Abstract
During viral infections, the host secretory pathway is crucial for both innate and acquired immune responses. For example, the export of most proinflammatory and antiviral cytokines, which recruit lymphocytes and initiate antiviral defenses, requires traffic through the host secretory pathway. To investigate potential effects of the known inhibition of cellular protein secretion during poliovirus infection on pathogenesis, cytokine secretion from cells infected with wild-type virus and with 3A-2, a mutant virus carrying an insertion in viral protein 3A which renders the virus defective in the inhibition of protein secretion, was tested. We show here that cells infected with 3A-2 mutant virus secrete greater amounts of cytokines interleukin-6 (IL-6), IL-8, and beta interferon than cells infected with wild-type poliovirus. Increased cytokine secretion from the mutant-infected cells can be attributed to the reduced inhibition of host protein secretion, because no significant differences between 3A-2- and wild-type-infected cells were observed in the inhibition of viral growth, host cell translation, or the ability of wild-type- or 3A-2-infected cells to support the transcriptional induction of beta interferon mRNA. We surmise that the wild-type function of 3A in inhibiting ER-to-Golgi traffic is not required for viral replication in tissue culture but, by altering the amount of secreted cytokines, could have substantial effects on pathogenesis within an infected host. The global inhibition of protein secretion by poliovirus may reflect a general mechanism by which pathogens that do not require a functional protein secretory apparatus can reduce the native immune response and inflammation associated with infection.
The functional integrity of the protein secretory apparatus is important for the cellular response to viral infection. For example, virus-infected cells can promote an antiviral state in neighboring uninfected cells through the secretion of alpha and beta interferons. Subsequent autocrine or paracrine signaling through the alpha/beta interferon receptor results in the induction of more than 50 genes that promote an antiviral cellular environment, including the double-stranded RNA protein kinase and the alpha and beta interferon genes themselves (reviewed in references 53 and 57 to 59). Another cellular response to viral infection in either fibroblast or endothelial cells is the secretion of cytokines such as interferon-inducible protein 10 (IP-10), granulocyte-macrophage colony-stimulating factor (GM-CSF), monocyte chemoattractant protein 1 (MCP-1), interleukin-1β (IL-1β), IL-8, and IL-6, which activate and attract cells of the immune system (reviewed in references 40 and 44). IL-8, for example, is a chemoattractant that recruits neutrophils as well as basophils and T cells to damaged and infected peripheral tissues. IL-6 is a proinflammatory cytokine that is believed to induce the terminal differentiation of proliferating B cells to plasma cells, stimulate antibody secretion from plasma cells, and enhance T-lymphocyte responses in secondary lymphoid organs. Infected cells can also present viral antigens in the context of MHC class I molecules to activate specific CD8+ cytotoxic T lymphocytes. All of the above responses require a functional protein secretory apparatus for the infected cell to communicate its status to surrounding cells and to the effector cells of the immune system.
Poliovirus is a nonenveloped positive-sense RNA virus that infects primate cells. Although the virus lacks envelope proteins or other proteins that require conventional anterograde traffic through the protein secretory pathway, the virus extensively alters and utilizes the membranes of the host secretory pathway. Some of these alterations are likely to be required for replication of the virus. For example, poliovirus RNA replication occurs on the cytosolic surface (6) of double-membraned vesicles derived from the endoplasmic reticulum (ER) via the combined action of viral proteins 2BC and 3A (10, 54, 60). Several different mutations in the 2BC and 3A coding regions impair or destroy viral viability, imparting specific defects in RNA replication (3, 4, 23, 24, 36).
We have shown that ER-to-Golgi transport is inhibited early in poliovirus infection (15, 16). Either viral protein 3A or 2B can reduce the rate of protein secretion in isolation, although 3A has the stronger effect and appears to be specific for ER-to-Golgi traffic (15, 16). Poliovirus 3A protein sequences associate tightly with membranes (62) and, when expressed in isolation, localize to the ER (15, 60). Previously, the inhibition of ER-to-Golgi traffic by 3A was shown for two different marker proteins expressed from transfected plasmids, the G protein from vesicular stomatitis virus (16) and human alpha-1 protease inhibitor (15). During infection with poliovirus, it is possible that all cargo of the secretory pathway is delayed in its transport. This could be significant in viral pathogenesis, because many of the well-known proteins that are induced and secreted during viral infection are cytokines that aid in the antiviral response by the infected cell.
