Abstract
Progress in developing a small animal model of human immunodeficiency virus type 1 (HIV-1) disease would greatly facilitate studies of transmission, pathogenesis, host immune responses, and antiviral strategies. In this study, we have explored the potential of rats as a susceptible host. In a single replication cycle, rat cell lines Rat2 and Nb2 produced infectious virus at levels 10- to 60-fold lower than those produced by human cells. Rat-derived cells supported substantial levels of early HIV-1 gene expression, which was further enhanced by overexpression of human cyclin T1. Rat cells displayed quantitative, qualitative, and cell-type-specific limitations in the late phase of the HIV-1 replication cycle including relative expression levels of HIV-1 Gag proteins, intracellular Gag processing, and viral egress. Nb2 cells were rendered permissive to HIV-1 R5 viruses by coexpression of human CD4 and CCR5, indicating that the major restriction on HIV-1 replication was at the level of cellular entry. We also found that primary rat lymphocytes, macrophages, and microglia expressed considerable levels of early HIV-1 gene products following infection with pseudotyped HIV-1. Importantly, primary rat macrophages and microglia, but not lymphocytes, also expressed substantial levels of HIV-1 p24 CA and produced infectious virions. Collectively, these results identify the rat as a promising candidate for a transgenic small animal model of HIV-1 infection and highlight pertinent cell-type-specific restrictions that are features of this species.
It has long been recognized that human immunodeficiency virus type 1 (HIV-1) replicates efficiently in human cells but is subject to potent species-specific restrictions in cells from many nonprimate species (25, 33, 34, 44). Several advances have been made recently in elucidating the molecular bases of such blocks to HIV-1 replication in nonprimate cells. These discoveries have recharged efforts to develop small animal models of infection for use in the study of pathogenesis, the screening of drugs, and the testing of vaccines.
A first important advance was made with the understanding of the requirements for human chemokine receptors as cofactors with human CD4 (hCD4) for cell surface fusion and entry of HIV-1 (reviewed in references 28 and 43). The discovery of the primary roles played by human CCR5 (hCCR5) and human CXCR4 (hCXCR4) renewed hopes for the development of a transgenic mouse model. However, while coexpression of hCD4 and a human chemokine receptor largely overcame the entry block in mouse NIH 3T3 cells (5, 20, 40) as well as in primary T cells from mice transgenic for either hCD4 and hCCR5 (11) or hCD4 and hCXCR4 (49), these mouse cells exhibited little or no productive infection.
A second advance has been in understanding postentry blocks to HIV-1 replication in some rodent cell lines. At least one restriction in cell lines from mice and hamsters is the inability of the viral transcription regulator, Tat, to activate transcription from the long terminal repeat (LTR) of HIV-1, which is normally a crucial step for vigorous viral replication (24). Recently, a novel Tat-interacting protein, human cyclin T1 (CycT1), was discovered (58), which is a component of the pTEFb transcription factor complex (38, 60). Human CycT1, in association with the cyclin-dependent kinase CDK9, interacts with Tat to form a heterodimer with very high affinity for binding the TAR stem-loop near the 5′ end of nascent HIV-1 mRNA transcripts. This complex mediates hyperphosphorylation of RNA polymerase II, thereby increasing its processivity (20). Mouse CycT1 differs from its human homologue at several amino acids, and the change from tyrosine to cysteine at position 261 prevents it from interacting with Tat (6, 20, 31). Expression of human CycT1 in NIH 3T3 cells reverses the block to Tat function and allows the HIV-1 replication cycle to proceed through entry, reverse transcription, integration, and proviral gene expression in cells coexpressing hCD4 and an appropriate coreceptor but is not sufficient to reconstitute the full HIV-1 replication cycle (5, 20, 40). There has been some controversy over the existence of an additional block in mouse and hamster cells that limits the function of HIV-1 Rev (37, 55), and more recent studies suggest a relative, rather than an absolute, limitation in the function of this regulatory protein in rodent cells (5, 39). Moreover, a viral assembly block was reported for NIH 3T3 cells (40) that could be partially complemented by human-mouse heterokaryon fusions (5, 39). Also, conflicting data have been published regarding the infectivity of virions released from murine cell lines (5, 20, 39–41).
With regard to the rat species, the rat fibroblast cell line Rat2 and neuroblastoma cell line B50 have been demonstrated to support robust levels of HIV-1 LTR activity (5, 40, 42), and persistently infected Rat2 cells that secrete considerable levels of p24 CA were reported elsewhere (42). Furthermore, Rattus norvegicus offers several advantages for the development of an animal model, including small size, convenience in breeding, and well-characterized immune and central nervous systems. In addition, rat transgenesis can be performed with relative ease, thereby enabling the selective expression of human genes that may be essential for full realization of the HIV-1 replication cycle.
Therefore, in the present work we sought to extend these provocative cell culture data as a further exploration of the rat as a potential small animal model of HIV disease. These experiments used both cell lines and primary rat-derived cell cultures to define in greater detail the extent to which the HIV-1 replication cycle is intact in this species context. The results reveal the integrity of the early phase of the replication cycle but also expose cell-type-specific limitations in the late phase. These features are likely to dictate patterns of productive infection, and probably pathogenesis, in a transgenic model based on rats.
MATERIALS AND METHODS
Plasmids.
pCMV4hygro is a version of the mammalian expression vector pCMV4 (2), which has the neomycin resistance gene replaced by a hygromycin resistance gene. pCD4neo, which expresses hCD4 and a neomycin resistance gene, has been described previously (23). pCMVFCCR5 contains an epitope-tagged version of hCCR5 and has been previously described (3). pCCR5hygro was developed by excising the epitope-tagged version of hCCR5 as a HindIII-XbaI fragment from pCMVFCCR5 and inserting it into these sites of pCMVhygro. Human CycT1 was expressed under the control of the cytomegalovirus (CMV) promoter by transfection of a plasmid (pCICycT) kindly provided by Katherine Jones. pVSV-G, the mammalian expression vector for vesicular stomatitis virus G (VSV-G) protein (17), was kindly provided by Jane Burns. The molecular clone pNL-4-3 Luc E−R− (15), an NL4-3 provirus (along with mutations in env, nef, and vpr) carrying a luciferase gene within the nef locus driven by the 5′ LTR, was a gift of Nathaniel Landau via the AIDS Research and Reference Reagent Program. A three-plasmid lentivirus expression system was used to generate HIV-derived retrovirus vector particles that express firefly luciferase under the control of the CMV intermediate-early promoter, including the packaging construct pCMVΔR8.91 (62), the transfer vector construct pHR′CMV-Luc-SIN-W (16, 61) (kindly provided by Didier Trono), and pVSV-G.
Cell lines and transfectants.
Rat2 cells (rat fibroblast-like cell line [ATCC CRL-1764]) and C58 cells (rat T-cell lymphoma [ATCC TIB-236]) were obtained from the American Type Culture Collection, Manassas, Va., and cultivated as recommended. Nb2 cells (rat T-cell lymphoma) (18) were a kind gift from Jay Ryan. Nb2 hCD4 cells were generated by electroporation of Nb2 cells with pCD4neo and neomycin selection (Life Technologies). Nb2 hCD4/hCCR5 cells were generated by electroporation of Nb2 hCD4 cells with pCCR5hygro and selection in medium containing both neomycin and hygromycin B (Roche Diagnostics). Pretreatment of Nb2 hCD4/hCCR5 cells with sodium butyrate (Sigma) (1 mM, 16 h) was required to induce expression of hCCR5 at detectable levels by flow cytometry. SUP-T1 hCCR5 cells were generated by transfection of SUP-T1 cells with pCCR5hygro and hygromycin B selection. Jurkat hCCR5 cells were a kind gift from Paula Pitha. The rat T-cell line SD1 was established by human T-cell leukemia virus type 1 transformation of primary hCD4-transgenic T lymphocytes from outbred Sprague-Dawley (SD) rats following an established protocol (1).
Primary cells.
