Abstract
Influenza A virus expresses three viral polymerase (P) subunits—PB1, PB2, and PA—all of which are essential for RNA and viral replication. The functions of P proteins in transcription and replication have been partially elucidated, yet some of these functions seem to be dependent on the formation of a heterotrimer for optimal viral RNA transcription and replication. Although it is conceivable that heterotrimer subunit interactions may allow a more efficient catalysis, direct evidence of their essentiality for viral replication is lacking. Biochemical studies addressing the molecular anatomy of the P complexes have revealed direct interactions between PB1 and PB2 as well as between PB1 and PA. Previous studies have shown that the N-terminal 48 amino acids of PB1, termed domain α, contain the residues required for binding PA. We report here the refined mapping of the amino acid sequences within this small region of PB1 that are indispensable for binding PA by deletion mutagenesis of PB1 in a two-hybrid assay. Subsequently, we used site-directed mutagenesis to identify the critical amino acid residues of PB1 for interaction with PA in vivo. The first 12 amino acids of PB1 were found to constitute the core of the interaction interface, thus narrowing the previous boundaries of domain α. The role of the minimal PB1 domain α in influenza virus gene expression and genome replication was subsequently analyzed by evaluating the activity of a set of PB1 mutants in a model reporter minigenome system. A strong correlation was observed between a functional PA binding site on PB1 and P activity. Influenza viruses bearing mutant PB1 genes were recovered using a plasmid-based influenza virus reverse genetics system. Interestingly, mutations that rendered PB1 unable to bind PA were either nonviable or severely growth impaired. These data are consistent with an essential role for the N terminus of PB1 in binding PA, P activity, and virus growth.
The influenza A virus genome consists of eight single-stranded RNA segments of negative polarity (viral RNAs [vRNAs]) (26, 36). In virions, each vRNA segment is assembled into a ribonucleoprotein particle (RNP) by association with the nucleoprotein and the three viral polymerase subunits (PB1, PB2, and PA; herein referred to as P proteins) (30). Other viral proteins mediate morphogenesis of virions by budding at the plasma membrane. Upon entry into a permissive cell by endocytosis and envelope fusion, viral RNPs travel to the nucleus, where transcription and replication of vRNA segments takes place (7, 21). The viral RNPs constitute the active transcription and replication competent unit (25). During infection, incoming vRNAs are initially transcribed into viral mRNAs. Nascent host cell polymerase II pre-mRNA transcripts provide capped oligonucleotides to initiate viral transcription (6). Polyadenylation of viral transcripts to yield mRNA occurs through a polymerase stuttering mechanism involving an oligo(U) signal adjacent to the RNA panhandle structure at the 5′ termini of the vRNA genes (40, 45). During replication, the P proteins switch to a primer-independent mode of RNA synthesis, generating full-length copies of cRNA which are used as intermediates for the production of progeny vRNAs.
The P proteins are found largely as heterotrimeric complexes within virions or the nuclei of infected cells (12, 33). Of the three P proteins, PB1 is the best characterized functionally. Biochemical and structural analyses recognize PB1 as responsible for RNA chain elongation. PB1 contains amino acid motifs common to all RNA-dependent RNA polymerases and RNA-dependent DNA polymerases (32). Mutations within these motifs render the complex inactive for transcription and replication in tissue culture cells (5). In cell-free systems, e.g., nuclear extracts of insect cells expressing PB1 alone, the protein can catalyze the synthesis of RNA using synthetic short minigenome model RNAs as templates, in a primer-dependent mode. PB2 activity appears essential for transcription (27). PB2 binds to methylated cap-1 structures at the 5′ termini of actively transcribed cellular mRNAs which are subsequently endonucleolytically cleaved by the P complex, producing 10- to 13-mers used for priming of viral mRNA transcription (6). On the other hand, PA seems required for vRNA replication, although its role in this process remains obscure. Temperature-sensitive influenza PA mutants are severely impaired in vRNA replication with normal viral mRNA synthesis (29).
At least one essential role has been tentatively assigned to each P protein in viral replication, yet some of these functions seem to be dependent on the formation of the heterotrimer for optimal viral RNA transcription and replication (24, 25). Although it is conceivable that heterotrimer subunit interactions may allow more efficient catalysis, direct evidence of their essentiality for viral replication is lacking. Elucidating the P protein interactions will help understand the integration of P protein catalytic activities in viral RNA replication and mRNA transcription. Using a mammalian two-hybrid system, we have previously identified the N-terminal 48-amino-acid region of PB1, termed domain α, as a determinant of interaction with PA (39). The interaction between PA and domain α appeared neither modulated nor altered by other regions of the PB1 polypeptide. This observation is supported by the findings of Toyoda et al. and González et al. (19, 42). Interestingly, a high degree of sequence conservation was found within the domain α of homologous PB1 polymerases from influenza virus types B and C, the salmon orthomyxovirus, and the orthomyxovirus-like insect viruses Thogoto and Dhori (39). Here we report the high-resolution functional map of the PB1 domain α boundaries: only the N-terminal 12 amino acids of PB1 are required for interaction with PA. We used a model influenza virus reporter minigenome (RG) to analyze the role of specific PB1 regions in viral gene expression and genome replication (25, 38). Efficient transcription of viral RG into translatable mRNA requires the PA protein and a functional PA binding α domain on PB1. Influenza viruses bearing these mutant PB1 genes were either nonviable or displayed severely impaired growth. These results indicate that the PB1-PA interaction is essential for virus viability in cell culture.
MATERIALS AND METHODS
Enzymes reagents and software.
DNA restriction and modification enzymes were purchased from Promega (Madison, Wis.), New England Biolabs (Beverly, Mass.), and Roche Molecular Biochemicals (Indianapolis, Ind.). Reagents for DNA sequencing were obtained from United States Biochemical (Cleveland, Ohio). Fetal bovine serum (FBS), antibiotic solution (containing penicillin and streptomycin), l-glutamine, sodium butyrate, and butiryl coenzyme A (butiryl-CoA) were from Sigma (St. Louis, Mo.). Dulbecco's modified essential medium (DMEM) and Lipofectamine reagent were bought from Gibco/BRL (Gaithersburg, Md.), and [14C]chloramphenicol and Tran35S-label were bought from ICN (Costa Mesa, Calif.). Oligonucleotides were purchased from Ransom Hill (Ramona, Calif.). Sequence alignments were performed using the Wisconsin Genetics Computer Group package (13) and influenza virus type A, A/WSN/33 (Swissprot accession no. P03430); influenza virus type B, B/Lee/40 (Swissprot accession no. P07832); and influenza virus type C, C/JJ/50 (Swissprot accession no. P19703). Secondary-structure prediction analyses were done with the Jpred multiple approach consensus algorithm of Cuff and colleagues (8, 9). Rabbit monspecific polyclonal antisera against PB1, PB2, and PA proteins were kindly provided by D. Nayak (1).
Cells and vaccinia T7 recombinant virus.
CV-1 and Cos-7 African green monkey cells (ATCC CCL-70 and CRL-1651, respectively; American Type Culture Collection, Manassas, Ua.) were maintained in DMEM supplemented with 10% (vol/vol) FBS. MDCK and 293T cells were kindly provided by E. Hoffmann and maintained in DMEM–5% FBS. Cloning and mutagenesis procedures were carried out in Escherichia coli mutS and DH10B strains (2). Recombinant vaccinia virus encoding T7 RNA polymerase (vaccinia-T7 virus) strain vTF7-3 was obtained through the AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (catalog no. 356) (18).
