight the importance of following proper contact precautions when handling IPD patients, especially in critical areas more prone to hospital-acquired infections.
Currently, Vancomycin and Teicoplanin are viable treatment options for these Gram-positive infections, but it is essential to follow proper guidelines regarding antibiotic usage to limit the spread of resistance. Only 50% of the Enterococci isolates exhibited susceptibility to doxycycline and Linezolid. This finding aligns with a series of investigations undertaken in Ethiopia and India.[31,32] Four of the Enterococcus species isolates were resistant to critical antibiotics such as Vancomycin and Teicoplanin, thus posing a challenge as effective antibiotics against Enterococci are limited, highlighting the need for continuous surveillance and developing novel therapeutic strategies.
Only 23.1% of isolates were Gram-negative bacteria, with Escherichia coli and Acinetobacter baumannii complex being the most prevalent (n = 15 each), closely followed by Klebsiella spp. (n = 12). This observation is in line with other studies.[1,5]
Upon analysing cumulative GNB data, the most significant susceptibility is to Colistin (90%), followed by Carbapenems (56%) and Aminoglycosides (50%) in general. Cephalosporins proved effective only against certain bacterial strains (16%). The cephalosporin resistance pattern of the Gram-negative isolates was similar to that reported by Agnihotri Bhat et al.[33] Pseudomonas species exhibited high sensitivity to Netilmicin (75%) and Aztreonam (50%), in addition to Colistin (90%). Such a high resistance rate to Carbapenems could be attributed to the indiscrete use of this antibiotic class in our region. The increasing prevalence of MDR in Gram-negative bacterial blood isolates is a severe issue, with the current rate at 68%. This is in agreement with other studies.[1,31,34] Klebsiella sp., Escherichia coli, and Acinetobacter baumannii complex isolates are particularly MDROs, highlighting the need for formulation of antibiotic policy and strict implementation of antibiotic stewardship practices to prevent the proliferation of resistant strains further.
Due to the paucity of isolates, individual analysis of isolates according to wards and Intensive Care Units (ICUs) was not done. The percentage of MDR organisms was slightly higher in the ICUs, but it was not statistically significant. This highlights the importance of implementing infection control measures and antibiotic policies at all levels and not just for critically ill or vulnerable patients of the ICUs.
Limitations
While the study provides valuable observations, certain limitations should be acknowledged.
As the study is retrospective, causation or temporality cannot be established. Moreover, the results of this single-centre study cannot be generalized to the whole Indian population. Multicentric studies with a larger sample size would increase the robustness of the findings.
This study’s antibiotic sensitivity testing method was restricted to disc diffusion sensitivity testing for all antibiotics except Vancomycin and Colistin.
The number of individual Gram-negative organisms was too low to be analyzed separately.
Conclusion
In conclusion, this study contributes valuable data on the epidemiology of BSIs and antibiotic resistance patterns among bacterial isolates. The findings underscore the complexity of BSIs and the importance of tailoring clinical practices based on local epidemiological data. The identification of isolates obtained from patients can aid primary healthcare physicians in the selection of appropriate antibiotics for early empirical introduction, thus preventing patients from progressing to sepsis and other complications. Further multicentric trials are necessary to expand our knowledge and database. These trials will enhance our understanding of the antibiotics required to combat infections caused by these isolates, in addition to providing a more comprehensive database to support clinical decision-making. The study also highlights the need for a multifaceted approach to infection control, which includes strict adherence to aseptic practices during invasive procedures, judicious use of indwelling devices, and effective management of underlying medical conditions like DM. The demographic diversity of our study population strengthens the generalizability of the study’s findings to a broader patient population.