Rat strains were obtained from Charles River Laboratories (inbred strains Fischer F344 [Fischer], Brown-Norway [BN], Noble, Copenhagen, Lewis, and WKY) or from Taconics (SD). Primary rat lymphocytes and macrophages were prepared from spleens, which had been removed aseptically from euthanatized rats. Single-cell suspensions were prepared by pushing tissue pieces through a nylon mesh screen (Falcon cell strainer; 70-μm-pore-size nylon; Becton Dickinson). The cell suspension was washed once with phosphate-buffered saline (PBS), and erythrocytes were then lysed by incubation in ACK lysis buffer (BioWhittaker) for 5 min, followed by another PBS wash.
For lymphocyte cultures, splenocytes were cultivated at 2 × 106/ml in RPMI 1640 containing 15% fetal bovine serum (FBS) (Gemini BioProducts, catalog no. 900-108, lot no. A25305R), 1% penicillin-streptomycin, 2 mM l-glutamine, 5 × 10−5 M 2-mercaptoethanol (GIBCO-BRL), nonessential amino acids (Cellgro), 1 mM sodium pyruvate, and minimum essential medium vitamin solution (GIBCO-BRL). To activate rat lymphocytes, concanavalin A (Sigma) was added at a final concentration of 1 μg/ml. After overnight incubation, cells were washed twice with PBS and then resuspended in fresh medium supplemented with 20 nM human recombinant interleukin 2 (IL-2) (a gift of Chiron Corporation).
For macrophage cultures, rat splenocytes were plated at 7 × 106/ml in 24-well plastic tissue culture dishes in the above medium with the addition of 5% complement-inactivated rat serum. Five days later, nonadherent cells were vigorously washed off with PBS, and the adherent macrophage-enriched population was cultivated in fresh medium for another 2 to 5 days before infection. At infection, the purity of rat macrophage cultures was approximately 80 to 90%.
Human lymphocytes from random, multiple donors were prepared and activated with phytohemagglutinin P (PHA-P) (Sigma) as described previously (51). Human macrophage cultures were derived from human CD14 magnetic bead-selected monocytes according to the manufacturer's protocol (Miltenyi Biotec) from Ficoll gradient-purified human peripheral blood mononuclear cells (PBMC) from a single donor; cultivated in Dulbecco's modified Eagle medium containing 20% FBS, 10% human AB serum (GemCell), 1% penicillin-streptomycin, and 2 mM l-glutamine; and plated at 106/ml in 48-well plastic tissue culture dishes. The purity of human macrophage cultures routinely exceeded 98%.
Primary glial cell cultures were prepared as follows. One- to three-day-old SD rat pups were euthanatized using CO2 and decapitated using scissors. Brains were gently removed from the skull and placed in a PBS-filled petri dish on ice. Meninges, blood vessels, and brain stem were removed. Brains were then minced with scissors and dissociated by trituration. The brain homogenate was filtered through a 70-μm-pore-size cell strainer and then washed three times with PBS. The brain homogenate was then resuspended in microglial growth medium (Dulbecco's modified Eagle medium supplemented with 10% FBS [Gemini BioProducts, catalog no. 100-107, lot no. A1923N; lipopolysaccharide concentration, <10.0 EU/ml], 2 mM l-glutamine, 100 μg of gentamicin sulfate/ml, 100 μg of ampicillin/ml, and 15% colony-stimulating factor 1-conditioned medium. Colony-stimulating factor 1-conditioned medium was obtained as the sterile-filtered supernatant of the LAD-MAC mouse producer cell line (ATCC CRL-2420). These mixed glial cultures were maintained in humidified 37°C, 5% CO2 incubators. Culture medium was replaced after 48 h. After 1 to 2 weeks, numerous astrocytes and microglia as well as rare neurons and oligodendrocytes were present in cultures, based on morphology. Microglia were subcultured 1 to 2 weeks after plating the mixed glial cultures based on adhesion (7). Briefly, microglia were detached from mixed glial cultures by shaking. Supernatants were collected and centrifuged at 600 × g for 5 min. Microglia-enriched pellets were then resuspended in microglia growth medium and plated at a density of 5,000 cells/well in 96-well flat-bottomed tissue culture plates.
Viruses and infections.
NL4-3 or 49.5/(VSV-G) replication-competent pseudotypes were produced by calcium phosphate transfection of 293T cells with an equal amount of proviral DNA plasmid and pVSV-G. Culture supernatants were harvested 48 h posttransfection, filtered, and frozen in aliquots at −80°C. Virus titers were determined on GHOST.R5 and GHOST.X4 cells (12), respectively, and a multiplicity of infection (MOI) of 1 was used for subsequent infections. Molecular clones of pYU-2, pJR-CSF, pNL4-3, and pLAI.2 were obtained through the AIDS Research and Reference Reagent Program and were prepared as described previously (52). p49.5 (14) was a gift from Bruce Chesebro. Infections were carried out at the indicated MOIs (YU-2 and JR-CSF) or at a p24 CA inoculum of 100 ng/ml (LAI.2). For experiments shown in Fig. 4, T-cell lines were pretreated with sodium butyrate at 1 mM for 16 h. The preparation of NL4-3 Luc E−R− pseudotype viruses with autologous or heterologous envelopes has been described previously (13).
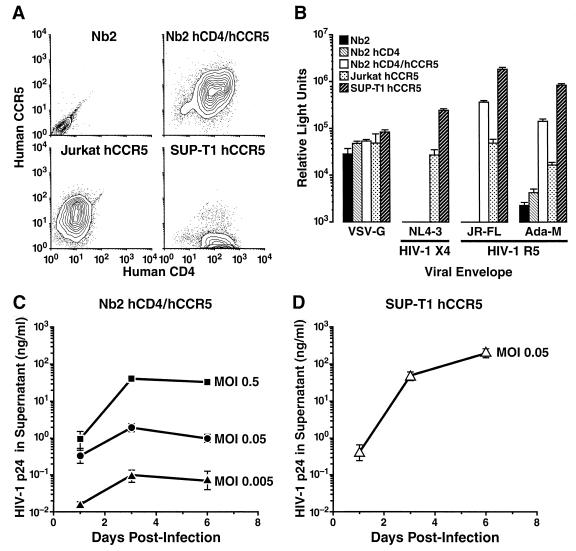
FIG. 4.
The rat T-cell line Nb2 can be rendered permissive to HIV-1 R5 viruses by coexpression of hCD4 and hCCR5. (A) Flow cytometry analysis of human and rat T-cell lines for expression of hCD4 and hCCR5. Data are shown as contour plots, with outlier dots representing less than 5% of the live cell population. (B) Single-round infections of HIV-1 pseudotypes carrying different autologous or heterologous envelopes. Cells were infected with NL4-3 luciferase reporter viruses pseudotyped with HIV-1 R5 (JR-FL and Ada-M), HIV-1 X4 (NL4-3), and VSV-G envelopes. Luciferase activity in cell lysates was quantified as relative light units 4 days postinfection. (C and D) Rat Nb2 hCD4/hCCR5 and human SUP-T1 hCCR5 cells were infected overnight with the indicated MOIs of YU-2. The p24 CA concentration was determined at days 1 (postwash), 3, and 6. All values given are arithmetic means ± standard deviations of triplicates.
Antibodies and flow cytometry.
Fluorescence-activated cell sorting analyses were performed as previously described (3), using phycoerythrin (PE)-conjugated anti-hCD4 (monoclonal antibody [MAb] Leu-3a); fluorescein isothiocyanate (FITC)-conjugated anti-hCCR5 (MAb 2D7); PE-conjugated anti-human HLA-A, -B, and -C (MAb G46-2.6); FITC-conjugated anti-human CD3 (MAb Leu-4); PE-conjugated anti-rat major histocompatibility complex class I (MAb OX-18); FITC-conjugated anti-rat CD3 (MAb G4.18); PE-conjugated anti-rat macrophage subset (ED2-like antigen; MAb HIS36); FITC-conjugated anti-rat CD11b (MAb WT.5); or PE-conjugated anti-rat CD11b/c (MAb OX-42). All antibodies were from BD Pharmingen.
Viral infectivity assay.