Plasmid constructions.
Plasmids pSG424, pAASVVP16, and pG5EC for the two-hybrid system in mammalian cells were provided by H. A. Vasavada and G. N. Nallur (Yale University, New Haven, Conn.) (43). pSGPB1t, pSGPB1c, and pVP16PA have been previously described (39). Plasmids pcDNA762/PB2, pcDNA787/PA, pcDNA693/NP, and p2HHCATHdR for the influenza virus minigenome replication (P5) system have been described previously (38). pCMV/SEAP was provided by B. Cullen (3). pGEMT7Luc was acquired from Promega. Plasmids pHW181 through pHW188 for rescue of influenza viruses were kindly provided by E. Hoffman (23). A SmaI/BamHI fragment from pSGPB1t encoding the first 654 amino acids of the PB1 gene from A/WSN/33 was ligated into pSP72 (Promega), previously digested with BglII, blunt ended with Klenow fragment, and subsequently digested with BamHI. The resulting intermediate plasmid was further manipulated to generate p72PB1c by incorporation of a BamHI/HindIII fragment encoding the last 103 amino acids of PB1 from A/PR/8/34 (in pcDNA774/PB1) (38).
Internal deletions in pSGPB1c were created by inverse PCR using appropriate primer sets with half BamHI at the 5′ ends and the XL-PCR extender kit containing rTth DNA polymerase (Perkin-Elmer, Norwalk, Conn.). Conditions for PCR were 94°C for 1 min, 58°C for 2 min, and 72°C for 5 min for 15 cycles. The PCR strategy led to the introduction of a new BamHI site in the PB1 coding sequence that was used for screening positive clones. The insertion of the extra BamHI site resulted in the substitution of one or two amino acids at the site of the deletion (glycine and serine) except in pSGPB1PCR8, in which the deletion does not change the PB1 coding sequence.
Single amino acid substitutions were incorporated into pSGPB1t or p72PB1c following the method of unique-site elimination described by Deng and Nickoloff (11). pSGPB1 mutants engineered by this method had a single XbaI site eliminated outside the PB1 coding region and contain one or two (depending on the mutagenic primer used) amino acid substitutions within the PB1 sequence. p72PB1c mutants had a unique PvuI site eliminated within the β-lactamase gene (with no change in the amino acid coding sequence) and a single amino acid substitution within the PB1 coding sequence. A PB1 mutant lacking amino acids 2 through 12 (PB1/[−11]) in p72PB1c was also prepared by site-directed mutagenesis using a specific oligonucleotide carrying the deletion mentioned. PB1 mutants containing the entire PB1 open reading frame (ORF) fused to GAL4 were produced by subcloning a BamHI/FspI fragment from pSGPB1c into the pSGPB1 mutant indicated (see Results). All mutations within the PB1 coding sequence were confirmed by sequencing using the dideoxynucleotide chain termination method described by Sanger et al. (41). At least three independent clones for each particular mutation were tested in the two-hybrid and minigenome replication assays. A complete list of oligonucleotides used for the generation of PB1 mutants is available upon request.
p72PB1HA constructs were prepared by subcloning a HindIII/XhoI fragment encoding a hemagglutinin (HA) epitope tag (YPYDVPDYA) from pcDNAHA (D. R. Perez, unpublished data) into p72PB1cs. Further restriction enzyme manipulation rendered a plasmid encoding a PB1 fusion protein in which the last 6 C-terminal amino acids of the wild-type (WT) protein were replaced by a 29-amino-acid region carrying the HA epitope.
pDPPB1 mutants for the influenza virus rescue system were generated using two different approaches. Replacing a BsaBI/EcoRI fragment of pHW182 with the corresponding mutant version from pSGPB1 produced mutations at positions 3 through 14. Replacing a SalI/BsaBI fragment of pHW182 with an oligonucleotide encoding a substitution at either D2 or V3 of PB1's ORF generated PB1 mutants at positions 2 and 3, respectively.
Two-hybrid assay.
CV-1 cells (5 × 105 cells per well) were plated on 35-mm-diameter dishes (Costar, Cambridge, Mass.) 2 to 4 h before transfection. Cells were transfected with 3 μg of total DNA (1 μg of each two-hybrid plasmid and 0.5 μg of each reporter, pG5EC and pCMV/SEAP) and 20 μg of Lipofectamine reagent (Gibco/BRL, Grand Island, N.Y.) per well in a final volume of 1.2 ml of DMEM. DNA-Lipofectamine complexes were left in contact with cells for 6 h at 37°C in 5% CO2 according to the directions of the manufacturer (Gibco/BRL). After transfection, cells were supplemented with 1 ml of DMEM containing 10% (vol/vol) FBS and 10 mM sodium butyrate. Eighteen hours after the start of transfection, DNA-Lipofectamine complexes were removed and replaced with fresh 10% FBS–DMEM.
P5 influenza virus RG assay.
The P5 influenza virus RG assay was performed as previously described with minor modifications (38). CV-1 cells were inoculated with vaccinia-T7 at an input multiplicity of infection (MOI) of 5 and incubated at 37°C in a moist atmosphere containing 5% CO2 for 45 min. Lipofectamine (12 μg) in 100 μl of DMEM without serum was mixed with 0.020 μg of pGEMT7Luc and 0.100 μg of each P5 plasmid (pcDNA774/PB1 in the original P5 system was replaced by p72PB1c), and complexes were allowed to form at room temperature (37). Immediately after infection, cells were washed three times with DMEM–serum-free medium, transfected with the DNA-lipid complexes in a final volume of 1.2 ml in DMEM without serum, and incubated for 4 h at 37°C and 5% CO2. Once transfection was completed, DNA-lipid complexes were removed and cells were incubated for an additional 13 h in the presence of 2 ml of 10% FBS–DMEM. Control replication experiments were performed by substituting the PA encoding plasmid with DNA from pcDNA3neo in the transfection (Invitrogen).
Radiolabeling and coimmunoprecipitation assay.
For metabolic radiolabeling of proteins, Cos-7 cells were infected with vaccinia-T7 at an MOI of 10, as explained above, and transfected with a mixture of DNAs, including 1 μg of pcDNA787/PA or pcDNA762/PB2 and 1 μg of the WT or a mutant version of PB1 in p72PB1c (see Fig. 4). At 7 h postinfection (hpi), cells were washed twice with methionine-free medium and incubated in this medium for 1 h. Subsequently, cells were supplemented with Tran35S-label to reach a final concentration of 50 μCi/ml. Labeling was carried out for 2 h at 37°C in a 5% CO2 moist chamber. After labeling, cells were rinsed twice with 1× phosphate-buffered saline (PBS) and lysed in RIPA buffer (10 mM Tris-HCl, pH 7.5; 2 mM EDTA; 100 mM NaCl; 1% NP-40; 0.5% Na deoxycholate; 0.1% sodium dodecyl sulfate [SDS]; 1% aprotinin). Samples were subdivided into two aliquots and used for coimmunoprecipitation using monospecific polyclonal antibodies against either PB1, PB2, or PA as described (1).
