Abstract
An investigation of ×19 soil samples collected under the auspices of the Australian citizen science initiative, Soils for Science, returned ×559 chemically dereplicated microbial isolates, of which ×54 exhibited noteworthy anthelmintic activity against either the heartworm Dirofilaria immitis microfilaria and/or the gastrointestinal parasite Haemonchus contortus L1–L3 larvae. Chemical (GNPS and UPLC-DAD) and cultivation (MATRIX) profiling prompted a detailed chemical investigation of Streptomyces sp. S4S-00196A10, which yielded new anthelmintic polyketide goondapyrones A–J (1–10), together with the known actinopyrones A (11) and C (12). Structures for 1–12 were assigned on the basis of detailed spectroscopic and chemical analysis, with preliminary structure activity relationship analysis revealing selected γ-pyrones >50-fold and >13-fold more potent than isomeric α-pyrones against D. immitis mf motility (e.g., EC50 0.05 μM for 1; EC50 2.7 μM for 5) and H. contortus L1–L3 larvae development (e.g., EC50 0.58 μM for 1; EC50 8.2 μM for 5), respectively.
Keywords: anthelmintic, citizen science, Dirofilaria immitis, Haemonchus contortus, microbial natural product, polyketide, pyrones, Soils for Science, Streptomyces
1. Introduction
As part of the ongoing investigations at the University of Queensland—Institute of Molecular Bioscience into the bioactive natural products of soil-derived microbes, we recently launched a first of its kind Australian citizen science initiative, Soils for Science (S4S) [1]. S4S engages with and enables the public to collect and share soil samples from urban backyards, rural properties, and native ecosystems across the nation as a valuable resource from which pure microbial (bacterial and fungal) isolates are recovered. Natural products of these isolates hold the prospect of new scientific knowledge that can inspire the development of new and improved pharmaceuticals and agrochemicals.
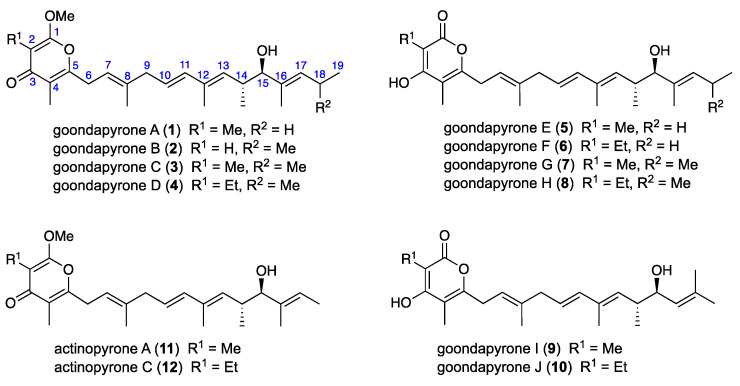
To illustrate this potential, this report describes a proof-of-concept search for microbial natural products capable of inspiring new treatments for animals infected by life-threatening nematodes, including the heartworm Dirofilaria immitis that afflicts companion animals (e.g., dogs and cats,) and gastrointestinal parasite Haemonchus contortus that infects ruminant livestock (e.g., sheep). In this study, an assessment of ×19 pasture soils collected from Goondicum Station, a cattle property situated in an extinct volcanic crater near the headwaters of the Burnett River, Queensland, Australia, returned a library of microbial isolates which were dereplicated and prioritized based on chemical profiling (UPLC-DAD and UPLC-QTOF-GNPS) and anthelmintic potential against either D. immitis and/or H. contortus. This report provides an account of a detailed chemical analysis of just one of these isolates, Streptomyces sp. S4S-00196A10, that yielded the new anthelmintic polyketide goondapyrones A–J (1–10), along with the known actinopyrones A (11) and C (12) (Figure 1).
Figure 1.
Streptomyces sp. S4S-00196A10 metabolites 1–12.
2. Results
M1 and ISP2 agar mother plates prepared from each of ×19 Goondicum Station soil samples were incubated for 14 d at 27 °C, after which manual colony picking returned ×704 pure microbial (bacterial and fungal) isolates. Each isolate was cultured on a fresh agar plate (ISP2 or M1, based on the mother plate media) to confirm purity and provide cells for cryopreservation, with residue mycelia/agar biomass per plate used to generate analytical scale EtOAc extracts. These extracts were dried in vacuo, resuspended in DMSO and dereplicated by chemical analysis (GNPS and UPLC-DAD) to ×559 unique extracts/isolates. Subsequent anthelmintic screening prioritized ×54 (10%) of these for their ability to inhibit the motility of D. immitis microfilaria (mf) and/or the development of H. contortus L1–L3 larvae (>75% inhibition at 25 μg/mL).
With a global natural product social (GNPS) [2] molecular network analysis of the prioritized extracts revealing a wealth of chemical diversity (Figure S3), attention was focused on those extracts/isolates that were the sole source/producer of a GNPS molecular family (i.e., cluster of MS/MS-related natural products), reasoning that targeting both anthelmintic activity and molecular rarity would enhance discovery prospects. Following this strategy, attention was drawn to Streptomyces sp. S4S-00196A10 as the sole producer of a GNPS molecular family that in a UPLC-DAD analysis aligned with major and minor metabolites exhibiting different UV–vis (DAD) chromophores (Figure S4). In an effort to determine how rare this class of chemistry may be, the GNPS molecular family was matched against a larger in-house library of ×1957 microbial extracts, derived from single agar plate cultivations of additional Goondicum (56%) and S4S (25%) soils, Australian sheep pasture plants (8%) and venomous snakes (1.8%), scorpions (1.5%), cone snails (2.5%), spiders (0.2%), centipedes (2%) and wasps (3%). This search revealed only a single replicate of S4S-00196A10, the Goondicum soil-derived Streptomyces sp. S4S-00246A11, an EtOAc extract of which also displayed activity against D. immitis mf.
Prior to scaled up cultivation, with a view to accessing both major and minor metabolites, we employed miniaturized 24-well plate cultivation profiling (MATRIX) [3,4] using ×11 different media compositions (Table S1) under solid phase as well as static and shaken conditions, supported by quantitative UPLC-DAD analysis (Figure S4). This process revealed solid phase glucose yeast extract agar (GYA) as optimal for the major co-metabolites, and a shaken broth comprised of glucose, malt and yeast extract, with peptone and soluble starch agar (D400) as optimal for the minor metabolites (as detected in ISP2).
An EtOAc extract prepared from a ×100 plate GYA 14 d, 30 °C cultivation was subjected to sequential solvent trituration and reversed phase fractionation to yield the new (γ-pyrone) goondapyrones A–D (1–4) and known (γ-pyrone) actinopyrones A (11) and C (12). By contrast, comparable fractionation of an EtOAc extract prepared from a D400 shaken broth cultivation (2 × 500 mL) yielded (α-pyrone) goondapyrones E–J (5–10). Structures were assigned to 1–12 by detailed spectroscopic analysis as summarized below, with the difference in UV–vis (DAD) spectra for major versus minor metabolites attributed to γ-pyrone versus α-pyrone chromophores, respectively.