The infectious titer (50% tissue culture infective dose [TCID50]) of HIV-1 in cell culture supernatants was determined by endpoint limiting dilution on PHA-P-activated human PBMC (pool of human PBMC from three or four HIV-1-negative donors) 5 days after inoculation. The p24 CA concentration in supernatants was quantified by the NEN/DuPont p24 enzyme-linked immunosorbent assay (NEN Life Sciences), which selectively measures p24 CA but not unprocessed p55 Gag (5). In some analyses, supernatant from a hybridoma producing the neutralizing anti-VSV-G MAb I1 (8) (a kind gift from Leo Lefrancois) was added at a final volume of 1:100 [a 1:1,000 dilution was still neutralizing for the HIV-1/(VSV-G) input stock].
cDNA sequencing of rat CycT1.
Total RNA was prepared from rat Nb2 using the Qiagen RNeasy kit. First-strand cDNA for PCR was carried out with avian myeloblastosis virus reverse transcriptase reagents from Roche Diagnostics (catalog no. 1483-188) according to the manufacturer's instructions, using oligo(dT) as a primer. The 5′ half of rat CycT1 was amplified using primer RATTAT 1 (AAGCTTGCCGCCATGGAGGGAGAGAGG), containing the start codon, and RATTAT 3 (CAAGTGCAGTGCCTTCCCTGC). The 3′ half was amplified using RATTAT 4 (GCAGGGAAGGCACTGCACTTG), the inverse complement of RATTAT 3, and RATTAT 2 (GGATCCTYACTTAGGAAGRGGTGG), with the stop codon. These primers were chosen based upon sequence similarities between the mouse and human sequences, and RATTAT 2 contains two degeneracies. The two halves were amplified using AmpliTaq and standard conditions. Amplified DNA was TA cloned into pCR2.1, and a TA clone of each half was obtained and sequenced. This preliminary sequence of 94% of the entire rat CycT1 clearly identified the amplified sequence as CycT1 and no other known cyclin. The identity of residue 261 was confirmed by preparing another set of RNA samples from Nb2, Rat2, and C58 cells and amplifying the region around residue 261 using another set of primers.
Western blot analysis.
The cell pellets were lysed in buffer A (50 mM HEPES, 135 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, and 1× protease inhibitor cocktail [Calbiochem]), pH 7.2, for 1 h at 4°C. The cell lysate was collected after centrifugation at 13,200 × g for 20 min at 4°C. The lysate was analyzed for protein concentration using the bicinchoninic acid protein assay (Pierce), and 10 μg of cellular protein was run on sodium dodecyl sulfate–12% polyacrylamide gels (Bio-Rad) and transferred to nitrocellulose (Osmonics). The blots were sequentially probed with primary antibodies to HIV-1 Nef (1:1,000; GF-7 [9]), gp120 (1:500; no. 387; from the AIDS Research and Reference Reagent Program), and α-tubulin (3 μg/ml; Oncogene Sciences) or an anti-p24 CA ascites (1:2,000; a generous gift from Beckman Coulter). Following secondary antibody treatment, the blots were exposed to autoradiographic film using the ECL system (Amersham).
RESULTS
Early and late HIV-1 gene expression in cell lines from humans, rats, and mice.
As a first assessment of the postentry susceptibility of rat cells to HIV-1 replication, various rat, human, and mouse cell lines were infected with pseudotyped HIV-1 virions carrying VSV-G, which mediates entry into a broad range of mammalian cells. To determine the relative expression of an early, Rev-independent HIV-1 gene product, we used HIV-1 pseudotype virus based on the HIV-1 NL4-3 proviral backbone containing the firefly luciferase gene within the nef gene locus (NL4-3 Luc E−R−) (15), the expression of which provides a quantitative marker of successful entry, reverse transcription, integration, and early viral gene expression in a given target cell. To quantify the relative expression and egress of a Rev-dependent, fully processed structural HIV-1 gene product, cells were similarly challenged with a VSV-G-pseudotyped HIV-1 molecular clone based on NL4-3 (49.5) and the p24 CA antigen concentration was measured in culture supernatants. To assess quantitative differences in VSV-G-mediated entry among different cell types, parallel infections were performed with VSV-G pseudotypes containing a firefly luciferase gene under the control of the CMV promoter (16, 61, 62).
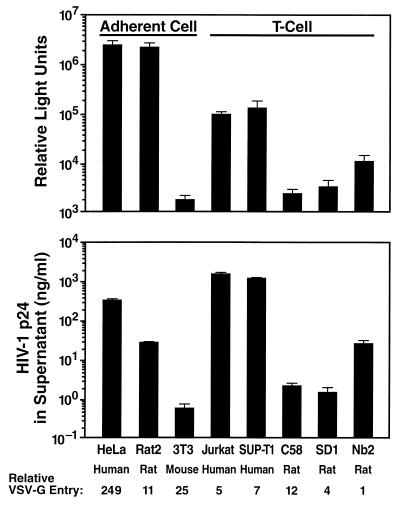
The species origin of all cell lines used in this study was confirmed by flow cytometry staining with major histocompatibility complex class I-specific antibodies (data not shown). Among adherent cell lines, HeLa and Rat2 cells exhibited very robust signals for both early and late HIV-1 gene expression following challenge with HIV-1/(VSV-G) pseudotypes, while NIH 3T3 cells yielded 100- to 3,000-fold-lower signals (Fig. 1). Among T-cell lines, both human cell lines expectedly yielded strong signals for early and late gene expression. Interestingly, the rat-derived T-cell lines displayed quite various relative expression levels. Early gene expression in rat Nb2 cells appeared to be 10- and 14-fold lower than that seen in human Jurkat and SUP-T1 cells, respectively (Fig. 1, top panel). However, taking into account the differences in relative VSV-G-mediated entry into these cell types, the relative early gene expression in Nb2 cells was actually only twofold lower than that determined for the two human T-cell lines. In contrast, rat T-cell lines SD1 and C58 yielded considerably lower signals, reflecting a transcriptional activity markedly reduced from that of the HIV-1 LTR. It should be noted that in both Rat2 and Nb2 cells there was a steeper drop in the expression levels from early (top panel) to late (lower panel) HIV-1 gene products than that seen for human reference cell lines. These results collectively indicate that rat cell lines can vary considerably in their ability to support specific steps in the HIV-1 replication cycle. More importantly, rat-derived cells, despite quantitative limitations, are not absolutely restricted in postentry steps of the HIV-1 replication cycle.
FIG. 1.
Relative expression of early and late HIV-1 gene products in cell lines from humans, rats, and mice following VSV-G pseudotype infections. (Top) Cells were infected with VSV-G pseudotyped NL4-3 Luc E−R− reporter viruses, and 3 days postinfection luciferase signals were measured as relative light units in cell lysates as a marker of early HIV-1 gene expression. (Bottom) The same cell types were infected overnight with VSV-G pseudotyped HIV-1 49.5. The postwash residual p24 CA concentration was below 0.12 ng/ml for all cultures. Three days postinfection, the p24 CA concentration in supernatants was determined as a marker for expression and egress of a late, fully processed HIV-1 gene product. The susceptibilities of cell lines for VSV-G-mediated entry were assessed using a VSV-G pseudotyped HIV-1-based vector containing a luciferase gene under CMV promoter control, and values representing the relative VSV-G entry are given. Bars represent the arithmetic means ± standard deviations of triplicates.