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Birru M, Woldemariam M, Manilal A, Aklilu A, Tsalla T, Mitiku A, et al. Bacterial profile, antimicrobial susceptibility patterns, and associated factors among bloodstream infection suspected patients attending Arba Minch General Hospital, Ethiopia. Sci Rep. 2021;11:15882. doi: 10.1038/s41598-021-95314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrei AI, Popescu GA, Popoiu MA, Mihai A, Tălăpan D. Changes in use of blood cultures in a covid-19-dedicated tertiary hospital. Antibiotics (Basel) 2022;11:1694. doi: 10.3390/antibiotics11121694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeganathan N. Burden of sepsis in India. Chest. 2022;161:1438–9. doi: 10.1016/j.chest.2022.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Marchello CS, Dale AP, Pisharody S, Rubach MP, Crump JA. A systematic review and meta-analysis of the prevalence of community-onset bloodstream infections among hospitalized patients in Africa and Asia. Antimicrob Agents Chemother. 2019;64:e1974–2019. doi: 10.1128/AAC.01974-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banik A, Lyngdoh VW, Durairaj E, Phukan AC, Kotal R. Ecology of bloodstream infections and temporal trends of their antibiograms with respect to source and duration of incubation: A 5-year retrospective observational analysis. J Lab Physicians. 2020;12:56–67. doi: 10.1055/s-0040-1714199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamba K, Lukwesa-Musyani C, Samutela MT, Kapesa C, Hang'ombe MB, Mpabalwani E, et al. Phenotypic and genotypic antibiotic susceptibility profiles of Gram-negative bacteria isolated from bloodstream infections at a referral hospital, Lusaka, Zambia. PLoS Glob Public Health. 2023;3:e0001414. doi: 10.1371/journal.pgph.0001414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timsit JF, Ruppe´ E, Barbier F, Tabah A, Bassetti M. Bloodstream infections in critically ill patients: An expert statement. Intensive Care Med. 2020;46:266. doi: 10.1007/s00134-020-05950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Neonatology Forum NNPD Network. National Neonatal Perinatal Database Report 2002–2003. New Delhi: National Neonatology Forum NNPD Network, India; 2005. p. 70. [Google Scholar]
- 9.Collee JG, Miles RS, Watt B. Tests for the identification of bacteria. In: Collee JG, Marmion BP, Fraser AG, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology. 14th ed. New York: Churchill Livingstone; 1996. pp. 131–51. [Google Scholar]
- 10.Winn WC, Allen SD, Janda WN, Koneman E, Procop G, Schreckenberger P, et al. 6th ed. Philadelphia: Lippincott; 2006. Koneman's Colour Atlas and Textbook of Diagnostic Microbiology. [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing;Thirty-Second Informational Supplement. CLSI Document M100-S25. 32nd ed. Wayne, PA, USA: Clinical and Laboratory Standard Institute; 2022. [Google Scholar]
- 12.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 13.Shrestha S, Amatya R, Shrestha RK, Shrestha R. Frequency of blood culture isolates and their antibiogram in a teaching hospital. J Nepal Med Assoc. 2014;52:692–6. [PubMed] [Google Scholar]
- 14.Sharma R, Sharma R, Gupta S. Bacteriological analysis of blood culture isolates with their antibiogram from a tertiary care hospital. Int J Pharm Sci Res. 2015;6:4847–51. [Google Scholar]
- 15.Pokhrel B, Koirala T, Shah G, Joshi S, Baral P. Bacteriological profile and antibiotic susceptibility of neonatal sepsis in neonatal intensive care unit of a tertiary hospital in Nepal. BMC Paediatr. 2018;18:208. doi: 10.1186/s12887-018-1176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ejaz A, Khawaja A, Arshad F, Tauseef A, Ullah R, Ahmad I. Etiological profile and antimicrobial patterns in blood culture specimens in a tertiary care setting. Cureus. 2020;12:e11000. doi: 10.7759/cureus.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajumbula H, Fujita AW, Mbabazi O, Najjuka C, Izale C, Akampurira A, et al. Antimicrobial drug resistance in blood culture isolates at a tertiary hospital, Uganda. Emerg Infect Dis. 2018;24:174. doi: 10.3201/eid2401.171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.York MK, Henry M, Gilligan PH. Blood Cultures: General Dectection and Interpretation. In: Isenberg H. D, editor. Clinical Microbiology Procedures Handbook: Volume 1. American Society for Microbiology. Washington, DC, USA: 2007. pp. 3.4.1.1–3.4.1.19. [Google Scholar]