HRESIMS measurements established molecular formula for 11 (C25H36O4, m/z M+Na ∆mmu +0.9) and 12 (C26H38O4, m/z M+Na ∆mmu −0.9), with the latter consistent with a higher homologue (+CH2) of the former. A literature comparison confirmed excellent concordance between the 1H NMR (CDCl3) data for 11 with those reported in 2006 [4] for actinopyrone A (Table S12, Figure S75). Likewise, the [α]D for 11 (+13.6 in CH2Cl2) compared well with that reported for both natural and synthetic 14R,15R actinopyrone A [4], although it is important to note that the [α]D in MeOH for 11 was of opposite sign (−14.6), highlighting the importance of making [α]D comparisons (experimental vs literature) in the same solvent. Comparison of the 1D NMR (DMSO-d6) data for 12 (Table S14, Figures S78 and S79) with those for 11 (Table S13, Figures S76 and S77) allowed the key difference to be attributed to replacement of the 2-Me in 11 (δH 1.68, s; δC 6.8) with a 2-Et in 12 (δH 2.23, q, J 7.4 Hz, CH2; 0.91, t, J 7.4 Hz, CH3; δC 14.8, CH2; 12.9, CH3), thereby identifying 12 as actinopyrone C, first reported in 1986 as a co-metabolite with actinopyrone A from the Japanese soil-derived Streptomyces pactum S12538 [5,6]. Co-metabolites 1–10 co-clustered in the GNPS with 11–12 suggested structural (biosynthetic) similarities, as evident in the structure assignments outlined below.
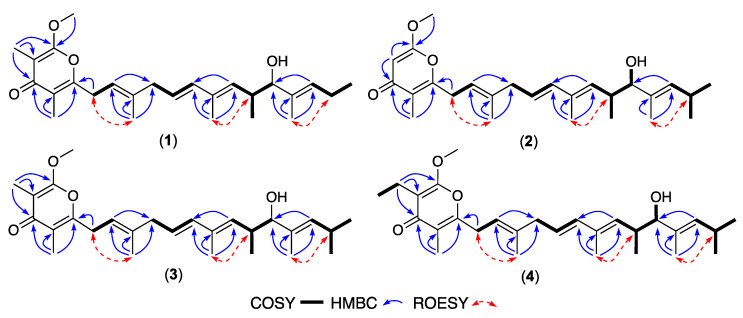
HRESIMS measurements established isomeric molecular formula for 1 (C26H38O4, m/z M+Na ∆mmu +1.1) and 2 (C26H38O4, m/z M+Na ∆mmu +1.1), consistent with lower homologues (-CH2) of 11. Comparison of the NMR (DMSO-d6) data for 1 (Table 1, Table 2 and Table S2, Figures S5–S10) with those for 11 revealed a high degree of concordance with the principle differences attributed to elongation of the polyketide chain (δH 0.89, t, J 7.5 Hz, H3-19; δC 14.1, C-19). Likewise, comparison of the NMR (DMSO-d6) data for 2 (Table 1, Table 2 and Table S3, Figures S12–S17) with those for 1 allowed the principle differences to be attributed to the absence of the 2-Me and inclusion of an 18-Me in 2 (δH 5.52, s, H-2; 0.87, d, J 6.1 Hz, H3-18; 0.89, d, J 5.9 Hz, H3-19; δC 87.7, C-2; 22.8, 18-CH3). Diagnostic 2D NMR (DMSO-d6) correlations supported the proposed planar structures for 1–2 (Figure 2), with a large J10,11 together with ROESY correlations between H2-6 and 8-CH3, H-14 and 12-CH3, and H2-18 and 16-CH3, allowing assignment of an all-E configuration. These observations, together with 1–2 and 11–12 sharing comparable [α]D (MeOH) and ECD spectra (Figure S84), along with biosynthetic considerations, allowed the structures for goondapyrones A (1) and B (2) to be assigned as shown.
Table 1.
1H NMR (DMSO-d6) data for goondapyrones A–D (1–4).
| Position | (1) δH, Mult (J in Hz) |
(2) δH, Mult (J in Hz) |
(3) δH, Mult (J in Hz) |
(4) δH, Mult (J in Hz) |
|---|---|---|---|---|
| 2 | - | 5.52, s | - | - |
| 6 | 3.35 A | 3.30, d (7.3) | 3.35, d (7.1) | 3.34, d (7.3) |
| 7 | 5.29, t (7.2) | 5.22, t (7.3) | 5.28, t (7.0) | 5.28, t (7.3) |
| 9 | 2.77, d (6.9) | 2.76, d (7.5) | 2.77, d (6.9) | 2.76, d (7.0) |
| 10 | 5.46, dt (15.5, 6.9) | 5.44, dt (15.6, 7.5) | 5.46, dt (15.5, 6.9) | 5.46, dt (15.5, 7.0) |
| 11 | 6.02, d (15.5) | 6.02, d (15.6) | 6.01, d (15.5) | 6.01, d (15.5) |
| 13 | 5.27, d (9.4) | 5.26, d (9.4) | 5.25, d (9.3) | 5.25, d (9.5) |
| 14 | 2.53, m | 2.53, m | 2.54, m | 2.53, m |
| 15 | 3.59, d (6.9) | 3.56, dd (7.2, 4.1) | 3.56, d (6.9) | 3.56, dd (6.8, 4.0) |
| 16 | - | - | - | - |
| 17 | 5.25, t (7.3) | 5.07, d (9.1) | 5.07, d (9.1) | 5.07, d (9.1) |
| 18 | 1.96, m | 2.47, m | 2.46, m | 2.46, m |
| 19 | 0.89, t (7.5) | 0.89, d (5.9) | 0.88, d (6.2) | 0.88, d (6.3) |
| 2-CH3 | 1.68, s | - | 1.68, s | - |
| 2-CH2CH3 | - | - | - | 2.23, q (7.5) |
| 2-CH2CH3 | - | - | - | 0.91, t (7.5) |
| 4-CH3 | 1.83, s | 1.80, s | 1.82, s | 1.82, s |
| 8-CH3 | 1.70, s | 1.66, s | 1.69, s | 1.69, s |
| 12-CH3 | 1.65, s | 1.64, s | 1.64, s | 1.64, s |
| 14-CH3 | 0.77, d (6.8) | 0.78, d (6.6) | 0.77, d (6.9) | 0.77, d (6.7) |
| 16-CH3 | 1.50, s | 1.51, s | 1.51, s | 1.51, s |
| 18-CH3 | - | 0.87, d (6.1) | 0.87, d (6.2) | 0.87, d (6.3) |
| 1-OCH3 | 3.91, s | 3.83, s | 3.91, s | 3.90, s |
| 15-OH | ND | 4.49, d (4.1) | ND | 4.50, d (4.0) |
A—obscured by solvent, ND—not detected.
Table 2.