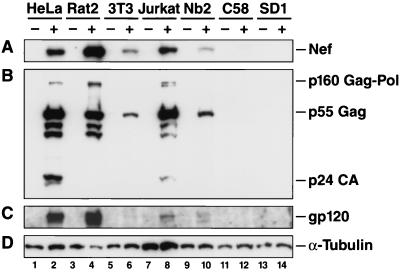
A Western blot analysis of lysates from the above-described HIV-1/(VSV-G)-infected cells revealed a complex picture regarding the expression of early and late gene products, the processing of Gag protein, and viral egress. Several interesting observations could be made. First, confirming our earlier analysis, Rat2 showed abundant expression of HIV-1 Nef (Fig. 2A, lane 4). Nb2 cells expressed low but clearly detectable levels of Nef (Fig. 2B, lane 10) despite their very low intrinsic susceptibility to VSV-G pseudotypes (Fig. 1). Second, expression levels of gp120 in human and rat cells paralleled those of Nef (Fig. 2A and C, lanes 2, 4, 8, and 10), suggesting that rat cells have a largely unimpaired expression of this late, Rev-dependent HIV-1 gene product that is translated off a partially spliced viral mRNA. In both Rat2 and Nb2 cells, a second band with higher electrophoretic mobility was detected, possibly representing a hypoglycosylated form of gp120. Third, levels of p55 Gag relative to Nef were reduced fourfold in Rat2 cells from that in human cells but were not reduced in Nb2 cells (Fig. 2A and B, lanes 2, 4, 8, and 10). Fourth, processing of p55 Gag to p24 CA was detectable in only human cell lysates, not in rat cell lysates (Fig. 2B, lanes 2, 4, 8, and 10). In this context, the overall levels of p55 Gag or p24 CA in lysates were not a good indicator of levels of secreted p24 CA within any given cell type. For example, p24 CA was 18-fold less abundant in Jurkat lysates than in HeLa lysates (Fig. 2B, lanes 2 and 8), and yet Jurkat cells secreted fivefold more p24 CA into the supernatant (Fig. 1). Similarly, levels of Gag products in Nb2 cells were approximately fivefold lower than those in Rat2 cells (Fig. 2B, lanes 10 and 4), and yet their levels of secreted p24 CA were almost identical (Fig. 1). Finally, p55 Gag levels in Nb2 lysates were only twofold higher than those in NIH 3T3 lysates (Fig. 2B, lanes 10 and 6), and yet concentrations of secreted p24 CA were 45-fold higher in Nb2 cells (Fig. 1). Collectively, these discrepancies indicate that virion assembly, maturation, and egress occur at quite different efficiencies that cannot be predicted from cell-associated levels of structural proteins. Judged from the limited selection of cell lines in this study, rat cells appear to be somewhat less efficient than human cells but more efficient than mouse cells in supporting these processes. Overall, these experiments highlight quantitative, qualitative, and cell-specific limitations rather than absolute blocks at various postentry steps in the HIV-1 replication cycle in rat cell lines, including LTR-driven transcription, relative levels of Gag products, the intracellular processing of p55 Gag, and viral egress.
FIG. 2.
Immunoblot analysis of early and late HIV-1 gene products following HIV-1/(VSV-G) pseudotype infections. 49.5/(VSV-G)-infected cells (+) (taken from the experiment shown in the bottom panel of Fig. 1) or uninfected control cells (−) were lysed and processed. Immunoblotting with anti-Nef (A), anti-p24 CA (B), anti-gp120 (C), and anti-α-tubulin (D) antibodies was performed consecutively.
The HIV-1 LTR exhibits significant activity in Rat2 cells which can be further enhanced by human CycT1.
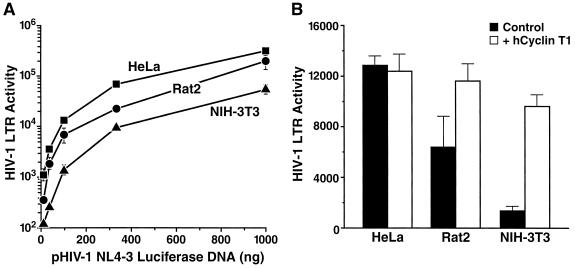
To characterize further the efficiency of HIV-1 LTR transactivation and transcript elongation in rat cells, increasing amounts of the proviral plasmid pNL4-3 Luc E−R− were transfected into Rat2, HeLa, and NIH 3T3 cells, and luciferase activities in cellular lysates were quantified 48 h later. The measured HIV-1 LTR activity was normalized for variability in transfection efficiency by cotransfection of an LTR-independent reporter construct (58). Over a range of pNL4-3 Luc E−R− concentrations, Rat2 cells yielded a substantial signal that was significantly higher than that found in NIH 3T3 cells but somewhat lower than that of HeLa cells (Fig. 3A). Interestingly, the substitution of a single amino residue (tyrosine-261) in mouse CycT1 compared to its human homologue was previously identified as the major determinant restricting Tat-mediated HIV-1 LTR transactivation in NIH 3T3 cells (6, 20, 31, 58). In view of the substantial transcriptional activity observed in Rat2 cells, we sought to establish the sequence of the rat CycT1 homologue. A partial sequence of rat CycT1 cloned from a rat cDNA library revealed that it shared a tyrosine residue at position 261 with mouse CycT1 (data not shown). This fact raises the possibility that this sequence change in CycT1 may not be essential for its interaction with Tat in rat cells or that viral transcription in rat cells is relatively Tat independent. In the same HIV-1 LTR assay system described above, cotransfection of an expression plasmid encoding human CycT1 nevertheless significantly augmented luciferase signals in both NIH 3T3 and Rat2 cells, achieving levels comparable to those of HeLa cells (Fig. 3B). Therefore, the activity of CycT1 may be a quantitatively limiting factor for Tat transactivation in rat cells, although human CycT1 is nonessential.
FIG. 3.
The HIV-1 LTR exhibits significant activity in Rat2 cells which can be further enhanced by overexpression of human CycT1. Cell lines HeLa, Rat2, and NIH 3T3 were transfected using the TransFast transfection reagent (Promega) with different amounts (10 to 1,000 ng) of pNL4-3 Luc E−R− (firefly luciferase), 10 ng of pRL-HSV-1-tk (Renilla luciferase control plasmid), and 0 to 990 ng of pcDNA3.1 (empty vector) (A) or 100 ng of pNL4-3 Luc E−R−, 10 ng of pRL-HSV-1-tk, 800 ng of pcDNA3.1, and 100 ng of pCICyT (expression plasmid for human CycT1) (B). Luciferase activities in cell lysates were determined 48 h after transfection, and the firefly luciferase activity was normalized to the Renilla luciferase activity as described previously (58). Values are arithmetic means ± standard deviations of triplicates.
Coexpression of hCD4 and hCCR5 in rat Nb2 cells renders them permissive for HIV-1 infection.
Cellular entry is a major restriction of HIV-1 replication in rodent cells (reviewed in reference 4). To test whether coexpression of hCD4 and the human chemokine receptor CCR5 could overcome this block, we generated stable transfectants of the rat T-cell line Nb2 that express hCD4 alone (Nb2 hCD4) or hCD4 and hCCR5 (Nb2 hCD4/hCCR5). Clones of the human CD4+ T-cell lines Jurkat (Jurkat hCCR5) (48) and SUP-T1 (SUP-T1 hCCR5), which had also been engineered to express hCCR5, served as controls. Cell surface expression of human receptors was quantified by flow cytometry, revealing various levels of hCD4 and hCCR5 on Nb2 hCD4/hCCR5, Jurkat hCCR5, and SUP-T1 hCCR5 cells (Fig. 4A). We first performed single-round infection studies with NL4-3 luciferase reporter viruses pseudotyped with different autologous or heterologous envelopes. VSV-G pseudotypes comparably infected Nb2, Nb2 hCD4, and Nb2 hCD4/hCCR5, as well as the human T-cell line clones (Fig. 4B). The HIV-1 R5 envelope pseudotypes efficiently infected Nb2 hCD4/hCCR5 cells but failed to produce significant signals in parental Nb2 cells. Interestingly, SUP-T1 hCCR5 cells yielded the highest signals for both R5 pseudotypes and yet expressed almost undetectable levels of hCCR5. As a confirmation of specificity, the HIV-1 X4 pseudotype did not show significant entry into any of the Nb2 cell lines but readily infected the human T-cell lines expressing endogenous hCXCR4.
