Abstract
The nef genes of human immunodeficiency virus and simian immunodeficiency virus (SIV) overlap about 80% of the U3 region of the 3′ long terminal repeat (LTR) and contain several essential cis-acting elements (here referred to as the TPI region): a T-rich region, the polypurine tract, and attachment (att) sequences required for integration. We inactivated the TPI region in the nef reading frame of the pathogenic SIVmac239 clone (239wt) by 13 silent point mutations. To restore viral infectivity, intact cis-regulatory elements were inserted just downstream of the mutated nef gene. The resulting SIV genome contains U3 regions that are 384 bp shorter than the 517-bp 239wt U3 region. Overall, elimination of the duplicated Nef coding sequences truncates the proviral genome by 350 bp. Nonetheless, it contains all known coding sequences and cis-acting elements. The TPI mutant virus expressed functional Nef and replicated like 239wt in all cell culture assays and in vivo in rhesus macaques. Notably, these SIVmac constructs allow us to study Nef function in the context of replication-competent viruses without the restrictions of overlapping LTR sequences and important cis-acting elements. The genomes of all known primate lentiviruses contain a large overlap between nef and the U3 region. We demonstrate that this conserved genomic organization is not obligatory for efficient viral replication and pathogenicity.
The nef gene is characteristic of primate lentiviruses and important for the full pathogenic potential of both human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) (11, 24, 27). A number of Nef functions that might increase virulence and allow the maintenance of high virus burdens have been described (reviewed in references 14 and 32). Nef downmodulates class I MHC and CD4 cell surface expression (3, 9, 15, 30, 35). Furthermore, Nef increases the infectivity of viral particles and stimulates viral replication in primary peripheral blood mononuclear cells, macrophages, and tonsillar histocultures (1, 8, 12, 16, 28, 31, 36). Recent studies suggest that most or all of these Nef activities are functionally independent and contribute to efficient viral replication and persistence in vivo (4, 6, 19, 20, 29, 37).
The use of replication-competent primate lentiviruses is obligatory to assess the effect of specific alterations in Nef on viral replication and pathogenicity. Also, the effects of Nef on cell surface expression of class I MHC and CD4 molecules and on signal transduction pathways are ideally investigated in infected cells. However, a detailed structure-function analysis is complicated because the nef open reading frame (ORF) overlaps several cis-acting elements and the U3 region in the viral long terminal repeat (LTR). A T-rich sequence (22), the polypurine tract (PPT), and the viral attachment (att) site at the 5′ end of the U3 region are essential for lentiviral replication (reviewed in reference 39). Mutations in the corresponding Nef region cannot be made without changing these critical cis-acting elements. Furthermore, the entire 3′ half of the nef gene overlaps the U3 region of the LTR. Therefore, after reverse transcription (RT), changes in the C-terminal half of Nef might have unexpected effects on transcriptional 5′ LTR activity, which could affect the results of replication and infectivity assays.
Previous findings suggest that 332 to 407 bp of U3 sequences upstream of the major core enhancer and promoter elements (US sequences) serve mainly as a nef coding sequence (21, 26, 27, 33). After infection of rhesus macaques with an SIVmac239 variant containing deletions in the nef unique region, additional deletions accumulate over time in the US region (26). Similar U3 deletions were selected in a long-term survivor of HIV-1 infection in whom only nef-deleted proviral sequences could be detected (27). Furthermore, an SIV variant containing a large number of nucleotide changes in the US region which did not affect the predicted nef coding sequence showed normal pathogenic potential in infected rhesus macaques (21). These SIVmac and HIV-1 variants did not contain most of the wild-type upstream U3 sequences. However, they were either attenuated, because of a deleted nef ORF (26, 27), or they maintained the nef-LTR overlap and the wild-type length of the U3 region as well as the essential cis-regulatory elements in the nef gene (21).
These previous studies showed that large parts of the US region serve mainly as nef coding sequence and do not contain important transcriptional elements. Therefore, it is unclear why the U3 regions of HIV and SIV are considerably longer than those of other lentiviruses (about 450 to 560 bp in length) and why they overlap the nef ORF by about 70%. To address these questions, we mutated the cis-acting elements in the SIVmac239 nef gene and introduced an intact TPI element downstream of the mutated nef ORF. The TPI region was either inserted just 14 bp upstream of the 5′ end of the single NF-κB site or 50 bp further upstream, because previous studies suggested that this US region might contain important enhancer elements (21, 26, 27, 33). These modifications had several consequences: (i) the nef gene of these proviral constructs does not overlap the LTR region; (ii) the nef ORF does not contain essential cis-acting elements; and (iii) these SIVmac variants possess short U3-LTR sequences. We found that SIVmac does not require long U3 regions for efficient replication in vitro and in vivo in rhesus macaques. Furthermore, we established a system to study nef function using infectious viruses without the limitations of overlapping cis-regulatory and LTR elements.
MATERIALS AND METHODS
Mutant construction.