13C NMR (DMSO-d6) data for goondapyrones A–H (1–8).
| Position | (1) δC, Type A |
(2) δC, Type A |
(3) δC, Type A |
(4) δC, Type A |
(5) δC, Type A |
(6) δC, Type A |
(7) δC, Type A |
(8) δC, Type A |
|---|---|---|---|---|---|---|---|---|
| 1 | 161.8, C | 167.0, C | 161.8, C | 161.8, C | 164.4, C | 164.0, C | 164.6, C | 164.0, C |
| 2 | 97.5, C | 87.7, CH | 97.5, C | 103.7, C | 97.3, C | 103.5, C | 97.1, C | 103.5, C |
| 3 | 179.4, C | 180.0, C | 179.4, C | 178.8, C | 164.7, C | 164.2, C | 165.2, C | 164.2, C |
| 4 | 116.8, C | 117.4, C | 116.8, C | 117.2, C | 106.4, C | 106.4, C | 106.6, C | 106.4, C |
| 5 | 157.2, C | 158.9, C | 157.2, C | 157.3, C | 156.7, C | 156.9, C | 156.6, C | 156.9, C |
| 6 | 29.4, CH2 | 29.5, CH2 | 29.4, CH2 | 29.4, CH2 | 29.7, CH2 | 29.7, CH2 | 29.7, CH2 | 29.7, CH2 |
| 7 | 117.8, CH | 117.9, CH | 117.8, CH | 117.8, CH | 118.6, CH | 118.6, CH | 118.7, CH | 118.6, CH |
| 8 | 137.7, C | 137.7, C | 137.7, C | 137.7, C | 136.9, C | 136.9, C | 136.9, C | 136.9, C |
| 9 | 42.1, CH2 | 42.1, CH2 | 42.1, CH2 | 42.1, CH2 | 42.2, CH2 | 42.2, CH2 | 42.3, CH2 | 42.2, CH2 |
| 10 | 124.0, CH | 123.9, CH | 123.9, CH | 124.0, CH | 124.0, CH | 124.0, CH | 124.0, CH | 124.0, CH |
| 11 | 136.6, CH | 136.6, CH | 136.6, CH | 136.6, CH | 136.6, CH | 136.6, CH | 136.6, CH | 136.6, CH |
| 12 | 132.0, C | 132.0, C | 132.0, C | 132.0, C | 132.1, C | 132.1, C | 132.1, C | 132.0, C |
| 13 | 135.4, CH | 135.3, CH | 135.3, CH | 135.3, CH | 135.4, CH | 135.4, CH | 135.4, CH | 135.3, CH |
| 14 | 36.0, CH | 36.1, CH | 36.1, CH | 36.1, CH | 36.1, CH | 36.1, CH | 36.2, CH | 36.1, CH |
| 15 | 80.4, CH | 80.2, CH | 80.2, CH | 80.2, CH | 80.4, CH | 80.4, CH | 80.3, CH | 80.2, CH |
| 16 | 136.2, C | 134.4, C | 134.4, C | 134.4, C | 136.2, C | 136.2, C | 134.4, C | 134.4, C |
| 17 | 127.1, CH | 133.0, CH | 133.0, CH | 133.0, CH | 127.1, CH | 127.1, CH | 133.1, CH | 133.0, CH |
| 18 | 20.2, CH2 | 26.2, CH | 26.1, CH | 26.2, CH | 20.2, CH2 | 20.2, CH | 26.2, CH | 26.1, CH |
| 19 | 14.1, CH3 | 23.0, CH3 | 23.0, CH3 | 23.0, CH3 | 14.1, CH3 | 14.1, CH3 | 23.0, CH3 | 23.0, CH3 |
| 2-CH3 | 6.8, CH3 | - | 6.8, CH3 | - | 9.2, CH3 | - | 9.3, CH3 | - |
| 2-CH2CH3 | - | - | - | 14.8, CH2 | - | 16.5, CH2 | - | 16.5, CH2 |
| 2-CH2CH3 | - | - | - | 12.9, CH3 | - | 12.8, CH3 | - | 12.8, CH3 |
| 4-CH3 | 9.5, CH3 | 9.1, CH3 | 9.5, CH3 | 9.5, CH3 | 9.8, CH3 | 9.8, CH3 | 9.9, CH3 | 9.8, CH3 |
| 8-CH3 | 16.3, CH3 | 16.3, CH3 | 16.3, CH3 | 16.3, CH3 | 16.3, CH3 | 16.3, CH3 | 16.4, CH3 | 16.3, CH3 |
| 12-CH3 | 12.7, CH3 | 12.7, CH3 | 12.7, CH3 | 12.7, CH3 | 12.8, CH3 | 12.8, CH3 | 12.8, CH3 | 12.8, CH3 |
| 14-CH3 | 17.9, CH3 | 17.9, CH3 | 17.9, CH3 | 17.9, CH3 | 17.9, CH3 | 17.9, CH3 | 17.9, CH3 | 17.9, CH3 |
| 16-CH3 | 11.4, CH3 | 11.6, CH3 | 11.6, CH3 | 11.6, CH3 | 11.4, CH3 | 11.4, CH3 | 11.6, CH3 | 11.6, CH3 |
| 18-CH3 | - | 22.8, CH3 | 22.8, CH3 | 22.8, CH3 | - | - | 22.9, CH3 | 22.8, CH3 |
| 1-OCH3 | 55.6, CH3 | 56.4, CH3 | 55.6, CH3 | 55.7, CH3 | - | - | - | - |
A—Resonance type assigned on the basis of 1D and 2D NMR analysis.
Figure 2.
Selected 2D NMR (DMSO-d6) correlations for 1–4.
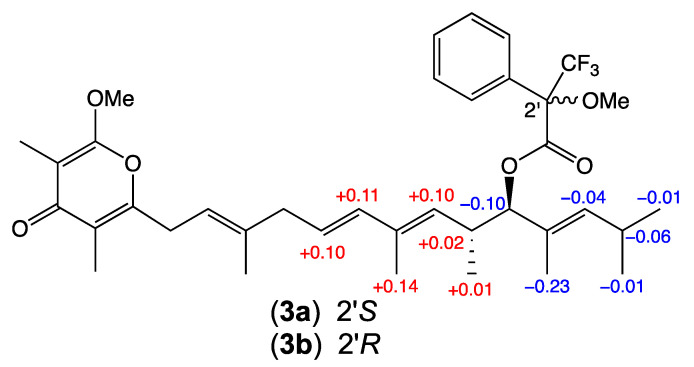
HRESIMS measurements established molecular formula for 3 (C27H40O4, m/z M+Na ∆mmu +0.5) and 4 (C28H42O4, m/z M+Na ∆mmu −0.4), with 3 consistent with a higher homologue (+CH2) of 2, and 4 consistent with a higher homologue (+CH2) of 3. Comparison of the NMR (DMSO-d6) data for 3 (Table 1, Table 2 and Table S4, Figures S19–S24) with those for 2 revealed a high degree of concordance with the principle differences attributed to inclusion of a 2-Me in 3 (δH 1.68, s, 2-CH3; δC 6.8, 2-CH3). Likewise, comparison of the NMR (DMSO-d6) data for 4 (Table 1, Table 2 and Table S5, Figures S26–S31) with those for 3 allowed the principle differences to be attributed to inclusion of a 2-Et in 4 (δH 0.91, t, J 7.5 Hz, CH3; 2.23, q, J 7.5 Hz, CH2; δC 12.9, CH3; 14.8, CH2). A large value for J10,11 together with diagnostic 2D NMR (DMSO-d6) correlations supported the proposed planar structures for 3–4 inclusive of an all-E configuration (Figure 2). In addition to 1–4 sharing comparable [α]D (MeOH) and ECD spectra (Figure S84), analysis of 1H NMR data for the Mosher esters 3a (S-MTPA) and 3b (R-MTPA) independently confirmed a 15R absolute configuration for 3 (Figure 3), consistent with data reported prior [4] for 11 and allowing the structures for goondapyrones C (3) and D (4) to be assigned as shown.