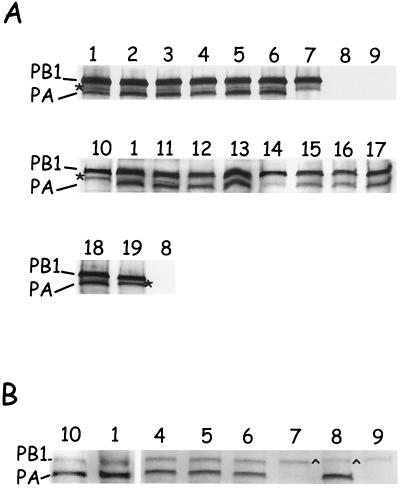
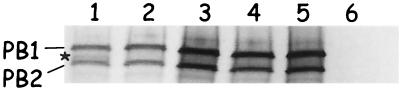
FIG. 4.
Coimmunoprecipitation of PA-PB1 complexes. (A) PA and PB1 proteins were coexpressed in Cos-7 cells using the vaccinia-T7 virus system, radiolabeled, and immunoprecipitated with an anti-PB1 monospecific antibody (see Materials and Methods). Lanes: 1, PA-PB1 WT; 2, PA-T6D; 3, PA-L7D; 4, PA-L8D; 5, PA-F9D; 6, PA-L10D; 7, PB1 WT alone; 8, PA alone; 9, mock-transfected control; 10, PA-PB1/[−11]; 11, PA-D2V; 12, PA-V3D; 13, PA-N4D; 14, PA-P5L; 15, PA-K11D; 16, PA-V12D; 17, PA-P13D; 18, PA-A14D; 19, PB1/[−11] alone. Proteins were separated on an SDS–7.5% PAGE gel, treated for fluorography, and exposed to X-ray film. Approximately 36 h after exposure, the films were developed, scanned, and digitized using Adobe Photoshop software. ∗, presence of the second PB1-specific band. (B) Immunoprecipitation as described in panel A except that anti-PA monospecific antibody was used. Proteins were separated on an SDS–6.0% PAGE gel and treated as described in panel A. are, presence of a vaccinia-expressed protein that coprecipitates with the anti-PA antibody and migrates on the polyacrylamide gel like the PB1 protein.
Recovery of influenza viruses with mutant PB1 genes.
Transfections with eight plasmids for rescue of influenza virus were performed essentially as described by Hoffman et al. (23). At 72 h posttransfection (hpt), virus in supernatant was removed and used to inoculate confluent MDCK cells (second blind passage). Plaque assays using agar overlay were performed with 10-fold serial dilutions of the second-blind-passage virus. Plaque-purified viruses were propagated on fresh MDCK cells and saved for future use. A 200-μl aliquot of the supernatant of the plaque-purified virus was subjected to RNA isolation using RNEasy (Qiagen, Inc., Valencia, Calif.) and reverse transcription-PCR using avian myeloblastosis virus reverse transcriptase and Taq polymerase (Roche Molecular Biochemicals) and a specific set of primers (available upon request). PCR products were purified using the Qiaquick PCR purification kit (Qiagen) and sequenced as described above. Positive plaques were further propagated at least two more times, and RNA was extracted for PCR and sequencing analysis as explained above. In all cases the predicted mutation was maintained in the virus progeny. To compare virus yield, MDCK cells in 75-cm2 flasks were infected with WT or mutant virus at an MOI of 0.1. Two days after infection (6 days in the case of mutants L7D and L10D starting with an MOI of 1 and 3 days in the case of mutant P5L), supernatants were separated from floating cells by low-speed centrifugation, and virus yield was analyzed by plaque assay as explained above. The strategy outlined above was repeated at least one more time with those mutants that produced virus. Virus rescue for PB1 mutants D2V, V3D, L8D, F9D, and A14D was attempted four times, with identical negative results.
CAT, alkaline phosphatase, luciferase, and Western blot analyses.
Cells were harvested 48 hpt (two-hybrid mammalian protein-protein interaction) or 18 hpi with vaccinia-T7 virus (influenza virus P5 replication system) in 70 μl of lysis buffer (100 mM KPO4 pH 7.6; 1 mM dithiothreitol). Chloramphenicol acetyltransferase (CAT) activity was analyzed as described (20) except that butiryl-CoA was used instead of acetyl-CoA and CAT activity was measured by either liquid scintillation counting using a Wallac counter from LKB-Pharmacia (Gaithersburg, Md.) or thin-layer chromatography. Conversion of [14C]chloramphenicol into the acetylated forms was allowed to progress for 1 h at 37°C and represented approximately 60% of maximal conversion rate for the most active constructs (expressing WT PB1). Thus, only experiments in which CAT activity was in the linear range are reported. Experiments with PB1 mutants that lacked CAT activity per standard assay conditions were subsequently carried out using an extended incubation period (10 h) to increase the sensitivity of the assays.
The secreted alkaline phosphatase (SEAP) assay was performed with supernatants of cells transfected with two-hybrid plasmids 48 hpt using the PhosphaLight kit from Tropix (PE Biosystems, Bedford, Mass.) according to the manufacturer's directions. The linear range of the assay was on the order of 106 relative light units/100 μl of tissue culture supernatant. A CAT/SEAP ratio normalized data to account for transfection efficiency variation among monolayers in different wells. Only data from experiments with variation of SEAP activity among transfections of ≤0.5-fold were considered. Standard error values were derived from the relative CAT/SEAP activity ratios calculated for each mutant in three different experiments. To this end, we calculated a quotient of mutant sample value to WT samples. The standard error for the quotient was calculated by standard methods (28). The interaction capacity (R) of each PB1 mutant relative to WT in the two-hybrid assay was calculated as follows: R = (CAT activity from mutant PB1/CAT activity from WT PB1)/(SEAP activity from mutant PB1/SEAP activity from WT PB1). Normalized CAT activity from two-hybrid transfections using the WT PB1 construct was arbitrarily set at R = 1.
Luciferase assays were performed using the luciferase assay system (Promega) following the manufacturer's directions. Luciferase activity was measured by light emission using the TopCount Luminescence Counter (Packard Instruments, Meriden, Conn.). Calculating a CAT/Luc ratio normalized transfection efficiency variation between wells. The replication activity (R) of each PB1 mutant relative to WT was calculated as follows: R = (CAT activity from mutant PB1/CAT activity from WT PB1)/(Luc activity from mutant PB1/Luc activity from WT PB1). The normalized CAT activity from P5 transfections with WT P proteins was set arbitrarily at R = 1.
For Western blots, PB1HA proteins contained in CV-1 cell lysates were separated on SDS–10% polyacrylamide gel electrophoresis (10% PAGE) gels and electro-transferred to nitrocellulose membranes (Hybond-C; Amersham, Arlington Heights, Ill.) using a Trans-Blot SD semidry transfer cell (Bio-Rad, Hercules, Calif.). Epitope-tagged proteins were detected with HA epitope-specific monoclonal antibody 12CA5 (44) and peroxidase-conjugated goat anti-mouse and ECL reagent (Amersham).
RESULTS
Refined map of the PA-binding site of PB1.