Nb2 hCD4/hCCR5 cells were next challenged with the replication-competent HIV-1 R5 virus YU-2 at various input doses (Fig. 4C). Following overnight infection, cells were washed and the p24 CA concentration in supernatants was monitored over the course of 6 days. Nb2 hCD4/hCCR5 supported a measurable level of HIV-1 replication that was clearly dependent on the initial MOI; approximately 95% of the p24 CA increase from day 1 to day 3 was sensitive to treatment with the reverse transcriptase inhibitor 3′-azido-3′deoxythymidine (AZT) (data not shown). Notably, the p24 CA concentration for Nb2 hCD4/hCCR5 cells 3 days postinfection was 30-fold lower than that seen for human SUP-T1 hCCR5 cells (Fig. 4C and D). Furthermore, the p24 CA kinetics observed for Nb2 hCD4/hCCR5 cells suggest that the infection was transient and did not spread efficiently through the culture. Similar results were obtained for the HIV-1 R5 virus JR-CSF (data not shown). The transfer of cell-free supernatant from HIV-1 YU-2-infected Nb2 hCD4/hCCR5 cells onto freshly sodium butyrate-induced Nb2 hCD4/hCCR5 cells resulted in a significant infection in these secondary target cells (data not shown), confirming that these cells support a productive HIV-1 infection leading to secretion of infectious virions. In contrast, the HIV-1 X4 virus LAI.2 did not productively infect Nb2 hCD4/hCCR5 cells but readily spread in the human T-cell lines (data not shown). Thus, Nb2 cells expressing hCD4 and hCCR5 were permissive for productive HIV-1 R5 virus infection but failed to support a robust spreading infection.
Primary rat cells can support substantial early HIV-1 gene expression, but only macrophages and microglia support significant expression of late, fully processed HIV-1 gene products.
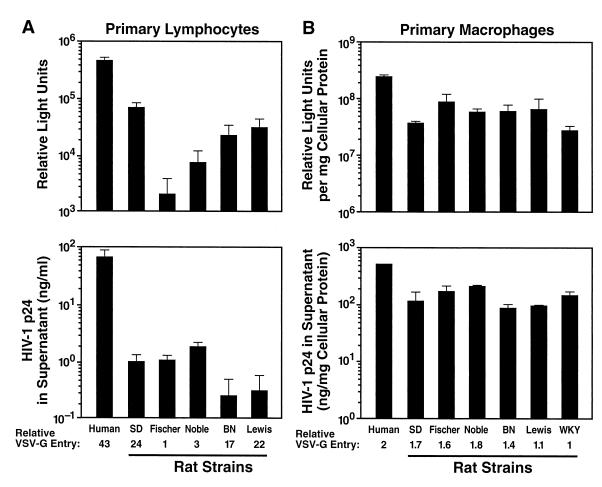
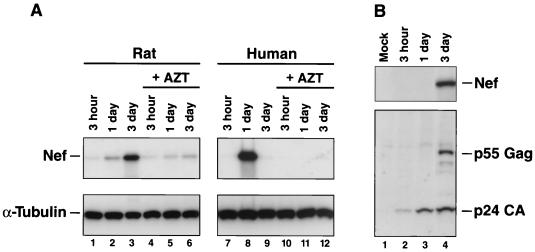
Tissue-specific differences in supporting particular steps in the HIV-1 replication cycle have been described previously (35, 45), which emphasizes the need to study the specific cell types that are expected to express human transgenes and thereby be rendered infectible by HIV-1 in a transgenic animal. Primary T lymphocytes and cells from the monocyte/macrophage lineage are considered to be the most important sites of HIV-1 replication in humans and are thus obvious targets for rodent transgenesis. We therefore investigated the ability of HIV-1 to replicate in primary lymphocytes, macrophages, and microglia from rats. The same VSV-G pseudotype infection strategies described above for cell lines were used to assess early and late events in the HIV-1 replication cycle in primary cells from a variety of different rat strains. Both primary rat lymphocytes and macrophages supported substantial LTR-driven early gene expression even in the absence of human cofactors for Tat (Fig. 5, top panels). When normalized for differences in VSV-G-mediated entry, levels for rat lymphocytes were 12 to 31% of that of human reference cultures, and levels for rat macrophages were 16 to 45%. Since luciferase activity is a surrogate marker of early gene expression that may be affected by other factors, we also used Nef expression in primary rat cells as an independent and direct measure. In agreement with our earlier findings, primary rat lymphocytes and macrophages expressed abundant levels of HIV-1 Nef following NL4-3/(VSV-G) pseudotype infections (Fig. 6A, lanes 3 and 8, and B, lane 4). AZT treatment inhibited Nef expression in both rat and human lymphocytes, confirming that this protein had been synthesized de novo. Of note, the kinetics of Nef expression in primary rat lymphocytes were somewhat delayed compared to those in human cultures, and human lymphocytes displayed massive cell death on day 3 postinfection.
FIG. 5.
Primary rat cells can express substantial levels of an early HIV-1 gene product, but only macrophages and microglia also secrete p24 CA. HIV-1 gene expression was assessed in spleen-derived lymphocytes and macrophages from different rat strains relative to comparable human control cultures as described in the legend to Fig. 1. Concanavalin A–IL-2-activated primary rat lymphocytes (A) or macrophages (B) were infected with either VSV-G pseudotyped NL4-3 Luc E−R− reporter pseudotypes (top panels) or NL4-3/(VSV-G) (bottom panels). Parallel infections were performed in human monocyte-derived macrophages or PHA-P–IL-2-activated human lymphocytes in the presence of the X4 entry inhibitor AMD3100 (250 nM) (50) to limit infections to a single round by preventing viral spread. The relative susceptibility of cells for VSV-G-mediated entry is given below each panel. All bars represent mean values of triplicate samples, with error bars representing standard deviations.
FIG. 6.
Immunoblot analysis of early and late HIV-1 gene products in primary lymphocytes and macrophages from rats and humans following HIV-1/(VSV-G) pseudotype infections. NL4-3/(VSV-G)-infected lymphocytes (A) and macrophages (B) were lysed and processed for immunoblotting at the indicated time points. (A) Human lymphocytes were infected and cultivated in the presence of the CXCR4 antagonist AMD3100 to limit infections to a single round. AZT (10 μM) was added to the indicated cultures 1 h prior to viral challenge.
Moreover, primary rat macrophages synthesized and secreted substantial amounts of p24 CA (Fig. 5B, bottom panel, and 6B). Macrophage lysates contained abundant levels of Gag products p55 Gag and p24 CA 3 days postinfection (Fig. 6B) and did not seem to have significant additional limitations on the HIV-1 replication cycle past early gene expression as indicated by levels of luciferase activities and secreted p24 CA concentrations comparable to those of human reference cultures (Fig. 5B). In contrast, primary rat lymphocytes secreted only very low levels of p24 CA (<2 ng/ml) (Fig. 5A, lower panel), and the de novo production of Gag proteins could also not be demonstrated for cellular lysates (data not shown). The results indicate a severe block in the transition to the late phase of the viral life cycle in primary rat lymphocytes but not in primary rat macrophages.
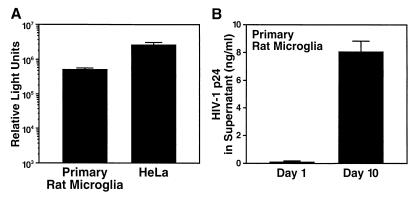
The susceptibility of primary rat microglia for the HIV-1 replication cycle was also studied using pseudotyped viruses as described above. Microglia were isolated from neonatal rat brains and enriched by subcultivation (see Materials and Methods), and the purity of cultures exceeded 95% as verified by positive staining for the rat microglial marker CD11b/c and an absence of staining for the astrocyte-specific glial fibrillary acidic protein (GFAP) marker (data not shown). As seen with primary macrophages, rat microglia supported robust early HIV-1 gene expression that was comparable to that observed for human HeLa cells (Fig. 7A). Moreover, late viral gene expression, Gag processing, and viral egress appeared intact in these microglia, as indicated by secretion of p24 CA (Fig. 7B). Thus, rat cells relating to the monocyte/macrophage lineage, including primary spleen-derived macrophages and brain-derived microglia, support substantial early and late HIV-1 gene expression.
FIG. 7.
Primary rat microglia support all steps in the HIV-1 replication cycle. Subcultured microglia were infected with the same VSV-G pseudotype stocks as described in the legend to Fig. 1 to assess early (A) or late (B) HIV-1 gene expression. HeLa cells served as a reference. The relative susceptibility of HeLa cells for VSV-G-mediated entry was 1.9-fold higher than that of primary rat microglia. All bars represent mean values of triplicate samples, with error bars representing standard deviations.
HIV-1 produced by human cells and that produced by rat cells are equally infectious.
