Sir,
We read with interest the findings of Gerber et al. (2017) detailing the observation that three large multi-generational families with dominant optic atrophy appear to have dominant segregating DNM1L missense alleles (Gerber et al., 2017). The phenotypic similarity to human subjects with OPA1 alleles is interesting given the role of both DNM1L and OPA1 in mitochondrial dynamics, although OPA1 and DNM1L are involved in opposed aspects, mitochondrial fusion and fission, respectively. Indeed monogenic examples of additional loci tying altered mitochondrial dynamics with optic atrophy have recently been uncovered, e.g. ATAD3A is implicated in optic atrophy as well as in severely altered mitochondrial dynamics (Harel et al., 2016). The observation of dominant optic atrophy and segregating DNM1L alleles represents the first observation of a dominant inherited disease due to DNM1L mutations as the other dominant cases have been due to de novo events or from parents with low-level mosaicism. Optic atrophy is a dramatically milder phenotype than all previously published DNM1L related cases. The authors provide a review of ‘all cases previously reported’, but fail to include an infant with a p.G350R variant, a patient that exhibited an early onset neurodegenerative disease with optic atrophy (Chao et al., 2016). The latter study used model organism studies to demonstrate that the p.G350R corresponds to a dominant negative allele and is one of a handful of heterozygous alleles experimentally demonstrated to be dominant negative (Fig. 1A).
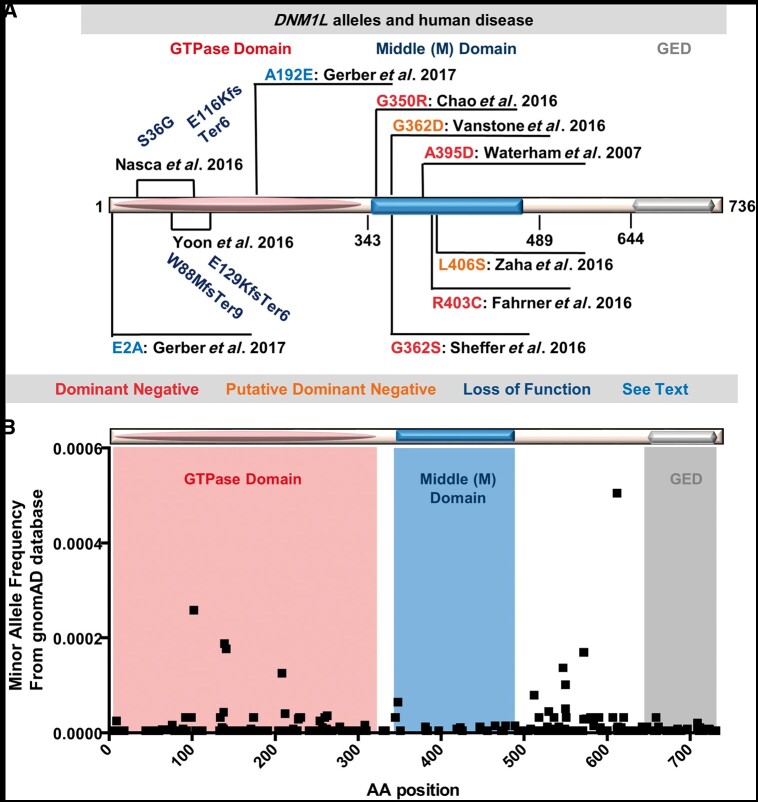
Figure 1.
DNM1L alleles in humans. (A) DNM1L alleles associated with human disease and their effects on gene function. Red: dominant negative heterozygous mutations associated with early onset disease such as encephalopathy due to defective mitochondrial and peroxisomal fission-1 (EMPF1) and experimentally validated in cells or animal models as dominant negative alleles. Orange: putative dominant negative heterozygous mutations associated with early disease. Dark blue: Compound heterozygous mutations resulting in loss of function in cases with bi-allelic variants in DNM1L. Light blue: heterozygous mutations associated with isolated optic atrophy reported by Gerber et al. (B) A graph of minor allele frequency on the y-axis versus the encoded amino acid position in the DNM1L protein on the x-axis for all the missense allele calls available in the gnomAD (gnomad.broadinstitute.org/) a database of >120 000 individuals. An approximate overlay of the domains encoded by the amino acids for the GTPase, middle domain and GED (effector domain) are shown in salmon, blue and grey, respectively.