Figure 3.
Mosher analysis of 3. Δδ (δS − δR) data for the S-MTPA (3a) and R-MTPA (3b).
Whereas goondapyrones 1–4 and actinopyrones 11–12 produced under GYA solid phase cultivation share a common γ-pyrone chromophore (as detailed above), the metabolites 5–10 produced under D400 shaken broth cultivation co-clustered (GNPS molecular network) with 1–4 and 11–12 but featured a different chromophore. This difference was further evident in the 1D NMR (DMSO-d6) data where the 1-OMe moiety common to all of 1–4 and 11–12 was absent in 5–10. Importantly, where the O-methylated γ-pyrone moieties in 1–4 and 11–12 cannot undergo tautomerisation, as demonstrated below, the pyrones in 5–10 can tautomerize and are expected to exist as the more thermodynamically stable α-pyrone tautomer, with a corresponding change in UV–vis (DAD) spectra.
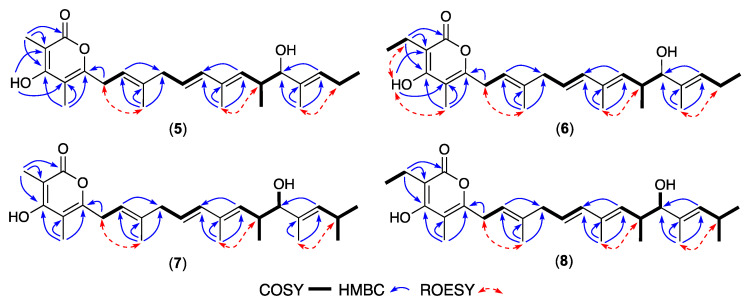
HRESIMS measurements established molecular formula for 5 (C25H36O4, m/z M+Na ∆mmu −1.7) and 6 (C26H38O4, m/z M+Na ∆mmu +1.5), with 5 consistent with a lower homologue (-CH2) of 1 and 6 consistent with a higher homologue (+CH2) of 5. Comparison of the NMR (DMSO-d6) data for 5 (Table 2, Table 3 and Table S6, Figures S33–S38) with 1 allowed the principle difference to be attributed to replacement of the 1-OMe in 1 with a chelated 3–OH in 5 (δH 10.48, s), with the alternate tautomeric regiochemistry evidenced by HMBC correlations from 3-OH to C-2 (δC 97.3) and C-4 (δC 106.4), and from 2-CH3 to C-1 (δC 164.4), C-2 and C-3 (δC 164.7) (Figure 4). Likewise, a comparison of the NMR (DMSO-d6) data for 6 (Table 2, Table 3 and Table S7, Figures S40–S45) with 5 attributed differences to the replacement of the 2-Me in 5 with a 2-Et in 6 (δH 0.93, t, J 7.4 Hz, CH3; 2.35, q, J 7.4 Hz, CH2; δC 12.8, CH3; 16.5, CH2). A large J10,11 value and diagnostic 2D NMR ROESY correlations supported the proposed planar structures for 5–6, inclusive of an all-E configuration (Figure 4). The above, together with comparable [α]D (MeOH) and ECD spectra (Figure S84) and biosynthetic considerations, permitted assignment of structures to goondapyrones E (5) and F (6).
Table 3.
1H NMR (DMSO-d6) data for goondapyrones E–H (5–8).
| Position | (5) δH, Mult (J in Hz) |
(6) δH, Mult (J in Hz) |
(7) δH, Mult (J in Hz) |
(8) δH, Mult (J in Hz) |
|---|---|---|---|---|
| 6 | 3.21, d (6.9) | 3.21, d (6.9) | 3.20, d (7.0) | 3.20, d (7.0) |
| 7 | 5.19, t (6.9) | 5.19, t (6.8) | 5.19, t (7.0) | 5.19, t (7.0) |
| 9 | 2.75, d (6.9) | 2.75, d (6.9) | 2.75, d (6.9) | 2.75, d (6.9) |
| 10 | 5.44, dt (15.5, 6.9) | 5.44, dt (15.5, 7.0) | 5.44, dt (15.6, 6.9) | 5.44, dt (15.6, 6.9) |
| 11 | 6.02, d (15.5) | 6.02, d (15.5) | 6.03, d (15.6) | 6.02, d (15.6) |
| 13 | 5.28, d (9.3) | 5.28, d (9.3) | 5.28, d (9.4) | 5.27, d (9.3) |
| 14 | 2.54, m | 2.53, m | 2.54, m | 2.53, m |
| 15 | 3.58, d (7.1) | 3.58, d (7.2) | 3.57, br d (7.1) | 3.56, dd (7.1, 4.3) |
| 17 | 5.25, t (7.1) | 5.25, t (7.2) | 5.08, d (9.1) | 5.07, t (9.0) |
| 18 | 1.96, m | 1.96, m | 2.48, m | 2.47, m |
| 19 | 0.89, t (7.3) | 0.89, t (7.5) | 0.89, d (5.3) | 0.89, d (5.5) |
| 2-CH3 | 1.82, s | - | 1.81, s | - |
| 2-CH2CH3 | - | 2.35, q (7.4) | - | 2.35, q (7.4) |
| 2-CH2CH3 | - | 0.93, t (7.4) | - | 0.93, t (7.4) |
| 4-CH3 | 1.88, s | 1.88, s | 1.87, s | 1.88, s |
| 8-CH3 | 1.66, s | 1.66, s | 1.66, s | 1.66, s |
| 12-CH3 | 1.65, s | 1.65, s | 1.65, s | 1.64, s |
| 14-CH3 | 0.77, d (6.8) | 0.77, d (6.8) | 0.78, d (6.8) | 0.77, d (6.9) |
| 16-CH3 | 1.50, s | 1.50, s | 1.52, s | 1.51, s |
| 18-CH3 | - | - | 0.88, d (5.3) | 0.88, d (5.5) |
| 3-OH | 10.48, s | 10.44, s | ND | ND |
| 15-OH | ND | ND | 4.48, br s | 4.48, d (4.3) |
ND—not detected.
Figure 4.
Selected 2D NMR (DMSO-d6) correlations for 5–8.
HRESIMS measurements established molecular formula for 7 (C26H38O4, m/z M+Na ∆mmu +1.4) and 8 (C27H40O4, m/z M+Na ∆mmu +1.2), consistent with higher homologues (+CH2) of 5 and 6, respectively. Comparison of the NMR (DMSO-d6) data for 7 and 8 (Table 2, Table 3, Tables S8 and S9, Figure 4, Figures S47–S52 and S54–S59) with 5 and 6 attributed the principle differences to the inclusion of an 18-Me moiety in both 7 and 8 (absent in both 5 and 6) while retaining a common all-E configuration. These observations together with comparable [α]D (MeOH), ECD spectra (Figure S84) and biosynthetic considerations allowed the structures for goondapyrones G (7) and H (8) to be assigned as shown.