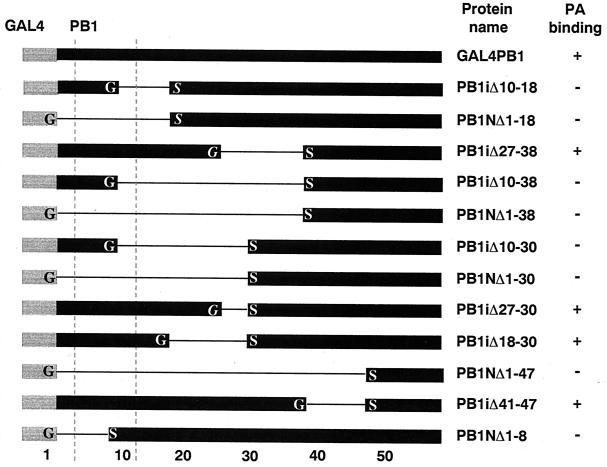
PB1 and PA interact to assemble influenza virus transcription and replication complexes, which also include PB2. A stretch of 48 amino acids at the N terminus of PB1 was shown previously to be sufficient for binding of PA in vivo (39). Our previous experiments also showed that the 709 amino acids outside of this 48-amino-acid N-terminal region of PB1 did not affect binding of PA. The first goal of this work was to refine the mapping of the domain α boundaries using a mammalian two-hybrid interaction assay. To this end, we engineered a set of nested deletion mutants within the PB1 N-terminal 48 amino acids, fused to the GAL4 DNA binding domain (GAL4 DBD), termed GAL4PB1Δ chimeras (Fig. 1). In some of these deletion mutants, the modification of restriction site-generated DNA ends resulted in the creation of glycine or serine amino acid codons flanking the deletion (Fig. 1). To assess the PA-binding properties of the mutant GAL4PB1Δ chimeras in two-hybrid assays, they were cotransfected into CV-1 cells with a mixture of the VP16PA transcription activation domain plasmid and the DNA encoding the CAT reporter controlled by the GAL4 promoter. In this system, the interaction of two chimeric proteins activates CAT mRNA transcription (39). The magnitude of two-hybrid reporter activity can be correlated with the strength of the interaction between the two chimeras, as reported by Estojak et al. (15). Efficient PA binding was observed with PB1 mutants carrying various deletions within the region spanning residues 18 to 47 (Fig. 1). The levels of CAT observed for these PB1 mutants were indistinguishable from that obtained with the WT PB1 chimeric protein. In contrast, deletions encompassing all or part of the N-terminal 17 amino acids of PB1 did not mediate PA binding. PB1iΔ18-30 shows interaction despite the alanine-to-glycine substitution at position 17, suggesting that this residue is not essential for binding PA. Taken together, the deletion mapping data suggested that the region responsible for binding PA is located within residues 1 to 17 of PB1.
FIG. 1.
Amino acid residues essential for the interaction of PB1 with PA in a mammalian two-hybrid system. Polymerase subunits PB1 and PA from influenza virus A/WSN/33 were fused to GAL4 DBD and VP16 AD, respectively, and tested for interaction in a mammalian two-hybrid assay as described (39). Small N-terminal (NΔ) and internal deletions (iΔ) were introduced within the first 48 amino acids of PB1 fused to GAL4. PB1 deletion mutants that interact with PA, such as the WT PB1 chimeric construct GAL4PB1, as revealed by CAT expression, are marked (+); lack of interaction is also shown (−). The amino acid positions of the deletion termini are reflected in the nomenclature (e.g., positions 10 to 18 in GAL4PB1iΔ10-18). Except when printed in italics, G (glycine) and S (serine) shown flanking the deletion were absent in wild-type PB1; they arose from codons created by modification of DNA overhangs prior to ligation. The numeric ruler indicates the amino acid positions within PB1 (domain α). Vertical dotted lines indicate the position of β-sheet structure boundaries predicted by Jpred analysis of the first 50 amino acids of PB1 (10).
Site-directed mutagenesis of PB1 domain α. (i) Effects on PA interaction.
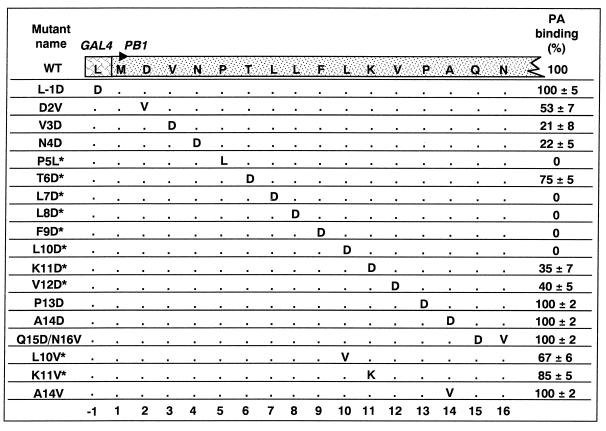
In order to identify the individual amino acids within the 16 N-terminal residues of PB1 that are indispensable for binding to PA we expressed mutants in which every residue in this region was replaced (Fig. 2). We engineered mutants carrying aspartic acid in place of the nonpolar residues in the WT sequence. Thus, valine (V) at positions 3 and 12; threonine (T) at position 6; leucine (L) at positions 7, 8, and 10; and phenylalanine (F) at position 9 were individually replaced with aspartic acid (D). Proline (P) residues 5 and 13 were replaced with leucine and aspartate, respectively. The basic residue lysine (K) 11 and the amide residue asparagine (N) 4 were also replaced with aspartate. The only aspartic acid residue in this region, at position 2, was mutated to valine. A double mutant was prepared in which glutamine (Q) 15 and asparagine 16 were supplanted by aspartate and valine.
FIG. 2.
Site-directed mutagenesis of the PA-binding domain in GAL4PB1. Single or double amino acid substitutions were introduced within PB1 and GAL4 DBD coding regions. Amino acids are indicated in single-letter code; dots denote unchanged amino acids. Mutant PB1 interaction efficiency with VP16PA, based on CAT transactivation in the two-hybrid assay, is indicated as a percentage relative to the WT GAL4PB1 (set arbitrarily at 100%). A minimum of three independent experiments were carried out for each mutant (the standard deviation for each mutant is shown). Two-hybrid assays were performed with pSGPB1t-derived PB1 mutants, lacking the C-terminal 103 amino acids of PB1. Asterisks indicate mutants that were tested also as GAL4PB1 chimeras encoding the entire ORF of PB1, with similar results (see Materials and Methods).
The two-hybrid assay results are subject to variability in the efficiency of transfection, precluding comparisons among mutants that reveal interaction strengths. To normalize these data, we cotransfected an additional plasmid encoding the SEAP gene under the control of the cytomegalovirus promoter. SEAP activity obtained from supernatants of transfected cells was used to normalize two-hybrid CAT expression (see Materials and Methods). The normalized CAT two-hybrid reporter activities produced by the mutants ranged from nil to WT (Fig. 2). The continuum of CAT values reflected a spectrum of interaction strengths (15). Mutants P13D and A14D, as well as the double mutant Q15D-N16V interacted efficiently with VP16PA, paralleling the WT GAL4PB1 interaction (Fig. 2). In contrast, the lack of CAT reporter activity observed with mutants L7D, L8D, F9D, and L10D indicated the absence of PA binding. Mutation of P5 to L also appeared to have lost PA interaction activity completely (Table 1). P5L mutant is interesting because the change to leucine maintains the hydrophobicity of the region but still prevents binding of PA to PB1 in this system. Other mutants, including D2V, V3D, N4D, T6D, K11D, and V12D retained various levels of PA binding ability (CAT reporter levels of 53, 21, 22, 75, 35, and 40%, respectively, relative to WT PB1). The amino acid at position 6 appeared to be less important for the interaction with PA (CAT reporter levels of 75% relative to WT PB1) although it is flanked by residues that appear to be essential for it (Fig. 2). Four substitutions downstream of position 12 were inconsequential for the interaction, suggesting that the C-terminal boundary of the domain required for PA binding is residue V12.