Site-directed mutagenesis to generate the SIVmac239 TPI variants was performed by spliced overlap extension PCR as described previously (28). Briefly, the env-nef region of SIVmac239 (23, 34) was amplified using primer pNheI (5′-GTACAAATGCTAGCTAAG-3′) and pPPT5 (5′-CTAAACCACCTTTCTCCTTAATGAAGTGAGACATGTCTATTGC-3′). The nef-LTR region was amplified using primer pPPT3 (5′-AAGGAGAAAGGTGGTTTAGAGGGTATCTATTACAGTGCAAGAAG-3′) and p3nefSmaI (5′-TCCCCCCGGGGGAAAGTCCCTGCTGTT-3′). Mutated positions are underlined. The left- and right-half PCR products were gel purified, mixed in equimolar amounts, and subjected to a second PCR with primers pNhe1 and p3nefSmaI. To generate 239nefMTPIΔ5, the PCR product was cloned into the SIVmac 239ΔUS384 construct (33) by using the unique NheI and SmaI sites in the env gene and just upstream of the TPI region. An overview of the mutants analyzed is given in Fig. 1 and 2. To generate 239TPImut, the TPI region of wild-type SIVmac239 was replaced with the mutated sequence using the flanking BglII and NdeI sites in nef. Conversely, the mutated region in the 239nefMTPIΔ5 construct was replaced with the corresponding 239wt and nef* sequences to generate the 239nef+TPIΔ5 and 239nef*TPIΔ5 variants. SIVmac239 nef* contains a premature in-frame TAA stop signal at the 93rd codon of nef (23). Three additional variants (239nefMTPI, 239nef+TPI, and 239nef*TPI) were generated the same way except that the mutated nef alleles were inserted into the proviral 239ΔNU clone, in which 65 bp upstream of the core enhancer elements are maintained (17). The U3 deletions were present in both LTRs to prevent recombination. All PCR-derived inserts were sequenced to confirmed that only the intended changes were present.
FIG. 1.
Schematic representation of modifications introduced into the SIVmac239 genome to eliminate the nef-LTR overlap. The 239wt clone (top) contains a 517-bp U3 region that overlaps the 792-bp nef ORF by 407 bp (79%) (34). The TPI region in nef was mutated, and an intact TPI element was inserted upstream of the single NF-κB site. Furthermore, 384 bp of upstream U3 sequences were deleted from the 5′ LTR to prevent recombination with the modified 3′ LTR region. The nefMTPIΔ5 provirus (bottom) contains a U3 region of 133 bp and a nef gene that neither overlaps the 3′ LTR nor contains essential cis-regulatory elements. The black bar indicates the position of the deletion, and the arrows indicate the positions of the mutated and functional TPI regions. Abbreviations: x, vpx; r, vpr.
FIG. 2.
Mutant construction and SIVmac239 nef-LTR variants analyzed. (A) The ΔUS384 construct, which contains a deletion of 182 bp in the nef unique region and 384 bp in the U3 region (33), was used to generate nefMTPIΔ5. As indicated below, a PCR restriction fragment containing 13 silent point mutations in the TPI region was cloned into ΔUS384 to generate nefMTPIΔ5. (B) Schematic presentation of the SIVmac constructs analyzed (left) and the deduced 5′ LTR promoter region (right). TPImut differs from 239wt only by the specific point mutations in the TPI region; nef+TPI and nef+TPIΔ5 contain wild-type nef genes; nefMTPI and nefMTPIΔ5 contain TPI-mutated nef alleles. Otherwise isogenic forms with a premature stop signal at the 93rd codon of the nef ORF were also generated. The TPIΔ5 forms contain 14 bp and the TPI forms contain 65 bp of US sequences. Abbreviations: E-CP, enhancer-core promoter; Int, U3-terminal sequences required for integration.
Virus stocks, cells, and infectivity assays.
For virus production, 293T cells were transfected by the calcium phosphate method, and virus production was quantitated as described previously (4, 5). 293T, CEMx174 cells, the herpesvirus saimiri-transformed T-cell line 221 (1), and rhesus peripheral blood mononuclear cells (rPBMC) were isolated and cultured as described previously (4, 5). The cells were infected, and virus replication was measured by reverse transcriptase assay as described (5). SIVmac infectivity was determined using sMAGI cells as described previously (7) and quantitated using the Galacto-Light Plus chemiluminescence reporter assay kit (Tropix, Bedford, Mass.), as recommended by the manufacturer.
Animal studies.
Three juvenile rhesus macaques of Indian origin were infected by intravenous inoculation of SIVmac239nefMTPIΔ5 containing 5 ng of p27 produced by transfected 293T cells. The animals were healthy and seronegative for SIV, type D retroviruses, and simian T-cell lymphotropic virus type 1 at the time of infection. Blood was collected at regular intervals, and serological, virological, and immunological analysis was performed as described previously (5, 28, 38).
PCR analysis.
SIV sequences spanning the entire nef-U3 region were amplified from rPBMC DNA with a nested PCR approach or from DNA isolated from positive PBMC-CEMx174 bulk cocultivation or infected CEMx174 cells by one round of amplification essentially as described (5, 28). Viral plasma RNA was isolated with the QIAamp RNA kit (Diagen, Basel, Switzerland), reverse transcribed with Superscript RT (Gibco-BRL, Eggenstein, Germany), and subjected to a standard nested PCR approach. PCR fragments were sequenced directly or following subcloning into the pCRII vector (Invitrogen Corp., San Diego, Calif.). Sequencing was performed as described previously (28). The following primers were used to analyze the proviral LTR sequences: pP1 (5′-GATCCAACTCTGGCCTACAC-3′; 260 to 278 and 9722 to 9741); pP2 (5′-CCGTCGTGGTTGGTTCCTGCC-3′; 891 to 912); pP3 (5′-TCGCTGAAACAGCAGGGACT-3; 400 to 420 and 9862 to 9882); and pP4 (5′-GATTTTCCTGCTTCGGTTTCCC-3′; 790 to 808 and 10252 to 10270). Numbers refer to positions in the proviral SIVmac 239wt sequence (34).
Western blot analysis.
