SUMMARY
Here we describe an approach to correct the genetic defect in Fragile X syndrome (FXS) via recruitment of endogenous repair mechanisms. A leading cause of autism spectrum disorders, FXS results from epigenetic silencing of FMR1 due to a congenital trinucleotide (CGG) repeat expansion. By investigating conditions favorable to FMR1 reactivation, we find MEK and BRAF inhibitors that induce a strong repeat contraction and full FMR1 reactivation in cellular models. We trace the mechanism to DNA demethylation and site-specific R-loops, which are necessary and sufficient for repeat contraction. A positive feedback cycle comprising demethylation, de novo FMR1 transcription, and R-loop formation results in recruitment of endogenous DNA repair mechanisms that then drive excision of the long CGG repeat. Repeat contraction is specific to FMR1 and restores production of FMRP protein. Our study therefore identifies a potential method of treating FXS in the future.
INTRODUCTION
Fragile X syndrome (FXS) is an X-linked neurodevelopmental disorder and one of the most common monogenic causes of inherited intellectual disability1,2. FXS has a higher incidence among males (~1:3000) than females (~1:6000) and is observed worldwide at similar frequencies. Approximately 60% of FXS individuals demonstrate autistic features, 86% have an anxiety disorder, and almost all exhibit cognitive, motor, and developmental delays. Disease-modifying treatments have been of major pharmaceutical interest. A phosphodiesterase-4D (PDE4D) allosteric inhibitor was recently shown to improve cognitive function3, but there remains a dearth of disease-specific treatments, despite intensive efforts to better understand the etiology4–6. FXS derives its name from a chromosomal fragile site at Xq27.3 manifested in cultured patient cells7,8. Underlying the cytogenetic fragility is a trinucleotide repeat expansion located in the 5’-UTR of FMR1, the causal gene that produces Fragile X messenger ribonucleoprotein (FMRP)9–13. FMRP is a polyribosome-associated RNA-binding protein that concentrates in the brain, especially at neuronal synapses where it associates with ~4% of all brain mRNAs to regulate their transport and translation14,15, thereby accounting for the broad neurologic deficits in the FXS presentation.
Interestingly, the FMR1 coding sequence is almost always normal in patients. Failed FMRP expression is instead caused by aberrant expansion of a CGG trinucleotide repeat in the 5’-UTR. Wildtype (WT) FMR1 alleles typically carry <50 CGG repeats. Once the repeat number exceeds 50, one of two aberrant states results. Alleles with 55–200 repeats fall in the “pre-mutation” (PM) range. PM alleles can still be transcribed to make FMRP, do not cause FXS, and display no obvious symptoms before age 5016. These FXS “carriers”, however, hypertranscribe and accumulate FMR1 mRNA to toxic levels17,18, leading to a different disorder known as “fragile X-associated tremor/ataxia syndrome” (FXTAS) after age 5019. The PM allele is also a precursor of the FM allele and carries the risk of expansion when passed through the maternal germ line20. Once CGG repeats expand beyond 200 copies, these “Full mutation” alleles (FM) become hypermethylated at CpG dinucleotides in the FMR1 promoter, with resulting gene silencing and loss of FMRP production.
Repeat expansion is believed to occur during oogenesis and early embryogenesis, but underlying mechanisms are not well understood. Current models suggest several distinct mechanisms (see reviews21–23). For example, CGG expansion could occur during DNA replication when DNA polymerase slips or when Okazaki fragments become displaced. Misalignment of repeat tracts during double-strand break repair could also underlie repeat expansion24. Understanding how CGG repeats expand could lead the way to clinical intervention — if the process could be reversed and CGG repeats could be induced to contract for a restoration of FMRP expression. Indeed, the proposed slippage mechanisms that result in repeat expansions should (equally likely) yield contractions, depending on whether the daughter or parent strand undergoes slippage. Repeat contraction would address the root cause of the disorder and provide a potential life-long “cure”. However, contractions are only occasionally observed in vivo and in natural populations25,26. Here we hypothesize that conditions favorable to repeat contraction can be developed using endogenous means — i.e., without introduction of exogenous nucleases such as in CRISPR/Cas9. Our investigation leads to a site-specific editing method for FMR1 via formation of R-loops.
RESULTS
FMR1 reactivation by cellular reprogramming
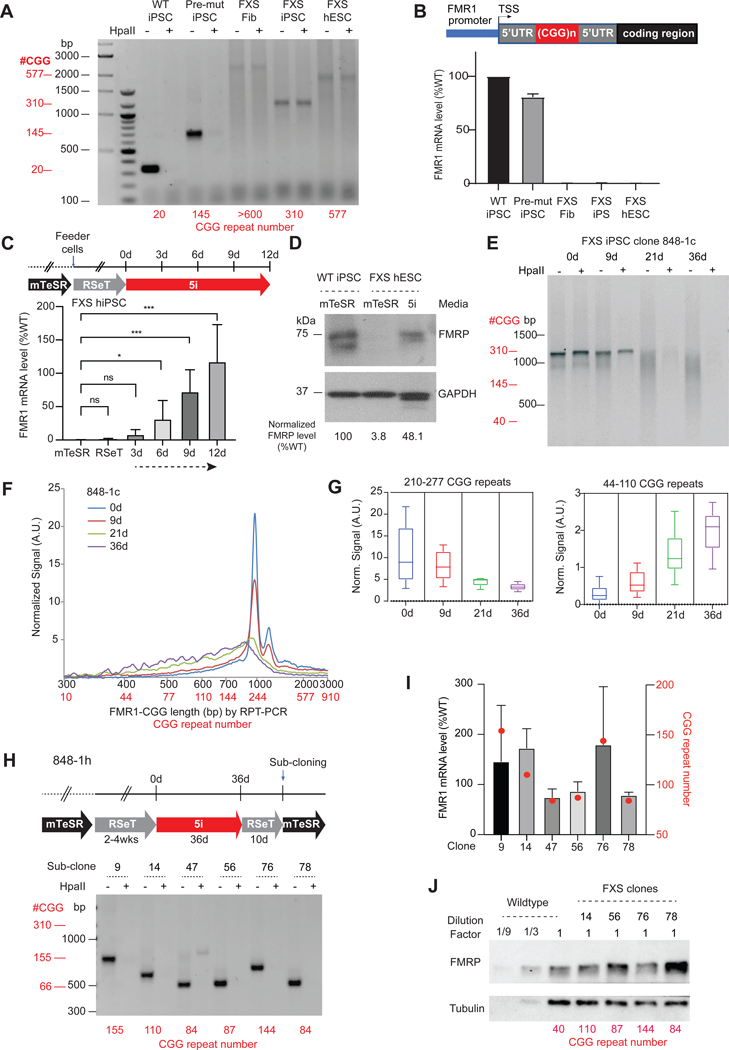
We used two established cellular models derived from FXS patients: (i) a human induced pluripotent stem cell (hiPSC) line from a FXS FM-male (848-iPS1)27, and (ii) a human embryonic stem cell (hESC) line outgrown from an IVF-cultured FM-blastocyst (WCMC37)28. Using established repeat-PCR (RPT-PCR) assays29, we confirmed repeat lengths in these cell lines (Figure 1A). Previous studies showed that FMR1 is silent in FXS hiPSCs27,30, but can be either expressed in early passage FXS hESCs28 or hypermethylated and silenced in other hESC lines30. Our hESCs and hiPSCs showed FMR1 silencing (Figure 1B). Its hypermethylated state was revealed by resistance to the methylation-sensitive HpaII restriction enzyme (Figure 1A). As early passage FXS hESCs showed FMR1 activity28, we surmised that prolonged passage of cells in culture could cause partial cellular differentiation and change the epigenetic state of FMR1. We therefore asked if returning the hESC’s to the so-called ‘naïve state’ could reactivate FMR131,32. Indeed, one previous work showed that culturing FXS hiPSCs in Naïve Human Stem cell Media (NHSM) resulted in FMR1 reactivation31. However, such reactivation was not observed in another study33.
Figure 1. CGG repeat contraction and FMR1 reactivation by cellular reprogramming.
See also Figure S1.
(A) Gel electrophoresis of PCR products from RPT-PCR to examine repeat length and DNA methylation status in indicated patient cell lines. Addition (+) of HpaII to genomic DNA prior to RPT-PCR tests methylation status by degrading non-methylated templates. RPT-PCR product lengths (black) and corresponding CGG repeat copy numbers (red, below each cell line) are shown.
(B) FMR1 mRNA quantitation from the cells shown in (A). Expression levels normalized to WT hiPSC levels. Schematic map of FMR1 gene depicted above.
(C) Experimental timeline showing periods of acclimation to mTeSR, RSeT, and 5i media, along with accompanying changes in FMR1 mRNA levels after 12 days of 5i treatment. P values determined by the Student t-test. ns, not significant, *, P<0.05, ***, P<0.001.
(D) Western blot analysis of FMRP in WT hiPSC versus FXS hESC grown in indicated media. GAPDH, loading control. FMRP levels quantified by imageJ67 and normalized to GAPDH.
(E) Time course analysis of CGG repeat length and DNA methylation status (HpaII +/−) in FXS full mutation clone, 848–1c, between 0–36 days of 5i treatment. Gel electrophoresis of RPT-PCR products is shown, with RPT-PCR product lengths (black) and corresponding repeat copy numbers (red).
(F) Bioanalyzer quantitation of CGG repeat lengths in a 0–36 day time course. RPT-PCR product lengths (black) and corresponding repeat copy numbers (red) are shown.
(G) Box plots for quantitation of high copy number (210–277x CGGs) and low copy number (44–110x CGGs) repeats after 0–36 days of 5i treatment.
(H) RPT-PCR assays of 6 subclones derived from 848–1h after 5i treatment to copy number of shortened CGG repeats (indicated in red).
(I) Quantitation of FMR1 mRNA levels in subclones of 848–1h in (H) by RT-qPCR. Red dots, repeat numbers as indicated in the alternate y-axis (red axis).
(J) Western blot quantitation of FMRP levels in indicated 848–1h subclones. Tubulin, loading control. CGG repeat numbers shown below each subclone.
We reasoned that differences in the culture conditions could cause the disparate observations. To find conditions permissive for reactivation in our hands, we tested RSeT media, a modified commercial version of NHSM, but observed no reactivation (Figure 1C). We tested another naïve state formulation dubbed “5i”32. To avoid cell shock caused by a sudden change in media, we transitioned cells by co-culturing with feeder cells in RSeT and then gradually titrating in 5i media (Figure 1C). Under 5i condition, we observed a progressive increase in FMR1 mRNA expression in FXS iPSCs by 3–6 days, with full reactivation by day 12, whereas WT iPSCs showed no significant changes (Figure S1A). Western blot analysis indicated that RNA upregulation was accompanied by FMRP production, restoring protein levels to ~50% of WT levels (Figure 1D). These data identify a specific media formulation in which FMR1 can be reactivated robustly.
FMR1 reactivation is accompanied by CGG repeat contraction
To ask if FMR1 reactivation were accompanied by CGG hypomethylation, we derived single-cell clones from the FXS hESC and hiPSC lines to obtain a homogeneous cell population (Figure S1B). RPT-PCR analysis showed that hiPSC clone 848–1c, initially carried ~310 repeats and was fully methylated. To our surprise, culturing in 5i media resulted in a progressive shortening of the CGG repeats, as evidenced by smearing of the CGG band in an electrophoretic gel (Figure 1E). Bioanalyzer quantitation indicated a progressive contraction over a 36 day time course (Figure 1F). As the subpopulation of long repeats (210–277 CGG copies: denoted as 210–277x) decreased, the subpopulation of shorter repeats (44–110x) increased (Figure 1G). By 36 days, CGG repeats showed a range from ~200x to <100x (Figures 1E-G) —in the pre-mutation range. Because the Ethidium Bromide intercalation assay intrinsically underestimates the stoichiometry of shorter repeats, repeat lengths in the normal range (<50x) could be present and substantially underestimated. Altogether, these data demonstrate that FRM1 reactivation is accompanied by CGG contraction.
Interestingly, contraction was not evident at day 6 (Figure S1C) when FMR1 reactivation was already detectable (Figure 1C), suggesting that some degree of FMR1 expression might precede the contraction phenomenon. Significantly, reactivation was also accompanied by DNA demethylation, as shown by a progressive digestion by the methylation-sensitive restriction enzyme, HpaII, beginning at day 9 (Figures 1C,E). Additional hiPSC and hESC clones demonstrated a similarly strong demethylation and repeat contraction after 27–36 days in 5i media (Figures S1B,D). hESC clones 37–1b and 37–1d initially had 450–500 repeats, whereas hiPSC clones 848–1a and 848–1h had 650–700 and ~310 repeats, respectively, and each contracted to <200 repeats. When the 5i-treated (and contracted) 848–1h clone was further subcloned after 36 days, we observed demethylated repeats as short as 80x (subclones 47, 56, 78; Figure 1H). Notably, PM carriers express normal FMRP levels but have elevated mRNA levels16,27,34,35. Our CGG-contracted subclones also exhibited high-level mRNA expression (Figure 1I) and fully restored FMRP levels (Fig 1J). There was a correlation between repeat number and mRNA expression: Shorter repeats showed nearly normal mRNA levels (subclones 47, 56, 78), whereas longer repeats showed higher levels (subclones 9, 14, 76).