TABLE 1.
Functional assessment of PA-binding mutants of PB1a
| Strain or mutant | % PA binding (two-hybrid)b | P5 polymerase activity (CAT) (%)b | PB1 mutant virus yield (PFU/ml) | Plaque purified and sequenced |
|---|---|---|---|---|
| WT | 100 | 100 | 4.0 × 107 | + |
| D2V | 53 ± 7 | 0 | 0 | − |
| V3D | 21 ± 8 | 0 | 0 | − |
| N4D | 22 ± 5 | 24 ± 1 | 4.0 × 104 | + |
| P5Lc | NDd | 1.0 × 103 | + | |
| T6D | 75 ± 5 | 56 ± 4 | 5.1 × 106 | + |
| L7De | 0 | 21 ± 2 | 2.0 × 103e | +e |
| L8D | 0 | 0 | − | |
| F9D | 0 | 3 ± 1 | 0 | − |
| L10De | 0 | 15 ± 5 | 2.0 × 103e | +e |
| K11D | 35 ± 7 | 48 ± 2 | 3.0 × 103 | + |
| V12D | 40 ± 5 | 54 ± 7 | 4.5 × 103 | + |
| P13D | 100 ± 2 | 78 ± 2 | 1.9 × 105 | + |
| A14D | 100 ± 2 | 0 | 0 | − |
Single amino acid substitutions were introduced within the first 14 N-terminal residues of PB1, and the effect of these mutations was tested in two functional assays: the influenza virus RG system (38), and the rescue of mutant influenza virus (23). The polymerase activity of each mutant is expressed as a percentage relative to the WT PB1 protein in the P5 RG system. Rescued PB1 mutant viruses were plaque purified and stocks were prepared. Sequence analysis was performed with three independent plaques from each mutant (except L7D and L10D, from which 10 plaques were used in sequence analysis). Plaque-purified mutant viruses were propagated on MDCK cells for 2 days (P5L mutant virus was grown for 3 days), and their yields (in PFU per milliliter) were analyzed by plaque assay (two independent experiments).
Means ± standard deviations.
Mutant P5L polymerase activity was demonstrated using a green fluorescent protein RG (not shown).
ND, not determined.
Mutants L7D and L10D were propagated on MDCK cells for 6 days before analysis of infectious virus yield by plaque assay.
The low reporter activity generated by mutant D2V suggested that this hydrophilic residue is important. D2V reduces the interaction activity twofold, as a consequence of replacing aspartate with a hydrophobic residue. Proline at position 5 appears important for interaction given the results obtained with P5L. Position 11 (lysine) is more permissive and tolerates a more extreme change than in the previous two cases (mutant K11V, retained ∼85% of the interaction with VP16PA).
Since our observations were based on the interaction between two fusion proteins, GAL4PB1 and VP16PA, in which the majority of critical residues (I, V, and L) were aliphatic, we analyzed the potentially spurious contribution of the leucine residue present at the junction between the GAL4 and PB1 protein chimera. The GAL4PB1 L-1D mutant (Fig. 2) yielded WT level of CAT (100%) in two-hybrid assays, ruling out a possible contribution of this position to the interaction with PA. Taken together, these results suggest that the region encompassing amino acids 2 to 12 of the PB1 α domain constitutes the core of a PB1 interface for specific interaction with PA.
For technical reasons, PB1 mutant constructs using a GAL4PB1 fusion backbone carry a deletion of the C-terminal 103 amino acids of PB1. Therefore, it was paramount to establish if there was a small contribution of these C-terminal residues to the interaction with PA. We engineered some of the mutations described above also in the context of a full-length PB1 gene fused to GAL4 (Fig. 2). CAT activity from lysates of CV-1 cells transfected with this new set of plasmids suggests that the C terminus of PB1 does not contribute to the binding to PA (Fig. 1 and reference 39).
In summary, domain α spans an 11-amino-acid region that participates in the interaction with PA. Each residue in this region seems to make its unique contribution to PA binding: some of them are important for binding PA, such as V3, N4, P5, L7, L8, F9, and L10, because they result in the loss of more than two-thirds of their PA-interacting capacity. In contrast, other positions tolerate nonconservative substitutions while retaining more than two-thirds of PA binding activity; e.g., D2, K11, and V12.
(ii) Effects on polymerase activity.
To assess the role of the PB1-PA interaction in influenza virus genome replication and gene expression in vivo, we utilized a modified plasmid-based influenza virus transcription-replication system. CV-1 cells are infected with vaccinia-T7 virus and transfected with a mixture of five plasmids with transcription units under the control of the bacteriophage T7 promoter. One of the plasmids in the set encodes a model influenza virus reporter gene flanked by two cis-acting ribozymes. Transcription of the precursor reporter gene RNA and subsequent cis cleavage at specific locations result in a mature RNA whose termini are identical to authentic influenza virus A genomic RNA segment 8 and flank the reporter CAT, mimicking an influenza virus vRNA (38). The remaining four plasmids in the set express the influenza virus nucleoprotein and the three-polymerase subunits. In this vaccinia-T7-driven influenza virus RG system, transfection of the five-plasmid set (P5) is necessary and sufficient for CAT expression in cells, as reported previously (38). Omission of a plasmid expressing any of the four proteins causes a >99% reduction in RG CAT accumulation (reference and data not shown). Because total reporter activity from the P5 RG system is the result of transcription and replication, it is important to emphasize that no RG expression is observed in the absence of PA (data not shown), consistent with previous observations (25).
Each of the site-directed PB1 mutations analyzed in the two-hybrid system was introduced into a PB1 mammalian expression plasmid used for the functional evaluation of polymerase function with the RG system. With proper normalization, CAT expression levels can be an indicator of the transcription-replication activity of the RNP complex. In order to account for variability in the P5 transfection efficiency, we incorporated an additional plasmid encoding the luciferase gene under the control of T7 promoter to the DNA mixture. The luciferase (Luc) activity level obtained for each lysate was used to normalize P5 RG CAT expression (see Materials and Methods). Normalized CAT values from transfected cells revealed the impact of PB1 domain α mutations on the function of the polymerase complex (Table 1). Some PB1 mutations led to a severe or even complete loss of polymerase function, while others retained biological activity despite considerable loss of PA binding efficiency in the two-hybrid assay. From a total of six mutants that maintain a >33% level of interaction between PB1 and PA, four mutants, namely, T6, K11, V12, and P13, allow substantial transcription-replication of RG in the P5 system, although often at reduced levels (RG expression levels of 56, 48, 54, and 78% relative to WT, respectively). The remaining two mutants, D2V and A14D, showed no polymerase activity despite moderate to maximal interaction, respectively, suggesting that these two residues have other crucial functions besides PA binding. A cluster of PB1 mutants with aspartate substitutions in residues L7, L8, F9, and L10, which failed to bind PA, showed a severe reduction or complete loss of polymerase activity. Interestingly, L7 and L10 expressed a residual low level of RG mRNA synthesis (21 and 15% of WT, respectively). Similarly, the V3 residue of PB1 appears important for PA binding and essential for polymerase activity because abrogation of PA binding caused a loss of polymerase activity.