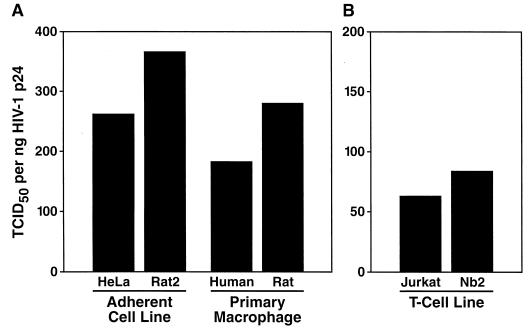
Finally, we sought to assess the relative infectivity of virions released from comparable human and rat cell cultures. We challenged adherent cell lines HeLa and Rat2, T-cell lines Jurkat and Nb2, and a set of primary macrophage cultures with VSV-G pseudotyped virus containing an intact, replication-competent HIV-1 genome. Five days postinfection, culture supernatants were collected and analyzed for both the concentration of p24 CA and infectious titer (TCID50 per milliliter) on human PBMC. The relative infectivity of released HIV-1, defined as the ratio of the TCID50 per milliliter to nanograms of p24 CA per milliliter, was comparable for the respective cell pairs (Fig. 8). Importantly, in earlier experiments of this type, TCID50 values determined in the presence or absence of a neutralizing concentration of the anti-VSV-G MAb I1 (8) had not revealed significant differences, indicating that the washing procedures had completely removed the initial VSV-G pseudotyped inoculum (data not shown). Thus, the comparable infectivities of human and rat cell-derived virions demonstrate that fully mature, infectious HIV-1 particles can be efficiently synthesized, assembled, and released from rat cells.
FIG. 8.
HIV-1 produced by cells from humans and that produced by cells from rats are equally infectious. The relative infectivity of HIV-1 released by adherent cell lines, T-cell lines (49.5), or primary macrophages (NL4-3) of human or rat origin was determined as the ratio of TCID50 measurements (determined on a PBMC pool from four [A] or three [B] donors) and p24 CA concentrations. Cells were infected overnight with HIV-1/(VSV-G). The postwash residual p24 CA concentration was below 0.3 ng/ml for all cultures. Five days postinfection, the supernatant was harvested and analyzed. The absolute p24 CA concentrations for the respective cell lines are as follows (in nanograms per milliliter): HeLa, 324; Rat2, 49; Jurkat, 1,681; Nb2, 27; human macrophages, 217; and rat macrophages, 25.
DISCUSSION
The urgent need for enhanced model systems to study HIV-1 transmission, pathogenesis, and candidate vaccines or therapeutic strategies necessitates a deeper understanding of quantitative limitations and blocks in the viral replication cycle in animals that may be used as hosts. Here we show that cellular entry constitutes the only absolute block to HIV-1 replication in certain rat cell lines and that this restriction can be overcome by coexpression of hCD4 and hCCR5. Our study revealed quantitative, qualitative, and cell-type-specific limitations in various aspects of the HIV-1 replication cycle in rat cells. Despite such limitations, primary rat cells from the monocyte/macrophage lineage supported all postentry steps in the viral life cycle and secreted substantial levels of infectious virions.
A first indication that rat-derived cells are permissive for postentry steps in the HIV-1 replication cycle resulted from infection studies of established cell lines using HIV-1 pseudotyped with a VSV-G envelope. Rat cell lines Rat2 (fibroblast) and Nb2 (T cell) supported considerable levels of productive infection, yielding p24 CA concentrations of approximately 30 ng/ml. Such high levels of HIV-1 replication have never been reported for mouse cell lines, even in the presence of human cofactors for Tat (4, 5, 40). Consistent with our findings, Rat2 clones stably transfected with HIV-1 provirus that secreted significant levels of virus have been reported previously (42). In contrast, one recent report found infection levels in Rat2 cells that were 10-fold lower than those seen in our study (5). The explanation for this discrepancy is unclear but may relate to different clones of Rat2 cells used in different laboratories. Nevertheless, these results prompted us to explore in subsequent experiments the efficiency of specific steps in the HIV-1 replication cycle in rat cells.
In these experiments, the postentry steps including uncoating, reverse transcription, nuclear importation, and proviral integration appeared to be intact in many rat-derived cells. Furthermore, rat-derived cells exhibited robust HIV-1 LTR activity as determined by HIV-1 NL4-3 pseudotype reporter virus infections, HIV-1 immunoblot analyses, HIV-1 LTR reporter assays, and infection studies with replication-competent HIV-1. The remarkably high HIV-1 LTR activity in rat cells seen in our study and previously reported by others (5, 39, 42) is in clear contrast to the previously reported inability of native mouse and hamster cells to support Tat-dependent HIV-1 transcription (5, 40, 58). The change of amino residue 261 from cysteine to tyrosine in CycT1 had previously been shown to be the major determinant restricting Tat-mediated HIV-1 LTR transactivation in NIH 3T3 cells (6, 20, 31, 58). Surprisingly, we found that rat and mouse CycT1 shared this amino acid substitution. Our experiments did not directly address whether the robust LTR activity in rat cells is indeed CycT1 dependent, and it is possible that viral transcription is at least partially CycT1 independent in this context. Alternatively, it can be speculated that this specific tyrosine residue is nonessential for rat CycT1 in supporting HIV-1 LTR transactivation. Specifically, one might hypothesize that other sequence changes in rat CycT1 may augment its interaction with Tat despite tyrosine-261, and indeed the sequence of a rat CycT1 clone that we obtained diverged at other positions from its mouse counterpart (data not shown). Furthermore, our results suggest that CycT1 may nonetheless be a quantitatively limiting factor for Tat transactivation in rat cells. Importantly, we also extended our analyses to primary rat cells, including lymphocytes and macrophages from different rat strains, all of which supported substantial early HIV-1 gene expression in the range of 12 to 45% of that of human reference cultures. We also found marked HIV-1 LTR activity in primary rat microglia, another cell type of the macrophage/monocyte lineage. Collectively, these data suggest that the ability to support robust HIV-1 LTR activity is a universal property of R. norvegicus.
We also found that coexpression of hCD4 and hCCR5 in rat Nb2 cells mediated substantial and specific entry of HIV-1. This finding is consistent with a number of studies demonstrating that the coexpression of hCD4 and hCCR5 or hCXCR4 on rodent cell lines is sufficient to overcome the HIV-1 entry block (5, 11, 39, 40, 49, 51), although there has been some controversy over the efficiency of this process (5, 40). Although Nb2 hCD4/hCCR5 cells could be productively infected and secrete infectious HIV-1, they failed to support an efficiently spreading infection. The reason for this inefficiency is unclear. One possible explanation is the lower absolute number of infectious particles being released from rat cells; compared to Nb2 cells, human T-cell lines produced approximately 40- to 60-fold-more infectious particles per cell in a single replication cycle. Also, Nb2 hCD4/hCCR5 cells only transiently express detectable levels of hCCR5 on the surface following sodium butyrate pretreatment, which conceivably limited a spreading infection by loss of potential secondary target cells for newly released virions in these experiments. Nevertheless, our data show that the major block to HIV-1 replication in rat cells can be overcome by expression of the HIV-1 receptor complex.
Quantitative and qualitative limitations in the late phase of the viral life cycle that appeared to be cell type specific were also found, although these relationships were complex. Intracellular p55 Gag processing, virion assembly, and egress occurred at various efficiencies that could not be predicted from cell-associated levels of structural proteins; in general, rat cell lines appeared to be less efficient than human cells but more efficient than mouse cells in supporting these steps. Absolute levels of secreted p24 CA were 10- to 60-fold lower in Rat2 and Nb2 cells than in human cell lines. However, there was no single step in the late phase of the HIV-1 replication cycle in these rat cell lines that could be pinpointed as being rate limiting for virus production. First, there has been some controversy over the existence of a profound block in Rev function in rodent cells (5, 37, 40, 55). This regulatory HIV-1 protein is thought to control the transition into the late phase of the viral replication cycle by orchestrating the ordered expression of the various gene products through effects on splicing, stability, and nuclear export of the unspliced genomic transcripts that depend on interactions with cellular factors (reviewed in reference 47). Our data do not support the existence of a rat species-specific defect in the function of HIV-1 Rev. In cell lines Rat2 and Nb2, as well as primary rat macrophages, we detected abundant levels of p55 Gag, a protein that derives from the Rev-dependent p160 Gag-Pol precursor. Furthermore, Rev function was conserved in rat T-cell lines C58 and SD1 (data not shown), as assessed with the chloramphenicol acetyltransferase reporter construct pDM128 and Rev supplied in trans by an expression vector (26). Second, there were clear quantitative differences in terms of the intracellular levels of fully processed p24 CA in rat and human cell lysates, as has been noted by others (5, 39, 40), but intracellular protein content was not a good indicator of levels of secreted p24 CA. Although difficult to quantify, the efficiency of viral assembly and egress may also be partly impaired in rat cells. Taking the data together, we speculate that several minor limitations at different steps of the HIV-1 replication cycle in rat cell lines in aggregate account for the quantitative reduction in virus production that we observed.