We conclude that FMR1 reactivation of FXS cells by 5i is accompanied by strong CGG contraction and DNA hypomethylation down to a PM state. Given these results, we were curious if naturally occurring PM or WT CGG lengths could respond to 5i. We derived clones from PM iPSC line, 131, and examined subclone 131–1a with ~150 CGG repeats. No obvious repeat contraction was observed after 27 days of 5i treatment (Figure S1E). Similarly, no repeat contraction was observed from WT iPSCs after 12 days of 5i treatment (Figure S1F). These data suggest a copy number threshold of ~150 CGG below which 5i cannot initiate contraction.
Repeat contraction attributed to MEK and BRAF inhibition
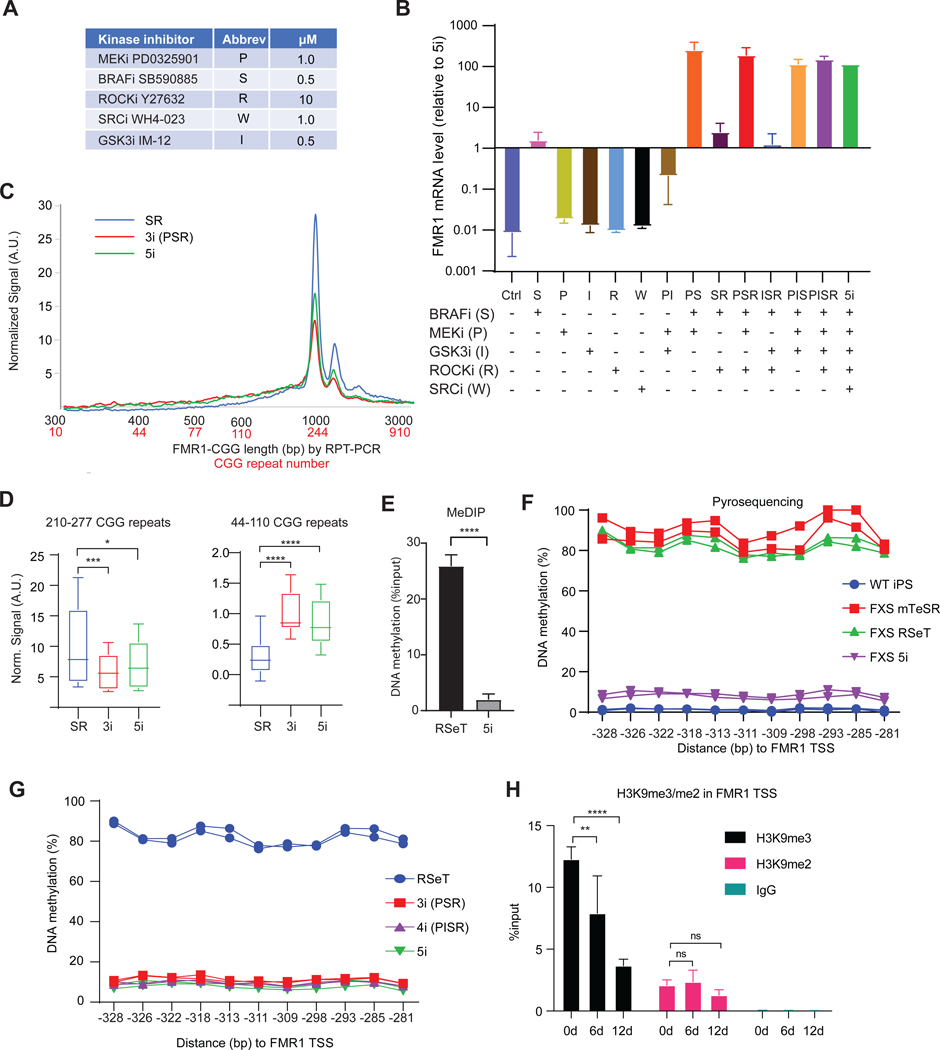
5i media contains a mixture of 5 inhibitors (“i”) of various kinases (Figure 2A): PD0325901 (“P”, MEKi), IM-12 (“I”, GSK3i), SB590885 (“S”, BRAFi), Y27632 (“R”, ROCKi), and WH4–023 (“W”, SRCi)32. To ask which is responsible for reactivating FMR1, we treated 848–1c hiPSCs with one or various combinations of the 5 inhibitors without changing the base media. While no single inhibitor alone was sufficient, the combination of MEKi and BRAFi (P+S) triggered full FMR1 expression (Figure 2B). The fact that MEK and BRAF kinases belong to the same MAPK pathway but MEKi and BRAFi alone could not reactivate FMR1 suggested that MEK and BRAF have downstream targets that do not overlap and that are necessary for reactivation. While the ROCKi was not essential for FMR1 reactivation, we noticed that its addition promoted hiPSC viability during cell passaging and was therefore beneficial to the formulation. P+S+R treatment (henceforth “3i”) resulted in CGG repeat contraction as robust as that observed with 5i (Figures 2C,D). Other combinations, including S+R, S+R+I, and P+I did not substantially increase FMR1 expression. We conclude that MEKi + BRAFi are sufficient to trigger CGG contraction and FMR1 reactivation.
Figure 2. MEKi and BRAFi are sufficient to drive CGG contraction.
See also Figure S2.
(A) Table of kinase inhibitors, abbreviations, and concentrations used in 5i formulation.
(B) Determination of active compounds in 5i media for FMR1 reactivation by testing single, double, and triple combinations of inhibitors in 848–1c cells. FMR1 RNA levels determined by RT-qPCR, normalized to levels in 5i.
(C) Bioanalyzer quantitation of CGG repeat length/copy number after indicated treatment for 12 days.
(D) Box plot quantitation of high (210–277x CGGs) versus low (44–110x CGGs) copy numbers after 12 days of indicated treatment. P determined by Student t-test. *, P<0.05, ***, P<0.001, ****, P<0.0001.
(E) MeDIP-qPCR assay using anti-5mC antibodies measures DNA methylation levels at FMR1 in 848–1c grown in RSeT versus 9 days of 5i. ****, P<0.0001 (Student t-test).
(F) Pyrosequencing analysis of DNA methylation at CpG islands in FMR1 promoter in WT versus FXS cells grown in indicated media.
(G) Pyrosequencing analysis of DNA methylation levels at FMR1 CpG islands after treating 848–1c cells with RSeT, 3i (PSR), 4i (PISR), or 5i.
(H) ChIP-qPCR assay for H3K9me3 and H3K9me2 at FMR1 TSS in 848–1c cells in RSeT versus 6 or 12 days in 5i. Student t-tests: ns, not significant, **, P<0.01, ****, P<0.0001. IgG ChIP, negative control.
Kinase inhibition upregulates TET enzymes and demethylates FMR1
The 5i media is known to trigger DNA demethylation32. To quantitate the observed demethylation in 5i-treated FXS cells (Figure 1E), we performed MeDIP, an immunoprecipitation method that captures methylated DNA using 5-methyl-C (5mC) antibodies, and confirmed a strong loss of DNA methylation in the FMR1 promoter after 9 days of 5i (Figure 2E). Pyrosequencing further confirmed the demethylation with single-nucleotide precision. After 5i treatment for 12 days, the unmethylated state resembled that of WT iPSCs (Figure 2F). Demethylation did not occur in FXS cells grown in mTeSR or RseT (Figures 2F,S2A). Strong DNA demethylation also occurred in FXS cells under 3i or 4i (P+I+S+R) treatment (Figures 2G,S2B). In contrast, reactivation did not seem to require loss of all H3K9me2 and H3K9me3 — two repressive histone methylation marks associated the FM allele36,37. Our ChIP-qPCR analysis revealed H3K9me2 and H3K9me3 enrichment at FMR1 in FM cells grown in RseT media (Figure 2H). There was no significant decrease in H3K9me2 after 5i treatment. However, there was a decrease in H3K9me3, though it was delayed relative to FMR1 reactivation (Figure 2H). Exposure to 5i only modestly reduced H3K9me3 levels in 6 days, despite clear FMR1 reactivation after 6 days (Figure 1C). At 12 days, a drop to ~25% of H3K9me3 was observed, but only after FMR1 was fully reactivated (Figure 1C).
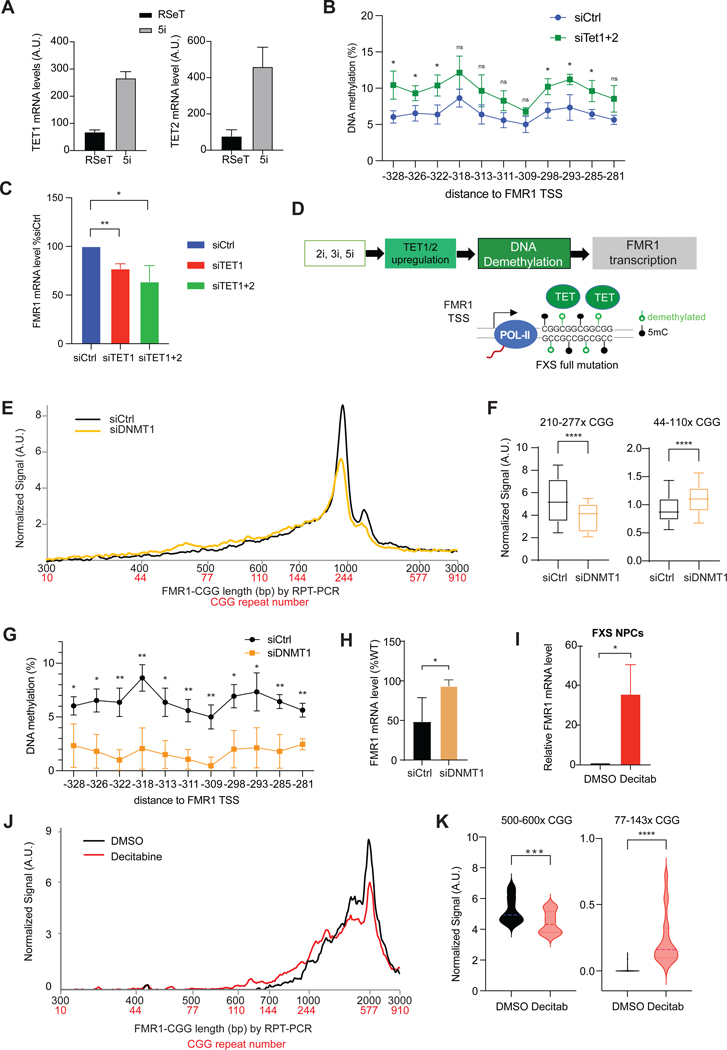
Thus, 5i induces a promoter CpG-demethylation concomitant with FMR1 reactivation. Several DNA demethylases are active in pluripotent stem cells, including TET1 and TET238. Interestingly, 5i treatment of FXS cells resulted in a 4-to 6-fold increase in endogenous TET1 and TET2 mRNA levels and a 2.5-to 3-fold increase in protein levels (Figures 3A,S3A), whereas various control genes did not show a significant change (Figure S3B). To ask if the 5i mechanism could be attributed to upregulation of TET1 and TET2, we partially knocked down TET1/TET2 using siRNA (Figures S3A,C) and observed loss of FMR1 demethylation and reactivation (Figure 3B-C,S3D). These results suggest that TET1 and TET2 mediate the effect of 5i by removing DNA methylation at the FMR1 promoter (Figure 3D).
Figure 3. DNA demethylation potentiates CGG contraction and FMR1 reactivation.
See also Figure S3.
(A) TET1 and TET2 RT-qPCR quantitation in FXS iPSCs treated with RSeT versus 5i for 6 days.
(B) Pyrosequencing analysis of DNA methylation at FMR1 promoter after 12 days of TET1/TET2 versus control knockdown in 848–1c cells grown in 5i. Student t-test: * P<0.05. ns, not significant.
(C) RT-qPCR of FMR1 in 848–1c cells treated with siCtrl, siTET1, and siTET1/siTET2 double knockdown in 5i for 6 days. Student t-test: *, P<0.05. **, P<0.01.
(D) Proposed epistatic pathway depicting effects of 5i, 3i, or 2i treatment on TET1/2 expression, DNA methylation, and de novo FM FMR1 transcription. 5mC, methylated C.
(E) Bioanalyzer analysis of CGG repeat length distribution after DNMT1 knockdown in 5i.
(F) Box plot quantitation of high (210–277x CGGs) versus low (44–110x CGGs) copy numbers after 12 days of siDNMT1 knockdown in 5i. Student t-test: ****, P<0.0001.
G) Pyrosequencing analysis of DNA methylation at FMR1 CpG islands after siDNMT1 (or siCtrl) treatment for 12 days in 848–1c grown in 5i. Student t-test: *, P<0.05. **, P<0.01.
(H) RT-qPCR of FMR1 expression in 848–1c cells treated with siCtrl versus siDNMT1 in 5i for 6 days. Student t-test: *, P<0.05.
(I) RT-qPCR of FMR1 expression in FXS NPC treated with decitabine versus DMSO vehicle for 3 days and grown for an additional 12 days. Student t-test: *, P<0.05.
(J) Bioanalyzer analysis of CGG repeat length distribution after DMSO or decitabine treatment with NPC per (I).
(K) Violin plots quantitation of high copy number (500–600x CGGs) versus low copy number (77–143x CGGs) repeats after DMSO or decitabine treatment per (I). t-test: ***, P<0.001. ****, P<0.0001.