To determine if the observed RG transcription and replication differences of the PB1 mutants result from changes in their catalytic activity rather than alterations in expression level, we analyzed the intracellular concentration of P proteins. To monitor expression by Western blotting, we engineered an epitope tag at the C terminus of WT and mutant PB1 used in the P5 system. Western blot analyses using a monoclonal antibody against the HA epitope tag (Fig. 3 and data not shown) revealed no differences between the expression levels of WT PB1 and any of its mutants. Interestingly, PB1 expressed in vaccinia virus-infected cells appears as a double band (Fig. 3). This effect seems to correlate with the use of vaccinia virus infection in the expression system; PB1 expressed in influenza virus-infected cells appears as a single band (not shown). We speculate that the second PB1 band corresponds to initiation of translation from a second ATG located in the PB1 ORF at nucleotide position 142 (codon 48). For the purposes of interpreting our results, it is important to emphasize that the relative proportions of the two forms of PB1 remain constant for all mutants expressed.
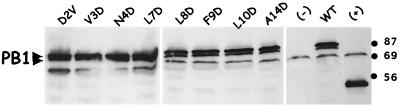
FIG. 3.
Expression of PB1 mutants in primate cells. WT and mutant PB1 were expressed in CV-1 cells infected with vaccinia-T7 and transfected with expression plasmids. Cells were harvested at 18 hpi, and lysates were analyzed by Western blotting with a monoclonal antibody to the HA epitope tag (12CA5). WT and representative mutants of PB1 are shown (D2V, V3D, N4D, L7D, L8D, F9D, L10D, and A14D). A lysate from nontransfected cells was used as a negative control (−), while a lysate expressing an unrelated HA-tagged protein of 56 kDa served as positive control (+). Proteins were separated on an SDS–10% PAGE gel and transferred to a Hybond-C membrane (Amersham). The luminescent signal (ECL; Amersham) was recorded by exposure to X-ray film, which was subsequently developed, scanned, and digitized using Adobe Photoshop software. Small arrowheads indicate the two PB1 polypeptides that reacted with the anti-HA antibody.
(iii) Effects on virus viability.
A correlation between the PA interaction capability of the PB1 mutants and their ability to function in the RG system was observed. These mutants included some with intermediate polymerase function relative to WT (e.g., 30 to 60% activity). The biological significance of intermediate polymerase activities could be best interpreted by evaluating the viability of influenza viruses bearing mutant PB1 genes. New developments in reverse genetics of influenza virus allow the recovery of recombinant or mutant influenza A viruses without the use of a helper virus, extending the application of influenza virus to the engineering of PB1 (16, 23, 35). We engineered PB1 mutants using the method recently described by Hoffman et al. in which eight-plasmid transfections into cells lead to the production of infectious influenza virus particles with high efficiency (106 PFU) (23). Transfection of plasmids with single amino acid substitutions in PB1 along with seven other plasmids encoding the remaining influenza virus genes of the influenza virus A/WSN/33 strain into 293T-MDCK cocultured cells resulted in the production of recombinant viruses for the mutants without lethal mutations. In direct correlation with our polymerase activity data, we were able to rescue mutant viruses that displayed detectable and significant polymerase activity. The exception is mutant F9D, which had only a 3% polymerase activity of the WT (Table 1) and thus might be too low for virus rescue. Supernatants of rescued mutant viruses were subjected to plaque purification and subsequent RNA isolation, reverse transcription-PCR, and sequence analysis of the PB1 gene. All rescued viruses contained the introduced mutation in PB1 and were stable after successive passages in tissue culture (data not shown). An initial characterization of this panel of mutant viruses indicates that their growth characteristics are different from the WT A/WSN/33 virus; all display very small plaque morphology (diameters from 0.5 to 1.0 mm, compared to 2.5 to 3.0 mm in the WT) and low yields of infectious progeny in cell culture. The entire process of virus rescue, from the transfection of eight plasmids to plaque purification and growth phenotype determination was repeated at least one more time for each mutant. The same results were obtained for each mutant in the repetitions, including their growth characteristics, suggesting the absence of spurious substitutions that could arise with these manipulations. However, a detailed molecular characterization of these viruses will be the subject of a future report.
Analysis of PA binding to PB1 mutants by coimmunoprecipitation.
The observation that PB1 mutants P5L, L7D, and L10D failed to interact with PA in two-hybrid assays but displayed low polymerase activity and were viable upon plasmid rescue from transfected cells was intriguing (Table 1). Influenza virus mutants L7D and L10D replicated very poorly, requiring 6 days in culture to reach titers on the order of 103 PFU/ml starting from an MOI of 1. Mutant P5L was only marginally viable, with a titer of <103 PFU/ml achieved after 3 days in MDCK cells (Table 1). This partial conflict between the two-hybrid interaction data, polymerase acitivity, and virus viability upon rescue was explored further using a biochemical assay. PA was coexpressed in cultured cells with each of the PB1 domain α mutants, and the formation of radiolabeled heteromeric complexes was analyzed by coimmunoprecipitation with monospecific anti-PB1 or anti-PA antibody (1). Interestingly, PA was found to bind to all PB1 mutants, albeit with different efficiencies (Fig. 4). PA binding to these PB1 mutants is specific; PA was not immunoprecipitated in the absence of PB1, ruling out the existence of artifacts caused by the anti-PB1 antibody (Fig 4, lane 8). Although this assay did not allow quantitation of interaction efficiencies it shows that defective binding rather than absence of PA binding is likely responsible for lower polymerase activity. As such, these results explain the discrepancy between the two-hybrid and polymerase data observed for mutants L7D and L10D (Fig. 4A, lanes 3 and 6). We also tested PA binding to D2V and P5L mutants by coimmunoprecipitation. PA binding to the D2V mutant was readily detected, in agreement with our two-hybrid data (Fig. 4A, lane 11). Only a weak binding between PA and P5L was detected (Fig. 4A, lane 14), in general agreement with our two-hybrid data. However, considering that the P5L mutant possesses polymerase activity and a viable virus was rescued, it is conceivable that the interaction between this mutant and PA is unstable and difficult to detect by these methodologies.
Since we observed interaction of all PB1 mutants with PA, it was important to establish that the N-terminus of PB1 was indeed responsible for all of the PA binding activity of PB1, as the two-hybrid data had shown. We investigated the interaction of a deletion mutant (PB1/[−11]) that shows no interaction with PA in the two-hybrid assay. PB1/[−11] expressed using the vaccinia virus system and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) showed a faster migration than the WT PB1 protein, coincident with a deletion that reduces the molecular mass of this polypeptide by approximately 1 kDa (not shown). Like the WT PB1 expressed under the same conditions, PB1/[−11] also showed a double-band pattern upon Western blotting (not shown). The double-band pattern was more prominent for mutant PB1/[−11] than for the WT PB1, maybe because the translation efficiency from the first methionine is reduced (Fig. 4A, lane 10 and 19). PB1/[−11] coexpressed with PA did not bind PA by coimmunoprecipitation under the same conditions under which all other PB1 mutants interacted with PA (Fig. 4A lane 10). Coimmunoprecipitation of PA and mutants of PB1 were also observed when the anti-PA antibody was used. Unfortunately, the anti-PA antibody also precipitated a vaccinia virus polypeptide that migrated almost identically to WT PB1, blocking the full interpretation of our data (Fig. 4B). However, we confirmed the lack of binding between PB1/[−11] and PA, because a band corresponding to the size of PB1/[−11] was not observed under these conditions (Fig. 5B, lane 10). From these experiments, it can be assumed that with PB1 as a full-length protein, there is sufficient interaction between the PB1 mutants and PA to allow polymerase activity for some of them. Combined with our two-hybrid data these results indicate that the N terminus of PB1 (12 amino acids) is sufficient and necessary for binding PA.