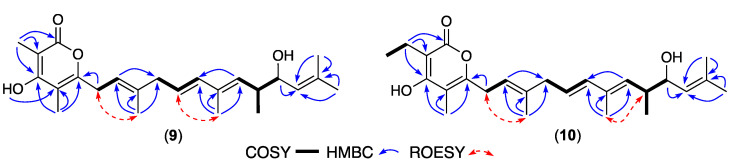
HRESIMS measurements established molecular formula for 9 (C24H34O4, m/z M+Na ∆mmu +0.6) and 10 (C25H36O4, m/z M+Na ∆mmu +1.4), consistent with lower homologues (-CH2) of 5 and 6, respectively. Comparison of the NMR (DMSO-d6) data for 9 and 10 (Table 4, Tables S10 and S11, Figure 5, Figures S61–S66 and S68–S73) with those for 5 and 6 revealed a common C-1 to C-15 scaffold inclusive of an a-pyrone moiety, alkyl branching and all-E configuration, with the principle differences attributed to the loss of the 16-Me and C-19 and incorporation of a 17-Me. These observations, together with comparable [α]D (MeOH), ECD spectra (Figure S84) and biosynthetic considerations, allowed the structures for goondapyrones I (9) and J (10) to be assigned as shown.
Table 4.
1H and 13C NMR (DMSO-d6) data for goondapyrones I–J (9–10).
| Position | (9) δC, Type A |
δH, Mult (J in Hz) |
(10) δC, Type A |
δH, Mult (J in Hz) |
|---|---|---|---|---|
| 1 | 164.4, C | - | 164.0, C | - |
| 2 | 97.3, C | - | 103.5, C | - |
| 3 | 164.7, C | - | 164.2, C | - |
| 4 | 106.4, C | - | 106.4, C | - |
| 5 | 156.6, C | - | 156.9, C | - |
| 6 | 29.6, CH2 | 3.21, d (7.0) | 29.7, CH2 | 3.21, d (7.0) |
| 7 | 118.6, CH | 5.19, t (7.0) | 118.6, CH | 5.19, t (7.0) |
| 8 | 136.9, C | - | 136.9, C | - |
| 9 | 42.2, CH2 | 2.75, d (6.9) | 42.2, CH2 | 2.75, d (6.9) |
| 10 | 124.0, CH | 5.45, dt (15.6, 6.9) | 124.0, CH | 5.45, dt (15.6, 6.9) |
| 11 | 136.6, CH | 6.02, d (15.6) | 136.6, CH | 6.02, d (15.6) |
| 12 | 132.1, C | - | 132.1, C | - |
| 13 | 135.4, CH | 5.29, d (9.3) | 135.4, CH | 5.29, d (9.3) |
| 14 | 36.1, CH | 2.53, m | 36.1, CH | 2.54, m |
| 15 | 80.5, CH | 3.59, br d (6.6) | 80.5, CH | 3.59, br s |
| 16 | 119.4, CH | 5.33, q (6.6) | 119.4, CH | 5.32, q (6.6) |
| 17 | 137.7, C | - | 137.8, C | - |
| 18 | 11.3, CH3 | 1.50, s | 11.3, CH3 | 1.50, s |
| 2-CH3 | 9.2, CH3 | 1.82, s | - | - |
| 2-CH2CH3 | - | - | 16.5, CH2 | - |
| 2-CH2CH3 | - | - | 12.8, CH3 | - |
| 4-CH3 | 9.8, CH3 | 1.88, s | 9.8, CH3 | 1.88, s |
| 8-CH3 | 16.3, CH3 | 1.66, s | 16.3, CH3 | 1.66, s |
| 12-CH3 | 12.7, CH3 | 1.65, s | 12.7, CH3 | 1.65, s |
| 14-CH3 | 17.9, CH3 | 0.76, d (6.8) | 17.9, CH3 | 0.75, d (6.7) |
| 17-CH3 | 12.8, CH3 | 1.54, d (6.7) | 12.8, CH3 | 1.54, d (6.6) |
| 3-OH | - | 10.46, s | - | 10.44, br s |
| 15-OH | - | 4.46, d (3.6) | - | 4.47, d (3.9) |
A—Resonance type assigned on the basis of 1D and 2D NMR analysis.
Figure 5.
Selected 2D NMR (DMSO-d6) correlations for 9–10.
3. Discussion
The polyketide pyrones 1–12 did not exhibit significant antibacterial activity against either the Gram-positive Staphylococcus aureus ATCC 25923 or Gram-negative Escherichia coli ATCC 11775 or antifungal activity against Candida albicans ATCC 10231 or cytotoxic activity against human colorectal (SW620) or lung (NCI-H460) carcinoma cells (EC50 > 30 μM) (Figures S80–S82). On the other hand, goondapyrones A–B (1–2) and actinopyrones A and C (11 and 12) were exceptionally potent inhibitors of the motility of D. immitis microfilaria (EC50 0.01 to 0.07 μM), but exhibited marginal activity against the L4 larvae, and along with goondapyrone C (3) also significantly inhibited the development of H. contortus L1–L3 larvae (EC50 0.15 to 0.66 μM) (Table 5). D. immitis, commonly known as heartworm, is a significant pathogen affecting animals such as dogs and cats [7]. It poses serious health risks, often leading to heart failure if untreated. Similarly, H. contortus is a major gastrointestinal parasite that infects ruminant livestock, including sheep and goats, leading to severe economic losses in the agricultural sector due to reduced productivity and increased mortality [8]. The discovery of natural products that exhibit potent anthelmintic activity against these parasites is crucial due to increasing resistance to existing anthelmintics such as ivermectins [9]. There is an urgent need to identify new bioactive compounds for managing and controlling these parasitic infections effectively.
Table 5.
Summary of anthelmintic properties of 1–13 (plus positive controls)—EC50 (μM).