To test this idea, we depleted DNMT1, the enzyme that maintains CpG methylation — reasoning that failure to maintain CpG methylation would have the same effect as upregulating TET1/2. We used a DNMT1-specific siRNA and compared results to a control scramble siRNA (siCtrl) in 848–1c hiPSCs. After 6 days in RSeT media, DNMT1 depletion did not reactivate FMR1 (Figures S3E,F). However, in 5i media (Figure S3G), the DNMT1 knockdown potentiated the CGG contraction beyond that of 5i alone (Figure 3E-F), When siDNMT1 and 5i were combined, there was also a significantly stronger DNA demethylation at the FMR1 promoter and a ~2-fold increase in FMR1 mRNA (Figure 3G-H). Thus, perturbing DNMT1 and TET1/2 have opposite effects. We conclude that CpG demethylation of FMR1 is a crucial step towards CGG contraction and FMR1 reactivation.
Similar effects were observed in cells in the neuronal lineage. In neuronal progenitor cells (NPCs) derived from the same FXS iPSCs27, we applied the demethylating agent, decitabine39 at 0.5 μM for 3 days and followed FMR1 expression over 15-days. Significantly, a ~40-fold increase in FMR1 levels was observed over DMSO-treatd controls (Figure 3I). A clear CGG repeat contraction accompanied this reactivation (Figure 3J,K). We conclude that demethylation causes CGG contraction and FMR1 reactivation in both FXS iPSCs and neuronal cells (NPCs).
Site-specific R-loops trigger CGG repeat contraction
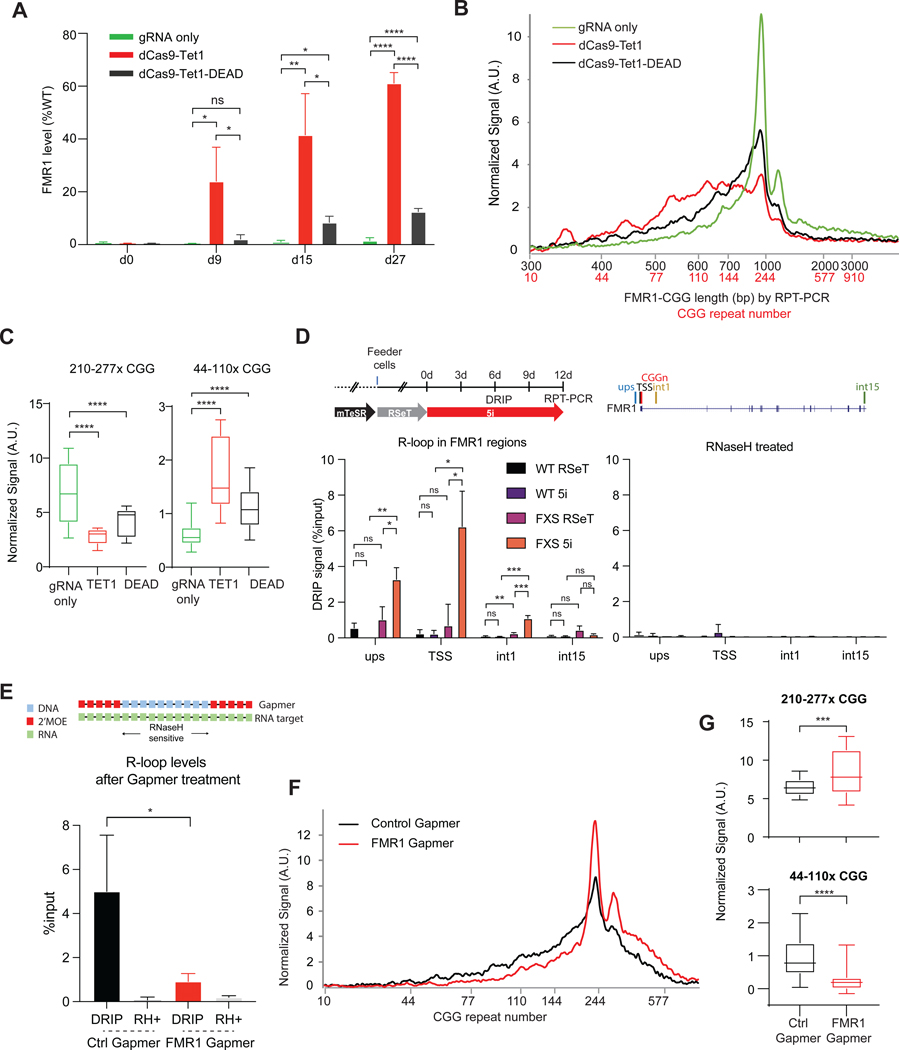
The data indicate that DNA demethylation may be a first step towards CGG contraction. To determine if CpG demethylation at the FMR1 promoter is sufficient to induce contraction, we targeted TET1 to FMR1 using a catalytically dead Cas9 (dCas9-Tet1) and CGG-specific gRNAs. In 848–1c cells grown in mTeSR, we observed a strong FMR1 reactivation (Figure 4A), consistent with a prior study in which targeting TET1 yielded FMR1 reactivation40. In contrast to the prior study, however, we found that dCas9-Tet1 targeting contracted the CGG repeats (Figures 4B,C). Furthermore, to our great surprise, the “control” harboring a catalytically dead TET1 (dCas9-Tet1-DEAD)40 also contracted the CGG repeat (Figures 4B,C) and showed FMR1 reactivation (Figure 4A, p<0.0001 in 27d). The degree of contraction and reactivation was smaller but still significant. As dCas-Tet1-DEAD lacks a demethylase activity, this finding indicates that the initial DNA demethylation can be bypassed by a separate activity in dCas-Tet1-DEAD.
Figure 4. R-loop formation in FMR1 locus.
(A) FMR1 reactivation after 848–1c cells were exposed to dCas9-Tet1, dCas9-Tet1-DEAD, versus gRNA only for 0–27 days in mTeSR. t-tests: ns, not significant. *, P<0.05. **, P<0.01. ****, P<0.0001.
(B) Bioanalyzer traces of repeat length (black) and copy number (red) after 27 days of treatment.
(C) Bioanalyzer signals for ranges of 210–277x CGGs versus 44–110x CGGs for indicated conditions. P<0.0001, t-test.
(D) Left panel: DRIP assay at the FMR1 regions including upstream (ups), transcription start site (TSS), first intron (int1) and 15th intron (int15) in WT iPSC and 848–1c FXS iPSC after 6 days of 5i. Right panel: Treatment with RNaseH (RH+) abolished DRIP signals. t-tests: ns, not significant. *, P<0.05. **, P<0.01. ***, P<0.001.
(E) Top panel: Gapmer design based on 2’MOE chemistry. Bottom panel: DRIP assay at the FMR1 TSS after FMR1 downregulation by gapmer treatment in 5i for 6 days.
(F) Bioanalyzer profiles for RPT-PCR product length and estimated CGG repeat copy number in 848–1c iPSC after 12 days treatment with FMR1 versus control Gapmer.
(G) Box plot quantitation of high (210–277x CGGs) versus low (44–110x CGGs) copy numbers after indicated treatment for 12 days. t-test: ***, P<0.001. ****, P<0.0001.
To identify the activity, we reasoned that dCas9-Tet1-DEAD can still target FMR1 via the CGG gRNA, resulting in a three-stranded nucleic acid structure known as an “R-loop” comprising an annealed DNA:RNA hybrid at the CGG repeat and a complementary single-stranded DNA. Previous structural and biophysical studies had shown that CRISPR-Cas9 and dCas9 both form R-loops at target DNA sites41–43. When not properly cleared or shielded, R-loops can cause DNA breaks and induce recruitment of the DNA repair machinery24,44–46. Notably, previous studies observed R-loops at the FMR1 locus of normal, PM, and FM FXS cells28,37,47.
We asked if enhanced R-loops could explain the CGG contraction under either 5i and/or dCas9 conditions. By performing DRIP (DNA-RNA hybrid immunoprecipitation) using S9.6 antibodies that recognizes RNA-DNA hybrids48, we observed a significant enrichment of RNA-DNA hybrids at FMR1 in 848–1c cells grown in 5i media for 6 days (Figure 4D). Consistent with previous observations28,37,47, R-loops were strongly enriched at transcription start site (TSS) of FMR1 but not in the distal gene body (Figure 4D, left panel). Pre-treatment with RNaseH (RH+) abolished DRIP signals, indicating a specific detection of R-loops (Figure 4D, right panel). To test if R-loops were necessary, we targeted the CGG-containing nascent FMR1 mRNA with a gapmer to degrade the transcript (Figure 4E, top panel). Gapmers contain an internal stretch of unmodified DNA nucleotides which, when base-paired with RNA, degrade the RNA strand of RNA-DNA hybrid by recruiting endogenous RNaseH49,50, thereby destroying the R-loop. Indeed, R-loop formation at FMR1 was significantly reduced by the FMR1-specific gapmer relative to control (Ctrl) gapmer (Figure 4E, bottom). RPT-PCR showed a concurrent attenuation of CGG repeat contraction (Figures 4F,G). Thus, the RNA-DNA hybrid within the presumptive R-loop is crucial for CGG contraction.
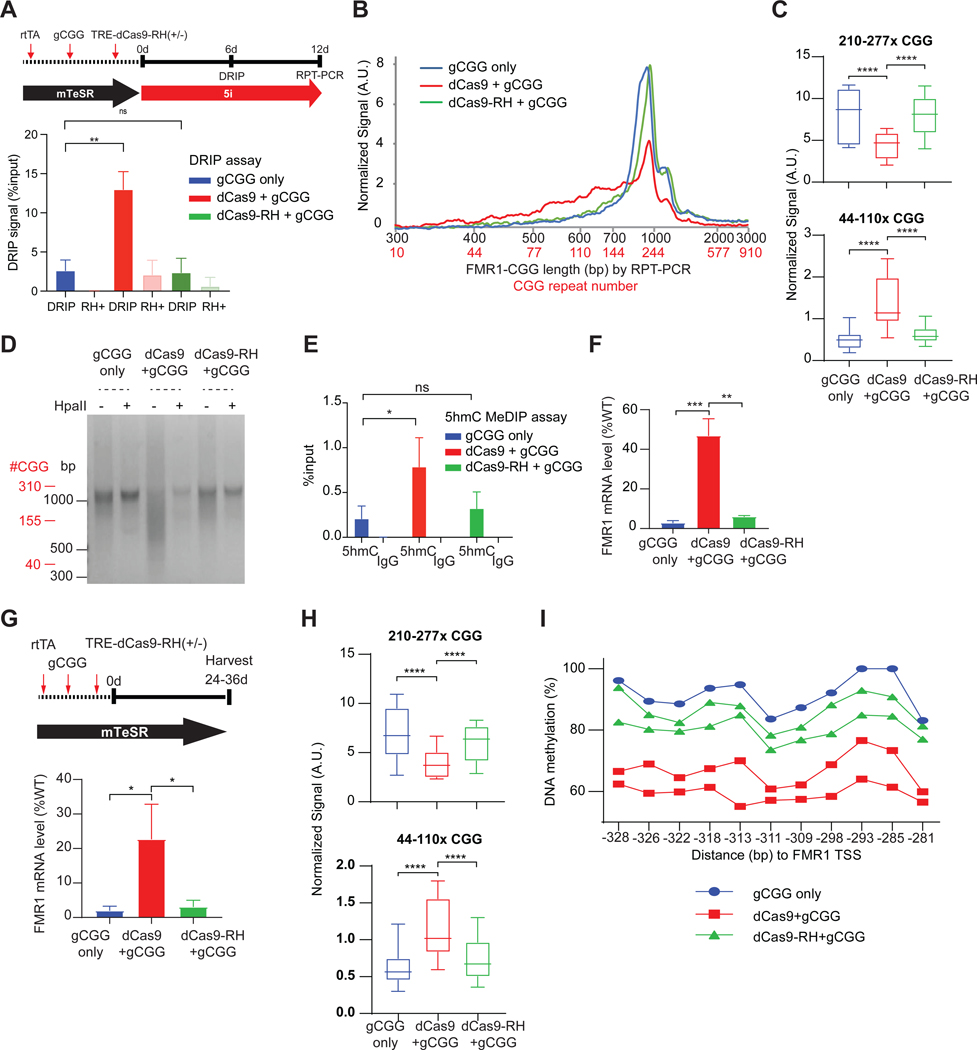
We then asked if dCas9 + CGG gRNA (gCGG) potentiates the effect of 5i by enhancing R-loops. We generally grow iPSCs in RSeT with feeder cell support, as RSeT helps iPSCs adapt to 5i (e.g., Figure 4D). However, dCas9 and gRNA introduction required blasticidin and puromycin selection markers which affected feeder survival in RSeT media. We therefore eliminated the RSeT transition and shifted FXS iPSCs from mTeSR (which does not require feeder support) to 5i directly (Figure 5A, timeline). Without the more favorable RSeT and feeder condition, the FXS iPSCs showed less robust R-loop formation (compare Figure 5A “gCGG-only,” to Figure 4D). To determine whether dCas9+gCGG could indeed enhance R-loop formation and accelerate the contraction associated with 5i treatment, we performed DRIP analysis and observed a stronger enrichment of R-loops after 6 days of treatment with dCas9+gCGG relative to gCGG alone (Figure 5A). The increase in R-loop formation was observed only in FXS iPSCs and not in WT cells (Figure S4A). By 12 days, dCas9+gCGG together induced a stronger CGG contraction at FMR1 compared to the gCGG-only control in FXS iPSCs (Figure 5B,C). In contrast, WT iPSCs did not exhibit any contraction (Figure S4B). The HpaII assay also showed a stronger FMR1 promoter demethylation in dCas9+gCGG cells (Figure 5D).
