Abstract
We examined the population structure and genetic variation of four genomic regions within and between 30 Citrus tristeza virus (CTV) isolates from Spain and California. Our analyses showed that most isolates contained a population of sequence variants, with one being predominant. Four isolates showed two major sequence variants in some genomic regions. The two major variants of three of these isolates showed very low nucleotide identity to each other but were very similar to those of other isolates, suggesting the possibility of mixed infections with two divergent isolates. Incongruencies of phylogenetic relationships in the different genomic regions and statistical analyses suggested that the genomes of some CTV sequence variants originated by recombination events between diverged sequence variants. No correlation was observed between geographic origin and nucleotide distance, and thus from a genetic view, the Spanish and Californian isolates analyzed here could be considered members of the same population.
Citrus tristeza virus (CTV) is distributed worldwide and is the causal agent of one of the most economically important diseases of citrus. CTV, a member of the genus Closterovirus within the family Closteroviridae, is phloem limited and is transmitted by aphids in a semipersistent manner. CTV virions are filamentous flexuous particles about 2,000 nm long, with two coat proteins (CP and CPm) covering 95 and 5% of the particle length, respectively (8). The CTV genome is a single-stranded, positive-sense RNA of 19,226 to 19,296 nucleotides (nt) (18, 27, 48, 51) organized in 12 open reading frames encoding at least 19 proteins. These include two papain-like proteases, replication-associated proteins (RNA polymerase, helicase, and methyltransferase), a homologue of the HSP70 protein, two coat proteins (CP and CPm), RNA-binding protein p23 (23), a p20 protein that accumulates in the amorphous inclusion bodies (14), and other proteins of so far unknown function (p61, p13, and p18) (Fig. 1). CTV-infected plants contain, in addition to the genomic RNA, 3′-coterminal subgenomic RNAs (15) and defective RNAs (D RNAs), the latter resulting from extensive internal deletions of the genomic RNA (2, 26, 28, 50).
FIG. 1.
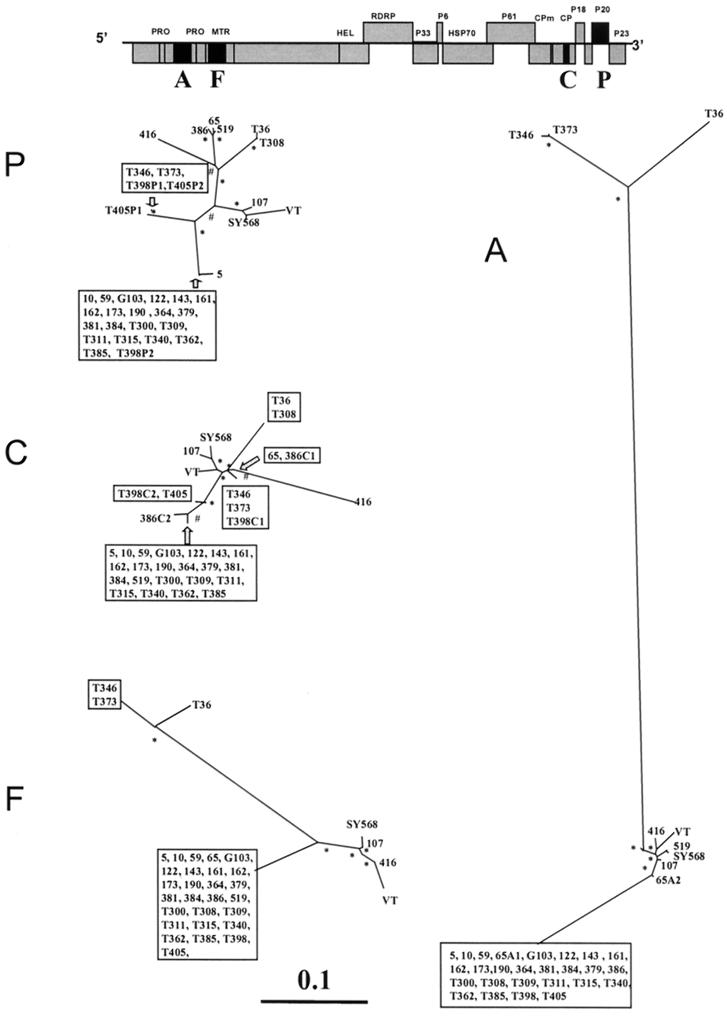
Unrooted maximum-likelihood phylogenetic trees of genomic regions A, F, C, and P (see also Table 2) of 34 CTV isolates (Table 1), constructed using the PHYLIP program DNAML. Bootstrap values of between 600 and 800 for 1,000 replicates are indicated by #, and values greater than 800 are indicated by ∗. Branch lengths are proportional to the genetic distances. Boxes include sequences with a nucleotide identity of greater than 99%. Above is a layout of the CTV genome, with the regions analyzed in black boxes. When an isolate contained more than one major sequence, these are indicated by a letter (corresponding to the genomic region) and a number (corresponding to the sequence variant), e.g., 65A1 and 65A2 occurred in genomic region A of isolate 65.
CTV isolates differing in the type and intensity of symptoms induced in different citrus species and cultivars and in their aphid transmissibility have been reported worldwide (38). In the last two decades, efforts have been taken to develop molecular techniques for rapid differentiation of CTV isolates and identification of molecular markers related to CTV-induced symptoms. Variation in serological reactivity, peptide maps of the coat protein, double-stranded RNA (dsRNA) patterns, hybridization with cDNA probes, restriction fragment length polymorphism, and single-strand conformation polymorphism (SSCP) have been described in attempts to differentiate CTV isolates (29).
Nucleotide sequence analysis is the most accurate procedure for CTV differentiation and estimation of molecular or genetic variation. To date, the complete genome nucleotide sequences of the five CTV isolates T36 and T30 from Florida (1, 18, 34), VT from Israel (27), SY568 from California (51), and T385 from Spain (48) have been reported. Also the partial nucleotide sequences of several CTV isolates have been reported (1, 17, 22, 25, 35, 36). Recently, it has been shown that individual CTV isolates are composed of a population of sequence variants (3, 19, 22). These reports showed the genetic differences between CTV isolates but did not estimate the genetic diversity of natural populations of CTV. Only Moya and García-Arenal (31) estimated the genetic diversity of CTV in Spain based on the number and position of D RNAs associated with Spanish CTV isolates (14). However, for CTV, nucleotide identity seems not to be correlated with the similarity of D RNA patterns (2).