CEMx174 cells were infected with virus containing 10 ng of p27 core antigen derived from transfected 293 T cells. When cytopathic effects were observed, cells were pelleted, and lysates were generated as described previously (5). Expression of Nef proteins in whole cellular lysates was analyzed by immunoblot using a rabbit anti-Nef serum (13). For detection of p27 core protein, an anti-Gag monoclonal antibody derived from SIVmac p27 hybridoma cells (55-2F12) was used (18). For enhanced chemiluminescent detection, horseradish peroxidase-conjugated secondary antibodies were used as described by the manufacturer of the ECL detection system (Amersham, Chicago, Ill.).
RESULTS
Construction of SIV TPI mutants.
The 792-bp SIVmac239 nef ORF overlaps about 80% (407 bp) of the 517-bp U3 region of the LTR and contains several essential cis-acting elements (named the TPI region in this study): a T-rich region (bp 363 to 368); the polypurine tract (bp 369 to 383); and sequences required for integration (bp 389 to 397) (numbers refer to the positions in the 239wt nef ORF). The TPI region in nef was inactivated, and intact cis-regulatory elements were inserted upstream of the single NF-κB site in the U3 region of the SIVmac239 LTR to eliminate the nef-LTR overlap (Fig. 1). A total of 13 point mutations were introduced to render the TPI region dysfunctional (Fig. 2A). These nucleotide substitutions did not alter the predicted Nef amino acid sequence. The following SIVmac239 variants were generated: (i) 239-TPImut is isogenic to 239wt except for the specific changes in the TPI region shown in Fig. 2A; (ii) 239-nef+TPI contains an insertion encompassing the TPI region and 65 bp of upstream U3 sequences, downstream of the nef gene; (iii) in 239-nefMTPI, the TPI region in the nef ORF of 239-nef+TPI is mutated; (iv) 239-nef+TPIΔ5 contains an insertion encompassing the TPI region just downstream of the nef gene and 14 bp upstream of the NF-κB binding site; and (v) in 239-nefMTPIΔ5, the TPI region in the nef ORF of 239-nef+TPIΔ5 is mutated (Fig. 2B). The right panel of Fig. 2B shows the 5′ LTR regions predicted after RT. The TPImut mutant is predicted to be inactive because it lacks sequences required for RT and integration. In contrast, the SIVmac239 nef+TPI and nef+TPIΔ5 variants contain two copies of these cis-regulatory elements. Therefore, RT and integration can result in two different forms of the proviral 5′ LTR. In comparison, the nefMTPI and nefMTPIΔ5 proviruses are predicted to contain only short U3 regions at the 5′ end of the genome. In addition to the mutants shown in Fig. 2B, nef-defective variants of the 239-nefMTPI and 239-nefMTPIΔ5 clones, containing a stop signal at the 93rd codon of the nef ORF, were also generated (239-nef*TPI and 239-nef*TPIΔ5, respectively).
SIV variants containing TPI elements downstream of nef are replication competent and express functional Nef.
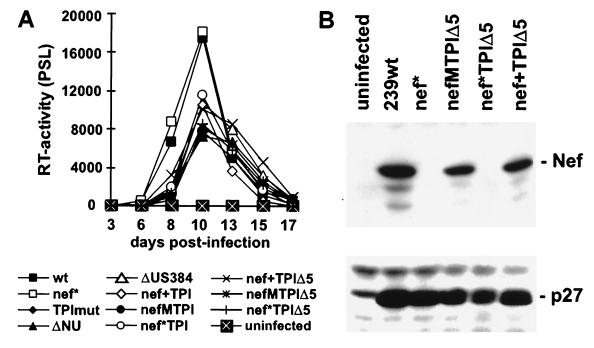
CEMx174 cells were infected with virus stocks derived from transiently transfected 293T cells to investigate the replicative potential of the SIVmac TPI variants. No RT activities above background levels were measured after infection with the control 239-TPImut virus (Fig. 3A). This variant, which does not contain intact TPI sequences at the 3′ end of the proviral genome, also did not replicate when very high doses of virus (up to 100 ng of p27 antigen) were used for infection (data not shown). In agreement with previous studies (24, 28), SIVmac 239wt and 239nef* replicated with comparable efficiency in CEMx174 cells. With the exception of 239-TPImut, all other SIVmac239 TPI variants replicated only slightly less efficiently than the parental 239wt clone (Fig. 3A). Thus, the presence of an additional TPI region or truncation of US sequences did not impair SIVmac replication in CEMx174 cells. PCR amplification and sequence analysis of nef-LTR and LTR-gag sequences at the end of culture demonstrated that all proviral sequences contained the predicted U3 sequences at the 5′ end of the viral genome (Fig. 2B) and showed that no reversions in the mutated nef alleles were selected during in vitro culture (data not shown). Western blot analysis revealed that SIVmac forms containing the 239wt or the TPI-mutated nef genes, but not the nef* variants, expressed the Nef protein (Fig. 3B and data not shown).
FIG. 3.
SIVmac239 TPI variants are replication competent and express Nef. (A) Replication of SIVmac239 variants in CEMx174 cells. Virus containing 10 ng of p27 was used for infection. RT activity was determined using a phosphorimager. PSL, photon-stimulated light emission. (B) Nef and p27 antigen expression in infected CEMx174 cells was verified by immunoblot using rabbit anti-Nef antiserum or a monoclonal anti-p27 antibody as described previously (5). Similar results were obtained in two independent experiments.