- 19.Willems E, Smismans A, Cartuyvels R, Coppens G, Van Vaerenbergh K, Van den Abeele AM, et al. The preanalytical optimization of blood cultures: A review and the clinical importance of benchmarking in 5 Belgian hospitals. Diagn Microbiol Infect Dis. 2012;73:1–8. doi: 10.1016/j.diagmicrobio.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Lee A, Mirrett S, Reller LB, Weinstein MP. Detection of bloodstream infections in adults: How many blood cultures are needed? J Clin Microbiol. 2007;45:3546–8. doi: 10.1128/JCM.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acheampong EN, Tsiase JA, Afriyie DK, Amponsah SK. Neonatal Sepsis in a resource-limited setting: Causative microorganisms and antimicrobial susceptibility profile. Interdiscip Perspect Infect Dis 2022. 2022 doi: 10.1155/2022/7905727. 7905727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ombelet S, Kpossou G, Kotchare C, Agbobli E, Sogbo F, Massou F, et al. Blood culture surveillance in a secondary care hospital in Benin: Epidemiology of bloodstream infection pathogens and antimicrobial resistance. BMC Infect Dis. 2022;22:1–5. doi: 10.1186/s12879-022-07077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banik A, Bhat SH, Kumar A, Palit A, Snehaa K. Bloodstream infections and trends of antimicrobial sensitivity patterns at Port Blair. J Lab Physicians. 2018;10:332–7. doi: 10.4103/JLP.JLP_50_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarai B, Jain D, Das P, Budhiraja S. Paired blood cultures increase the sensitivity for detecting pathogens in both inpatients and outpatients. Eur J Clin Microbiol Infect Dis. 2018;37:435–41. doi: 10.1007/s10096-018-3188-8. [DOI] [PubMed] [Google Scholar]
- 25.Kaur M, Gupta V, Gombar S, Chander J, Sahoo T. Incidence, risk factors, microbiology of venous catheter associated bloodstream infections-A prospective study from a tertiary care hospital. Indian J Med Microbiol. 2015;33:248–54. doi: 10.4103/0255-0857.153572. [DOI] [PubMed] [Google Scholar]
- 26.Lona-Reyes JC, López-Barragán B, Pérez-Molina JJ, Ascencio-Esparza EP. Central venous-catheter related bacteremia: Incidence and risk factors in a hospital in western Mexico. Bol Med Hosp Infant Mex. 2016;73:105–10. doi: 10.1016/j.bmhimx.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Stoeckle M, Kaech C, Trampuz A, Zimmerli W. The role of diabetes mellitus in patients with bloodstream infections. Swiss Med Wkly. 2008;138:512–9. doi: 10.4414/smw.2008.12228. doi: 10.4414/smw.2008.12228. PMID: 18792825. [DOI] [PubMed] [Google Scholar]
- 28.Ghonim EM, El Hindawy GR, Abd El Motelb TM, Labib AZ, Ahmady I, Salem EH. Study of bacteremia in diabetic patients. Menoufia Med J. 2016;29:846. [Google Scholar]
- 29.Hall KK, Lyman JA. Updated review of blood culture contamination. Clin Microbiol Rev. 2006;19:788–802. doi: 10.1128/CMR.00062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasudeva N, Nirwan PS, Shrivastava P. Bloodstream infections and antimicrobial sensitivity patterns in a tertiary care hospital of India. Ther Adv Infect Dis. 2016;3:119–27. doi: 10.1177/2049936116666983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wasihun AG, Wlekidan LN, Gebremariam SA, Dejene TA, Welderufael AL, Haile TD, et al. Bacteriological profle and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle Hospital Northern Ethiopia. Springerplus. 2015;4:314. doi: 10.1186/s40064-015-1056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gohel K, Jojera A, Soni S, Gang S, Sabnis R, Desai M. Bacteriological profle and drug resistance patterns of blood culture isolates in a tertiary care nephrourology teaching institute. Biomed Res Int 2014. 2014 doi: 10.1155/2014/153747. 153747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhat YR, Lewis LE, Ke V. Bacterial isolates of early-onset neonatal sepsis and their antibiotic susceptibility pattern between 1998 and 2004: An audit from a center in India. Ital J Pediatr. 2011;37:32. doi: 10.1186/1824-7288-37-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thakur S, Thakur K, Sood A, Chaudhary S. Bacteriological profile and antibiotic sensitivity pattern of neonatal septicaemia in a rural tertiary care hospital in North India. Indian J Med Microbiol. 2016;34:67–71. doi: 10.4103/0255-0857.174108. [DOI] [PubMed] [Google Scholar]