In this study, we assessed the structure and genetic diversity of four genomic regions from two natural CTV populations located in two important citrus-growing areas, Spain and California. By SSCP analysis we estimated the population structure of sequence variants within individual isolates. Nucleotide analysis was used to estimate the genetic distance between sequence variants. These analyses open new insights about mixed infections, recombination, and spatial population structure in the effort to understand CTV complexity.
MATERIALS AND METHODS
Virus isolates.
Nineteen Californian and 11 Spanish CTV isolates were obtained from field trees and maintained in sweet orange (Citrus sinensis) plants in insect-proof greenhouses. Symptom evaluation was performed for these isolates in sweet orange plants and in graft-inoculated Mexican lime (Citrus aurantiifolia) and grapefruit (Citrus paradisi) plants. Geographic origin and symptoms induced by these CTV isolates are summarized in Table 1. More detailed information about the biological characteristics of CTV Spanish isolates can be found in Ballester-Olmos et al. (4).
TABLE 1.
Origin and biological characterization of CTV isolates
| Origin | Isolate no. | Specific origin (city, county or province) | Symptoms induceda
|
||
|---|---|---|---|---|---|
| Mexican lime | Sweet orange | SY | |||
| California | 5 | Lindsay, Tulare | 2 | 0 | − |
| 10 | Exeter, Tulare | 3 | 0 | − | |
| 59 | Edison, Kern | 2 | 0 | + | |
| 65 | Exeter, Tulare | 0 | 0 | − | |
| G103 | Ventura | N | 2 | + | |
| 107 | North Exeter, Tulare | 2 | 2 | − | |
| 122 | Exeter, Tulare | 2 | 1 | − | |
| 143 | Mc Farland, Kern | N | 0 | + | |
| 161 | Fresno | N | 0 | + | |
| 162 | McFarland, Kern | N | 0 | + | |
| 173 | Delano, Kern | N | 0 | + | |
| 190 | McFarland, Kern | 1 | 0 | − | |
| 364 | Delano, Kern | N | 0 | + | |
| 379 | Lindsay, Tulare | N | 0 | + | |
| 381 | Lindsay, Tulare | N | 0 | − | |
| 384 | Hemet, Riverside | N | 0 | + | |
| 386 | Riverside City, Riverside | N | 0 | + | |
| 416 | Fresno | N | N | N | |
| 519 | Riverside | N | 0 | − | |
| Spain | T300 | Puebla Larga, Valencia | 1 | 0 | − |
| T308 | Burjasot, Valencia | 3 | 0 | + | |
| T309 | Almenara, Castellon | 3 | 0 | − | |
| T311 | Xeresa, Valencia | 1 | 0 | − | |
| T315 | Huercal, Almeria | 1 | 0 | − | |
| T340 | Godella, Valencia | 2 | 0 | − | |
| T346 | Brenes, Sevilla | 2 | 0 | − | |
| T362 | Tortosa, Tarragona | 3 | 0 | − | |
| T373 | Alhama, Murcia | 2 | 0 | − | |
| T398 | Alcira, Valencia | 2 | 2 | − | |
| T405 | Alcira, Valencia | 2 | 0 | − | |
Symptoms induced by CTV isolates in different hosts: Mexican lime (vein clearing and stem pitting) and sweet orange (stunting and stem pitting). Symptom intensity is scored from 1 to 3, with 1 being mild and 3 very severe. SY, seedling yellows reaction in sour orange and grapefruit; − and +, absence and presence of symptoms, respectively; N, characterization not done.
Purification of CTV dsRNA.
CTV-infected sweet orange bark was pulverized with nitrogen liquid, and total nucleic acids were extracted with phenol-detergent buffer. The dsRNAs were then purified by column chromatography on nonionic cellulose and precipitated as previously described (30).
cDNA synthesis and PCR amplification.
Four pairs of primers (A, F, C, and P) were designed for performing reverse transcription (RT)-PCR of different regions of the CTV genomic RNA (Table 2). Approximately 200 ng of dsRNA was denatured at 95°C for 5 min and reverse transcribed by incubation at 42°C for 45 min in a reaction mixture (20 μl) containing 1× avian myeloblastosis virus (AMV) buffer, 200 μM each of the four deoxynucleoside triphosphates (dNTPs), 40 ng of reverse primer, 2 U of RNasin (Promega Corp.), and 0.3 U of AMV reverse transcriptase. An aliquot (1/10) of cDNA product was PCR amplified in a 20-μl reaction mixture containing 1× PCR buffer, 1.5 mM MgCl2, 200 μM each dNTPs, 20 ng of each primer, and Taq DNA polymerase (Promega). The following PCR conditions were used: 94°C for 2 min; 30 cycles each of 94°C for 30 s, 50°C for 30 s, and 72°C 40 s; and 72°C for 5 min. The resulting RT-PCR products were separated by electrophoresis in a 2% agarose gel and detected by ethidium bromide staining.
TABLE 2.
Primers designed for PCR amplification of four regions of CTV genomic RNA
| Primera | Nucleotide sequence | Genomic region | Positionsb | Sizec (nt) |
|---|---|---|---|---|
| A-forward | 5′-ACGTGTTCGTGAAACGCGG-3′ | A | 2021–2039 | 528 |
| A-reverse | 5′-GTCGATAACTCGACAAACGAGC-3′ | 2527–2548 | ||
| F-forward | 5′-GTGTTATCATGCGTCTGAAGCG-3′ | F | 3561–3582 | 438 |
| F-reverse | 5′-GGAATCTTAATCCTAATCAAG-3′ | 3978–3998 | ||
| CP-forward | 5′-AACGCCCTTCGAGTCTGGGGTAGGA-3′ | C | 16515–16539 | 273 |
| CP-reverse | 5′-TCAACGTGTGTTGAATTTCCCAAGC-3′ | 16763–16787 | ||
| P20-forward | 5′-ACAATATGCGAGCTTACTTTA-3′ | P | 17720–17740 | 561 |
| P20-reverse | 5′-AACCTACACGCAAGATGGA-3′ | 18262–18280 |
Oligonucleotide primers used. Forward indicates primers complementary to the negative strand, and reverse indicates those complementary to the positive strand.
Nucleotide positions of the primers in the genomic RNA of CTV isolate T385 (48).
Expected size of the RT-PCR products.
Cloning and SSCP analysis.