| Compounds |
D. immitis mf Motility |
D. immitis L4 Motility |
H. contortus L1–L3 Larval Development |
|---|---|---|---|
| 1 | 0.05 | 34 | 0.58 |
| 2 | 0.07 | 31 | 0.63 |
| 3 | 22.3 | 48 | 0.66 |
| 4 | 3.2 | 25 | 15.3 |
| 5 | 2.7 | >60 | 8.2 |
| 6 | 16.7 | >60 | >60 |
| 7 | 3.2 | >60 | 11.9 |
| 8 | 3.2 | >60 | 31.9 |
| 9 | 6.5 | >60 | 17.4 |
| 10 | 1.4 | 45 | 23.8 |
| 11 | 0.01 | 27 | 0.15 |
| 12 | 0.02 | 34 | 0.65 |
| 13 | 1.77 | >60 | 0.72 |
| gramicidin | 0.10 | - | - |
| monensin | - | 0.4 | - |
| ivermectin | - | - | 0.0005 |
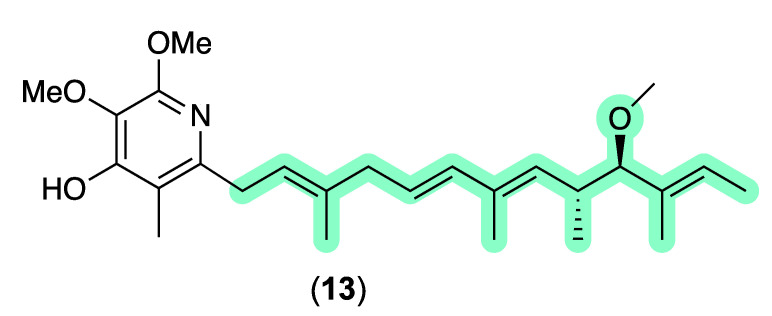
An anthelmintic structure activity relationship (SAR) assessment of 1–12 revealed that γ-pyrones 1–2 and 11–12 are up to 1000-fold more active than α-pyrones 5–10, and up to 10-fold more effective against D. immitis mf than H. contortus L1–L3 larvae. As an aside, γ-pyrones 1–2 and 11–12 share common structural features with the well-known piericidin family of Streptomyces polyketides, with an authentic sample of piericidin B (13) active in our assays against D. immitis mf and H. contortus L1–L3 larvae (Figure 6, Table 5). Of note, piericidins can be detected across our microbe libraries with >4-fold higher frequency than the actinopyrones/goondapyrones, and typically also register as hits in our anthelmintic assays. Notwithstanding, as piericidins are known neurotoxins that inhibit NADH-ubiquinone oxidoreductase (Complex I) in the mitochondrial electron transport, anthelmintic hit extracts containing piericidins are typically deprioritized against further investigation. By contrast, pyrones 1–12 do not exhibit mammalian cell cytotoxicity (or antibiotic activity), with goondapyrone A (1) and actinopyrone A (11) being 35- and 170-fold more active against D. immitis mf, respectively, than authentic piericidin B (13).
Figure 6.
Piericidin B (13). Colour highlights—similarities with actinopyrone A (11) (green).
4. Materials and Methods
For general experimental details, see Supplementary Materials.
4.1. Collection of Soils and Isolation of Microbes
Soil samples collected under the auspices of the Australian Soils for Science (S4S) citizen science initiative [1] from Goondicum Station situated in an extinct volcanic crater near the headwaters of the Burnett River, Queensland, Australia were used to inoculate ISP2 and M1 agar mother plates. After incubation at 27 °C for 14 days, manual colony picking yielded isolates were cultivated on fresh ISP2 or M1 agar plates (media choice based on that of the source mother plate) before being (i) photographed and the image uploaded to the S4S Gallery [1], (ii) cryopreserved at −80 °C, and (iii) an EtOAc extract was prepared, dried, resuspended in DMSO and archived at −20 °C.
4.2. Chemical Profiling (UPLC-DAD and UPLC-QTOF-GNPS)
EtOAc extracts prepared from agar plate cultivations of S4S soil-derived microbes were subjected to both UPLC-DAD and UPLC-QTOF-GNPS chemical profiling. UPLC-DAD chemical profiling involved injection of an aliquot of each analyte (2 μL at ~1 mg/mL in MeOH) through an Agilent 1290 infinity UPLC system (Zorbax SB-C8 RRHD 1.8 μm, 2.1 × 50 mm column, eluting at 0.417 mL/min, 2.50 min gradient elution from 90% H2O/MeCN to 100% MeCN with a constant 0.01% TFA modifier) equipped with a diode array multiple wavelength detector (DAD). UPLC-QTOF-GNPS chemical profiling involved injection of an aliquot of each analyte (1 μL at ~100 μg/mL in MeOH) through an Agilent 1290 Infinity II UPLC (Zorbax SB-C8 RRHD 1.8 μm, 2.1 × 50 mm column, eluting with 0.417 mL/min, 2.50 min gradient elution from 90% H2O/MeCN to 100% MeCN with a constant 0.1% formic acid/MeCN modifier) equipped with Agilent 6545 QTOF detector. UPLC-QTOF-(+) MS/MS data for GNPS analysis were acquired for all samples at a collision energy of 35 eV, transferred to the GNPS server [10], and resulting spectral network visualized using Cytoscape version 3.8.0 [11] (see Supplementary Materials for details).
4.3. Taxonomic Identification of S4S-00196A10 (and S4S-00246A11)
Genomic DNA was extracted from an ISP2 agar plate cultivation of target bacteria using the DNeasy Blood and Tissue Kit (Qiagen) as per the manufacturer’s protocol (see Supplementary Materials for details). BLAST analysis (NCBI database) showed that the amplified 16S rRNA sequence for S4S-00196A10 (accession number MG597592) has 98.9% identity with Streptomyces parvisporogenes strain MJM12043 (Figures S1a and S2). A comparable analysis of S4S-00246A11 revealed it to be a replicate of S4S-00196A10 (Figures S1b and S2).
4.4. Cultivation Profiling (MATRIX) of Streptomyces sp. S4S-00196A10
S4S-00196A10 was subjected to cultivation profiling in a 24-well plate (MATRIX) [3] system using 11 different media compositions (Table S1) under solid agar (1.5 g) as well as static (1.5 mL) and shaken (1.5 mL, 190 rpm) broth formats at 27 °C for 10 days. An additional set of control incubations were prepared from 11 different media solid-phase cultivations without inoculation. Individual MATRIX wells (Figure S4) were extracted in situ with EtOAc (2 mL) with the organic phases dried at 40 °C under a stream of N2, re-suspended in MeOH (100 mL). A portion of the analyte was then subjected to GNPS chemical profiling (as described above), while a second portion was treated with an internal calibrant (1-decyloxy-2,4-dinitrobenzene, 50 μg/mL) and subjected to UPLC-DAD-MS chemical profiling (as described above) (Figure S4).
4.5. Scale-Up Cultivation and Fractionation of Streptomyces sp. S4S-00196A10
Cultivation in GY agar media: A seed culture of S4S-00196A10 was prepared by inoculating D400 broth medium (70 mL) and shaking at 190 rpm at 30 °C for 5 days. Aliquots of the seed culture (100 μL) were used to inoculate glucose yeast (GY) agar plates (×100), and after incubation at 30 °C for 14 days, the combined agar was extracted with EtOAc (2 × 500 mL) and the organic phase concentrated in vacuo to yield an extract (585.4 mg). This extract was then subjected to sequential trituration to afford, after drying under nitrogen at 40 °C, n-hexane (120.5 mg), CH2Cl2 (298.6 mg) and MeOH (166.3 mg) solubles. The combined CH2Cl2 and MeOH solubles were fractionated by preparative reverse-phase HPLC (Zorbax RX-C8 7 μm, 21.2 × 250 mm column, 20 mL/min gradient elution over 20 min from 90% H2O/MeCN to 100% MeCN, with a constant 0.01% TFA/MeCN modifier) to result in 40 fractions. Fractions 30–33 were combined and subjected to semi-preparative reverse-phase HPLC (Zorbax SB-C18 5 μm, 9.4×250 mm column, 3 mL/min gradient elution over 20 min from 25% H2O/MeCN to 15% H2O/MeCN, with a constant 0.01% TFA/MeCN modifier) to produce pure goondapyrones A (1) (2.0 mg, 0.25%), B (2) (1.7 mg, 0.21%), C (3) (10.3 mg, 1.30%) and D (4) (1.5 mg, 0.19%), and actinopyrones A (11) (2.5 mg, 0.31%) and C (12) (4.0 mg, 0.50%).