The genetic mechanism as to how these novel DNM1L missense alleles reported by Gerber et al. cause human disease remains to be explored. The authors propose a ‘restricted dominant negative’ effect based on fibroblast analyses, and they state that they base this on the observation that the protein levels are not affected by the variants, and the ability of the mutant to polymerize with wild-type or mutant DNM1L appears intact. However, it is not obvious to us why these mutations are not loss- or gain-of-function mutations, as the function of the protein can be affected without impacting the levels. Given that there are three general mechanisms to explain dominant inheritance, haploinsufficiency, dominant negative, and gain of function, these three possibilities should be explored further. The parallels between the human subjects and the haploinsufficient mouse model provide support for the haploinsuffiency model rather than the dominant negative model proposed by the authors. If haploinsufficiency is indeed the cause, we would predict that the parents of the reported cases in Nasca et al. (2016) and Yoon et al. (2016) would be at risk of developing similar optic atrophy and could become symptomatic. Unfortunately, this information is not currently available.
A subtle gain of function could be considered as the phenotypes are consistent with what has been reported with OPA1. In the case of OPA1, haploinsuffiency of a gene required for mitochondrial fusion would be consistent with a gain of function of DNM1L as that would lead to increased fission of mitochondria. However, the observed increased mitochondrial network in the patients' fibroblasts from these optic atrophy families would argue against a gain of function mutation.
Third, a dominant negative function is also possible, and as noted this is what the authors favour. However, it should be noted that these cases are much more subtle than the severe human phenotypes that have been observed with the already reported DNM1L dominant negative alleles (Fahrner et al., 2016; Sheffer et al., 2016; Vanstone et al., 2016; Zaha et al., 2016) (Fig. 1A). These possibilities have not yet been explored sufficiently to conclude that the mutations described here are subtle dominant negative mutations.
Several genetic mechanisms are currently known for DNM1L in human disease. In four reported individuals from two families, a progressive infantile disorder resulting from biallelic loss-of-function pathogenic variants in DNM1L has been observed. In these cases the parents were reportedly heterozygous carriers without known disease at the time of the reports (Nasca et al., 2016; Yoon et al., 2016). In a number of other cases de novo variants (Waterham et al., 2007; Fahrner et al., 2016; Sheffer et al., 2016; Vanstone et al., 2016; Zaha et al., 2016), or variants inherited from a mosaic parent (Chao et al., 2016) have been reported in individuals with a range of severe metabolic and neurological disease. The study by Gerber et al. is the first report of dominant inherited disease due to DNM1L. If the optic atrophy in these individuals were in fact due to DNM1L loss-of-function then the implication is that the parents of individuals with biallelic disease could have milder disease as well. Indeed, the individuals Gerber reports were often asymptomatic when ascertained earlier in life (Gerber et al., 2017).
One possible explanation is that the authors are correct and these missense alleles are all dominant negative but have different functional impact depending on what protein domain is affected. We had previously observed that for DNM1L the middle domain appears to have a paucity of missense variation in genomic databases from more general human populations (Chao et al., 2016) suggesting this region is relatively intolerant to missense change. With the availability of genomic data on over 120 000 individuals in the gnomAD database (Lek et al., 2016) a similar picture emerges of rare missense variation with few alleles and in particular the middle domain and GED domain appear intolerant to missense changes in the population at least based on the allele frequencies in comparison to the protein domain where missense changes occur (Fig. 1B). The middle domain is responsible for protein assembly and the molecular mechanism for how missense alleles in the middle domain can produce dominant negative effects has been established and relates to the assembly of DNM1L oligomers (Chang et al., 2010).
The impact on mitochondrial and peroxisomal morphology has also been explored in vivo and the dominant negative middle domain variants affect these organelles dramatically (Waterham et al., 2007; Chao et al., 2016). In contrast, the GTPase domain is not implicated in oligomerization or assembly per se and dominant negative GTPase alleles have not been previously observed in humans. Dominant negative GTPase alleles for DNM1L have been generated (Li and Gould, 2003). A simple overexpression of the variants found in the patients in cell culture or in model organisms would resolve the dominant negative versus haploinsufficiency question.