The cDNAs obtained by RT-PCR were cloned into PGEM-T (Promega) using T4 DNA ligase (Promega) according to the manufacturer's instructions, followed by transformation into Eschericia coli DH5α (44). Ten clones obtained from each cDNA product were selected and PCR amplified using the same conditions as the PCR described above. SSCP analysis was performed on the resulting PCR products as previously described (19, 40). Denatured samples were electrophoresed in nondenaturing 8% polyacrylamide gels at 200 V for 3 h (genomic regions A and F), 2 h (region P), or 1 h (region C).
Nucleotide sequences and statistical analysis.
Nucleotide sequences of cDNAs were determined in both directions by means of an ABI Prism DNA sequencer 377 (Perkin-Elmer, Foster City, Calif.). Multiple alignments of the nucleotide sequences were realized with the program CLUSTAL W (47). Numbers of synonymous and nonsynonymous nucleotide substitutions were determined using the program DIVERGE in the Genetics Computer Group (GCG) package (7), which is based on the method described by Pamilo and Bianchi (33) and Li (21) (PBL method). Nucleotide identity of pairs of sequences over the entire sequence length was plotted using GCG's program PLOTSIMILARITY (7). Nucleotide distances were estimated by using the DNADIST program of PHYLIP package version 3.573 (9) using the Jukes and Cantor method for correction of superimposed substitutions. Phylogenetic relationships were inferred using the PHYLIP programs DNAML, based on the maximum-likelihood method, and NEIGHBOR, which implements the neighbor-joining method from a nucleotide distance matrix. SEQBOOT (1,000 repetitions) and CONSENSE were used for bootstrap analysis. Genetic diversity (average number and standard errors of the number of nucleotide substitutions for each pair of sequence variants) and the degree of population subdivision were calculated by the method of Lynch and Crease (24). Recombination events between diverged nucleotide sequences were explored with the programs GENECONV (45) and PHYLPRO (49).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited in the GenBank database under accession numbers AF356226 to AF356329. Other CTV nucleotide sequences used in our analyses were obtained from the indicated GenBank entries: isolate T36 (U16034), isolate VT (U56902), isolate T385 (Y18420), isolate SY568 (AF001623), isolate T30 (AF260651), Japanese isolates (AB011185 to AB011197), Portuguese isolates (AF184113 to 184118), and Californian isolates 65, 107, 122, 173, and 190 (AF203035 to AF203037, AF203040, AF203043, AF203047, AF203048, AF203051, AF203053, AF203055, AF203057, AF203060, AF203062, AF203063, AF203066, AF203070, AF203073, AF203075, AF203077, AF203079, and AF203081).
RESULTS AND DISCUSSION
Genetic variation within CTV isolates: evidence of mixed infections.
RNA viruses have a great potential for genetic variation due to their error-prone RNA replication, large populations, and short replication times. As a consequence, each isolate of RNA virus is expected to consist of a population of genetically related variants, termed quasispecies (16). To estimate the within-isolate population structure, we performed RT-PCR of four genomic regions (Table 2) of 11 Spanish and 19 Californian CTV isolates and analyzed the SSCP patterns of 10 clones obtained from each genomic region and isolate. Previously we assessed the accuracy of these SSCP analyses (19). The average number of nucleotide differences per site between pairs of CTV sequence variant clones was only 0.001346 ± 0.002126, supporting SSCP analysis as a precise tool for population studies. To minimize primer-directed selection of sequence variants within a isolate, the primers used here (Table 2) were designed from nucleotide sequences conserved for the five CTV isolates whose complete genome sequences are known (1, 18, 27, 34, 48, 51). Previously, we also found that the intensities of the DNA bands in SSCP profiles reflected the relative proportion of CTV RNA variants within an isolate, suggesting that these primers do not bind preferentially to some CTV sequence variants (42). Furthermore, to minimize the possibility that nucleotide incorporation errors in the initial phases of the RT-PCR give rise to a detectable subpopulation of false mutants (20), we used as a template a large amount of dsRNAs (∼200 ng). Nonetheless, because we analyzed only the major sequence variants, RT-PCR-induced errors, if present, did not affect our analysis. Finally, to minimize possible variations due to irregular distribution of sequence variants in infected tissues, the dsRNAs were extracted from different branches of a large plant and pooled.
Our results and the results reported previously (19) showed that 26 of 30 field CTV isolates, in the four genomic regions analyzed, had a within-isolate population structure consisting of one major variant (frequency greater than 0.7) and other sequence variants of lower frequencies (minor variants), a typical quasispecies structure (16). In contrast, genomic region A of isolate 65 (19), genomic region C of isolates T398 and 386, and genomic region P of isolates T398 and T405 had two major variants with frequencies of 0.4 or greater. SSCP frequencies and nucleotide distances for all variants from Californian isolates 65, 107, 122, 173, and 190 have been described in detail by Kong et al. (19). We determined the nucleotide sequences of all CTV major variants and estimated the nucleotide distances between them. The nucleotide distances between sequences are represented in Fig. 1 as branch lengths of phylogenetic trees (see next section). Interestingly, for 65 A, T398 and 386 C, and T398 P, the nucleotide distances between the two major variants within the same isolate were high (Fig. 1). For example, the nucleotide distance of genomic region P between the two major variants of isolate T398 (T398P1 and T398P2) was 0.0795. Furthermore, within these isolate regions, each major variant was significantly more similar to major variants of other isolates than to the other major variant from the same isolate (Fig. 1). For example, the major variant T398P2 and that of isolate T300 showed a nucleotide distance of 0.0019, whereas T398P1 and T346 had a nucleotide distance of 0.0039. These data suggest that 65 A, T398 and 386 C, and T398 P could have originated from mixed infections of two CTV isolates with diverged sequence variants. Also, the fact that some genomic regions in the same isolate have two diverged major variants whereas other genomic regions have only one suggests the possibility of recombination events between sequence variants of the original coinfecting CTV isolates.
Mixed infections are possible, as CTV hosts are long-lived perennial plants (some living 100 years or more), allowing the possibility of repeated inoculations of CTV by viruliferous aphids. Although we have evidence for mixed infections in a small proportion of CTV isolates here analyzed (3 of 30), it is probable that mixed CTV infections occur more frequently in nature. With the approaches used here, mixed infections could only be detected between two isolates with diverged sequence variants, and both sequences should be in relatively high proportion when they were sampled. The coexistence of two quasispecies in the same host is probably not common, as differences in fitness would likely cause the displacement of one quasispecies.