FIG. 5.
Coimmunoprecipitation of PB2-PB1 complexes. PB2 and PB1 proteins were coexpressed in Cos-7 cells using the vaccinia-T7 virus system, radiolabeled, and immunoprecipitated with an anti-PB1 monospecific antibody. Lanes: 1, PB2-D2V; 2, PB2-V3D; 3, PB2-P5L; 4, PB2-A14D; 5, PB2-PB1 WT; 6, PB2 alone. ∗, presence of the second PB1 specific band. After coimmunoprecipitation, proteins were treated as described in Fig. 4A.
Mutants D2V, V3D, and A14D showed specific binding to PA as observed using two different methods but lacked polymerase activity, suggesting that this phenotype was due to failure to bind PB2, the other viral polypeptide in the heterotrimeric complex. Following coexpression of PB2 and mutants of PB1 we found that PB2 was coimmunoprecipitated with all three PB1 mutants tested (Fig. 5, lanes 1, 2, and 4). PB2 binding was dependent upon the presence of the PB1 mutant proteins when the anti-PB1 antibody was used (Fig. 5, lane 6). Likewise, PB1 mutants were specifically coimmunoprecipitated with PB2 when an anti-PB2 antibody was used (not shown). Thus, lack of polymerase activity in mutants D2V, V3D, and A14D cannot be explained by either lack of binding to PA or PB2 or poor stability (Fig. 5, 4, and 3, respectively). Therefore, it is reasonable to speculate that these PB1 mutants have alterations in folding that prevent polymerase activity. Similar considerations apply to the interaction between mutant P5L and PA (Fig. 4, lane 14), and P5L and PB2 (Fig. 5, lane 3), although in this particular case the mutation does preserve polymerase activity and is able to rescue virus.
DISCUSSION
A region of 48 amino acids at the N terminus of PB1 was previously reported to include the interface for recruitment of PA to the polymerase complex (39). Similarly, coimmunoprecipitation of PA protein with deletion mutants of PB1 transiently expressed in mammalian cells pointed to the importance of the N terminus of PB1 to bind PA (this report and reference 42). The alignment of the PB1 genes from influenza virus types A, B, and C, as well as those from the insect and fish orthomyxoviruses, revealed a clustering of amino acid sequence conservation towards the C terminus of this 48-amino-acid region (residues 17 to 48). Because of the putative functional importance of P complex formation, we favored the notion that the PB1 domain that binds PA would be conserved among members of the family. Thus, the interaction was proposed to lie between residues 17 and 48 (39). Contrary to that prediction, the high-resolution mapping of the PB1 domain α entailing functional analyses of two sets of PB1 mutants revealed that the interaction of domain α with PA actually occurs through the N-terminal 12 amino acids. The presence of a β-sheet structure spanning residues 6 to 12 is predicted by the Jpred algorithm of Cuff and Burton (data not shown) (8, 9). The loss of interaction resulting from substitutions at proline 5 indicates that the integrity of this structure may be required. Although this N-terminal region is absolutely conserved among all known type A influenza virus PB1 sequences, PB1 genes from different influenza virus types and from more-distant orthomyxoviruses from insects and fish reveal that the N-terminal 12 amino acids are not conserved (data not shown). There are significant amino acid differences between the PB1 domain α of influenza virus type A and those of types B and C virus (e.g., B/Lee/40 and C/JJ/50) within the region that appears to be important for interaction and replication. For example, type B PB1 has F7 in place of the L7 found in influenza virus type A. Another nonconservative substitution can be found at T6, which has been replaced by Y6 in types B and C PB1 orthologs. This substitution for a bulky aromatic amino acid in a region predicted to fold as a β-sheet could preclude the interaction with the influenza virus type A PA. Thus, domain α divergence during influenza virus type speciation may have made subsequent intertypic PB1 gene exchange among members of the family impossible, by disrupting the PB1-PA interaction interface. Nevertheless, PB1 proteins from influenza type A, B, and C viruses maintain an overall similarity within this region, including the high content of aliphatic amino acids, the presence of charged amino acids at exactly the same locations, and a predicted secondary β-sheet structure (data not shown).
To determine if PA binding is important for the polymerase activity of PB1 complexed with PB2, we measured the reporter expression activity of PB1 mutants with known PA interaction abilities. Polymerase activity in vivo was assessed using an influenza virus plasmid-based transcription-replication system, which revealed that there was a general correlation between PB1-PA interaction ability and polymerase activity. This was especially true for the region extending from N4 to P13. Within this region, regression analysis of relative interaction on polymerase activity yielded values of 0.93 (data not shown). Mutant D2V had a limited impact on PA interaction efficiency yet showed no polymerase activity. We contemplate two major possibilities to explain this result. First, D2V could be binding PA with an abnormal spatial geometry that interferes with the normal function of the P complex. Alternatively, this substitution may have no impact on PA binding but instead may abolish polymerase activity by altering the conformation of a discontinuous PB1 domain important for PA function. The first interpretation would be consistent with the notion that D2 contributes to orient PA in an optimal functional conformation. A similar interpretation can be made about position V3. The A14D mutation that lies outside the region for interaction with PA has no polymerase activity but binds PA with WT efficiency as observed by two-hybrid and coimmunoprecipitation assays. The A14D mutation also appears to possess intact PB2 binding. No role has been assigned to this position for either interaction with other viral components and/or intrinsic polynucleotide extension ability of PB1. The presence of a hydrophobic amino acid at this position appears important since it is conserved within the three influenza virus types (isoleucine in type B and valine in type C). Deleterious effects on the activity of PB1 could result from substitution of a single highly conserved amino acid, as previously reported (5).
We have used reverse genetics to attempt the introduction of representative mutant PB1 genes, bearing mutations within the 12 N-terminal amino acids, into the influenza virus genome. Interestingly, the recovered mutant viruses displayed altered phenotypes and growth characteristics relative to the WT. It would be of interest to establish if any of these PB1 mutants with altered PA interaction and/or polymerase function display novel replication phenotypes and/or modified disease pathogenesis in laboratory animal hosts (35). To the best of our knowledge, the PB1 mutant set described in this report is the first to include a number of mutants that display partial polymerase activity that has been rescued in fully competent viruses. We attribute the failure to recover viable virus from certain PB1 mutant genomes such as F9D to RNA synthesis levels below the threshold required for progeny virion assembly. This conclusion rests on the improvements to the virus rescue system described by Hoffmann et al. (23), which render it comparable to the one described by Neumann et al. in terms of virus yield from a given amount of transfected DNA (35).
Interestingly, our results suggest that the polymerase activity of PB1 is stimulated by PA even in the α domain mutants failing to bind PA efficiently, as evidenced by PB1 L7D and L10D mutants (Fig. 2). However, the RG expression level in these mutants is five- to sixfold below WT levels, suggesting that the PB2-PB1-PA interaction is essential for maximum influenza virus PB1 transcriptase activity. It is possible that PA is mediating this enhancement by way of its ability to induce proteolysis of many cellular proteins as reported by Perales et al. (37). Our experiments also provided additional evidence for a role of PA in transcription and are consistent with previous findings on the direct role of PA in genome transcription and replication by the P complex. Future studies are needed to evaluate the functional properties of PA mutants deficient in their ability to interact with PB1.