Figure 5. Site-specific R-loops underlie repeat contraction.
See also Figure S4.
(A) DRIP assay at the FMR1 TSS in 848–1c iPSC treated as shown in schematic (top panel), with gCGG alone, dCas9+gCGG, or dCas9-RH+gCGG for 6 days. t-test: **, P<0.01. ns, not significant.
(B) Bioanalyzer analysis of RPT-PCR product length and estimated CGG repeat copy number for the samples depicted in (A).
(C) Box plot quantitation of high (210–277x CGGs) versus low (44–110x CGGs) copy numbers after indicated treatment for 12 days. t-test: ****, P<0.0001
(D) Gel electrophoresis of CGG RPT-PCR products +/− HpaII digestion in 848–1c iPSC, treated for 12 days using the scheme shown in (A).
(E) 5hmC MeDIP assay at the FMR1 5’UTR in 848–1c iPSC treated, with gCGG alone, dCas9+gCGG, or dCas9-RH+gCGG for 6 days. t-test: *, P<0.05. ns, not significant.
(F) RT-qPCR of FMR1 mRNA levels for corresponding samples shown in (A-E). t-test: **, P<0.01; ***, P<0.001.
(G) Top: Timeline to test sufficiency of dCas9-targeted R-loops in 848–1c iPSC without 5i treatment. Bottom: RT-qPCR of FMR1 mRNA levels following 24-day exposure to gCGG alone, dCas9+gCGG, or dCas9-RH+gCGG. t-test: *, P<0.05.
(H) Box plot quantitation of high (210–277x CGGs) versus low (44–110x CGGs) copy numbers after indicated treatment in (G). t-test: ****, P<0.0001.
(I) Pyrosequencing to examine DNA methylation at FMR1 promoter for samples in (G,H).
Because R-loop formation can bypass the requirement for 5i and targeted TET1/TET2 (Figure 4A), we suspected that R-loops might be able to induce demethylation in FMR1. Indeed, we detected a significant increase of the 5hmC intermediate (Figure 5E,S4C), indicating the R-loop mediated active DNA demethylation51. These effects were abolished by tethering RNaseH to dCas9 (dCas9-RH + gCGG) to destroy R-loops. The 6 day treatment with dCas9 + gCGG also resulted in strong FMR1 reactivation to ~50% of WT levels (Figure 5F), while the same treatment in WT iPSCs did not have significant changes (Figure S4D). The accelerated reactivation with dCas9+gCGG was also abolished by tethering RNaseH (dCas9-RH + gCGG). Thus, promoting R-loop formation using dCas9 + gCGG yields a stronger reactivation relative to 5i alone or 5i + gCGG.
If R-loop formation were causal to the contraction and reactivation, treating with dCas9 + gCGG alone — without any 5i exposure — should be sufficient to induce the effect. To test this, we repeated the experiment in mTeSR media without 5i (Figure 5G). Indeed, dCas9 + gCGG in FXS iPSCs yielded 20–25% FMR1 reactivation after 24 days, while WT iPSCs showed no changes (Figure 5G,S4E). Reactivation was accompanied by CGG contraction (Figure 5H,S4F) and significant promoter demethylation (Figure 5I,S4G). Tethering RNaseH to dCas9 significantly reduced contraction, demethylation, and FMR1 reactivation (Figure 5G-I). We conclude that R-loop formation is both necessary and sufficient for CGG repeat contraction and FMR1 restoration in FXS. As these changes occurred independently of 5i and targeted TET1/TET2, we conclude that R-loop formation at the long CGG repeat is central to the contraction phenomenon.
On-target effects
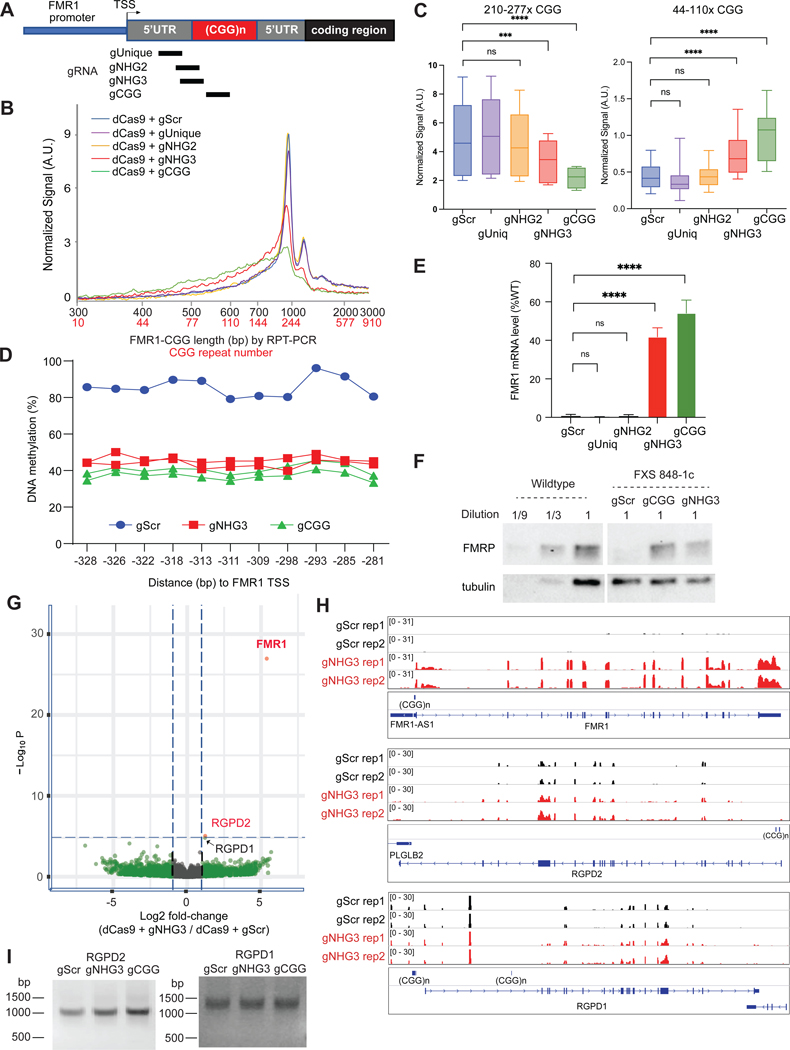
Because CGG repeats occur throughout the human genome, we considered the potential for gCGG to have “off-target” effects. For more specific targeting to FMR1, we designed FMR1-specific gRNAs by including unique sequences flanking the repeat (Figure 6A). gUnique binds unique sequences in the 5’UTR, whereas gNHG2 and gNHG3 target the junction between unique and repeat sequences, with gNHG3 including three CGG repeats and gNHG2 including only 2. Interestingly, neither gUnique nor gNHG2 resulted in CGG contraction in 848–1c FXS cells grown in mTeSR (Figures 6B,C). In contrast, gNHG3 showed a CGG repeat contraction (Figure 6B,C), DNA demethylation, and FMR1 reactivation as robust as with gCGG (Figure 6D-F,S5A). gNHG3-mediated reactivation was also observed at the protein level (Figure 6F). Thus, at least three CGGs are required in the gRNA.
Figure 6. Repeat contraction and gene reactivation are specific to FMR1.
See also Figure S5.
(A) Map of FMR1 and location of FMR1-specific gRNAs.
(B) Bioanalyzer profiles for CGG length and repeat copy number for 848–1c iPSC targeted as shown in (A) for 36 days in mTeSR regular media. Scrambled gRNA (gScr), negative control.
(C) Box plot quantitation of high (210–277x CGGs) versus low (44–110x CGGs) copy numbers after indicated treatment per (A) above. t-test: ***, P<0.001. ****, P<0.0001.
(D) Pyrosequencing to examine DNA methylation at FMR1 promoter for samples targeted as indicated per (A-B).
(E) RT-qPCR of FMR1 mRNA for samples shown in (A-D). t-test: ****, P<0.0001. ns, not significant.
(F) Western blot analysis of FMRP levels for the experiments in (A-E). Tubulin, loading control.
(G) Volcano plot of transcriptomic analysis of 848–1c iPSC targeted by dCas9+gNHG3 versus dCas9+gScr. Log2 fold-change for differentially expressed (DE) genes are plotted against statistical significance (-Log10P). Red dots, significantly changed genes.
(H) Integrated Genome Viewer (IGV) screenshots for transcriptomic analysis of two biological replicates (rep1, rep2) of cells exposed to either dCas9 + gScr or dCas9 + gNHG3. Three representative genes with CGG tracts are shown. Scale is indicated in brackets.
(I) RPT-PCR suggests absence of off-target CGG contraction at RGPD2 and RGPD1 following dCas9 targeting by gScr, gNHG3, and gCGG in 848–1c FXS hiPSCs.
We investigated whether gNHG3 could have off-target effects at other CGGs in the genome. Intriguingly, after 36 days of treatment in FXS cells, differential gene expression analysis of two biological replicates showed only one dominant change —FMR1 (Fig 6G,H). FMR1 was upregulated >40-fold compared to gScr controls. One other gene showed a small, though significant change: RGPD2 was upregulated by ~2-fold (Fig 6G,H). RGPD2 contains two relatively long CGG repeat tracts of 174bp and 129bp separated by a 621bp spacer (Figure 6H). These CGG repeats could have been targeted by dCas9 + gNHG3. However, RPT-PCR analysis revealed no obvious change in its CGG length after treatment (Figure 6I). Interestingly, a related gene, RGPD1, did show a minor, but insignificant change in expression (Figures 6G,H). Notably, RGPD1 carries 6 short CGG repeats (177, 75, 191, 174, 202, and 55 bp) in its promoter region, but they did not obviously contract after dCas9 + gNHG3 treatment (Figures 6H,I,S5B). Other CGG repeat-containing genes, including AFF2 and SIRT1, also showed no significant changes in gene expression or CGG length (Figures S5C,D). Lastly we asked whether WT hiPSCs respond to dCas9 + gCGG or gNHG3. No CGG contraction was observed in FMR1 (Figure S5E). Altogether, our data indicate that dCas9 + gNHG3 elicit strong site-specific effects at FMR1. They also suggest that contraction requires a minimum CGG copy number of >200.
R-loop recruits the DNA mismatch repair mechanism to correct the CGG expansion
POL-II transcription naturally and transiently forms R-loops during ribonucleotide polymerization on the DNA template, but R-loops can be aberrantly stabilized over long CG-rich regions, including FMR128,37,47. When not properly regulated, R-loops can cause DNA damage, trigger DNA repair24, and induce repeat instability52,53 (Figure 7A). CGG copy number fluidity is known to depend on endogenous DNA repair mechanisms21–23. Several DNA repair pathways exist in mammalian cells to correct R-loop-mediated DNA damage (Figure 7A), including the transcription-coupled nucleotide excision repair (TC-NER) involving CSB, XPG, and XPF54,55. TC-NER plays a critical role in R-loop-induced DNA damage repair54,55 and trinucleotide repeat instability56,57. The mismatch repair (MMR) pathway is also implicated in trinucleotide repeat instability. The MMR recognition factor, MSH2, binds to slipped-strand DNA structures formed within trinucleotide repeat tracts58,59, causing either repeat expansion or contraction in various trinucleotide repeat disorders52,60,61.
Figure 7. A positive feedback loop of DNA demethylation, de novo nascent transcription, R-loop formation, and recruitment of DNA mismatch repair mechanisms drives FMR1 reactivation.
See also Figure S6.
(A) R-loop formation can trigger two major DNA repair pathways. MMR and TC-NER are candidate mechanisms for CGG repeat contraction.
(B) ChIP-qPCR for γ-H2AX at four FMR1 positions (upstream, TSS, first intron, and 15th intron) in 848–1c cells grown in RSeT or 5i media. t-test: *, P<0.05. **, P<0.01.
(C) Bioanalyzer profiles for RPT-PCR product length and estimated CGG repeat copy number in 848–1c iPSC after 12 days treatment with siMSH2, siXPG, or siCSB, as compared to control siRNA treatment. Cells are grown in 5i media.
(D) Box plot quantitation of high (210–277x CGGs) versus low (44–110x CGGs) copy numbers after indicated treatment for 12 days. t-test: ***, P<0.001, ****, P<0.0001.
(E) Positive feedback cycle of DNA demethylation, transcription, R-loop formation, and repeat contraction driving reactivation of FMR1.
We therefore asked if the observed CGG contraction invokes either DNA damage response. ChIP analysis showed that γ-H2AX, a histone mark for DNA damage response, was significantly elevated in 5i-treated FXS cells at FMR1 (Figure 7B), consistent with R-loop-mediated DNA damage. To ask if the damage is repaired by the TC-NER/CSB-XPG pathway, we depleted CSB or XPG but did not observe a major effect on CGG contraction in 5i-treated FXS cells after 12 days (Figures 7C,D; S6). By contrast, depleting MSH2 in the MMR pathway significantly attenuated CGG contraction (Figures 7C,D; S6). Although we cannot rule out CSB/XPG participation, these data indicate that CGG contraction is at least dependent on the MSH2/MMR pathway. We conclude that R-loops recruit endogenous DNA repair mechanisms to FMR1 to correct the aberrant repeat length in FXS cells.