Genetic variation in different genomic regions: evidence for natural recombination events.
The major variant sequences from the CTV isolates analyzed here and the sequences of CTV isolates T36, VT, T385, and SY568 (18, 27, 34, 48, 51) were used to estimate the nucleotide diversity (average number of nucleotide substitutions per site in each pair of sequence variants) for each genomic region. These analyses showed appreciable differences between the different genomic regions. Genomic region A showed the greatest diversity, twice that for F and P and three times that for the C genomic region (Table 3). To estimate the degree of selective constraints on each genomic region, nonsynonymous and synonymous substitutions were computed separately. The number of synonymous substitutions per synonymous site (dS) was similar over the four genomic regions analyzed (Table 3). However, the number of nonsynonymous substitutions per nonsynonymous site (dN) was smaller than dS and varied considerably between genomic regions (Table 3). This suggests a negative selective pressure for most amino acid changes (functional constraints) and that the degree of the functional constraints varies for the different genomic regions. Genomic regions F and P had a ratio dN/dS in the range of most DNA and plant RNA virus protein-coding sequences (12, 32). F corresponds to part of the methyltransferase domain, involved in virus replication, and P corresponds to gene p20, coding for a protein of unknown function that accumulates in amorphous inclusion bodies (13). Genomic region A, which so far is not known to be part of any known functional domain, showed the greatest dN/dS ratio. Finally, C, corresponding to a portion of the coat protein gene, showed the greatest functional constraints. The CTV coat protein, in addition to constraints related to virion structure and stability, might play a critical role in interactions with its aphid vector and/or plant host. The dN/dS ratio of CTV genomic region C was very low in comparison with that of other plant virus coat protein genes (12).
TABLE 3.
Average number of nucleotide substitutions between CTV isolates in different genomic regions (A, F, CP, and P20)a
| Region | D | dN | dS | dN/dS |
|---|---|---|---|---|
| A | 0.13656 (0.04209) | 0.11125 | 0.20472 | 0.56323 |
| F | 0.05993 (0.01601) | 0.03455 | 0.29849 | 0.12302 |
| C | 0.03792 (0.00626) | 0.00511 | 0.25128 | 0.02739 |
| P | 0.06029 (0.00763) | 0.03221 | 0.25093 | 0.13424 |
D, nucleotide diversity: average number of nucleotide substitutions per site between pairs of sequences. Standard errors are indicated in parentheses. dN, average number of nonsynonymous substitutions per nonsynonymous site. dS, average of synonymous substitutions per synonymous site. dN/dS, average of the ratio between nonsynonymous and synonymous substitutions for each pair of comparisons. dN, dS, and dN/dS were estimated by the PBL method (21, 33).
Phylogenetic relationships between the CTV isolates were inferred using neighbor-joining and maximum-likelihood methods (9). Both gave basically the same relative phylogenetic grouping and showed bootstrap values greater than 80% in the main nodes (Fig. 1). Most CTV isolate major variants showed the same grouping in the phylogenetic trees obtained from the four genomic regions analyzed here. However, some isolate major variants showed sharp differences in their genetic relationships with other CTV sequences in different genomic regions. For example, Spanish isolate T308 was genetically very close to Florida isolate T36 and very different from isolate T385 in the 3′-terminal genomic regions (C and P), but in the 5′-terminal regions (A and F) the genetic relationships between these three isolates were opposite (Fig. 1). Incongruencies in the phylogenetic relationships of different genomic regions were also observed in CTV isolates 65, 386, 519, T398, and T405 (Fig. 1). These phylogenetic incongruencies between different genomic regions for the same CTV isolate major variants suggest that some sequence variants might have originated by recombination events between diverged sequence variants.
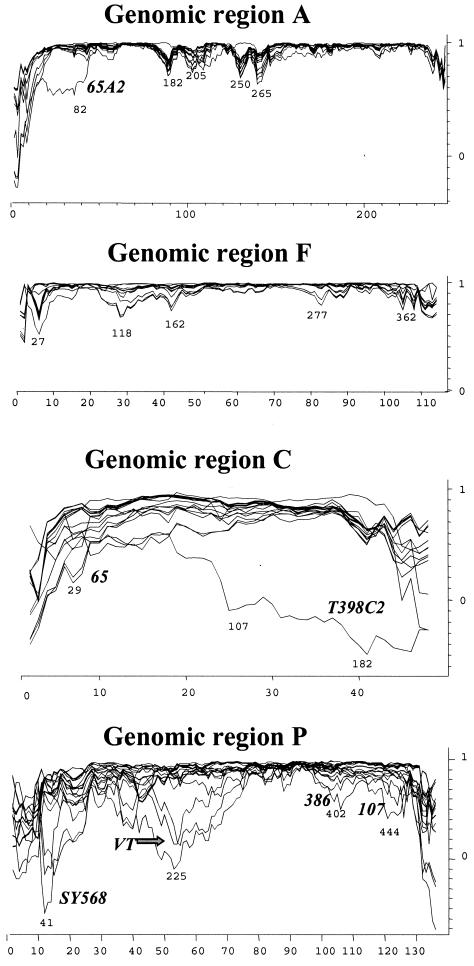
To attempt to identify possible recombination events located within each genomic region, we used the program PHYLPRO (49). PHYLPRO displays graphically the coherence of sequence relationships (phylogenetic correlation) over the entire length of a set of aligned homologous sequences. Recombination signals appear as areas of low phylogenetic correlation, visualized by single sharp-pointed downward peaks in the graph. In Fig. 2, the phylogenetic correlation profiles of all individual CTV sequences corresponding to the four genomic regions are shown. Genomic regions A and F showed several weak recombination signals, suggesting possible recombination events that might be blurred by mutations and other recombination events that have accumulated over time. Genomic regions C and P, however, gave very complex phylogenetic correlation profiles, suggesting the possibility of multiple recombination events in different sequence positions, hindering the identification of specific recombination points between CTV sequences (Fig. 2). Because strong recombination signals might obscure the weaker ones, to improve the detection of sequences with low recombination signals, new analyses excluding sequences with regions of low phylogenetic correlation were performed. Subsequently, the removed sequences were reintroduced individually into the set of sequences. By using this procedure and by modifying the program parameters, we could locate more likely recombination events for some CTV sequences (not shown). For example, in genomic region P, several possible recombination points between an ancestor of SY568, 107, and VT and an ancestor of 519, 65, 386, and 416 (see Fig. 1) were detected.