goondapyrone A (1). Light yellow oil; [α]24D −15.0 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6) (Table 1, Table 2 and Table S2, Figures S5–S10); HRESIMS m/z 437.2679 [M+Na]+ (calculated for C26H38NaO4, 437.2662).
goondapyrone B (2). Light yellow oil; [α]24D −13.7 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6) (Table 1, Table 2 and Table S3, Figures S12–S17); HRESIMS m/z 437.2679 [M+Na]+ (calculated for C26H38NaO4, 437.2662).
goondapyrone C (3). Light yellow oil; [α]24D −21.2 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6) (Table 1, Table 2 and Table S4, Figures S19–S24); HRESIMS m/z 451.2824 [M+Na]+ (calculated for C27H40NaO4, 451.2819).
goondapyrone D (4). Light yellow oil; [α]24D −14.2 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6) (Table 1, Table 2 and Table S5, Figures S26–S31); HRESIMS m/z 465.2971 [M+Na]+ (calculated for C28H42NaO4, 465.2975).
actinopyrone A (11). Light yellow oil; [α]24D −13.0 (c 0.1, MeOH) and [α]24D +13.6 (c 0.1, CH2Cl2); ECD (MeOH) (Figure S84); 1H NMR (CDCl3) (Table S12 and Figure S75) and 1H and 13C NMR (DMSO-d6) (Table S13 and Figures S76 and S77); HRESIMS m/z 423.2520 [M+Na]+ (calculated for C25H36NaO4, 423.2511).
actinopyrone C (12). Light yellow oil; [α]24D −14.6 (c 0.1, MeOH) and [α]24D +13.6 (c 0.1, CH2Cl2); ECD (MeOH) (Figure S84); 1D NMR (DMSO-d6) (Table S14 and Figures S78 and S79); HRESIMS m/z 437.2657 [M+Na]+ (calculated for C26H38NaO4, 437.2668).
Cultivation in D400 shaking broth media: A seed culture of S4S-00196A10 was prepared by inoculating D400 broth medium (70 mL) and shaking at 190 rpm at 30 °C for 5 days. Aliquots of the seed culture (100 μL) were used to inoculate D400 broth media (2 × 500 mL), and after incubation at 190 rpm and 30°C for 14 days, the cultures were each extracted with EtOAc (2 × 500 mL) and the combined organic phase concentrated in vacuo to yield an extract (676.7 mg). This extract was then subjected to sequential trituration to afford after drying under nitrogen at 40 °C n-hexane (127.7 mg), CH2Cl2 (346.5 mg) and MeOH (202.5 mg) solubles. The combined CH2Cl2 and MeOH solubles were fractionated by reverse-phase HPLC (preparative: Zorbax RX-C8 7 μm, 21.2 × 250 mm column, 20 mL/min gradient elution over 20 min from 90% H2O/MeCN to 100% MeCN, with a constant 0.01% TFA/MeCN modifier) to result in 40 fractions, which were further resolved by semi-preparative HPLC (Zorbax SB-C18 5 μm, 250 × 9.4 mm column, 3 mL/min gradient elution over 20 min from 30% H2O/MeCN to 20% MeCN, with a constant 0.01% TFA/MeCN modifier) to yield goondapyrones E (5) (2.0 mg, 0.33%), F (6) (1.5 mg, 0.25%), G (7) (3.6 mg, 0.60%), H (8) (1.2 mg, 0.20%), I (9) (3.0 mg, 0.50%) and J (10) (1.1 mg, 0.18%).
goondapyrone E (5). Light yellow oil; [α]24D −13.1 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6) (Table 2, Table 3 and Table S6, Figures S33–S38); HRESIMS m/z 423.2523 [M+Na]+ (calculated for C25H36NaO4, 423.2506).
goondapyrone F (6). Light yellow oil; [α]24D −12.1 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6), (Table 2, Table 3 and Table S7, Figures S40–S45); HRESIMS m/z 437.2677 [M+Na]+ (calculated for C26H38NaO4, 437.2662).
goondapyrone G (7). Light yellow oil; [α]24D −15.5 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6), (Table 2, Table 3 and Table S8, Figures S47–S52); HRESIMS m/z 437.2679 [M+Na]+ (calculated for C26H38NaO4, 437.2662).
goondapyrone H (8). Light yellow oil; [α]24D −13.6 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6), (Table 2, Table 3 and Table S9, Figures S54–S59); HRESIMS m/z 451.2831 [M+Na]+ (calculated for C27H40NaO4, 451.2819).
goondapyrone I (9). Light yellow oil; [α]24D −13.0 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6), (Table 4 and Table S10, Figures S61–S66); HRESIMS m/z 409.2355 [M+Na]+ (calculated for C24H34NaO4, 409.2349).
goondapyrone J (10). Light yellow oil; [α]24D −8.4 (c 0.1, MeOH); ECD (MeOH) (Figure S84); 1D and 2D NMR (DMSO-d6), (Table 4 and Table S11, Figures S68–S73); HRESIMS m/z 423.2520 [M+Na]+ (calculated for C25H36NaO4, 423.2506).
4.6. Mosher Analysis of Goondapyrone C (3)
A solution of goondapyrone C (3) (1 mg) and S-α-methoxy-α-trifluoromethylphenylacetic acid [(S)-MTPA, 2.3 µL, 14.3 µmol, 6.2 eq.] in anhydrous CH2Cl2 (100 μL) was treated with dicyclocarbodiimide (DCC, 3.0 mg, 14.3 µmol, 6.2 eq.) and 4-dimethylaminopyridine (DMAP, 1.7 mg, 14.3 µmol, 6.2 eq.) at room temperature for 24 h, after which the reaction mixture was dried under N2 at 40 °C and the was residue extracted with EtOAc, which was in turn dried, dissolved in MeOH and purified by semi-preparative HPLC (Zorbax C18 5 μm, 250 × 9.4 mm column, 3 mL/min gradient elution over 20 min from 10% H2O/MeCN to 100% MeCN, with a constant 0.01% TFA modifier) to yield the (S)-MTPA ester 3a (0.76 mg, 51%). The procedure as outlined above was repeated using (R)-MTPA to yield the (R)-MTPA ester 3b (0.71 mg, 48%).
goondapyrone C (S)-MTPA ester 3a. 1H NMR (DMSO-d6), (Figure S83); HRESIMS m/z 667.3245 [M+Na]+ (calculated for C37H47F3NaO6, 667.3217).
goondapyrone C (R)-MTPA ester 3b. 1H NMR (DMSO-d6), (Figure S83); HRESIMS m/z 667.3251 [M+Na]+ (calculated for C37H47F3NaO6, 667.3217).