Next, we investigated if the TPI-mutated nef alleles are able to enhance SIVmac infectivity. As shown in Fig. 4, SIVmac 239wt infected sMAGI with about eightfold higher efficiency than the nef-defective 239nef* variant. Insertion of TPI-US65 (nef+TPI) or the TPI region (nef+TPIΔ5) downstream of the nef ORF did not reduce viral infectivity. Mutation of the TPI sequences in nef in the presence of a second downstream TPI region (nefMTPI and nefMTPIΔ5) resulted in slightly enhanced infectivity compared to the corresponding constructs with duplicated TPI regions (nef+TPI and nef+TPIΔ5). A premature stop codon reduced the infectivity of the nefMTPI and nefMTPIΔ5 variants to a level comparable to that of 239nef* (see nef*TPI and nef*TPIΔ5, Fig. 4). These results show that the TPI-mutated nef alleles are functional in enhancing viral infectivity.
FIG. 4.
SIV nef alleles containing changes in the TPI region enhance viral infectivity and replication. sMAGI cells were infected with the indicated SIVmac239 TPI-nef variants containing 50 ng of p27 antigen. Infections were performed in triplicate with three different virus stocks.
It has been shown previously that a functional nef gene enhances SIVmac replication in the rhesus macaque T-lymphoid cell line 221, particularly in the absence of interleukin-2 (IL-2) (1). We investigated the replicative capacity of the SIVmac TPI variants in 221 cells to clarify if the mutated nef alleles are able to cause lymphoid cell activation. With the exception of the TPImut form, all variants replicated in 221 cells in the presence of IL-2 (Fig. 5A). However, intact nef genes resulted in faster growth kinetics and increased replication relative to the forms containing disrupted nef genes. In the absence of IL-2, only forms containing wild-type or TPI-mutated nef alleles showed marked levels of replication (Fig. 5B). The nef+TPIΔ5 and nefMTPIΔ5 variants, in which essentially the entire U3 region between the sequences required for integration and the core enhancer elements are deleted, were more active than the forms containing the 65 bp upstream of the NF-κB binding sites (Fig. 5). Thus, the TPI-mutated nef alleles stimulated SIVmac replication with an efficiency comparable to that of 239wt nef, and short U3 regions accelerated rather than reduced viral replication in 221 cells. In agreement with the results obtained using 221 cells, the 239nef+TPI, 239nefMTPI, 239nef+TPIΔ5, and 239nefMTPIΔ5 variants replicated efficiently in rPBMC which were infected immediately after isolation and stimulated with phytohemagglutinin (PHA) 3 days later (Fig. 6A). The nef-defective SIVmac variants 239nef*, 239ΔNU, 239ΔUS384, 239nef*TPI, and 239nef*TPIΔ5 were inactive under these experimental conditions. In comparison, only the TPImut variant did not show appreciable levels of replication in stimulated PBMC, although the forms expressing functional Nef showed a higher replicative capacity than the nef* or nef-deleted forms (Fig. 6B).
FIG. 5.
SIVmac239 variants without overlapping nef-U3 sequences express functional Nef and replicate efficiently in 221 cells. Replication in of the indicated 239 mutants in 221 cells was tested in the presence (A) and absence (B) of IL-2. Similar results were obtained in two independent experiments using different virus stocks.
FIG. 6.
Replication of SIVmac TPI-nef variants in rPBMC. (A) Unstimulated rPBMC were infected immediately after isolation and stimulated with PHA 3 days postinfection. (B) rPBMC were PHA stimulated for 3 days prior to infection. The results shown were derived from a single experiment using rPBMC derived from the same animal. Similar results were obtained with rPBMC from two different rhesus macaques.
These results demonstrate that the TPI-mutated nef alleles are expressed in infected cells and enhance SIVmac infectivity and replication with an efficiency comparable to that of the 239wt nef allele. In agreement with previous studies (21, 26, 33), the U3 region upstream of the core enhancer element was dispensable for efficient viral replication.
Amino acid residues encoded by TPI region are important for Nef function.
One rationale for the construction of the SIVmac TPI mutants was to establish a system that allows investigation of Nef function without the complications of essential overlapping cis-regulatory sequences. We introduced mutations in the nef gene of the nefMTPIΔ5 variant, predicting changes of two lysines, K124/K127→A124/A127 (MTPI-KK-AA), and two tyrosine residues, Y133/Y134→F133/F134 (MTPI-YY-FF). Some of the nucleotide substitutions are present at positions corresponding to important cis-regulatory elements, like the polypurine tract in 239wt (Fig. 2A). Changes in both the lysine and tyrosine residues reduced the ability of SIV-Nef to increase virion infectivity for sMAGI cells (Fig. 7A). Furthermore, the mutated nef alleles did not stimulate SIVmac replication in PBMC culture (Fig. 7B). Our results demonstrate that these amino acid residues in Nef are not only highly conserved, because they are encoded by essential cis-regulatory RNA elements, but are also important for Nef function.
FIG. 7.
cis-Regulatory elements in nef encode amino acids residues important for Nef function. (A) sMAGI cells were infected in triplicate with 293T cell-derived virus stocks containing 50 ng of p27. Infectivity is shown relative to that of 239wt virus. (B) rPBMC were infected immediately after isolation and stimulated with PHA at day 3 postinfection. Similar results were obtained in an independent experiment.
SIVmac239 nefMTPIΔ5 variant replicates efficiently in rhesus macaques.