FIG. 2.
Phylogenetic correlation profiles (graphic representation of the coherence of sequence relationships) of 34 CTV isolates (Table 1) for genomic regions A, F, C, and P (Table 2). Only the variable sites (x axis) are represented in the graph. Phylogenetic correlation (y axis) was obtained at each variable site from pairwise distance analysis of all aligned sequences by using the program PHYLPRO, with a fixed window of 40 bp. Numbers under low phylogenetic correlation areas (possible recombination signals) indicate nucleotide positions. Some CTV sequences are indicated near their individual correlation profiles.
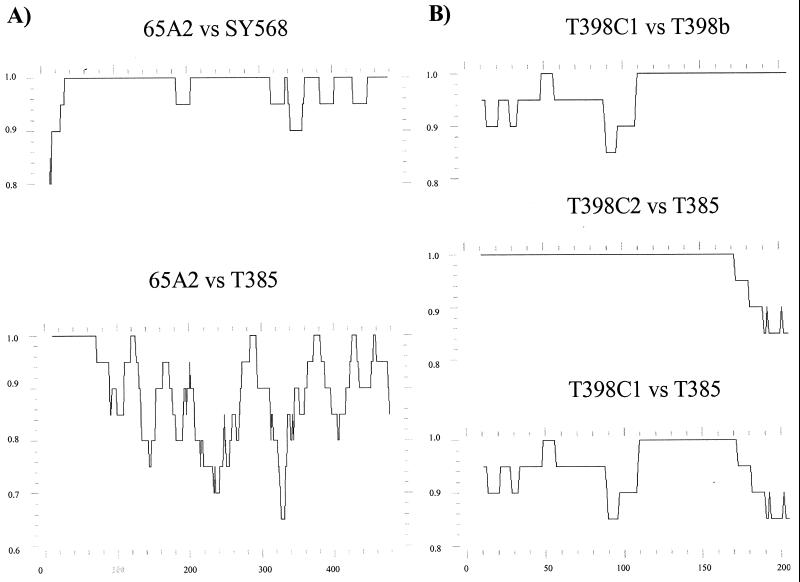
In cases with clear recombination signals, probably reflecting recent recombination events, the display of nucleotide identity between pairs of sequences over their length provided more precise information. For example, genomic region A of the major variant 65A2 seems to have originated by recombination between SY568-like and T385-like isolates in a position between nt 40 and 80 (Fig. 3). Another clear example would be genomic region C of the major variant T398C2. When the identity profiles of T398C2 with respect to T398C1 and T385 were displayed, it was observed that from positions 110 to 170, the three sequences show 100% nucleotide identity; T398a and T398b have 100% nucleotide identity in the segment between positions 110 and 273; and T398C2 and T385 have 100% identity between positions 1 and 170. All this suggests that the T398C2 sequence might have resulted from recombination between T398C1-like and T385-like sequences at some point between positions 110 and 170. It is possible that the complete identity between nucleotides 110 and 170 resulted from a previous recombination of the ancestor of T385 and T398C1 with another sequence variant (Fig. 3). In all cases, the recombination events inferred from the nucleotide identity profiles (Fig. 3) confirmed those obtained from the phylogenetic correlation profiles (Fig. 2). It seems unlikely that the possible recombination sites could have arisen in vitro during RT-PCR, as in our previous work no recombinant variants were observed when dsRNAs from two isolates, each with one major variant, were mixed and amplified by RT-PCR (42).
FIG. 3.
Nucleotide identity profiles between pairs of CTV sequences. (A) Genomic region A, sequence variants 65A2, SY568, and T385; (B) genomic region C, sequence variants T398C1, T398C2, and T385. The profiles were constructed using the PLOTSIMILARITY program, with a window of 20 bp. The y axis corresponds to nucleotide identity and the x axis to nucleotide positions.
We also performed conversion analyses using the program GENECONV (45) on the complete genome nucleotide sequences of isolates T36, VT, T385, and SY568. This program is based on the analysis of whether some regions of a pair of sequences have more consecutive identical silent polymorphic sites in common than would be expected by chance. GENECONV finds and ranks fragments with the highest score (number of matches for pairs of sequences). To avoid the selection of high-scoring fragments by chance, the polymorphic sites are permuted randomly among themselves 10,000 times and scored each time. Statistical significance is evaluated by the parameter P, the proportion of permuted alignments for which the maximal fragment score for that pair of sequences is greater than or equal to the original fragment score. Because mutations could accumulate after the recombination event, we used the program option to allow mismatches (penalty was set for a gscale of 2). Our GENECONV analyses suggested recombination between isolates T385 and SY568, which showed a nucleotide identity of greater than 99% between positions 9304 and 16107 (P = 0.0000), whereas the rest was below 93%, which corroborates the results obtained by Vives et al. (48). Also, we found a possible double recombination between VT and SY568: SY568 and VT showed a nucleotide identity of about 96% in positions 1309 to 2781 (P = 0.0027) and 6546 to 9302 (P = 0.0000), whereas the rest was below 92%.
Recombination in natural populations has been reported for other plant viruses (5, 10). It has been proposed that recombination can be advantageous for RNA viruses. The high mutation rates of RNA viruses cause accumulation of deleterious mutations, limiting the RNA genome size. In large populations, fitness can be maintained or increased by natural selection, but in small populations, genetic drift will lead to progressive loss of replicative fitness. Recombination of viral genomes with deleterious mutations can regenerate functional genomes (6, 39, 46). For CTV, having the largest genome among the known single-stranded plus-sense plant RNA viruses, recombination could act as a compensatory mechanism to offset accumulation of deleterious mutations in bottleneck episodes, such as aphid transmission. Also, coinfection and recombination of different genomic regions between diverged virus genomes allow greater genome diversity and adaptability to new environments (39, 46). Thus, recombination might explain in part the great number of CTV isolates with different biological characteristics described worldwide (38). Recombination among CTV isolates also has important practical implications. For example, for application of disease control measures, such as cross-protection or transgenic plant resistance, caution must be taken to avoid the introduction of exotic CTV sequences that might recombine and give rise to CTV isolates with new biological properties.
Genetic variation with respect to geographic distribution: absence of correlation between genetic and geographic proximity.