4.7. Antiparasitic Assays
D. immitis microfilariae motility inhibition assay. D. immitis microfilariae were resuspended in RPMI 1640 media (Hyclone) and ~250 suspended parasites (in 100 µL) were delivered to microtiter plate wells containing test compounds 1–12 dissolved in 100% DMSO. Compounds were delivered to generate 10 doses (3.2-fold serial dilutions) covering the range of 25–0.0007 µg/mL. Microtiter plates were then incubated at 37 °C and 5% CO2 for ~72 h, after which they were imaged in a camera-based system and quantitative descriptors of parasite motility were calculated. Values were normalized using positive (1.0 µM gramicidin) and negative (DMSO) controls and motility inhibition calculated for each well. For each dose–response curve, an EC50 value was calculated using a four-parametric logistic model in Boehringer Ingelheim’s MegaLab application.
H. contortus L1–L3 larvae development assay (LDA). ~20 L1 stage H. contortus were delivered to microtiter plate wells containing compounds 1–12 dissolved in 100% DMSO and nutrient medium. Compounds were delivered to generate 10 doses (3.2-fold serial dilutions) in the range of 25–0.0007 µg/mL. Microtiter plates were then incubated at 27 °C and 85% relative humidity. After incubating for ~96 h to allow development from the L1 to the L3 stage, plates were imaged on a camera-based system and quantitative motility descriptors were calculated on resulting developed worms. Values were normalized using positive (1.0 µM ivermectin) and negative (DMSO) controls and motility inhibition calculated for each well. For each dose–response curve, an EC50 value was calculated using a four-parametric logistic model in Boehringer Ingelheim’s MegaLab application.
Inhibition of motility of D. immitis L4 larvae. Four to six L4-stage D. immitis worms suspended in a 1:1 mixture of NCTC-109 (Thermo Fisher, Waltham, MA, USA) and iMDM (with sodium bicarbonate and 25 mM HEPES (Sigma-Aldrich, Burlington, NJ, USA)) media supplemented with antibiotics/antimycotics and 1% Fetal Bovine Serum (Cytiva Hyclone, Logan, UT, USA) were added to wells of a microtiter plate. Compounds diluted in 100% DMSO were prediluted in culture media and added to the wells containing the L4s for a final assay volume of 100 μL. Compounds were delivered to generate 5 doses (4-fold serial dilution) covering the range of 25–0.098 µg/mL. Plates were held for ~72 h at 37 °C and 5% CO2 and imaged for calculation of motility descriptors as described above. Percentage motility inhibition was calculated by normalization of the motility descriptor values of a test compound well with the average motility of positive (5.0 μM monensin) and negative (DMSO) controls on each plate. EC50 values were calculated with Boehringer Ingelheim’s MegaLab application using a four-parametric logistic model.
5. Conclusions
This study demonstrates that there is still much to learn from the defensive natural products that have evolved and are encoded within the global microbiome while validating a citizen science-based approach to acquiring microbial diversity and an integrated platform of biological, chemical and cultivation profiling as an efficient strategy for exploring bioactive microbial natural products. In particular, this study demonstrates that a selection of polyketide pyrones isolated from a pasture soil-derived Streptomyces can exhibit promising anthelmintic activity against both the dog heartworm D. immitis mf and the livestock gastrointestinal parasite H. contortus L1–L3 larvae, without displaying toxicity towards bacterial, fungal or mammalian (human carcinoma) cells.
Acknowledgments
We thank Rob and Nadia Campbell (the pastoralists) for generous access to Goondicum Station; K. Agnew for industry liaison; J. Han and T. Wu for technical advice; Boehringer Ingelheim Animal Health and the Australian Research Council (ARC) for project funding (LP190100083); the ARC for funding NMR instrumentation (LE210100148) and colony picking robotics (LE230100128); The University of Queensland, Institute for Molecular Bioscience; and the staff, donors, sponsors and members of the public, and students, that have supported Soils for Science. Z.G.K acknowledges support through the award of an ARC Future Fellowship (FT230100468).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13100989/s1, Bacterial taxonomy and phylogenetic analysis for Streptomyces spp. S4S-00196A10, together with NMR, HRESIMS, ECD and biological assay data for 1–12.
Author Contributions
Conceptualization, R.J.C.; investigation, S.J., D.F.B., C.T.C., E.B., Y.M., Z.G.K. and A.A.S.; resources, R.J.C. and Y.M.; data curation, S.J.; writing, R.J.C.; supervision, R.J.C., Z.G.K., A.A.S. and Y.M.; project administration, R.J.C. and Y.M.; funding acquisition, R.J.C. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
NMR and mass spectrometric data are available on request.
Conflicts of Interest
David F. Bruhn, Erica Burkman, Cynthia T. Childs and Yovany Moreno are employees of Boehringer Ingelheim Animal Health, USA Inc. All remaining authors declare no conflicts of interest.
Funding Statement
Australian Research Council (LE210100148, LE230100128, LP190100083 and FT230100468).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Soils for Science. [(accessed on 9 September 2020)]. Available online: https://imb.uq.edu.au/soilsforscience.
- 2.Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T., et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salim A.A., Khalil Z.G., Elbanna A.H., Wu T., Capon R.J. Methods in Microbial Biodiscovery. Mar. Drugs. 2021;19:503. doi: 10.3390/md19090503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosokawa S., Yokota K., Imamura K., Suzuki Y., Kawarasaki M., Tatsuta K. The first total synthesis and structural determination of actinopyrone A. Tetrahedron Lett. 2006;47:5415–5418. doi: 10.1016/j.tetlet.2006.05.028. [DOI] [Google Scholar]
- 5.Yano K., Yokoi K., Sato J., Oono J., Kouda T., Ogawa Y., Nakashima T. Actinopyrones A, B and C, new physiologically active substances. I. Producing organism, fermentation, isolation and biological properties. J. Antibiot. 1986;39:32–37. doi: 10.7164/antibiotics.39.32. [DOI] [PubMed] [Google Scholar]
- 6.Yano K., Yokoi K., Sato J., Oono J., Kouda T., Ogawa Y., Nakashima T. Actinopyrones A, B and C, new physiologically active substances. II. Physico-chemical properties and chemical structures. J. Antibiot. 1986;39:38–43. doi: 10.7164/antibiotics.39.38. [DOI] [PubMed] [Google Scholar]
- 7.Noack S., Harrington J., Carithers D.S., Kaminsky R., Selzer P.M. Heartworm disease—Overview, intervention, and industry perspective. Int. J. Parasitol. Drugs Drug Resist. 2021;16:65–89. doi: 10.1016/j.ijpddr.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delano M.L., Mischler S.A., Underwood W.J. Chapter 14—Biology and Diseases of Ruminants: Sheep, Goats, and Cattle. In: Fox J.G., Anderson L.C., Loew F.M., Quimby F.W., editors. Laboratory Animal Medicine. 2nd ed. Academic Press; Burlington, MA, USA: 2002. pp. 519–614. [DOI] [Google Scholar]
- 9.Baiak B.H.B., Lehnen C.R., da Rocha R.A. Anthelmintic resistance in cattle: A systematic review and meta-analysis. Livest. Sci. 2018;217:127–135. doi: 10.1016/j.livsci.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 10.GNPS: Global Natural Products Social Molecular Networking. [(accessed on 20 August 2021)]. Available online: https://gnps.ucsd.edu.
- 11.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NMR and mass spectrometric data are available on request.