Importantly, we also found that HIV-1 produced by rat cells and that produced by human cells had comparable levels of infectivity. HIV-1 derived from adherent cell lines and that derived from T-cell lines of human or rat origin had similar ratios of TCID50 to p24 CA, indicating that assembly and maturation of virions can proceed normally in rat cells. A similar finding was recently reported for NIH 3T3 (40) and CHO and Rat2 (39) cells, while another study reported rat and mouse cell-derived HIV-1 to be largely noninfectious (5). The basis for these discrepancies is not known but could be related to different methods for measuring infectious titers. We also found comparable infectivities for HIV-1 released from primary macrophages of rat origin and for that from macrophages of human origin. Future infection studies with HIV-1 Δvif mutants in primary rat macrophages may help to clarify the functionality of the HIV-1 Vif protein in this species context, since the vif gene is known to greatly enhance the infectivity of HIV-1 that is released from human cells classified as nonpermissive, which include primary macrophages (36, 54, 56).
An important aspect of our current study was to evaluate postentry steps in the HIV-1 replication cycle in primary rat cells, and we found that primary rat macrophages and microglia are remarkably susceptible. To our knowledge, this analysis is the first report describing primary rodent cells supporting HIV-1 replication at such high levels; the p24 CA concentrations secreted by rat macrophages were only two- to sixfold lower than those secreted by human monocyte-derived macrophages in a single replication cycle. In humans, macrophages and microglia in the central nervous system have long been known to be targets of HIV-1 that support virus replication in vivo (10, 19, 21, 29, 30, 53, 57, 59). Also, macrophages have been implicated as a source of increasing viremia in the latter stages of HIV disease in humans, in particular during opportunistic infections (46). A recent study suggested macrophages as the principal reservoir for sustained high virus load in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus–HIV-1 chimera (27). Interestingly, macrophages are the evolutionarily conserved host cell type for all animal lentiviruses (reviewed in reference 32). Furthermore, the productive infection of microglia and the presence of multinucleated giant cells were found in one study to correlate roughly with the antemortem diagnosis of HIV-associated dementia (22). We speculate that reconstituting the HIV-1 viral receptor complex in transgenic rats may be sufficient to permit recapitulation of the full viral replication cycle in cells of the macrophage/monocyte lineage in vitro and in vivo.
Primary rat T lymphocytes were found to harbor a major posttranscriptional block to HIV-1 replication. Despite considerable expression of Nef protein or a reporter gene, primary T cells failed to synthesize Gag proteins and secrete significant concentrations of p24 CA. This phenotype is consistent with an impaired function of Rev in promoting the nuclear export of unspliced genomic transcripts, and future studies will be needed to address this possibility. On one hand, the failure of HIV-1 replication to proceed beyond early gene expression in primary rat T lymphocytes may help to dissect mechanisms of CD4+ T-cell depletion in an in vivo model. On the other hand, the future identification of human cellular factors that participate in HIV-1 replication and that may surmount restrictions in primary rat T lymphocytes would both provide a better understanding of the molecular aspects of HIV-1 replication and increase the range of uses of a transgenic rat model for the study of HIV-related pathogenesis. The results of the present study employing rat cell lines and primary rat cells serve as a sound basis for our current efforts to develop a transgenic small animal model using Rattus norvegicus, in which hCD4 and human chemokine receptors are expressed on macrophages, microglia, and T lymphocytes. It will be important to determine the impact in vivo of the cell-specific viral replication described here, which may dictate additional modifications that should be incorporated in subsequent generations of such a model.
ACKNOWLEDGMENTS
We thank Jane Burns, Kathy Jones, Matija Peterlin, Ned Landau, Didier Trono, Malcolm Martin, Leo Lefrancois, Jay Ryan, Paula Pitha, and Kathleen Page for reagents. We thank Warner C. Greene for his support. We acknowledge the excellent administrative assistance of Heather Gravois in the preparation of the manuscript and of John Carroll and Jack Hull for graphics preparation.
This work was supported in part by the J. David Gladstone Institutes (M.A.G.), NIH grant R21-AI46258 (M.A.G.), and the UCSF AIDS Clinical Research Center (R.F.S.). Oliver T. Keppler is a Howard Hughes Medical Institute Physician Postdoctoral Fellow. Frank J. Welte is a Howard Hughes Medical Institute Medical Student Research Training Fellow.
REFERENCES
- 1.Akagi T, Takata H, Yoshino T, Teramoto N, Yano S, Oka T. Immortalization of rat spleen and thymus T-cells by human T-cell leukemia virus type-I. Acta Med Okayama. 1989;43:143–151. doi: 10.18926/AMO/30886. [DOI] [PubMed] [Google Scholar]
- 2.Andersson S, Davis D L, Dahlbäck H, Jörnvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 3.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 4.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz P D, Cullen B R. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J Virol. 2000;74:9868–9877. doi: 10.1128/jvi.74.21.9868-9877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boddeke E, Meigel I, Frentzel S, Gourmala N G, Harrison J K, Buttini M, Spleiss O, Gebicke-Harter P. Cultured rat microglia express functional beta-chemokine receptors. J Neuroimmunol. 1999;98:176–184. doi: 10.1016/s0165-5728(99)00096-x. [DOI] [PubMed] [Google Scholar]
- 8.Boritz E, Gerlach J, Johnson J E, Rose J K. Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J Virol. 1999;73:6937–6945. doi: 10.1128/jvi.73.8.6937-6945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bresnahan P A, Yonemoto W, Ferrell S, Williams-Herman D, Geleziunas R, Greene W C. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr Biol. 1998;8:1235–1238. doi: 10.1016/s0960-9822(07)00517-9. [DOI] [PubMed] [Google Scholar]
- 10.Brew B J, Rosenblum M, Cronin K, Price R W. AIDS dementia complex and HIV-1 brain infection: clinical-virological correlations. Ann Neurol. 1995;38:563–570. doi: 10.1002/ana.410380404. [DOI] [PubMed] [Google Scholar]
- 11.Browning J, Horner J W, Pettoello-Mantovani M, Raker C, Yurasov S, DePinho R A, Goldstein H. Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc Natl Acad Sci USA. 1997;94:14637–14641. doi: 10.1073/pnas.94.26.14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cecilia D, Kewalramani V N, O'Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman D R, Zolla-Pazner S. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan S Y, Speck R F, Power C, Gaffen S L, Chesebro B, Goldsmith M A. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connor R, Chen B, Choe S, Landau N. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 16.Deglon N, Tseng J L, Bensadoun J C, Zurn A D, Arsenijevic Y, De Almeida L P, Zufferey R, Trono D, Aebischer P. Self-inactivating lentiviral vectors with enhanced transgene expression as potential gene transfer system in Parkinson's disease. Hum Gene Ther. 2000;11:179–190. doi: 10.1089/10430340050016256. [DOI] [PubMed] [Google Scholar]
- 17.Emi N, Friedmann T, Yee J K. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming W H, Pettigrew N M, Matusik R J, Friesen H G. Thymic origin of the prolactin-dependent Nb2 lymphoma cell line. Cancer Res. 1982;42:3138–3141. [PubMed] [Google Scholar]
- 19.Gabuzda D H, Ho D D S, de la Monte M, Hirsch M S, Rota T R, Sobel R A. Immunohistochemical identification of HTLV-III antigen in brains of patients with AIDS. Ann Neurol. 1986;20:289–295. doi: 10.1002/ana.410200304. [DOI] [PubMed] [Google Scholar]