According to CTV genomic RNA 5′ untranslated region nucleotide identity, Lopez et al. (22) classified clones from 11 CTV isolates in three groups: I, represented by isolate T36; II, represented by VT; and III, represented by T385. According to our analyses, genomic regions A and F (both located in the 5′ half of the genome; Fig. 1) of all the Spanish and Californian isolates analyzed here can be included in these three groups. The major variants of Spanish isolates T346 and T373 are included in group I, three to five Californian isolates are in group II, and about 80% of the Spanish and California isolates belonged to group III (Fig. 1). For genomic regions C and P, the CTV isolates could not be readily assigned to these three discrete groups. Curiously, group III isolates showed a nucleotide identity of greater than 99% in the four genomic regions (Fig. 1). Group III isolates, with a very high nucleotide identity, has been also found in Florida, Taiwan, and Colombia (1).
For a better comparison of CTV populations, we estimated the nucleotide diversity (average nucleotide distance between two pairs of CTV major sequences chosen randomly) of Spanish and Californian CTV isolates (including T385 and SY568) for each genomic region (Table 4). We found that in each genomic region, the nucleotide diversity of CTV isolates from the same geographic population was remarkably higher than that assessed between the two geographic populations (Table 4). For example, the nucleotide diversity of CTV genomic region P within California and within Spain was about 0.050 for both, whereas the nucleotide diversity considering only nucleotide distances between Spanish and Californian isolates was less than 0.004 (Table 4). Application of the D statistic (24) showed that the Spanish and Californian CTV populations were not significantly genetically different (D = 0.0000), and hence, from a genetic view, the Spanish and Californian CTV isolates can be considered part of the same population. We wanted to know if this was also true for other CTV geographic populations. Unfortunately, not many CTV nucleotide sequences from the same geographic region are available. When genomic region C of six Portuguese isolates (GenBank accession nos. AF184113-AF84118) was included in the analysis, we found that the Portuguese CTV isolates were part of the same genetic population as the Spanish and Californian CTV isolates (Table 4). Analysis of the first 462 nt of the coat protein gene (C2) of 6 Portuguese and 13 Japanese CTV isolates (17) also showed higher diversity within a geographic population than between the two geographic populations (Table 4). Low genetic variation among geographically distant isolates has also been observed for Curcubit yellow stunting disorder virus and Beet pseudo-yellows virus in the genus Crinivirus, the other genus of the family Closteroviridae (41, 43), and for Tobacco mild green mosaic virus and Pepper mild mottle virus in the genus Tobamovirus (11, 37).
TABLE 4.
Nucleotide diversity of geographically different CTV populationsa
| Group | Region | Nucleotide diversity
|
Vb
|
Vw | Nst | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sp | Cal | Sp-Cal | ||||||||
| 1 | A | 0.17627 (0.08425) | 0.05809 (0.01306) | 0.00819 (0.01924) | 0.11718 (0.05909) | 0.06496 | ||||
| F | 0.06825 (0.02866) | 0.03328 (0.01094) | 0.00182 (0.00764) | 0.04846 (0.01979) | 0.03620 | |||||
| C | 0.03518 (0.00749) | 0.03264 (0.01025) | 0.00059 (0.00380) | 0.03391 (0.00582) | 0.01710 | |||||
| P | 0.05390 (0.01083) | 0.05349 (0.01174) | 0.00351 (0.00537) | 0.05369 (0.00784) | 0.06136 | |||||
| 2 | Sp | Cal | Por | Sp-Cal | Cal-Por | Sp-Por | Sp-Cal-Por | Vw | Nst | |
| C | 0.03518 | 0.03264 | 0.06946 | 0.00059 | 0.00432 | 0.00209 | 0.00233 | 0.04576 | 0.04845 | |
| (0.00749) | (0.01025) | (0.01535) | (0.00380) | (0.01103) | (0.00965) | (0.00722) | (0.01187) | |||
| 3 | Ja | Por | Ja-Por | VW | Nst | |||||
| C2 | 0.08262 (0.00652) | 0.07798 (0.01236) | 0.00544 (0.00798) | 0.0803 (0.00608) | 0.06344 | |||||
Genomic regions A, F, C (nt 424 to 647 of the coat protein gene), P (Table 1 and Fig. 1), and C2 (first 462 nt of coat protein gene) were analyzed. The first group shows diversities of regions A, F, C, and P using the sequences determined by us here. The second includes comparisons only of region C, using our sequences and additional nucleotide sequences obtained from GenBank (see Materials and Methods). The last shows nucleotide diversities of region C2 estimated from sequences obtained from GenBank. The CTV isolates analyzed were from Spain (Sp), California (Cal), Portugal (Por), and Japan (Ja). Vb, average nucleotide diversity between geographic populations (24); Vw, average nucleotide diversity within geographic populations (24); Nst = Vb/(Vw + Vb), proportion of the diversity between geographic populations with respect to total nucleotide diversity (24). Nucleotide diversity is the average number of nucleotide substitutions per site in each pair of sequence variants. Standard errors are indicated in parentheses.
The genetic structure observed here suggests migration of CTV isolates among geographically isolated CTV populations. It is well known that an intense traffic of CTV-infected propagative citrus material has occurred between distant regions in the world (38). Within each geographic region, local dispersion is then effected by aphids.
ACKNOWLEDGMENTS
Part of this work was supported by grants from the USDA special grants Citrus Tristeza Virus Research Program, the California Tristeza Research Coalition, and the University of California to B.W.F. Part of this work was supported by projects SC93–111 and SC97–098 (INIA). L.R. was supported in part by a postdoctoral fellowship from Ministerio de Educación y Ciencia, Spain.
We acknowledge the technical assistance of T. Olupona, M. Boil, E. Estela, and M. Martínez. We thank F. Garcia-Arenal for excellent critical review of the manuscript.