Three rhesus macaques were experimentally infected with the nefMTPIΔ5 variant to investigate whether SIVmac variants without overlapping nef-LTR sequences and with short U3 LTR sequences replicate efficiently in vivo. The nefMTPIΔ5 mutant was selected for in vivo analysis because it contained the shortest U3 region (133 bp) in conjunction with an intact nef gene and showed a phenotype similar to that of 239wt in in vitro infectivity and replication assays. The replicative capacity of the nefMTPIΔ5 mutant was compared with that of 239wt and ΔNU, which have been extensively analyzed in vivo (5, 17, 24). As shown in Fig. 8A, the p27 levels observed during the acute phase of infection by nefMTPIΔ5 (1,721 ± 1,017 pg/ml) were only 2-fold lower than those observed for 239wt infection (3,428 ± 2,648 pg/ml, n = 15) and about 25-fold higher than those observed in animals infected with the nef-deleted SIVmac ΔNU variant (67 ± 39 pg/ml, n = 4). In agreement with the high levels of plasma viremia, the levels of viral RNA (7.1 × 106 ± 6.2 × 106 copies/ml) were also more similar to infection with pathogenic nef-open forms of SIVmac239 (1.1 × 107 ± 7.5 × 106, n = 11), than to ΔNU infection (1.8 × 105 ± 1.2 × 105, n = 4) (Fig. 8B). The initial peak levels (days 11 to 15) of urinary neopterin, a marker of immune activation, were high (15.9 [± 2.0] times baseline), even compared to those measured in 239wt-infected animals (10.5 ± 3.1, n = 10) (Fig. 8C). All three animals that received the nefMTPIΔ5 variant became chronically infected and maintained high cell-associated viral loads throughout the course of infection, comparable to 239wt-infected macaques (Fig. 9A). Similarly, in monkeys Mm10032 and Mm10033, the RNA levels were comparable to those observed in 239wt infection (Fig. 9B). The remaining animal, Mm10034, showed RNA loads intermediate between those after 239wt and ΔNU infection. All animals showed a marked reduction in CD4+ cells during acute infection, which was most apparent for the CD4+ CD29+ memory T-cell subset (day 0, 199 ± 65/μl; 2 weeks postinfection [wpi], 101± 19/μl; 4 wpi, 35 ± 48/μl). Mm10032 and Mm10033 showed partial recovery and maintained relatively stable CD4+ cell counts (≥500/μl at 44 and 52 weeks of follow-up, respectively). The overall CD4+ T-cell count measured during chronic infection corresponded to about 50% of the preinfection values. Similarly, the number of CD4+ CD29+ cells remained at about 40% of the preinfection value (data not shown). Unexpectedly, Mm10034, which developed the lowest viral load (Fig. 9), had the greatest reduction in the number of circulating CD4+ T cells. The absolute number of CD4+ T cells dropped from 580/μl to 32/μl, and the CD4+ CD29+ cell count fell from 108/μl to 5/μl by 4 wpi. Thereafter, the CD4+ T-cell count of Mm10034 partially recovered (164/μl by 52 wpi), whereas the number of CD4+ CD29+ cells remained very low (<10/μl; data not shown). One animal, Mm10032, had to be euthanized at 44 wpi because of severe diarrhea. Hemolytic Escherichia coli, Klebsiella sp., Giardia sp., and Entamoeba sp. were isolated from the intestine, indicating a severe immunodeficiency associated with opportunistic infections. The remaining two animals are still alive 1 year postinfection. However, all animals infected with the nefMTPIΔ5 variant exhibited signs of immunodeficiency, indicated by either CD4+ cell loss or overt AIDS-like symptoms. The levels of replication and characteristics of infection were similar to those observed in nef-open SIVmac239 infection.
FIG. 8.
nefMTPIΔ5 variant behaves similar to 239wt in acutely infected rhesus macaques. Maximum levels of p27 plasma antigenemia (A), viral RNA (B), and urinary neopterin (C) in three animals infected with the nefMTPIΔ5 variant. For comparison, values obtained from macaques infected with ΔNU or 239wt are also indicated. Peak levels of p27 plasma antigenemia and of viral RNA were always observed at 2 wpi. The neopterin/creatinine ratio is expressed for each animal as the fold increase over the mean ratios determined prior to infection as described previously (20).
FIG. 9.
Replication of SIVmac239 nefMTPIΔ5 variant in vivo. (A) Number of infectious cells per 106 PBMC. (B) Viral RNA load. The detection limit for viral RNA is approximately 40 copies/ml (38). For comparison, curves for the average values obtained from rhesus macaques infected with 239wt and ΔNU are indicated. Standard deviations are not shown for clarity. Parameters were determined as described in Materials and Methods.