The clarification of the functional impact has implications for the individuals and family members, especially those that carry truncating alleles. In the previous reports of autosomal recessive disease due to DNM1L (Nasca et al., 2016; Yoon et al., 2016), healthy unaffected parents have been reported carrying heterozygous loss-of-function alleles. If haploinsufficiency of DNM1L can cause optic atrophy as suggested by the mouse model, these parents would be at risk for optic atrophy and should potentially be screened. In addition, a number of potential individuals in the general population may exhibit DNM1L heterozygous loss-of-function alleles. However, if the authors’ hypothesis of restricted dominant negative mechanisms is correct, there could be therapeutic implications. Advances in oligonucleotide-based therapies for neurodegenerative disease would theoretically allow the specific alleles to be targeted for degradation (Schoch and Miller, 2017). In either case, we propose that determining genetic mechanisms will require robust cellular and model organism studies that account for genetic mechanisms in the models and the specific variants in all the human cases.
Contributor Information
Michael F Wangler, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children's Hospital, Houston TX 77030, USA; Texas Children's Hospital, Houston TX 77030, USA.
Nurit Assia Batzir, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston TX 77030, USA.
Laurie A Robak, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston TX 77030, USA; Texas Children's Hospital, Houston TX 77030, USA.
Mary K Koenig, Department of Pediatric Neurology, University of Texas Medical School at Houston, Houston TX 77030, USA.
Carlos A Bacino, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston TX 77030, USA; Texas Children's Hospital, Houston TX 77030, USA.
Fernando Scaglia, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston TX 77030, USA; Texas Children's Hospital, Houston TX 77030, USA.
Hugo J Bellen, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children's Hospital, Houston TX 77030, USA; Howard Hughes Medical Institute, Baylor College of Medicine, Houston TX 77030, USA.
References
- Chang CR, Manlandro CM, Arnoult D, Stadler J, Posey AE, Hill RB, et al. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem 2010; 285: 32494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y-H, Robak LA, Xia F, Koenig MK, Adesina A, Bacino CA, et al. Missense variants in the middle domain of DNM1L in cases of infantile encephalopathy alter peroxisomes and mitochondria when assayed in Drosophila. Hum Mol Genet 2016; 25: 1846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrner JA, Liu R, Perry MS, Klein J, Chan DC. A novel de novo dominant negative mutation in DNM1L impairs mitochondrial fission and presents as childhood epileptic encephalopathy. Am J Med Genet A 2016; 170: 2002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S, Charif M, Chevrollier A, Chaumette T, Angebault C, Kane MS, et al. Mutations in DNM1L, as in OPA1, result indominant optic atrophy despite opposite effectson mitochondrial fusion and fission. Brain 2017; 140: 2586–96. [DOI] [PubMed] [Google Scholar]
- Harel T, Yoon WH, Garone C, Gu S, Coban-Akdemir Z, Eldomery MK, et al. Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am J Hum Genet 2016; 99: 831–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J Biol Chem 2003; 278: 17012–20. [DOI] [PubMed] [Google Scholar]
- Nasca A, Legati A, Baruffini E, Nolli C, Moroni I, Ardissone A, et al. Biallelic mutations in DNM1L are associated with a slowly progressive infantile encephalopathy. Hum Mutat 2016; 37: 898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch KM, Miller TM. Antisense Oligonucleotides: Translation from mouse models to human neurodegenerative diseases. Neuron 2017; 94: 1056–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffer R, Douiev L, Edvardson S, Shaag A, Tamimi K, Soiferman D, et al. Postnatal microcephaly and pain insensitivity due to a de novo heterozygous DNM1L mutation causing impaired mitochondrial fission and function. Am J Med Genet A 2016; 170: 1603–7. [DOI] [PubMed] [Google Scholar]
- Vanstone JR, Smith AM, McBride S, Naas T, Holcik M, Antoun G, et al. DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur J Hum Genet 2016; 24: 1084–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med 2007; 356: 1736–41. [DOI] [PubMed] [Google Scholar]
- Yoon G, Malam Z, Paton T, Marshall CR, Hyatt E, Ivakine Z, et al. Lethal disorder of mitochondrial fission caused by mutations in DNM1L. J Pediatr 2016; 171: 313–16.e1–2. [DOI] [PubMed] [Google Scholar]
- Zaha K, Matsumoto H, Itoh M, Saitsu H, Kato K, Kato M, et al. DNM1L-related encephalopathy in infancy with Leigh syndrome-like phenotype and suppression-burst. Clin Genet 2016; 90: 472–4. [DOI] [PubMed] [Google Scholar]