REFERENCES
- 1.Albiach-Martí M R, Mawassi M, Gowda S, Satyanarayana T, Hilf M E, Shanker S, Almira E C, Vives M C, López C, Guerri J, Flores R, Moreno P, Garnsey S M, Dawson W O. Sequences of citrus tristeza virus separated in time and space are essentially identical. J Virol. 2000;74:6856–6865. doi: 10.1128/jvi.74.15.6856-6865.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayllón M A, López C, Navas-Castillo J, Mawassi M, Dawson W O, Guerri J, Flores R, Moreno P. New defective RNAs from citrus tristeza virus: evidence for a replicase-driven template switching mechanism in their generation. J Gen Virol. 1999a;80:817–821. doi: 10.1099/0022-1317-80-3-817. [DOI] [PubMed] [Google Scholar]
- 3.Ayllon M A, Rubio L, Moya A, Guerri J, Moreno P. The haplotype distribution of two genes of citrus tristeza virus is altered after host change or aphid transmission. Virology. 1999b;255:32–39. doi: 10.1006/viro.1998.9566. [DOI] [PubMed] [Google Scholar]
- 4.Ballester-Olmos J F, Pina J A, Carbonell E A, Moreno P, Hermoso de Mendoza A, Cambra M, Navarro L. Biological diversity of citrus tristeza virus (CTV) isolates in Spain. Plant Pathol. 1988;42:219–229. [Google Scholar]
- 5.Bousalem M, Dizery E J P, Fargette D. High genetic diversity, distant phylogenetic relationships and intraspecies recombination events among natural populations of yam mosaic virus: a contribution to understanding potyvirus evolution. J Gen Virol. 2000;81:243–255. doi: 10.1099/0022-1317-81-1-243. [DOI] [PubMed] [Google Scholar]
- 6.Chao L. Fitness of RNA viruses decreased by Muller's ratchet. Nature. 1990;348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 7.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Febres V J, Ashoulin L, Mawasi M, Frank A, Bar-Joseph M, Manjunath K L, Lee R F, Niblett C L. The p27 protein is present at one end of citrus tristeza particles. Phytopathology. 1996;86:1331–1335. [Google Scholar]
- 9.Felsenstein J. PHYLIP—Phylogenetic Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 10.Fraile A, Alonso-Prados J L, Aranda M A, Bernal J J, Malpica J M, Garcia-Arenal F. Reports about recombination in virus natural populations. J Virol. 1997;71:934–940. doi: 10.1128/jvi.71.2.934-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraile A, Malpica J M, Aranda M A, Rodríguez-Cerezo E, García-Arenal F. Genetic diversity in tobacco mild green mosaic tobamovirus infecting the wild plant Nicotiana glauca. Virology. 1996;223:148–155. doi: 10.1006/viro.1996.0463. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Arenal F, Fraile A, Malpica J M. Genetic variability and evolution. In: Mandahar C L, editor. Molecular biology of plant viruses. Boston, Mass: Kluwer Academic Publishers; 1999. pp. 143–159. [Google Scholar]
- 13.Gowda S, Satyanarayana T, Davis C L, Navas-Castillo J, Albiach-Martí M R, Mawassi M, Valkov N, Bar-Joseph M, Moreno P, Dawson W O. The p20 gene product of Citrus tristeza closterovirus accumulates in the amorphous inclusion bodies. Virology. 2000;274:246–254. doi: 10.1006/viro.2000.0413. [DOI] [PubMed] [Google Scholar]
- 14.Guerri J, Moreno P, Muñoz N, Martinez E. Variability among Spanish citrus tristeza virus isolates revealed by double-stranded RNA analysis. Plant Pathol. 1991;40:38–44. [Google Scholar]
- 15.Hilf M E, Karasev A V, Pappu H R, Gumpf D J, Niblett C L, Garnsey S M. Characterization of citrus tristeza virus subgenomic RNAs in infected tissue. Virology. 1995;208:576–582. doi: 10.1006/viro.1995.1188. [DOI] [PubMed] [Google Scholar]
- 16.Holland J J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1582. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 17.Kano T, Hiyama T, Natsuaki T, Imanishi N, Okuda S, Ieki H. Comparative sequence analysis of biologically distinct isolates of citrus tristeza virus in Japan. Ann Phytopathol Soc Jpn. 1998;64:270–275. [Google Scholar]
- 18.Karasev A V, Boyko V P, Gowda S, Nikolaeva O V, Hilf M E, Koonin E V, Nibblet C L, Cline K, Gumpf D J, Lee R F, Garnsey S M, Dawson W O. Complete sequence of the citrus tristeza virus RNA genome. Virology. 1995;208:511–520. doi: 10.1006/viro.1995.1182. [DOI] [PubMed] [Google Scholar]
- 19.Kong P, Rubio L, Polek M, Falk B W. Population structure and genetic diversity of California citrus tristeza virus (CTV) field isolates. Virus Genes. 2000;21:139–145. doi: 10.1023/a:1008198311398. [DOI] [PubMed] [Google Scholar]
- 20.Krawczak M, Reiss J, Schmidtke J, Rösler U. Polymerase chain reaction: replication errors and reliability of gene diagnosis. Nucleic Acids Res. 1989;17:2197–2201. doi: 10.1093/nar/17.6.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W-H. Unbiased estimation of the rates of synonymous and nonsynonymous substitutions. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 22.López C, Ayllón M A, Navas J, Guerri J, Moreno P, Flores R. Molecular variability of the 5′ and 3′ terminal regions of the citrus tristeza virus RNA. Phytopathology. 1998;88:685–691. doi: 10.1094/PHYTO.1998.88.7.685. [DOI] [PubMed] [Google Scholar]
- 23.López C, Navas-Castillo J, Gowda S, Moreno P, Flores R. The 23-kDa protein coded by the 3′-terminal gene of citrus tristeza virus is an RNA-binding protein. Virology. 2000;269:462–470. doi: 10.1006/viro.2000.0235. [DOI] [PubMed] [Google Scholar]
- 24.Lynch M, Crease T J. The analysis of population survey data on DNA sequence variation. Mol Biol Evol. 1990;7:377–394. doi: 10.1093/oxfordjournals.molbev.a040607. [DOI] [PubMed] [Google Scholar]
- 25.Mawassi M, Gafny R, Bar-Joseph M. Nucleotide sequence of the coat protein gene of citrus tristeza virus: comparison of biologically diverse isolates collected in Israel. Virus Genes. 1993;7:265–275. doi: 10.1007/BF01702587. [DOI] [PubMed] [Google Scholar]
- 26.Mawassi M, Karasev A V, Mietkiewska E, Gafny R, Lee R F, Dawson W O, Bar-Joseph M. Defective RNA molecules associated with citrus tristeza virus. Virology. 1995;208:383–387. doi: 10.1006/viro.1995.1165. [DOI] [PubMed] [Google Scholar]