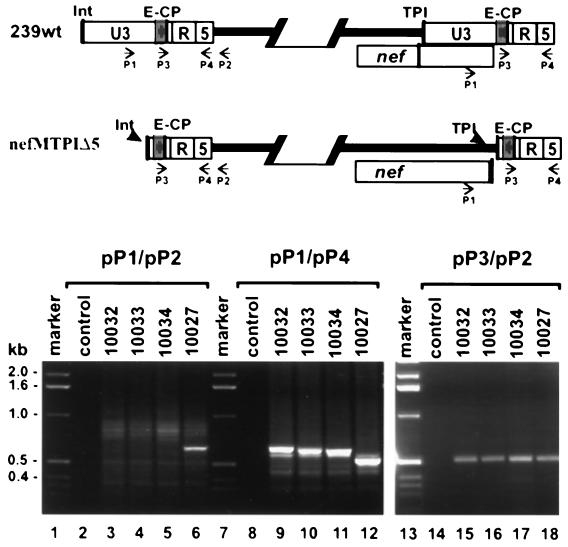
HIV and SIV are highly variable and point mutations in nef that attenuate viral replication can revert rapidly in vivo (24). Therefore, we investigated whether changes in the mutated nef-LTR region were selected in the animals infected with the nefMTPIΔ5 mutant. Sequence analysis of nef-LTR PCR fragments amplified directly from plasma RNA, PBMC, or virus-positive bulk cocultures revealed that no reversions in the mutated TPI region in the nef gene were detectable after 40 weeks of follow-up (data not shown). Furthermore, the TPI region downstream of nef and upstream of the NF-κB site was always maintained. We found that the TPImut variant did not show significant levels of replication (Fig. 5 and 6), suggesting that the mutated elements were nonfunctional. Next, we performed PCR analysis of nef-LTR sequences of virus reisolated from animals infected with SIVmac239 nefMTPIΔ5 to further confirm that the mutated TPI region was not used for reverse transcription and integration and that function was not restored in infected macaques. Amplification with primers pP1 and pP2, which bind to the US region and the noncoding region flanking the 5′ LTR, yielded a 632-bp fragment from DNA prepared from positive bulk cocultures derived from 239wt-infected animals (Fig. 10, lane 6, and data not shown). In contrast, no specific product was obtained with virus reisolated from the three animals infected with the nefMTPIΔ5 mutant (Fig. 10, lanes 3 to 5). This result confirms that the U3 region of the 5′ LTR is truncated and does not contain the binding site for primer pP1. In comparison, PCRs performed with primers p1 and p4 yielded products for both 239wt- and nefMTPIΔ5-infected animals (Fig. 10, lanes 9 to 12). The fragments obtained for the mutant form were larger than the products obtained for 239wt. This was expected because these primers span the region at the 3′ end of the viral genome that contains the inserted TPI region in the nefMTPIΔ5 genome (Fig. 10, upper panel). Finally, PCRs with primers pP3 and pP2, which bind to the 3′ end of the U3 region, which is maintained at the 5′ LTR of both the 239wt and the nefMTPIΔ5 proviruses, yielded products of 513 bp for DNA from all bulk cocultures (Fig. 10, lanes 15 to 18). Thus, the results of both the sequence and PCR analyses consistently show that the mutations in the nef-LTR region of nefMTPIΔ5 did not revert in infected animals and that the mutant virus maintained a truncated U3 region.
FIG. 10.
Analysis of nef-LTR sequences derived from nefMTPIΔ5-infected rhesus monkeys. (A) Schematic representation of the predicted 5′ and 3′ ends of the 239wt and nefMTPIΔ5 proviruses. Arrows indicate the positions of primers used for PCR amplification. The abbreviations are described in the legend to Fig. 2. (B) SIV LTR and nef-LTR sequences were amplified from positive PBMC-CEMx174 bulk cocultures. PBMC were derived at 40 wpi from three macaques infected with nefMTPIΔ5 (Mm10032, Mm10033, and Mm10034) and one animal that received 239wt (Mm10027). PCR products were separated by electrophoresis through 1.5% agarose gels.
DISCUSSION
We designed SIVmac239 mutants containing an intact nef gene which does not overlap the U3 region of the LTR and does not include essential cis-regulatory elements. The TPI-mutated nef alleles were functional in enhancing SIVmac infectivity and replication. Elimination of the U3 sequences upstream of the core enhancer elements had little if any effect on replicative capacity in cell culture or in rhesus macaques. Our observation that the upstream U3 sequences of SIVmac serve primarily or exclusively as nef coding sequence is consistent with previous studies. It has been shown that the US region is selectively deleted in vivo in the absence of an intact nef gene (26) and can be mutagenized extensively without loss of virulence (21).
We generated two sets of SIVmac239 TPI variants. One contained a U3 region of 183 bp, in which 65 bp of US sequences were maintained, and the other contained a U3 region of only 117 bp, in which essentially all US sequences are deleted. The 65 bp upstream of the single NF-κB binding site were always preserved in macaques infected with nef-deleted SIVmac239 (26) and were not altered in a previous study on the functional role of the US region (21). This region of the SIVmac LTR contains binding sites for Ets family transcription factors, which allow efficient viral replication in the absence of the entire core enhancer element (33). We found that the nefMTPIΔ5 variant, which does not contain the US65 region, was more active in infectivity and replication assays than the nefMTPI variant. Our results extend previous studies (21, 26, 33) and demonstrate that the 3′ end of the U3 region, encompassing the NF-κB and Sp1 binding sites and the TATA box is sufficient for efficient replication of SIVmac both in vitro and in vivo in rhesus macaques. It remains to be clarified why the genome of primate lentiviruses is always organized so that the nef gene overlaps the 3′ LTR. Certainly we cannot exclude that this genomic organization has some subtle advantage in certain cell types or tissues. Indeed, although the characteristics of infection with the nefMTPIΔ5 variant were much more similar to those of 239wt than to those of 239Δnef, it seemed that this mutant virus was relatively well controlled by the antiviral immune responses. Studies in large numbers of infected animals would be required to clarify if these minor differences are significant. A number of factors could explain how the TPI mutations affect viral replication in infected macaques. Nef expression levels might be slightly reduced, the mutations could affect the stability of the viral RNA, or the US sequences might have some effect on transcriptional activity in primary cells. However, the wild-type-like phenotype of the mutant virus in vivo and in vitro and the lack of reversions in infected macaques indicate that these attenuating effects may be very subtle.