DISCUSSION
Here we have identified methods of FMR1 gene editing without exogenous nucleases such as TALENS and CRISPR-Cas9. We demonstrate FMRP restoration can be induced by CpG demethylation and R-loop formation, which together recruit endogenous DNA repair mechanisms to correct the long CGG repeat length (Figure 7E). Initially, we observed that MEK and BRAF inhibitors together elicit CGG repeat contraction and full FMR1 reactivation in <12 days. While MEK and BRAF kinases belong to the same MAPK pathway, inhibiting either kinase alone was ineffective, suggesting that non-overlapping downstream targets of MEKi and BRAFi mediate the effect. Combined, the effects of kinase inhibition upregulate TET1/TET2 demethylases and demethylate FMR1 promoter and CCG repeat. This demethylation can be phenocopied by depleting DNMT1 or by tethering dCas9-TET1 to the CGG repeats. De-methylation induces de novo transcription from FMR1 and leads to formation of POL-II mediated R-loops that become aberrantly stabilized over the long CGG repeat due to the high GC content and high melting temperatures. The aberrant R-loops in turn trigger DNA damage signals, which are then resolved by the MSH2/MMR repair pathway (Figure 7) and possibly also the TC-NER pathway54–57. Resolution of the aberrant R-loops leads to CGG contraction, FMR1 reactivation, and restoration of FMRP. Thus, the central event in the contraction phenomenon is de novo transcription (caused by demethylation) and formation of site-specific R-loops at FMR1 (Figure 7E).
Our model posits a strong positive feedback loop involving DNA demethylation, de novo FMR1 transcription, and R-loop formation (Figure 7E). Within the loop, intervention could occur at any step. For instance, MEKi and BRAFi (2i, 3i, and 5i) can induce demethylation via TET1 and TET2 upregulation. DNMT1i also potentiates the effect. De novo transcription resulting from MEKi+BRAFi, TET1/TET2 upregulation, or DNMT1i can all create the aberrant R-loops that would then trigger DNA repair. Likewise, intervention at the step of R-loop formation would also trigger DNA repair — for example, by targeting dCas9 + gCGG. De novo R-loop formation would open up chromatin and promote new transcription, which in turn would further enhance R-loop formation in the positive feedback cycle. R-loops can indeed promote transcription62, possibly by recruiting transcription factors. For FXS, the initial transcription need not be robust, as even a single round of transcription could create a stable R-loop at the long CGG repeat. R-loops can reciprocally induce DNA demethylation through recruitment of TET151,63, further enhancing the positive feedback loop.
R-loops are both necessary and sufficient to achieve reactivation. dCas9 can initiate the cycle by melting the highly stable double-stranded CGG repeat. gCGG directs site-specific recruitment of dCas9. gCGG cannot work alone, as melting the long CGG repeat is energetically unfavorable and depends on the unwinding activity of dCas9. dCas9 enables the gRNA to anneal to the DNA to create the essential RNA-DNA hybrid41–43. Because dCas9 is catalytically dead, it lacks the endonuclease activity required to complete the programmed CRISPR reaction and becomes “stuck” at the CGG repeat. As the FXS repeat contains >200 CGG copies, the repeat could potentially host hundreds of such dCas9-mediated RNA-DNA hybrids — creating a long and stable R-loop stretch. The dependence on CGG repeat length indicates a copy number threshold and may explain why the wildtype FMR1 locus and other CGG-containing genes are not subject to contraction. The number of CGG repeats in the gRNA is also crucial factor. gCGG (with 7x repeats) and gNHG3 (with 3x repeats) were both effective at R-loop formation and CGG contraction, but gRNAs with only 2x repeats (gNHG2) or containing only unique FMR1 sequence (gUnique) were not. The inability of gUnique to induce contraction suggests that R-loops must be targeted to the repeats themselves.
Consistent with a copy number threshold, the method is surprisingly site-specific, as dCas9 + gNHG3 resulted only in a single major transcriptomic change — in FMR1 itself, which was upregulated >30-fold compared to control-treated cells. Other CGG repeat-containing genes (e.g., RGPD1, SIRT1, AFF2) were not affected significantly, except that RGPD2 (with two CGG tracts of 174 and 129 bp) and was upregulated by ~2-fold. RGPD2 repeats did not evidently contract, however. Wildtype cells with <50 CGG repeats in FMR1 and pre-mutation cells with ~150x CGG were also not affected (Figures S1E,F,S5E; 6I,S5B,D). We note that, while contraction does not initiate in PM cells, FM cells (>200 CGG repeats) can clearly contract down to PM levels and continue to contract below 200 copies (Figures 1E-G,5B-D,5H). It is worth noting that subclone 78, which only has 44–110x CGGs with full FMR1 reactivation (Fig. 1H-J), showed low R-loop levels compared to the full-mutation 848–1c FXS cell line in 5i (Fig. S7A). R-loop levels in clone 78 did not differ whether cultured in RSeT or 5i, in contrast to the full-mutation FXS line. Culturing clone 78 in 5i did not result in further FMR1 upregulation (Fig. S7B). These findings suggest a threshold of the CGG repeat length (between 44–110x CGG) below which contraction is not further observed. The reasons for a contraction threshold are currently unknown, but longer CGG repeats have the potential to form complex secondary structures in both the RNA and DNA21,64,65, and secondary structures formed in the non-template single-strand DNA strand could aid in R-loop stability and recruitment of additional mediators66.
In summary, our study demonstrates that R-loop formation in long CGG repeats in FXS cells can achieve up to 40–100% FMR1 reactivation. If FXS cellular phenotypes could be reversed with FMR1 restoration1,4,5, an approach to treating FXS in the future could involve targeting R-loops for trinucleotide repeat contraction and re-expression of the missing FMRP in neuronal cells. Here we have provided preliminary evidence that the mechanism strongly observed in iPSCs can also be seen in neuronal cells. For therapeutic applications, future work will focus on optimizing the contraction process in ex vivo neuronal and in vivo models.
LIMITATIONS OF THE STUDY
The two methods for estimating the CGG repeat sizes from RPT-PCR (agarose gel and bioanalyzer) yielded small differences in repeat sizes due to underlying inherent differences between in-gel and microfluidic dynamics. These small differences do not impact our conclusions, as there was clear CGG contraction using the two orthogonal methods. The RPT-PCR may not represent the full spectrum of repeat lengths, due to the inherent difficulties amplifying across longer repeats. On the other hand, because the fluorescence-based detection gives greater visual weight to longer repeat lengths, short repeats may be under-represented stoichiometrically. Finally, although our data implicate the MSH2-dependent MMR pathway, we cannot exclude CSB and XPG of the TC-NER pathway due to technical limitations and lack of reagent availability (e.g., ChIP-grade antibodies were not available). Both pathways may be involved under different circumstances. Whereas our present work establishes the phenomenon of R-loop-induced CGG contraction in FXS, future work will delve further into how MMR, as well as TC-NER pathways could mediate contraction under different conditions both in vitro and in vivo assays. Elucidating these mechanisms will aid in developing FXS therapeutics in the future.
RESOURCE AVAILABILITY
Lead contact
Please direct requests for resources and reagents to Lead Contact : Jeannie T. Lee (lee@molbio.mgh.harvard.edu)
Materials availability
Plasmids generated in this study are available upon request.
Data and code availability
Next-generation sequencing data reported in this study have been deposited at the Gene Expression Omnibus with accession number GSE184580.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE184580.
This paper does not report original codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture conditions for human embryonic stem (hES) cells and induced pluripotent (iPS) cells
FXS FM WCMC37 hES cells28, FXS FM 848-iPS127, Premutation 131-iPS127 and WT8330-iPS27 male human pluripotent cell lines were maintained in mTeSR media with Matrigel-treated plates at 37° C with 5% CO2 level. Cells were passaged either with Accutase or TrypLE Select (Gibco) by following the manufacturer’s manual. Feeder cells from UV-irradiated drug resistant mouse embryonic fibroblasts were used for the RSeT (Stem Cell Technology) or 5i32 media condition, and TrypLE Select or Accutase was treated in 5–10 minutes until the hPSC cells start to be detached by a gentle tapping without disturbing feeder cells. 5i medium was generated by following the protocol from Theunissen et al., 2014. Briefly, 240 ml DMEM/F12 (Gibco 10565–042), 240 ml Neurobasal media (Gibco 21103049), 1 mM L-glutamine (Gibco), 1% nonessential amino acids (Gibco), penicillin-streptomycin (Gibco), 50 mg/ml BSA (cell culture grade) were combined and used as a basal media. The following components were freshly added to 48.5ml of basal media: 0.5 ml N2 supplement (Gibco 17502048), 1 ml B27 supplement (Gibco 17504044), 20 ng/ml recombinant human LIF (Peprotech), 0.1 mM β-mercaptoethanol (Sigma), 10 ng/ml Activin A (Peprotech), and the following small molecules from Naive Stem Cell 5i inhibitor Set (Axon 5011) : PD0325901 (1 μM), IM-12 (1 μM), SB590885 (0.5 μM), WH-4–023 (1 μM), and Y-27632 (10 μM). For the composition optimization, the same basal media was used with different small molecule combinations. 40 μm cell strainer was used to collect the detached cells to minimize feeder contamination after TrypLE Select or Accutase is neutralized by DMEM media containing 10% FBS and 25mM HEPES pH 7.0. Cells were grown in 5% O2 incubator for 5i treatment.
Experimental conditions for small molecule treatments were optimized to minimize the toxicity while sustaining the effects of the small molecules. As the direct conversion from regular human ES/iPS media (mTeSR) to 5i media caused a massive cell death, we first adopted the hES/iPS cells in RSeT media with feeder condition and gradually introduced 5i media by using 50% mixture of RSeT and 5i media. While FMR1 gene expression was not significantly increased during the long-term cultivation in RSeT media, treating the FXS hES/iPS cells with 50% 5i media triggered a significant increase in FMR1 gene expression in 6 days (Figure 1C) without massive cell death.
Cell culture conditions for Neuronal Progenitor Cells (NPCs)
NPCs derived from 848–1 FXS iPSCs27 were grown in NPC media consisting of 35ml DMEM, 15ml Ham’s F12 medium, 1ml B27 (without Vitamin A; Cat# 12587010), 0.5ml penicillin-streptomycin (Gibco), 200ng/ml EGF (R & D SYSTEMS, Cat# 236-EG-200), 200ng/ml FGF (R & D SYSTEMS, Cat# 233-FB-025/CF) , and 5μg/ml Heparin (R & D SYSTEMS, Cat# 2812/100). For testing FMR1 reactivation in FXS neuronal cells, 0.5 μM Decitabine (Selleck Chemicals; S1200) was treated to Neuronal Progenitor Cells (NPCs) derived from FXS iPSCs27 for three consecutive days. Cells were grown in NPC media without Decitabine for 12 days and harvested for RNA and genomic DNA on d15.
METHOD DETAILS
Repeat-PCR (RPT-PCR)
Repeat-PCR (RPT-PCR), an assay for detecting the very long CGG repeats at the FMR1 locus, was performed as described in Hayward et al29. Due to the extremely high GC content and repetitive nature of the long CGG repeats, regular PCR protocols cannot be used for analyzing the length of the CGG repeats on full mutation FMR1 locus. Briefly, genomic DNA was isolated by ZYMO Quick-gDNA MiniPrep Plus kit (Zymogen), and 600ng of genomic DNA was treated with HindIII in 50mM Tris-Cl pH 8.8, 1.5mM MgCl2, 22mM (NH4)2SO4, 0.2% Triton X-100 overnight at 37°C. EcoNI was added to remove the RPT-PCR product from mouse feeder cells. HpaII, a methylation-sensitive enzyme, was additionally included in the half of the HindIII digestion mix for monitoring the methylation status of the target regions. RPT-PCR master mix containing 50 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, 22 mM (NH4)2SO4, 0.2% Triton X-100, 3.3 M betaine (Sigma, St. Louis, MO), 2.67% dimethyl sulfoxide, 0.27 mM dNTPs, 27 U/ml High-Fidelity Phusion DNA polymerase (New England Biolabs) and 0.67 μM of primers (Not-FraxC and Not-FraxR4 for FMR1) was prepared on ice. For the RPT-PCR reactions, 5 μl of the HindIII-digested genomic DNA was added to 15 μl of the PCR master mix on ice and mixed well before PCR reaction. The PCR was performed by following cycles : 98°C for 3 minutes, 30 × (98°C for 30 seconds, 64°C for 30 seconds, 72°C for 210 seconds), and 72°C for 10 minutes. Since the RPT-PCR product includes 269 base pair flanking regions, the number of CGG repeat length can be calculated by the following formula: number of CGG repeat = (fragment size −269)/3. At least three biological replicates were tested if not specified.
Bioanalyzer analysis
RPT-PCR products were purified by using PCR purification kit (Qiagen) and the purified samples were further analyzed by High sensitivity DNA kit with Bioanalyzer 2100 (Agilent) to quantify the proportion of long and short CGG fragments. Estimated fragment size was calculated by the migration time to size conversion based on the ladder migration. Raw signal data was normalized by the average intensity between 300bp to 3kb. Box plots are drawn to show the signal intensity distribution on the long (900–1100bp : 210–277 CGG repeats) and short (400–600bp : 44–110 CGG repeats) CGG repeat range.