- 27.Mawassi M, Mietkiewska E, Gofman R, Yang G, Bar-Joseph M. Unusual sequence relationship between two isolates of citrus tristeza virus. J Gen Virol. 1996;77:2359–2364. doi: 10.1099/0022-1317-77-9-2359. [DOI] [PubMed] [Google Scholar]
- 28.Mawassi M, Mietkiewska E, Hilf M E, Ashoulin L, Karasev A V, Gafny R, Lee R F, Garnsey S M, Dawson W O, Bar-Joseph M. Multiple species of defective RNAs in plants infected with citrus tristeza virus. Virology. 1995;214:264–268. doi: 10.1006/viro.1995.9930. [DOI] [PubMed] [Google Scholar]
- 29.Moreno P, Guerri J. Variability of citrus tristeza closterovirus (CTV): methods to differentiate isolates. In: Monette P, editor. Filamentous viruses of woody plants. Trivandrum, India: Research Signpost; 1997. pp. 97–107. [Google Scholar]
- 30.Moreno P, Guerri J, Muñoz N. Identification of Spanish strains of citrus tristeza virus (CTV) by analysis of double-stranded RNAs (dsRNA) Phytopathology. 1990;80:477–482. [Google Scholar]
- 31.Moya A, García-Arenal F. Populations genetics of viruses: an introduction. In: Gibbs A, Calisher C H, García-Arenal F, editors. Molecular basis of virus evolution. Cambridge, England: Cambridge University Press; 1995. pp. 213–223. [Google Scholar]
- 32.Nei M. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. [Google Scholar]
- 33.Pamilo P, Bianchi N O. Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol. 1993;10:271–281. doi: 10.1093/oxfordjournals.molbev.a040003. [DOI] [PubMed] [Google Scholar]
- 34.Pappu H R, Karasev A V, Anderson E J, Pappu S S, Hilf M E, Febres V J, Eckloff R M G, McCaffery M, Boyko V, Gowda S, Dolja V V, Koonin E V, Gumpf D J, Cline K C, Garnsey S M, Dawson W O, Lee R F, Niblett C L. Nucleotide sequence and organization of eight open reading frames of the citrus tristeza closterovirus genome. Virology. 1994;199:35–46. doi: 10.1006/viro.1994.1095. [DOI] [PubMed] [Google Scholar]
- 35.Pappu H R, Pappu S, Niblett C L, Lee R F, Civerolo E L. Comparative sequence analysis of the coat protein of biologically distinct citrus tristeza closterovirus isolates. Virus Genes. 1993;7:255–264. doi: 10.1007/BF01702586. [DOI] [PubMed] [Google Scholar]
- 36.Pappu S S, Febres V J, Pappu H R, Lee R F, Niblett C L. Characterization of the 3′ proximal gene of the citrus tristeza closterovirus genome. Virus Res. 1997;47:51–57. doi: 10.1016/s0168-1702(96)01405-0. [DOI] [PubMed] [Google Scholar]
- 37.Rodríguez-Cerezo E, Moya A, García-Arenal F. Variability and evolution of the plant RNA virus pepper mild mottle virus. J Virol. 1989;63:2198–2203. doi: 10.1128/jvi.63.5.2198-2203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roistacher R N, Moreno P. The worldwide threat from destructive isolates of citrus tristeza virus—a review. In: Brlansky R H, Lee R F, Timmer L W, editors. Proceedings of the 11th Conference of the International Organization of Citrus Virologists. Riverside, Calif: Department of Plant Pathology, University of California; 1991. pp. 7–19. [Google Scholar]
- 39.Roossinck M J. Mechanisms of plant virus evolution. Annu Rev Phytopathol. 1997;35:1953–1965. doi: 10.1146/annurev.phyto.35.1.191. [DOI] [PubMed] [Google Scholar]
- 40.Rubio L, Ayllón M A, Guerri J, Pappu H, Niblett C, Moreno P. Differentiation of citrus tristeza closterovirus (CTV) isolates by single-strand conformation polymorphism analysis of the coat protein gene. Ann Appl Biol. 1996;129:479–489. [Google Scholar]
- 41.Rubio L, Soong J, Kao J, Falk B W. Geographic and molecular variation of isolates of three whitefly-borne closteroviruses: lettuce infectious yellows virus, cucurbit yellow stunting disorder virus and beet pseudo-yellows virus. Phytopathology. 1999;89:707–711. doi: 10.1094/PHYTO.1999.89.8.707. [DOI] [PubMed] [Google Scholar]
- 42.Rubio L, Guerri J, Moreno P. Characterization of Citrus tristeza virus isolates by single-strand conformation polymorphism analysis of DNA complementary to their RNA population. In: Yokomi R H, da Graca J V, Lee R F, editors. Proceedings of the 14th Conference of the International Organization of Citrus Virologists. Riverside, Calif: Department of Plant Pathology, University of California; 2000. pp. 12–17. [Google Scholar]
- 43.Rubio L, Abou-Jawdah Y, Lin H, Falk B W. Geographically distant isolates of the Crinivirus cucurbit yellow stunting disorder virus (CYSDV) show very low genetic diversity in the coat protein gene. J Gen Virol. 2001;82:929–933. doi: 10.1099/0022-1317-82-4-929. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- 46.Simon A E, Bujarski J J. RNA-RNA recombination and evolution in virus-infected plants. Annu Rev Phytopathol. 1994;32:337–362. [Google Scholar]
- 47.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vives M C, Rubio L, Lopez C, Navas-Castillo J, Albiach-Martí M R, Dawson W O, Guerri J, Flores R, Moreno P. The complete genome sequence of the major component of a mild citrus tristeza isolate. J Gen Virol. 1999;80:811–816. doi: 10.1099/0022-1317-80-3-811. [DOI] [PubMed] [Google Scholar]
- 49.Weiller G F. Phylogenetic profiles: a graphical method for detecting genetic recombinations in homologous sequences. Mol Biol Evol. 1998;15:326–335. doi: 10.1093/oxfordjournals.molbev.a025929. [DOI] [PubMed] [Google Scholar]
- 50.Yang G, Mawassi M, Gofman R, Gafny R, Bar-Joseph M. Involvement of a subgenomic mRNA in the generation of a variable population of defective citrus tristeza virus molecules. J Virol. 1997;71:9800–9802. doi: 10.1128/jvi.71.12.9800-9802.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z-N, Mathews D H, Dodds J A, Mirkov T E. Molecular characterization of an isolate of citrus tristeza virus that causes severe symptoms in sweet orange. Virus Genes. 1999;19:131–142. doi: 10.1023/a:1008127224147. [DOI] [PubMed] [Google Scholar]