Previous results on the functional relevance of the upstream U3 region were derived from infections with nef-deleted viruses (26, 27) or from SIVmac mutants that still contained the overlap of the nef gene and the LTR (21). In contrast, the nefMTPIΔ5 variant contains an intact nef gene that neither overlaps the LTR nor contains any essential cis-regulatory elements. We demonstrate that the TPI-mutated nef allele increases SIVmac infectivity and replication with an efficiency indistinguishable from that of 239wt nef. Usually, about 60% of the nef gene is overlapped by the U3 region of the LTR or by important cis-regulatory elements. Detailed structure-function analysis of Nef is complicated because mutations in the central region or the 3′ half of the nef gene not only might alter Nef function but also could have unexpected effects on transcriptional activity, integration, or RT. The constructs generated in the present study extend the experimental possibilities for the analysis of nef function in infected cells. Such systems are obligatory to study the effect of Nef on viral replication and pathogenesis. However, it remains important to investigate other nef functions in primary infected cells because (i) effects of Nef on cellular signal transduction or activation are often not observed in immortalized tumor cell lines, (ii) cellular kinases or other factors might only be expressed in relevant primary cells and binding to Nef might be affected by the presence of other viral proteins, (iii) the influence of Nef on the cell surface expression of various molecules might depend on the coexpression of other viral factors, e.g., both Vpu and Env also reduce CD4 surface expression levels (10, 40), and (iv) essentially all activities of Nef strongly depend on protein expression levels. Thus, overexpression of Nef by expression constructs in transfected cells may generate misleading data. The constructs generated in this study should be useful for delineating the molecular mechanisms that underlie the various Nef functions and investigating their relevance to viral replication in vivo. We show that lysine and tyrosine residues, which are encoded by important cis-regulatory elements in the 239wt nef ORF, are important for the ability of Nef to increase infectivity and replication. Our results suggest that this region in the nef gene is highly conserved not only because it contains important regulatory elements, but also because it encodes residues that are important for Nef function.
We demonstrate that the conserved genomic organization of the 3′ end of the genome of primate lentiviruses, specifically the large overlap between the nef gene and the LTR, is not obligatory for efficient replication of SIVmac. We are currently investigating if analogous HIV-1 TPI variants can be generated. Our initial focus was on SIV rather than on HIV-1 in this study because SIVmac239 allows studies of viral pathogenicity in a well-characterized animal model. Pathogenic HIV-SIV hybrid Nefs have recently been described (2, 25). However, the current forms are of limited value for pathogenesis studies because they only induced disease in a subset of infected macaques. The SIV constructs described in this work should prove useful for the detailed structure-function analysis of Nef in infected cells.
ACKNOWLEDGMENTS
We thank Bernhard Fleckenstein for support and encouragement, Mandy Krumbiegel for excellent technical assistance, and Julie Overbaugh and Bryce Chackerian for sMAGI cells.
This work was supported by the Wilhelm-Sander Foundation, BMBF grant 01Ki9478, and the Deutsche Forschungsgesellschaft.
REFERENCES
- 1.Alexander L, Du Z, Rosenzweig M, Jung J J, Desrosiers R C. A role for natural SIV and HIV-1 nef alleles in lymphocyte activation. J Virol. 1997;71:6094–6099. doi: 10.1128/jvi.71.8.6094-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander L, Du Z, Howe A Y, Czajak S, Desrosiers R C. Induction of AIDS in rhesus monkeys by a recombinant simian immunodeficiency virus expressing nef of human immunodeficiency virus type 1. J Virol. 1999;73:5814–5825. doi: 10.1128/jvi.73.7.5814-5825.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson R E, Sanfridson A, Ottinger J S, Doyle C, Cullen B R. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J Exp Med. 1993;177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carl S, Iafrate A J, Stahl-Hennig C, Skowronski J, Kirchhoff F. Effect of the attenuating deletion and of sequence alterations evolving in vivo on simian immunodeficiency virus C8-Nef function. J Virol. 1999;73:2790–2797. doi: 10.1128/jvi.73.4.2790-2797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carl S, Iafrate A J, Lang S M, Stolte N, Matz-Rensing K, Fuchs D, Stahl-Hennig C, Skowronski J, Kirchhoff F. Simian immunodeficiency virus containing mutations in N-terminal tyrosine residues and in the PxxP motif in Nef replicates efficiently in rhesus macaques. J Virol. 2000;74:4155–4164. doi: 10.1128/jvi.74.9.4155-4164.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carl S, Greenough T C, Krumbiegel M, Greenberg M, Skowronski J, Sullivan J L, Kirchhoff F. Modulation of different human immunodeficiency virus type 1 Nef functions during progression to AIDS. J Virol. 2001;75:3657–3665. doi: 10.1128/JVI.75.8.3657-3665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chackerian B, Haigwood N L, Overbaugh J. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology. 1995;213:386–394. doi: 10.1006/viro.1995.0011. [DOI] [PubMed] [Google Scholar]
- 8.Chowers M Y, Spina C A, Kwoh T J, Fitch N J, Richman D D, Guatelli J C. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins K L, Chen B K, Kalams S A, Walker B D, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 10.Crise B, Buonocore L, Rose J K. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J Virol. 1990;64:5585–5593. doi: 10.1128/jvi.64.11.5585-5593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 12.deRonde A, Klaver B, Keulen W, Smit L, Goudsmit J. Natural HIV-1 NEF accelerates virus replication in primary human lymphocytes. Virology. 1992;188:391–395. doi: 10.1016/0042-6822(92)90772-h. [DOI] [PubMed] [Google Scholar]