To test whether 3A, in the context of the virus, would limit the amount of secretion of endogenous proteins known to be induced in the innate immune response, we used a virus with a mutation in 3A. The mutant virus was created by the molecular insertion of one codon, a serine, between amino acids 13 and 14 of 3A, into a wild-type poliovirus cDNA. Virus bearing the 3A-2 mutation, while slightly cold sensitive, is viable and does not show substantial growth defects at temperatures above 32.5oC. However, the 3A-2 protein when expressed in isolation displays little inhibition of cellular ER-to-Golgi traffic under all conditions examined in tissue culture (15).
The existence of the 3A-2 mutant poliovirus suggests that the function of 3A in inhibiting ER-to-Golgi traffic is not required for viral replication. We have recently shown that stimulation of antigen-specific CD8+ cytotoxic T cells is reduced in cells infected with wild-type poliovirus but not with the 3A-2 mutant virus. Thus, the wild-type function of 3A in inhibiting ER-to-Golgi traffic can serve to reduce the transport of newly synthesized MHC class I to the cell surface, and the difference in transport rate observed can have functional consequences for CD8+ T-cell recognition in tissue culture (14). Thus, we suggest that the ability of 3A to inhibit ER-to-Golgi traffic may serve as a virulence factor during infection of host animals.
Extensive interactions between poliovirus and the host take place during the infectious cycle, including the inhibition of cellular transcription and translation. To test the effect of inhibiting host protein secretion by viral protein 3A separately from other potential viral effects, such as the inhibition of host translation and transcription (2, 18, 55, 56, 67), we compared the effects of infection with wild-type and 3A-2 mutant polioviruses on the secretion of antiviral and proinflammatory cytokines during single-cycle infections. A significant increase in the secretion of beta interferon, IL-8, and IL-6 was observed in mutant-infected cells, arguing that the wild-type function of 3A acts to reduce the secretion of these cytokines during natural infection.
MATERIALS AND METHODS
Cells and viruses.
MG63 cells (kindly provided by the T. Maniatis laboratory, Harvard University) were grown as described (61). HEC-1B (American Type Culture Collection [ATCC]) cells (kindly provided by N. Reich, State University of New York, Stony Brook) were grown as described. COS-1 cells were grown as described (16). Type 1 Mahoney poliovirus was grown and counted on all cell types as described (32). Full-length poliovirus cDNA (49) containing the 3A-2 mutation (3) was transfected into COS cells using the DEAE-dextran method (43). Single plaques were picked and propagated to high-titer stocks on HeLa cell monolayers. Individual stocks were plaque assayed on HeLa cells at 32.5°C for reversion. Viral stocks with the lowest proportion of phenotypically revertant viruses (fewer than 2%) were used in subsequent experiments. Single-cycle growth curves in MG-63, HEC-1B, and COS-1 cells were all performed as described (39) after determination of the titers of the stocks on the relevant cell type.
Cellular protein synthesis.
MG63 cells (2 × 105) were washed with phosphate-buffered saline (PBS) containing 1 mg of MgCl2 and 1 mg CaCl2 per ml (PBS+) and infected with wild-type or 3A-2 mutant virus at a multiplicity of infection (MOI) of 20 PFU/cell for 30 min at 37°C. Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum was added. At the times indicated postinfection, cells were washed with PBS+. DMEM lacking methionine (Life Technologies, Gaithersburg, Md.) containing 55 μCi of [35S]methionine and [35S]cysteine per ml (Express Label; New England Nuclear, Beverly, Mass.) was added, incubation was continued for 15 min. at 37°C, and cells were washed in ice-cold PBS, collected into a total of 500 μl of PBS by scraping, and collected by centrifugation at 300 × g for 5 min at 4°C. Pelleted cells were resuspended in 50 ml of RSB+NP-40 (10 mM Tris [pH 7.5], 10 mM NaCl, 1.5 mM MgCl2, 1% NP-40 [pH 7.5]) and centrifuged at 2,000 × g for 10 min to remove the nuclei. Supernatants were collected, and radioactive proteins from equivalent numbers of cells were displayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described (16).