ACKNOWLEDGMENTS
We thank E. Hoffman and R. Webster for providing the plasmids for the influenza virus rescue system, B. Cullen for the plasmid pCMV/SEAP, and D. Nayak for the anti-P antibodies. We also thank I. H. Ansari for critical reading of the manuscript, C. M. Johnson for technical assistance, and N. Makarova for statistical analysis.
This work was supported in part by the Center for Biotechnology of the University of Nebraska—Lincoln. D.P. was supported in part by NIH contract N01 AI 95357, NIH grant AI29680, and the American Lebanese Syrian Associated Charities.
Footnotes
This is publication no. 12905 of the Agricultural Research Division, IANR, University of Nebraska—Lincoln.
REFERENCES
- 1.Akkina R K, Chambers T M, Londo D R, Nayak D P. Intracellular localization of the viral polymerase proteins in cells infected with influenza virus and cells expressing PB1 protein from cloned cDNA. J Virol. 1987;61:2217–2224. doi: 10.1128/jvi.61.7.2217-2224.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 2nd ed. Vol. 1. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. [Google Scholar]
- 3.Berger J, Hauber J, Hauber R, Geiger R, Cullen B. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 4.Biswas S, Boutz P, Nayak D. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J Virol. 1998;72:5493–5501. doi: 10.1128/jvi.72.7.5493-5501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas S K, Nayak D P. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J Virol. 1994;68:1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braam J, Ulmanen I, Krug R M. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell. 1983;34:609–618. doi: 10.1016/0092-8674(83)90393-8. [DOI] [PubMed] [Google Scholar]
- 7.Bukrinskaya A, Vorkunova G, Vorkunova N. Cytoplasmic and nuclear input virus RNPs in influenza virus-infected cells. J Gen Virol. 1979;45:557–567. doi: 10.1099/0022-1317-45-3-557. [DOI] [PubMed] [Google Scholar]
- 8.Cuff J A, Barton G J. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 9.Cuff J A, Clamp M E, Siddiqui A S, Finlay M, Barton G J. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 10.Cuff J A, Barton G J. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 11.Deng W, Nickoloff J. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 12.Detjen B M, St A C, Katze M G, Krug R M. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J Virol. 1987;61:16–22. doi: 10.1128/jvi.61.1.16-22.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Digard P, Blok V C, Inglis S C. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology. 1989;171:162–169. doi: 10.1016/0042-6822(89)90523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fodor E, Devenish L, Engelhardt O G, Palese P, Brownlee G G, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fodor E, Seong B L, Brownlee G G. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J Gen Virol. 1993;74:1327–1333. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]
- 18.Fuerst T, Niles E, Studier F, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez S, Zurcher T, Ortin J. Identification of two separate domains in the influenza virus PB1 protein involved in the interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 1996;24:4456–4463. doi: 10.1093/nar/24.22.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 22.Herget M, Scholtissek C. A temperature-sensitive mutation in the acidic polymerase gene of an influenza A virus alters the regulation of viral protein synthesis. J Gen Virol. 1993;74:1789–1794. doi: 10.1099/0022-1317-74-9-1789. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster R G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honda A, Mukaigawa J, Yokoiyama A, Kato A, Ueda S, Nagata K, Krystal M, Nayak D P, Ishihama A. Purification and molecular structure of RNA polymerase from influenza virus A/PR8. J Biochem. 1990;107:624–628. doi: 10.1093/oxfordjournals.jbchem.a123097. [DOI] [PubMed] [Google Scholar]
- 25.Huang T S, Palese P, Krystal M. Determination of influenza virus proteins required for genome replication. J Virol. 1990;64:5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inglis S C, Carroll A R, Lamb R A, Mahy B W. Polypeptides specified by the influenza virus genome. I. Evidence for eight distinct gene products specified by fowl plague virus. Virology. 1976;74:489–503. doi: 10.1016/0042-6822(76)90355-x. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi M, Toyoda T, Ishihama A. Influenza virus PB1 protein is the minimal and essential subunit of RNA polymerase. Arch Virol. 1996;141:525–539. doi: 10.1007/BF01718315. [DOI] [PubMed] [Google Scholar]
- 28.Korn G A, Korn T M. Mathematical handbook for scientists and engineers: definitions, theorems, and formulas for reference and review. Mineola, N.Y: Dover Publications; 2000. [Google Scholar]
- 29.Krug R M, Ueda M, Palese P. Temperature-sensitive mutants of influenza WSN virus defective in virus-specific RNA synthesis. J Virol. 1975;16:790–796. doi: 10.1128/jvi.16.4.790-796.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb R A. Genes and proteins of the influenza viruses. In: Krug R M, editor. The influenza viruses. 1st ed. New York, N.Y: Plenum Press; 1989. [Google Scholar]
- 31.Medcalf L, Poole E, Elton D, Digard P, Medcalf L, Poole E, Elton D, Digard P. Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J Virol. 1999;73:7349–7356. doi: 10.1128/jvi.73.9.7349-7356.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller R, Poch O, Delarue M, Bishop D H, Bouloy M. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol. 1994;75:1345–1352. doi: 10.1099/0022-1317-75-6-1345. [DOI] [PubMed] [Google Scholar]
- 33.Murti K G, Webster R G, Jones I M. Localization of RNA polymerases on influenza viral ribonucleoproteins by immunogold labeling. Virology. 1988;164:562–566. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa Y, Oda K, Nakada S. The PB1 subunit alone can catalyze cRNA synthesis, and the PA subunit in addition to the PB1 subunit is required for viral RNA synthesis in replication of the influenza virus genome. J Virol. 1996;70:6390–6394. doi: 10.1128/jvi.70.9.6390-6394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez D R, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palese P, Ritchey M B. Polyacrylamide gel electrophoresis of the RNAs of new influenza virus strains: an epidemiological tool. Dev Biol Stand. 1977;39:411–415. [PubMed] [Google Scholar]
- 37.Perales B, Sanz-Ezquerro J, Gastaminza P, Ortega J, Santaren J, Ortín J, Nieto A. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J Virol. 2000;74:1307–1312. doi: 10.1128/jvi.74.3.1307-1312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez D, Donis R. The matrix 1 protein of influenza A virus inhibits the transcriptase activity of a model influenza reporter genome in vivo. Virology. 1998;249:52–61. doi: 10.1006/viro.1998.9318. [DOI] [PubMed] [Google Scholar]
- 39.Perez D R, Donis R O. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J Virol. 1995;69:6932–6939. doi: 10.1128/jvi.69.11.6932-6939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poon L, Pritlove D, Fodor E, Brownlee G. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J Virol. 1999;73:3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toyoda T, Adyshev D, Kobayashi M, Iwata A, Ishihama A. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J Gen Virol. 1996;77:2149–2157. doi: 10.1099/0022-1317-77-9-2149. [DOI] [PubMed] [Google Scholar]
- 43.Vasavada H A, Ganguly S, Germino F J, Wang Z X, Weissman S M. A contingent replication assay for the detection of protein-protein interactions in animal cells. Proc Natl Acad Sci USA. 1991;88:10686–10690. doi: 10.1073/pnas.88.23.10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 45.Zheng H, Lee H, Palese P, Garcia-Sastre A. Influenza A virus RNA polymerase has the ability to stutter at the polyadenylation site of a viral RNA template during RNA replication. J Virol. 1999;73:5240–5243. doi: 10.1128/jvi.73.6.5240-5243.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]