- 13.Du Z, Lang S M, Sasseville V G, Lackner A A, Ilyinskii P O, Daniel M D, Jung J U, Desrosiers R C. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell. 1995;82:665–674. doi: 10.1016/0092-8674(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 14.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 15.Garcia J V, Miller A D. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 16.Glushakova S, Grivel J C, Suryanarayana K, Meylan P, Lifson J D, Desrosiers R C, Margolis L. Nef enhances human immunodeficiency virus replication and responsiveness to interleukin-2 in human lymphoid tissue ex vivo. J Virol. 1999;73:3968–3974. doi: 10.1128/jvi.73.5.3968-3974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gundlach B R, Linhart H, Dittmer U, Sopper S, Reiprich S, Fuchs D, Fleckenstein B, Hunsmann G, Stahl-Hennig C, Überla K. Construction, replication, and immunogenic properties of a simian immunodeficiency virus expressing interleukin 2. J Virol. 1997;71:2225–2232. doi: 10.1128/jvi.71.3.2225-2232.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins J R, Sutjipto S, Marx P A, Pedersen N C. Shared antigenic epitopes of the major core proteins of human and simian immunodeficiency virus isolates. J Med Primatol. 1992;21:265–269. [PubMed] [Google Scholar]
- 19.Iafrate A J, Bronson S, Skowronski J. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 1997;16:673–684. doi: 10.1093/emboj/16.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iafrate A J, Carl S, Bronson S, Stahl-Hennig C, Swigut T, Skowronski J, Kirchhoff F. Disrupting surfaces of Nef required for downregulation of CD4 and for enhancement of virion infectivity attenuates simian immunodeficiency virus replication in vivo. J Virol. 2000;74:9836–9844. doi: 10.1128/jvi.74.21.9836-9844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilyinskii P O, Daniel M D, Simon M A, Lackner A A, Desrosiers R C. The role of upstream U3 sequences in the pathogenesis of simian immunodeficiency virus-induced AIDS in rhesus monkeys. J Virol. 1994;68:5933–5944. doi: 10.1128/jvi.68.9.5933-5944.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilyinskii P O, Desrosiers R C. Identification of a sequence element immediately upstream of the polypurine tract that is essential for replication of simian immunodeficiency virus. EMBO J. 1998;17:3766–3774. doi: 10.1093/emboj/17.13.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kestler H W, Kodama T, Ringler D J, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P K, Daniel M D, Desrosiers R C. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 24.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 25.Kirchhoff F, Münch J, Carl S, Stolte N, Mätz-Rensing K, Fuchs D, Ten Haaft P, Heeney J L, Swigut T, Skowronski J, Stahl-Hennig C. The human immunodeficiency virus type 1 nef gene can to a large extent replace simian immunodeficiency virus nef in vivo. J Virol. 1999;73:8371–8383. doi: 10.1128/jvi.73.10.8371-8383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchhoff F, Kestler H W, Desrosiers R C. Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene. J Virol. 1994;68:2031–2037. doi: 10.1128/jvi.68.3.2031-2037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Absence of intact nef sequences in a long-term, nonprogressing survivor of HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 28.Lang S M, Iafrate A J, Stahl-Hennig C, Kuhn E M, Niβlein T, Haupt M, Hunsmann G, Skowronski J, Kirchhoff F. Association of simian immunodeficiency virus Nef with cellular serine/threonine kinases is dispensable for the development of AIDS in rhesus macaques. Nat Med. 1997;3:860–865. doi: 10.1038/nm0897-860. [DOI] [PubMed] [Google Scholar]
- 29.Lock M, Greenberg M E, Iafrate A J, Swigut T, Münch J, Kirchhoff F, Shohdy N, Skowronski J. Two elements target SIV Nef to the AP-2 clathrin adaptor complex, but only one is required for the induction of CD4 endocytosis. EMBO J. 1999;18:2722–2733. doi: 10.1093/emboj/18.10.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariani R, Skowronski J. CD4 down-regulation by nef alleles isolated from human immunodeficiency virus type 1-infected individuals. Proc Natl Acad Sci USA. 1993;90:5549–5553. doi: 10.1073/pnas.90.12.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M D, Warmerdam M T, Gaston I, Greene W C, Feinberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–114. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peter F. HIV nef: the mother of all evil? Immunity. 1998;9:433–437. doi: 10.1016/s1074-7613(00)80626-3. [DOI] [PubMed] [Google Scholar]
- 33.Pöhlmann S, Flöβ S, Ilyinskii P O, Stamminger T, Kirchhoff F. Sequences just upstream of the simian immunodeficiency virus core enhancer allow efficient replication in the absence of NF-κB and Sp1 binding elements. J Virol. 1998;72:5589–5598. doi: 10.1128/jvi.72.7.5589-5598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regier D A, Desrosiers R C. The complete nucleotide sequence of a pathogenic molecular clone of SIV. AIDS Res Hum Retrovir. 1989;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard J M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 36.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D D. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swigut T, Iafrate A J, Münch J, Kirchhoff F, Skowronski J. Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility antigen expression. J Virol. 2000;74:5691–5701. doi: 10.1128/jvi.74.12.5691-5701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ten Haaft P, Verstrepen B, Überla K, Rosenwirth B, Heeney J. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J Virol. 1998;72:10281–10285. doi: 10.1128/jvi.72.12.10281-10285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varmus H. Retroviruses. Science. 1988;240:1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- 40.Willey R L, Maldarelli F, Martin M A, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]