Quantitation of secreted beta interferon, IL-6, and IL-8.
MG63 cells were infected with wild-type or mutant 3A-2 virus at an MOI of 10 PFU/cell. For quantitation of beta interferon, duplicate samples of 5 × 106 cells were analyzed, and for IL-6 and IL-8, triplicate samples of 5 × 105 cells were studied. Cells were infected for 30 min at 37°C, washed with PBS+, and further incubated in the presence of DMEM containing 10% fetal bovine serum. At various times postinfection, samples of medium were collected and cleaned by centrifugation at 600 × g for 5 min at 4°C. For the quantitation of beta interferon, supernatants were lyophilized, resuspended in 150 μl of H2O (4°C), and subjected to enzyme-linked immunosorbent assay (ELISA) analysis as recommended by the manufacturer (Biosource International, Camarillo, Calif.). For IL-6 quantitation, supernatants were diluted threefold, and for IL-8 analysis, supernatants were diluted sixfold before performing ELISA analysis according to the manufacturer (Biosource International). Light absorbance at 405 nm was measured in a Bio-Rad model 550 microplate reader (Bio-Rad, Hercules, Calif.). The amount of cytokine in each sample was interpolated from duplicate standard curves performed for each assay.
RNase protection assay.
HEC-1B cells (8 × 106) on 100-mm plates were infected at an MOI of 20 PFU/cell with either wild-type or mutant 3A-2 virus and incubated for 2 h in 1.5 ml of minimal essential medium (Eagle) in Earle's balanced salt solution with nonessential amino acids and sodium pyruvate and without serum. After aspirating, 1.5 ml of medium containing 175 μg of poly(I)-poly(C) (Sigma, St. Louis, Mo.) + 800 μg of DEAE-dextran per ml was added. Total RNA was collected using RNEasy (Qiagen, Valencia, Calif.). Endogenous beta interferon mRNA was detected using a probe prepared from pSP65′IF (kindly provided by T. Maniatis, Harvard University) cleaved with EcoRI and transcribed with SP6 polymerase (19). Cyclophilin RNA was detected using pTRI-cyclophilin-Human (Ambion, Austin, Tex.) transcribed with SP6 polymerase. Hybridization and RNase digestion were done as described (68). Quantitation was performed using a Storm 860 (Molecular Dynamics, Sunnyvale, Calif.).
Electron microscopy.
COS-1 cells were infected at an MOI of 20 PFU/cell for 30 min. at 37°C. DMEM containing 10% calf serum was added, and cells were incubated at 37°C for 4.5 h. After trypsinization, cells were resuspended in PBS+ and spun at 240 × g for 3 min. The cell pellet was then resuspended in DMEM containing 10% calf serum and 150 mM mannitol and spun again at 240 × g for 3 min. The cell pellet was then high-pressure frozen in a BAL-TEC HPM-010, freeze-substituted in 0.1% tannic acid in acetone followed by 2% osmium tetroxide in acetone, embedded in Epon-Araldite epoxy resin, and thin sectioned for imaging in a Philips CM10 electron microscope as described (54).
RESULTS
Increased secretion of beta interferon, IL-6, and IL-8 from cells infected with 3A-2 mutant poliovirus.
To examine whether the ability of poliovirus 3A protein to inhibit ER-to-Golgi traffic has a role in inhibiting cytokines induced by poliovirus infection, we compared the secretion of cytokines known to be produced during poliovirus infection from cells infected with wild-type virus and with virus that contained a previously characterized mutation, 3A-2 (3). The secretion of beta interferon during multiple cycles of poliovirus infection had been published previously (29). IL-6 and IL-8 mRNAs were found to be induced and associated with polysomes during poliovirus infection (31), making it likely that these proteins would be produced. Several other cytokines, including RANTES, macrophage inflammatory protein 1β (MIP-1β), and IL-1β, remained undetectable after poliovirus infection in several cell lines (data not shown). Therefore, we focused our studies on the secretion of beta interferon, IL-6, and IL-8.