- 20.Garber M, Wei P, KewalRamani V, Mayall T, Herrmann C, Rice A, Littman D, Jones K. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 22.Glass J D, Fedor H, Wesselingh S L, McArthur J C. Immunocytochemical quantitation of human immunodeficiency virus in the brain—correlations with dementia. Ann Neurol. 1995;38:755–762. doi: 10.1002/ana.410380510. [DOI] [PubMed] [Google Scholar]
- 23.Goldsmith M A, Warmerdam M T, Atchison R E, Miller M D, Greene W C. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J Virol. 1995;69:4112–4121. doi: 10.1128/jvi.69.7.4112-4121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart C E, Ou C Y, Galphin J C, Moore J, Bacheler L T, Wasmuth J J, Petteway S R, Jr, Schochetman G. Human chromosome 12 is required for elevated HIV-1 expression in human-hamster hybrid cells. Science. 1989;246:488–491. doi: 10.1126/science.2683071. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hope T J, McDonald D, Huang X J, Low J, Parslow T G. Mutational analysis of the human immunodeficiency virus type 1 Rev transactivator: essential residues near the amino terminus. J Virol. 1990;64:5360–5366. doi: 10.1128/jvi.64.11.5360-5366.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Igarashi T, Brown C R, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin M A. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4(+) T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc Natl Acad Sci USA. 2001;98:658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinter A, Arthos J, Cicala C, Fauci A S. Chemokines, cytokines and HIV: a complex network of interactions that influence HIV pathogenesis. Immunol Rev. 2000;177:88–98. doi: 10.1034/j.1600-065x.2000.17708.x. [DOI] [PubMed] [Google Scholar]
- 29.Koenig S, Gendelman H, Orenstein J, Dal Canto M, Pezeshkpour G, Yungbluth M, Janotta F, Aksamit A, Martin M, Fauci A. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 30.Kolson D L, Gonzalez-Scarano F. HIV and HIV dementia. J Clin Investig. 2000;106:11–13. doi: 10.1172/JCI10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwak Y T, Ivanov D, Guo J, Nee E, Gaynor R B. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J Mol Biol. 1999;288:57–69. doi: 10.1006/jmbi.1999.2664. [DOI] [PubMed] [Google Scholar]
- 32.Levy J A. The value of primate models for studying human immunodeficiency virus pathogenesis. J Med Primatol. 1996;25:163–174. doi: 10.1111/j.1600-0684.1996.tb00013.x. [DOI] [PubMed] [Google Scholar]
- 33.Levy J A, Cheng-Mayer C, Dina D, Luciw P A. AIDS retrovirus (ARV-2) clone replicates in transfected human and animal fibroblasts. Science. 1986;232:998–1001. doi: 10.1126/science.3010461. [DOI] [PubMed] [Google Scholar]
- 34.Lewis A D, Johnson P R. Developing animal models for AIDS research—progress and problems. Trends Biotechnol. 1995;13:142–150. doi: 10.1016/S0167-7799(00)88925-8. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig E, Ceccherini-Silberstein F, van Empel J, Erfle V, Neumann M, Brack-Werner R. Diminished Rev-mediated stimulation of human immunodeficiency virus type 1 protein synthesis is a hallmark of human astrocytes. J Virol. 1999;73:8279–8289. doi: 10.1128/jvi.73.10.8279-8289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madani N, Kabat D. Cellular and viral specificities of human immunodeficiency virus type 1 Vif protein. J Virol. 2000;74:5982–5987. doi: 10.1128/jvi.74.13.5982-5987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral Rre—implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 38.Mancebo H S Y, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y R, Peng J M, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mariani R, Rasala B A, Rutter G, Wiegers K, Brandt S M, Kräusslich H G, Landau N R. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J Virol. 2001;75:3141–3151. doi: 10.1128/JVI.75.7.3141-3151.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mariani R, Rutter G, Harris M E, Hope T J, Kräusslich H G, Landau N R. A block to human immunodeficiency virus type 1 assembly in murine cells. J Virol. 2000;74:3859–3870. doi: 10.1128/jvi.74.8.3859-3870.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizrachi Y, Rubinstein A, Golodner M, Sakai K, Volsky D J. CD4 confers susceptibility to human immunodeficiency virus type 1 infection in a rat fibroblast cell line. AIDS Res Hum Retrovir. 1996;12:1315–1318. doi: 10.1089/aid.1996.12.1315. [DOI] [PubMed] [Google Scholar]
- 42.Mizrachi Y, Sternas L, Volsky D J. The establishment of rodent cell lines persistently producing HIV-1. Virology. 1992;186:167–174. doi: 10.1016/0042-6822(92)90071-v. [DOI] [PubMed] [Google Scholar]
- 43.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 44.Morrow W J, Wharton M, Lau D, Levy J A. Small animals are not susceptible to human immunodeficiency virus infection. J Gen Virol. 1987;68:2253–2257. doi: 10.1099/0022-1317-68-8-2253. [DOI] [PubMed] [Google Scholar]
- 45.Murphy K M, Sweet M J, Ross I L, Hume D A. Effects of the tat and nef gene products of human immunodeficiency virus type 1 (HIV-1) on transcription controlled by the HIV-1 long terminal repeat and on cell growth in macrophages. J Virol. 1993;67:6956–6964. doi: 10.1128/jvi.67.12.6956-6964.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orenstein J, Fox C, Wahl S. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276:1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 47.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 48.Popik W, Pitha P M. Early activation of mitogen-activated protein kinase kinase, extracellular signal-regulated kinase, p38 mitogen-activated protein kinase, and c-Jun N-terminal kinase in response to binding of simian immunodeficiency virus to Jurkat T cells expressing CCR5 receptor. Virology. 1998;252:210–217. doi: 10.1006/viro.1998.9466. [DOI] [PubMed] [Google Scholar]
- 49.Sawada S, Gowrishankar K, Kitamura R, Suzuki M, Suzuki G, Tahara S, Koito A. Disturbed CD4+ T cell homeostasis and in vitro HIV-1 susceptibility in transgenic mice expressing T cell line-tropic HIV-1 receptors. J Exp Med. 1998;187:1439–1449. doi: 10.1084/jem.187.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schramm B, Penn M L, Speck R F, Chan S Y, De Clercq E, Schols D, Connor R I, Goldsmith M A. Viral entry through CXCR4 is a pathogenic factor and therapeutic target in human immunodeficiency virus type 1 disease. J Virol. 2000;74:184–192. doi: 10.1128/jvi.74.1.184-192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speck R F, Penn M L, Wimmer J, Esser U, Hague B F, Kindt T J, Atchison R E, Goldsmith M A. Rabbit cells expressing human CD4 and human CCR5 are highly permissive for human immunodeficiency virus type 1 infection. J Virol. 1998;72:5728–5734. doi: 10.1128/jvi.72.7.5728-5734.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi K, Wesselingh S L, Griffin D E, McArthur J C, Johnson R T, Glass J D. Localization of HIV-1 in human brain using polymerase chain reaction/in situ hybridization and immunocytochemistry. Ann Neurol. 1996;39:705–711. doi: 10.1002/ana.410390606. [DOI] [PubMed] [Google Scholar]
- 54.Trono D. HIV accessory proteins—leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 55.Trono D, Baltimore D. A human cell factor is essential for HIV-1 Rev action. EMBO J. 1990;9:4155–4160. doi: 10.1002/j.1460-2075.1990.tb07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Schwedler U, Song J, Aiken C, Trono D. vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watkins B A, Dorn H H, Kelly W B, Armstrong R C, Potts B J, Michaels F, Kufta C V, Dubois-Dalcq M. Specific tropism of HIV-1 for microglial cells in primary human brain cultures. Science. 1990;249:549–553. doi: 10.1126/science.2200125. [DOI] [PubMed] [Google Scholar]
- 58.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 59.Wiley C A, Schrier R D, Nelson J A, Lampert P W, Oldstone M B. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc Natl Acad Sci USA. 1986;83:7089–7093. doi: 10.1073/pnas.83.18.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu Y R, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zufferey R, Donello J E, Trono D, Hope T J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]