DRIP (DNA-RNA hybrid immunoprecipitation)
R-loop formation (DNA-RNA hybrid) was monitored by DRIP. DRIP assay was performed by following the protocol from Loomis et al47 with slight modifications. Briefly, cell pellets were resuspended in 10 mM Tris-HCl, 10 mM EDTA, 100 mM NaCl pH 8, lysed with 0.5% SDS, and digested with 400 units of Proteinase K overnight at 37°C. Cell lysates were then extracted once with phenol pH 8 and twice with chloroform. DNA was precipitated with 1 volume of isopropanol and 300 mM sodium acetate, and the pellet was washed twice with 70% ethanol, and was resuspended in 10 mM Tris-HCl pH 8. Harvested nucleic acids (~10μg) were digested with EcoRI, HindIII, BsrGI, and XbaI (20 units each) overnight at 37°C in NEBuffer 2. RNaseH1, which degrades RNA-DNA hybrid, was treated to test the specificity of the DRIP-qPCR signals and served as negative control. After that, samples were purified by phenol and chloroform extraction followed by precipitation in isopropanol. The pellet was washed twice with 70% ethanol. Air-dried pellets were resuspended in 10 mM Tris-HCl pH 7.5, 1 mM EDTA.
Digested nucleic acids (2–5 μg) were diluted in 450 μL of TE, and 10 μL was reserved as input for qPCR. 10× IP buffer was added for a final buffer concentration of 10 mM sodium phosphate, 140 mM sodium chloride, 0.05% Triton X-100, and 4–10 μg of S9.6 antibody (Kerafast inc.; ENH001) was added and incubated at 4°C for 2 hours. After the antibody incubation, Dynabead Protein A/G magnetic beads (1:1 mix) were washed twice with 800 μL of 1× IP buffer for 5 minutes at room temperature and 40 μl of the beads were added to each sample and incubated for 2 hours at 4°C. Beads were washed three times with 700 μL 1× IP buffer for 10 minutes at room temperature. After the wash, the magnetic beads were resuspended in 250 μL of 1× IP buffer and incubated with 60 units of Proteinase K for 30 minutes at 50°C. Digested DRIP samples were purified by phenol/chloroform extraction and isopropanol precipitation. Pellets were resuspended in 80 μL of 10 mM Tris-HCl pH 8.0. Quantitative PCR was performed for four different loci in FMR1 – upstream promoter(ups), transcription start site (TSS), first intron(int1), and 15th intron (int15) (schematic description in Figure 4D).
Methylated DNA immunoprecipitation (MeDIP) and pyrosequencing
5mC and 5hmC levels on FMR1 locus were measured by qPCR assay with the pull-down material of methylated DNA by antibody recognizing 5mC (Active Motif monoclonal Ab clone 33D3) and 5hmC (Active Motif polyclonal Ab #39769). Briefly, genomic DNA was purified from cultured cells by overnight Proteinase K treatment, followed by phenol-chloroform extraction and ethanol precipitation. DNA was fragmented using HindIII, EcoRI, BsrGI, and XbaI at 37°C overnight. We used 2–4 μg of fragmented DNA for a standard MeDIP assay. After denaturing by incubating at 95°C for 10min, the sample was immunoprecipitated for 2h at 4°C with 10 μl of monoclonal antibody against 5-methylcytidine (Active motif; 33D3) in a final volume of 500 μl IP buffer (10 mM sodium phosphate (pH 7.0), 140 mM NaCl, 0.05% Triton X-100). We incubated the mixture with 30 μl of Dynabeads for 2 h at 4°C and washed it three times with 700 μl of IP buffer. Beads were treated with proteinase K for 3 h at 56°C and pull-downed methylated DNA was purified by phenol-chloroform extraction and ethanol precipitation. Pyrosequencing was performed by EpigenDx (ADS1451-FS2).
Chromatin immunoprecipitation (ChIP)
Harvested cells are fixed in 1% formaldehyde for 5 minutes at room temperature, and glycine was added to a final conc. of 0.125M and incubated for 5 minutes at room temperature to stop crosslinking. After washing twice with cold PBS, cross-linked cells were resuspended in Buffer 1 (50mM HEPES-KOH pH 7.5, 150mM NaCl, 1mM EDTA pH 8.0, 0.5% NP40, 0.25% Triton X-100 with freshly added protease inhibitor cocktail (PIC)) and rotated at 4°C for 10 min. After spin-down at 1700 rcf for 5 min in 4°C, cells were resuspended in Buffer 2 (10mM Tris-Cl pH 8.0, 200mM NaCl, 5mM EDTA pH 8.0, 2.5mM EGTA pH 8.0 with freshly added PIC) and rotated on a wheel for 10 min at 4°C. Finally, cells were incubated in Buffer 3 (10mM Tris-Cl pH 8.0, 5mM EDTA pH 8.0, 2.5mM EGTA pH 8.0 with freshly added PIC) with RNase A and rotated on a wheel for 30 min at 37°C. After adding N-lauroyl sarcosine to final conc. 0.5%, samples were sonicated by using Covaris system with 15×19mm tube, 5% duty cycle, intensity 140, and 200 burst per cycle for 10–25 min. Sonicated samples were centrifuged by 14000rpm for 10 min in 4°C and supernatant was used for immunoprecipitation and input samples. For immunoprecipitation, equal volume of 2x IP buffer (2% Triton X-100, 300mM NaCl, 30mM Tris-HCl pH 8.0, 1X PIC) was added to the sonicated samples and 2–5 μg of antibody was added per IP and incubated o/n at 4°C on a rotating wheel. 40 μl of Protein G Dynabead was washed twice with Buffer1 and added to each IP sample and incubated for 2hrs at 4C° on a rotating wheel. Beads were washed 3 times with 1/2x RIPA-1 buffer (50mM HEPES-KOH pH 7.5, 500mM NaCl, 10mM EDTA pH 8.0, 1% NP40, 0.5% Sodium Deoxycholate) and 3 times with RIPA-3 buffer (50mM HEPES-KOH pH 7.5, 50mM NaCl, 10mM EDTA pH 8.0, 0.2% NP40) at 4°C for 5 min on a rotating wheel. Washed beads were resuspended to 200 μl TES buffer (50mM Tris-Cl pH 8.0, 10mM EDTA pH 8.0, and 1% SDS) and incubated at 65°C for 15 min. After spin down, 40 μg of Proteinase K was added to the supernatant and incubated in 65°C for 4hrs to O/N for reversing the crosslinking. After phenol/chloroform extraction and ethanol precipitation with glycogen, resulted pellets were resuspended in ddH2O and analyzed by qPCR assay with purified input samples.
The following antibodies were used for ChIP experiment: H3K9me3 (Abcam; ab8898), H3K9me2 (Abcam, ab1220), gamma H2A.X (phospho S139) (Abcam; ab2893), IgG (Cell Signaling; 2729S).
Lentivirus production and molecular cloning
Lentiviruses were produced by transfecting HEK293T cells with various dCas9-containing constructs or pgRNA constructs together with psPAX2 and pMD2.G-VSVG packaging vectors using Lipofectamine 3000 (Invitrogen). Fuw-dCas9-Tet1CD-P2A-BFP, Fuw-dCas9-dead Tet1CD-P2A-BFP, pgRNA-CGG and pgRNA-modified (for the backbone of the sgRNA construct) were gifts from Rudolf Jaenisch (Addgene plasmid #108245, #108246, #108248, #84477). pGH125 dCas9-Blast was a gift from Michael Bassik (Addgene plasmid #85417). Inducible dCas9 and dCas9-RNaseH1 plasmids were generated by modifying the pGH125 dCas9-Blast backbone (Addgene plasmid # 85417). Briefly, EF1a promoter was swapped with TRE-tight Tet-responsive promoter of pTRE-Tight plasmid. The catalytic domain of human RNaseH1 (110–260 aa) was additionally subcloned into the 3’ end of dCas9 coding region as in-frame to create dCas9-RNaseH1 recombinant protein expressing cassette. One Shot Stbl3 chemically competent E. coli strain (Thermo Fisher) was used for the cloning. Puromycin and Blasticidin were used for the selection markers of gRNA and dCas9-related constructs integration, respectively. Produced lentiviruses were further concentrated by Lenti-concentrator (Origene) and stored in Lenti-Stabilizer (Origene). Lentivirus titer was measured by Lenti-X GoStix Plus (Takara Bio), and polybrene (10μg/ml) was treated to cells before lentiviral transduction.
RT-qPCR
RNAzol was treated to the collected cell pellets, and total RNA was extracted by following the manufacturer’s manual. Reverse transcription reaction was performed by using QuantiTect (Qiagen). Reverse transcribed cDNA was used for quantitative PCR (qPCR) with 2x iTaq SyBr Green qPCR mix (Biorad). GAPDH was used for normalization between samples.
Western blot analysis
Total whole cell extract was prepared by first lysing cells in Lysis buffer containing 50 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS. Samples were quantified by Bradford assay for even loading. Serial dilutions of cell lysates were loaded on a NuPAGE 4–12% or BioRad 4–20% gradient SDS polyacrylamide gel, separated by electrophoresis, transferred to a PVDF membrane. After 1hr blocking with 5% skim milk in PBST (1x PBS, and 0.1% Tween-20), the membrane was incubated with primary antibodies for overnight at 4˚C and then secondary antibodies (1:20,000) at room temperature for 30min with 1% skim milk in PBST followed by three washing steps. Primary antibodies were used with the following dilutions- 1:2000 for anti-FMRP 1c3 (Millipore-Sigma MAB2160), 1:1000 for anti-TET1 (Sigma-Aldrich 09–872), 1:1000 for anti-TET2 (Millipore-Sigma MABE462), 1:5000 for anti-tubulin (Sigma-Aldrich T5201) and anti-GAPDH (Invitrogen GA1R).
Knock-down assays
To degrade the nascent FMR1 transcript by RNase H1 activity, we used an FMR1 ASO gapmer with 5nt of flanking 2-methoxyethyl (2’ MOE) nucleotides. Control ASO is a scrambled gapmer ASO. For TET1, TET2, DNMT1, MSH2, XPG, and CSB (also known as ERCC6) knock-down, ON-TARGETplus human siRNA SmartPool (Dharmacon) sets were used. Lipofectamine 3000 was used for introducing ASOs (20nM final conc.) and siRNAs (45–90 nM final conc.) into the cells the day after splitting every 3 days.
Transcriptome analysis by RNA-seq
The whole transcriptome analysis was performed by making RNA-seq libraries from Poly(A) selected RNA samples using NEBNext Poly(A) mRNA Magnetic Isolation Module (#E7490) and NEBNext Ultra™ II Directional RNA Library Prep Kit for Illumina (#E7760S) by following the protocols from the manufacturer. NEBNext Multiplex Oligos for Illumina was used for barcoding the samples. Short-read sequencing by NovaSeq 6000 system (S4 flowcell) was performed by Novogene. The numbers of raw reads for gScr_rep1, gScr_rep2, gNHG3_rep1, gNHG3_rep2 are 80390332, 70236048, 84195678, 80627830, respectively. Trim-galore (https://github.com/FelixKrueger/TrimGalore) was used for preprocessing of the raw reads, and TopHat268 was used to align the RNA-seq reads to hg19 genome with default parameters. Generated bam files were further utilized by DEseq269, featureCounts70, and enhancedVolcano for analyzing the differentially expressed genes from total 28152 genes.
QUANTIFICATION AND STATISTICAL ANALYSIS
We conducted t-test for most of the statistical analysis if not specified. Statistical significance is indicated by the following notations: ns for P > 0.05, * for P ≤ 0.05, ** for P ≤ 0.01, *** for P ≤ 0.001, and **** for P ≤ 0.0001. At least three biological replicates were tested if not specified. Error bars in bar graphs or line graphs represent standard deviation of the data.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-FMRP (clone 1c3) | Millipore-Sigma | Cat# MAB2160; RRID:AB_2283007 |
| Rabbit polyclonal anti-TET1 (purified) | Millipore-Sigma | Cat# 09–872; RRID:AB_10806199 |
| Mouse monoclonal anti-TET2 (clone hT2H 21F11) | Sigma-Aldrich | Cat# MABE462; RRID:AB_2923169 |
| Mouse monoclonal anti-beta Tubulin (clone Tub2.1) | Sigma-Aldrich | Cat# T5201; RRID:AB_609915 |
| Rabbit monoclonal anti-GAPDH (GA1R) | Thermo Fisher Scientific | Cat# MA5-15738-D800; RRID:AB_2537658 |
| Mouse monoclonal anti-DNA-RNA Hybrid Antibody (clone S9.6) | Kerafast inc. | Cat# ENH001; RRID:AB_2861387 |
| Mouse monoclonal anti-Histone H3 (di methyl K9) antibody | ABCAM INC | Cat# ab1220; RRID:AB_449854 |
| Rabbit polyclonal anti-Histone H3 (tri methyl K9) antibody | ABCAM INC | Cat# ab8898; RRID:AB_306848 |
| Rabbit polyclonal 5-Hydroxymethylcytosine (5-hmC) antibody (pAb) | ACTIVE MOTIF | Cat# 39769; RRID:AB_10013602 |
| Mouse monoclonal 5-Methylcytosine (5-mC) antibody (mAb clone 33D3) | ACTIVE MOTIF | Cat# 39649; RRID:AB_2687950 |
| Rabbit polyclonal anti-gamma H2A.X (phospho S139) antibody | ABCAM INC | Cat# ab2893; RRID:AB_303388 |
| Rabbit normal IgG antibody | Cell Signaling | Cat # 2729; RRID:AB_1031062 |
| Bacterial and virus strains | ||
| One Shot Stbl3 Chemically Competent E. coli | Thermo Fisher Scientific | Cat# C737303 |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Naive Stem Cell 5i inhibitor Set | Axon Medi Chem | Cat# Axon5011 |
| recombinant human LIF | Peprotech | Cat# 300–05 |
| Activin A | Peprotech | Cat# 120–14E |
| Decitabine | SELLECK CHEMICALS LLC | Cat# S1200 |
| ROCKi Y-27632 | SELLECK CHEMICALS LLC | Cat# S1049 |
| Recombinant Human FGF basic/FGF2/bFGF | R & D SYSTEMS INC | Cat# 233-FB-025/CF |
| Recombinant Human EGF Protein | R & D SYSTEMS INC | Cat# 236-EG-200 |
| Heparin sodium salt | R & D SYSTEMS INC | Cat# 2812/100 |
| Blasticidin S HCl | FISHER SCIENTIFIC | Cat# R210–01 |
| Puromycin Dihydrochloride | LIFE TECHNOLOGIES CORPORATION | Cat# A1113803 |
| 5M Betaine solution | SIGMA-ALDRICH INC | Cat# B0300–5VL |
| Critical commercial assays | ||
| Agilent High Sensitivity DNA kit | Agilent | Cat# 5067–4626 |
| QuantiTect Reverse Transcription Kit | QIAGEN INC | Cat# 205311 |
| RNAzol RT | SIGMA-ALDRICH INC | Cat# R4533 |
| Lenti-X GoStix Plus | TAKARA BIO USA | Cat# 631280 |
| ZYMO Quick-gDNA MiniPrep Plus | ZYMOGEN | Cat# D4068 |
| NEBNext Poly(A) mRNA Magnetic Isolation Module | NEW ENGLAND BIOLABS | Cat# E7490S |
| NEBNext Ultra™ II Directional RNA Library Prep Kit for Illumina | NEW ENGLAND BIOLABS | Cat# E7760S |
| NEBNext Multiplex Oligos for Illumina | NEW ENGLAND BIOLABS | Cat# E7500S |
| QIAquick PCR Purification Kit | QIAGEN INC | Cat# 28104 |
| Pyrosequencing assays for FMR1 promoter | EpigenDx | Cat# ADS1451-FS2 |
| Deposited data | ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE184580 |
| Experimental models: Cell lines | ||
| Fragile X Patient Fibroblasts (GM05848) | CORIELL INSTITUTE | Cat# GM05848 |
| WCMC37 FXS human ES cells | Colak et al., 2014 1 | N.A. |
| FXS 848-iPS1 | Sheridan et al., 2011 2 | N.A. |
| WT 8330-iPS | Sheridan et al., 2011 2 | N.A. |
| Premutation 131-iPS | Sheridan et al., 2011 2 | N.A. |
| 848 FXS Neuronal Progenitor Cells | Sheridan et al., 2011 2 | N.A. |
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
| ON-TARGETplus Human TET1 siRNA – SMARTpool | Horizon Discovery | L-014635-03-0005 |
| ON-TARGETplus Human TET2 siRNA – SMARTpool | Horizon Discovery | L-013776-03-0005 |
| ON-TARGETplus DNMT1 siRNA SmartPool | Horizon Discovery | L-004605-00-0005 |
| ON-TARGETplus MSH2 siRNA SmartPool | Horizon Discovery | L-003909-00-0005 |
| ON-TARGETplus ERCC5(XPG) siRNA - SMARTpool | Horizon Discovery | L-006626-00-0005 |
| ON-TARGETplus ERCC6(CSB) siRNA SmartPool | Horizon Discovery | L-004888-00-0005 |
| ON-TARGETplus Non-targeting Pool | Horizon Discovery | D-001810-10-05 |
| FMR1-RT-5:CAGGGCTGAAGAGAAGATGG | Eiges et al., 2007 3 | N.A. |
| FMR1-RT-3:ACAGGAGGTGGGAATCTGA | Eiges et al., 2007 3 | N.A. |
| GAPDH.ex.F1:AGCCACATCGCTCAGACACC | Itskovitz-Eldor et al., 20004 | N.A. |
| GAPDH.ex.R1:GTACTCAGCGGCCAGCATCG | Itskovitz-Eldor et al., 20004 | N.A. |
| TET1.F1:CATCAGTCAAGACTTTAAGCCCT | Poole et al., 20195 | N.A. |
| TET1.R1:CGGGTGGTTTAGGTTCTGTTT | Poole et al., 20195 | N.A. |
| TET2.F1:GATAGAACCAACCATGTTGAGGG | Poole et al., 20195 | N.A. |
| TET2.R1:TGGAGCTTTGTAGCCAGAGGT | Poole et al., 20195 | N.A. |
| DNMT1.F1:CCATCAGGCATTCTACCA | Arosio et al., 2021 6 | N.A. |
| DNMT1.R1: CGTTCTCCTTGTCTTCTC | Arosio et al., 2021 6 | N.A. |
| MSH2.F1:GGAAAAGAAGATGCAGTCAACA | Ku et al., 2010 7 | N.A. |
| MSH2.R1:CAAACTGTGCACTGGAATCC | Ku et al., 2010 7 | N.A. |
| XPG.F1:CCCGGACTCTTTCAGCCATT | This study | N.A. |
| XPG.R1:ATTTCTTCTGCTCCTGGCCC | This study | N.A. |
| CSB.F1:AAATCTGTGCACTTTCCATAGAACTTC | Baas et al., 2010 8 | N.A. |
| CSB.R1:TATTCTGGCTTGAGTTTCCAAATTC | Baas et al., 2010 8 | N.A. |
| FMR1.TSS.F:gaacagcgttgatcacgtga | Colak et al., 2014 1 | N.A. |
| FMR1.TSS.R:accggaagtgaaaccgaaac | Colak et al., 2014 1 | N.A. |
| FMR1-int1-F:TTGCCCCTTAGTTCCCTGAG | Loomis et al., 2014 9 | N.A. |
| FMR1-int1-R:TCTTCCATCAGTGCAGACCA | Loomis et al., 2014 9 | N.A. |
| FMR1-ups-F:ACAGTGGAATGTAAAGGGTTG | Groh et al., 2014 10 | N.A. |
| FMR1-ups-R:GTGTTAAGCACTTGAGGTTCAT | Groh et al., 2014 10 | N.A. |
| FMR1-int15-F:GAACTTCCAGTAAGCATTTCAG | Groh et al., 2014 10 | N.A. |
| FMR1-ex16-R:CTGTTGTTCTTCCTTTAGCCTCTC | Groh et al., 2014 10 | N.A. |
| FMR1-RT-3:ACAGGAGGTGGGAATCTGA | Eiges et al., 2007 3 | N.A. |
| FMR1-RT-5:CAGGGCTGAAGAGAAGATGG | Eiges et al., 2007 3 | N.A. |
| FMR1-Not_FraxC:AGTTCAGCGGCCGCGCTCAGCTCCGTTTCGGTTTCACTTCCGGT | Hayward et al., 2016 11 | N.A. |
| FMR1-Not_FraxR4:CAAGTCGCGGCCGCCTTGTAGAAAGCGCCATTGGAGCCCCGCA | Hayward et al., 2016 11 | N.A. |
| AFF2-CCG-F1:CCCCCTGTGAGTGTGTAAGTG | This study | N.A. |
| AFF2-CCG-R1:CTGGAGGTGTCTGGTCCTGT | This study | N.A. |
| RGPD1-CGG.F1:TGCTGACGCAGTACACAAGT | This study | N.A. |
| RGPD1-CGG.R1:ACTGCACAATATTCCCCAACAG | This study | N.A. |
| RGPD1-CGG.F2:GTTGGAATTGGCGACTGCTG | This study | N.A. |
| RGPD1-CGG.R2:CTGCACAATATTCCCCAACAGA | This study | N.A. |
| RGPD2-CGGx2.F1:TTCCGTTGAAATCCCCAGGTT | This study | N.A. |
| RGPD2-CGGx2.R1:GTCGCCTGGAAAGGTGAGTG | This study | N.A. |
| Gapmer-FMR1:/52MOErC/*/i2MOErC/*/i2MOErG/*/i2MOErC/*/i2MOErC/*G*C*C*G*C*G*C*T*G*C*/i2MOErC/*/i2MOErG/*/i2MOErC/*/i2MOErA/*/32MOErC/ | This study | N.A. |
| Gapmer-Scramble:/52MOErG/*/i2MOErC/*/i2MOErG/*/i2MOErA/*/i2MOErC/*T*A*T*A*C*G*C*G*C*A*/i2MOErA/*/i2MOErU/*/i2MOErA/*/i2MOErU/*/32MOErG/ | This study | N.A. |
| scramble gRNA_For:TTGG cccccgggggaaaaattttt | Liu et al., 201812 | N.A. |
| scramble gRNA_Rev:AAAC aaaaatttttcccccggggg | Liu et al., 201812 | N.A. |
| CGG repeat targeting gRNA_For:TTGG GGCGGCGGCGGCGGCGGCGG | Liu et al., 201812 | N.A. |
| CGG repeat targeting gRNA_Rev:AAAC CCGCCGCCGCCGCCGCCGCC | Liu et al., 201812 | N.A. |
| NHG2-gRNA_For:TTGG GGCGTGCGGCAGCGCGGCGG | This study | N.A. |
| NHG2-gRNA_Rev:AAAC CCGCCGCGCTGCCGCACGCC | This study | N.A. |
| NHG3-gRNA_For:TTGG GTGCGGCAGCGCGGCGGCGG | This study | N.A. |
| NHG3-gRNA_Rev:AAAC CCGCCGCCGCGCTGCCGCAC | This study | N.A. |
| gUnique-gRNA_For:TTGG GTGACGGAGGCGCCGCTGCC | This study | N.A. |
| gUnique-gRNA_Rev:AAAC GGCAGCGGCGCCTCCGTCAC | This study | N.A. |
| Recombinant DNA | ||
| pgRNA-modified | ADDGENE | 84477 |
| pgRNA-CGG | ADDGENE | 108248 |
| pLV_hEF1a_rtTA3 (Plasmid #61472) | ADDGENE | 61472 |
| Fuw-dCas9-Tet1CD-P2A-BFP | ADDGENE | 108245 |
| Fuw-dCas9-dead Tet1CD-P2A-BFP | ADDGENE | 108246 |
| pGH125_dCas9-Blast | ADDGENE | 85417 |
| pTRE-dCas9-Blast | This paper | |
| pTRE-dCas9-RNaseH1-Blast | This paper | |
| psPAX2 | ADDGENE | 12260 |
| pMD2.G | ADDGENE | 12259 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 201213 | https://imagej.nih.gov/ij/ |
| TopHat2 | Kim et al., 201314 | https://ccb.jhu.edu/software/tophat/index.shtml |
| DEseq2 | Love et al., 201415 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| featureCounts | Liao et al., 201416 | https://subread.sourceforge.net/featureCounts.html |
| enhancedVolcano | Blighe et al., 2020 | https://github.com/kevinblighe/EnhancedVolcano |
| Prism | GraphPad | https://www.graphpad.com |
| Other | ||
| mTeSR™1 Medium for Human ES and iPS Cells | STEMCELL TECHNOLOGIES INC | Cat# 85850 |
| RSeT medium (2-components) | STEMCELL TECHNOLOGIES INC | Cat# 05978 |
| DMEM/F12 Media | LIFE TECHNOLOGIES CORPORATION | Cat# 11320-033 |
| Neurobasal medium | LIFE TECHNOLOGIES CORPORATION | Cat# 21103-049 |
| Corning Ham’s F12 Medium w L-Glutamine | FISHER SCIENTIFIC | Cat# MT10080CV |
| Accutase | STEMCELL TECHNOLOGIES INC | Cat# 07920 |
| Corning Matrigel hESC-Qualified Matrix | FISHER SCIENTIFIC | Cat# 08-774-552 |
| Geltrex LDEV-Free, hESC-Qualified, Reduced Growth Factor Basement Membrane Matrix | LIFE TECHNOLOGIES CORPORATION | Cat# A1413302 |
| L-Glutamine (200 mM) | LIFE TECHNOLOGIES CORPORATION | Cat# 25030-081 |
| TrypLE Select Enzyme (1X), no phenol red | LIFE TECHNOLOGIES CORPORATION | Cat# 12563011 |
| N2 supplement (100X) | LIFE TECHNOLOGIES CORPORATION | Cat# 17502048 |
| B-27 Supplement (50X), serum free | LIFE TECHNOLOGIES CORPORATION | Cat# 17504044 |
| B-27 Supplement (50X), minus vitamin A | LIFE TECHNOLOGIES CORPORATION | Cat# 12587010 |
| Lentivirus Stabilizer | ORIGENE TECH | Cat# TR30039 |
| Lenti Concentrator | ORIGENE TECH | Cat# TR30025 |
| Phusion High-Fidelity DNA Polymerase | NEW ENGLAND BIOLABS INC | Cat# M0530S |
| RNaseH | TAKARA BIO CORPORATION | Cat# 2150B |
| Polybrene Infection / Transfection Reagent | SIGMA-ALDRICH INC | Cat# TR-1003-G |
| HpaII restriction enzyme | NEW ENGLAND BIOLABS INC | Cat# R0171S |
| EcoNI | NEW ENGLAND BIOLABS INC | Cat# R0521L |
| HindIII HF (High Fidelity) | NEW ENGLAND BIOLABS INC | Cat# R3104M |
| BsrGI | NEW ENGLAND BIOLABS INC | Cat# R0575L |
| EcoRI | NEW ENGLAND BIOLABS INC | Cat# R0101L |
| XbaI | NEW ENGLAND BIOLABS INC | Cat# R0145L |
| Dynabeads Protein G | LIFE TECHNOLOGIES CORPORATION | Cat# 10003D |
| Dynabeads Protein A | LIFE TECHNOLOGIES CORPORATION | Cat# 10001D |
| Falcon 40μm Cell Strainer | FISHER SCIENTIFIC | Cat# 08-771-1 |
| iTaq UniverSYBR Green SMX 5000 | BIO-RAD LABS LIFE SCIENCES GROUP | Cat# 1725125 |
| Lipofectamine 3000 Transfection Reagent | LIFE TECHNOLOGIES CORPORATION | Cat# L3000008 |
ACKNOWLEDGMENTS
We thank SR Jaffrey, N Zaninovic, R Willemsen, and E Berry-Kravis for providing FXS cell lines; H Jacob for sharing information about naive state media; and B Bujisic, J Kim, Y Takeuchi, T Dial, J Wang, and T Fu for experimental suggestions and critical feedback. This study was supported by grants from the FRAXA Research Foundation to J.T.L. and H.-G.L., and MGH Sundry Funds to J.T.L. We are also grateful to the Pierce Family Fragile X Foundation for their longstanding support.
Footnotes
DECLARATION OF INTERESTS
J.T.L. is a founder of Translate Bio and Fulcrum Therapeutics, and is an Advisor to Skyhawk Therapeutics.
REFERENCES
- 1.Hagerman RJ, Des-Portes V, Gasparini F, Jacquemont S, and Gomez-Mancilla B. (2014). Translating molecular advances in fragile X syndrome into therapy: a review. J Clin Psychiatry 75, e294–307. 10.4088/JCP.13r08714. [DOI] [PubMed] [Google Scholar]
- 2.Hagerman RJ, Berry-Kravis E, Hazlett HC, Bailey DB Jr., Moine H, Kooy RF, Tassone F, Gantois I, Sonenberg N, Mandel JL, and Hagerman PJ (2017). Fragile X syndrome. Nat Rev Dis Primers 3, 17065. 10.1038/nrdp.2017.65. [DOI] [PubMed] [Google Scholar]
- 3.Berry-Kravis EM, Harnett MD, Reines SA, Reese MA, Ethridge LE, Outterson AH, Michalak C, Furman J, and Gurney ME (2021). Inhibition of phosphodiesterase-4D in adults with fragile X syndrome: a randomized, placebo-controlled, phase 2 clinical trial. Nat Med 27, 862–870. 10.1038/s41591-021-01321-w. [DOI] [PubMed] [Google Scholar]
- 4.Berry-Kravis EM, Lindemann L, Jonch AE, Apostol G, Bear MF, Carpenter RL, Crawley JN, Curie A, Des Portes V, Hossain F, et al. (2018). Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nat Rev Drug Discov 17, 280–299. 10.1038/nrd.2017.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagerman RJ, and Hagerman PJ (2021). Fragile X Syndrome: Lessons Learned and What New Treatment Avenues Are on the Horizon. Annu Rev Pharmacol Toxicol. 10.1146/annurev-pharmtox-052120-090147. [DOI] [PubMed] [Google Scholar]
- 6.Kang Y, Zhou Y, Li Y, Han Y, Xu J, Niu W, Li Z, Liu S, Feng H, Huang W, et al. (2021). A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies. Nat Neurosci. 10.1038/s41593-021-00913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lubs HA (1969). A marker X chromosome. Am J Hum Genet 21, 231–244. [PMC free article] [PubMed] [Google Scholar]
- 8.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, and et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914. 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 9.Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, and Nelson DL (1991). Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66, 817–822. 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 10.Kremer EJ, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren ST, Schlessinger D, Sutherland GR, and Richards RI (1991). Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science 252, 1711–1714. 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 11.Yu S, Pritchard M, Kremer E, Lynch M, Nancarrow J, Baker E, Holman K, Mulley JC, Warren ST, Schlessinger D, and et al. (1991). Fragile X genotype characterized by an unstable region of DNA. Science 252, 1179–1181. 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]
- 12.Nelson DL, Orr HT, and Warren ST (2013). The unstable repeats--three evolving faces of neurological disease. Neuron 77, 825–843. 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidorov MS, Auerbach BD, and Bear MF (2013). Fragile X mental retardation protein and synaptic plasticity. Mol Brain 6, 15. 10.1186/1756-6606-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomazeau A, Bosch M, Essayan-Perez S, Barnes SA, De Jesus-Cortes H, and Bear MF (2020). Dissociation of functional and structural plasticity of dendritic spines during NMDAR and mGluR-dependent long-term synaptic depression in wild-type and fragile X model mice. Mol Psychiatry. 10.1038/s41380-020-0821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tassone F, Beilina A, Carosi C, Albertosi S, Bagni C, Li L, Glover K, Bentley D, and Hagerman PJ (2007). Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA 13, 555–562. 10.1261/rna.280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, et al. (2010). Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J 29, 1248–1261. 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V, et al. (2013). CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455. 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik I, Kelley CP, Wang ET, and Todd PK (2021). Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat Rev Mol Cell Biol 22, 589–607. 10.1038/s41580-021-00382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rousseau F, Heitz D, Biancalana V, Blumenfeld S, Kretz C, Boue J, Tommerup N, Van Der Hagen C, DeLozier-Blanchet C, Croquette MF, and et al. (1991). Direct diagnosis by DNA analysis of the fragile X syndrome of mental retardation. N Engl J Med 325, 1673–1681. 10.1056/NEJM199112123252401. [DOI] [PubMed] [Google Scholar]
- 21.Mirkin SM (2007). Expandable DNA repeats and human disease. Nature 447, 932–940. 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 22.Moore H, Greenwell PW, Liu CP, Arnheim N, and Petes TD (1999). Triplet repeats form secondary structures that escape DNA repair in yeast. Proc Natl Acad Sci U S A 96, 1504–1509. 10.1073/pnas.96.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usdin K, House NC, and Freudenreich CH (2015). Repeat instability during DNA repair: Insights from model systems. Crit Rev Biochem Mol Biol 50, 142–167. 10.3109/10409238.2014.999192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinaldi C, Pizzul P, Longhese MP, and Bonetti D. (2020). Sensing R-Loop-Associated DNA Damage to Safeguard Genome Stability. Front Cell Dev Biol 8, 618157. 10.3389/fcell.2020.618157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiraanont P, Kumar M, Tang HT, Espinal G, Hagerman PJ, Hagerman RJ, Chutabhakdikul N, and Tassone F. (2017). Size and methylation mosaicism in males with Fragile X syndrome. Expert Rev Mol Diagn 17, 1023–1032. 10.1080/14737159.2017.1377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Kumari D, Sciascia N, and Usdin K. (2016). CGG-repeat dynamics and FMR1 gene silencing in fragile X syndrome stem cells and stem cell-derived neurons. Mol Autism 7, 42. 10.1186/s13229-016-0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, and Haggarty SJ (2011). Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One 6, e26203. 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD, and Jaffrey SR (2014). Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 343, 1002–1005. 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayward BE, Zhou Y, Kumari D, and Usdin K. (2016). A Set of Assays for the Comprehensive Analysis of FMR1 Alleles in the Fragile X-Related Disorders. J Mol Diagn 18, 762–774. 10.1016/j.jmoldx.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mor-Shaked H, and Eiges R. (2016). Modeling Fragile X Syndrome Using Human Pluripotent Stem Cells. Genes (Basel) 7. 10.3390/genes7100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. (2013). Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286. 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 32.Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. (2014). Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency. Cell Stem Cell 15, 524–526. 10.1016/j.stem.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Esch CE, Ghazvini M, Loos F, Schelling-Kazaryan N, Widagdo W, Munshi ST, van der Wal E, Douben H, Gunhanlar N, Kushner SA, et al. (2014). Epigenetic characterization of the FMR1 promoter in induced pluripotent stem cells from human fibroblasts carrying an unmethylated full mutation. Stem Cell Reports 3, 548–555. 10.1016/j.stemcr.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig AL, Espinal GM, Pretto DI, Jamal AL, Arque G, Tassone F, Berman RF, and Hagerman PJ (2014). CNS expression of murine fragile X protein (FMRP) as a function of CGG-repeat size. Hum Mol Genet 23, 3228–3238. 10.1093/hmg/ddu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Primerano B, Tassone F, Hagerman RJ, Hagerman P, Amaldi F, and Bagni C. (2002). Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA 8, 1482–1488. [PMC free article] [PubMed] [Google Scholar]
- 36.Coffee B, Zhang F, Ceman S, Warren ST, and Reines D. (2002). Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am J Hum Genet 71, 923–932. 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groh M, Lufino MM, Wade-Martins R, and Gromak N. (2014). R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet 10, e1004318. 10.1371/journal.pgen.1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213. 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saba HI (2007). Decitabine in the treatment of myelodysplastic syndromes. Ther Clin Risk Manag 3, 807–817. [PMC free article] [PubMed] [Google Scholar]
- 40.Liu XS, Wu H, Krzisch M, Wu X, Graef J, Muffat J, Hnisz D, Li CH, Yuan B, Xu C, et al. (2018). Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 172, 979–992 e976. 10.1016/j.cell.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, and Doudna JA (2016). Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871. 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang F, and Doudna JA (2017). CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys 46, 505–529. 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 43.Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, and Seidel R. (2014). Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A 111, 9798–9803. 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Rodriguez Y, Ross RL, Zhao R, Watts JA, Grunseich C, Bruzel A, Li D, Burdick JT, Prasad R, et al. (2020). RNA abasic sites in yeast and human cells. Proc Natl Acad Sci U S A 117, 20689–20695. 10.1073/pnas.2011511117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niehrs C, and Luke B. (2020). Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol 21, 167–178. 10.1038/s41580-019-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hegazy YA, Fernando CM, and Tran EJ (2020). The balancing act of R-loop biology: The good, the bad, and the ugly. J Biol Chem 295, 905–913. 10.1074/jbc.REV119.011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loomis EW, Sanz LA, Chedin F, and Hagerman PJ (2014). Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet 10, e1004294. 10.1371/journal.pgen.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips DD, Garboczi DN, Singh K, Hu Z, Leppla SH, and Leysath CE (2013). The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit 26, 376–381. 10.1002/jmr.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deleavey GF, and Damha MJ (2012). Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol 19, 937–954. 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 50.Bennett CF, and Swayze EE (2010). RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol 50, 259–293. 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 51.Arab K, Karaulanov E, Musheev M, Trnka P, Schafer A, Grummt I, and Niehrs C. (2019). GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat Genet 51, 217–223. 10.1038/s41588-018-0306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Y, Dent SY, Wilson JH, Wells RD, and Napierala M. (2010). R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci U S A 107, 692–697. 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freudenreich CH (2018). R-loops: targets for nuclease cleavage and repeat instability. Curr Genet 64, 789–794. 10.1007/s00294-018-0806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, and Cimprich KA (2014). Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 56, 777–785. 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cristini A, Ricci G, Britton S, Salimbeni S, Huang SN, Marinello J, Calsou P, Pommier Y, Favre G, Capranico G, et al. (2019). Dual Processing of R-Loops and Topoisomerase I Induces Transcription-Dependent DNA Double-Strand Breaks. Cell Rep 28, 3167–3181 e3166. 10.1016/j.celrep.2019.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin Y, and Wilson JH (2007). Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol 27, 6209–6217. 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin Y, and Wilson JH (2012). Nucleotide excision repair, mismatch repair, and R-loops modulate convergent transcription-induced cell death and repeat instability. PLoS One 7, e46807. 10.1371/journal.pone.0046807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pearson CE, Ewel A, Acharya S, Fishel RA, and Sinden RR (1997). Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum Mol Genet 6, 1117–1123. 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 59.Owen BA, Yang Z, Lai M, Gajec M, Badger JD 2nd, Hayes JJ, Edelmann W, Kucherlapati R, Wilson TM, and McMurray CT (2005). (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol 12, 663–670. 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 60.Lin Y, and Wilson JH (2009). Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair (Amst) 8, 878–885. 10.1016/j.dnarep.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakatani R, Nakamori M, Fujimura H, Mochizuki H, and Takahashi MP (2015). Large expansion of CTG*CAG repeats is exacerbated by MutSbeta in human cells. Sci Rep 5, 11020. 10.1038/srep11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan-Wong SM, Dhir S, and Proudfoot NJ (2019). R-Loops Promote Antisense Transcription across the Mammalian Genome. Mol Cell 76, 600–616 e606. 10.1016/j.molcel.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ginno PA, Lott PL, Christensen HC, Korf I, and Chedin F. (2012). R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45, 814–825. 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pearson CE, and Sinden RR (1996). Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry 35, 5041–5053. 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 65.Gacy AM, Goellner G, Juranic N, Macura S, and McMurray CT (1995). Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81, 533–540. 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 66.Hirst MC, and White PJ (1998). Cloned human FMR1 trinucleotide repeats exhibit a length- and orientation-dependent instability suggestive of in vivo lagging strand secondary structure. Nucleic Acids Res 26, 2353–2358. 10.1093/nar/26.10.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y, Smyth GK, and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing data reported in this study have been deposited at the Gene Expression Omnibus with accession number GSE184580.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE184580.
This paper does not report original codes.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.