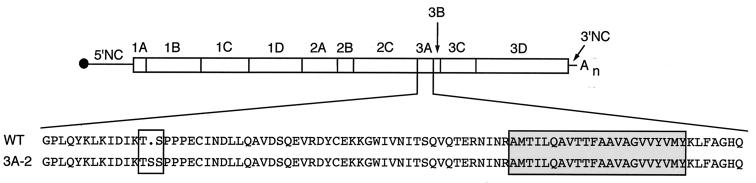
The 3A-2 mutant virus differs from wild-type virus only by the presence of a three-nucleotide insertion in the 3A coding region, introducing a serine codon between Thr13 and Ser14 (Fig. 1). Although the 3A-2 mutant virus displayed a growth defect in many cell types at low temperatures, its rate of growth at 37°C was indistinguishable from that of wild-type virus in several cell lines (3). When expressed in isolation, 3A-2 mutant protein was not as effective in inhibiting ER-to-Golgi traffic as wild-type 3A protein at 32.5, 37, or 39.5°C (15). Our working hypothesis was, therefore, that comparison of wild-type and 3A-2 mutant poliovirus infections at 37°C, at which temperature the viruses display similar growth, should test the effects of inhibiting host protein secretion by wild-type 3A protein.
FIG. 1.
Sequence changes in 3A-2 mutant virus. The sequences of the 87-amino-acid 3A protein coding region from wild-type Mahoney type I poliovirus and of the 3A-2 mutant protein are shown. The wild-type sequence and the sequence of the 3A-2 mutant protein are aligned, and the serine insertion at position 14 in 3A-2 is boxed. The hydrophobic C-terminal region is boxed and shaded. NC, noncoding.
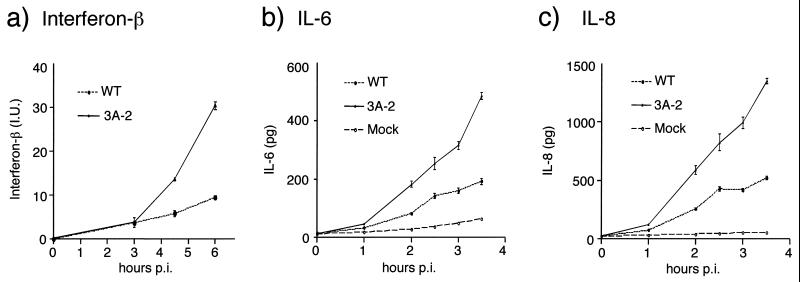
To test whether the inhibition of host protein secretion by poliovirus 3A protein affected the amount of beta interferon secreted, human MG63 cells, known to be highly inducible for beta interferon synthesis (8), were infected with either wild-type or 3A-2 mutant virus. The amount of beta interferon released into the medium during a single cycle of infection with 3A-2 mutant poliovirus was threefold greater than from cells infected with wild-type virus (Fig. 2a), consistent with the hypothesis that the wild-type function of 3A protein served to limit beta interferon secretion during infection.
FIG. 2.
(a) Amount of beta interferon secreted at various times after infection with wild-type (WT) poliovirus and with 3A-2 mutant poliovirus. MG63 cells were infected at 20 PFU/cell, and the amounts of beta interferon were determined by ELISA in conjunction with a standard curve. Standard error from replicate experiments is shown. Amounts of IL-6 (b) and IL-8 (c) secreted from MG63 cells as a function of time after mock infection or infection with wild-type or 3A-2 mutant poliovirus was also determined by ELISA in conjunction with standard curves. Standard error from replicate experiments is shown. At later time points there was a decrease in the total amount of IL-6 and IL-8 in the medium, leading to highly variable measurements and suggesting that the IL-6 and IL-8 proteins are unstable (data not shown).
Although the transcriptional and translational regulation of IL-6 and IL-8 differs from that of beta interferon (reviewed in references 28, 37, and 52), all of these cytokines are secreted in higher abundance from 3A-2 mutant-infected cells than from wild-type-infected cells. Approximately three times more IL-6 (Fig. 2b) and the chemokine IL-8 (Fig. 2c) were secreted from MG63 cells infected with 3A-2 mutant poliovirus than from cells infected with wild-type poliovirus. Overall, these findings support the hypothesis that the block of ER-to-Golgi trafficking by 3A is a general phenomenon that will inhibit any protein routed for export from the cell through the ER-to-Golgi pathway.
Increased secretion of cytokines from 3A-2 mutant virus-infected MG63 cells is not due to a difference in viral yield or delayed inhibition of host translation.
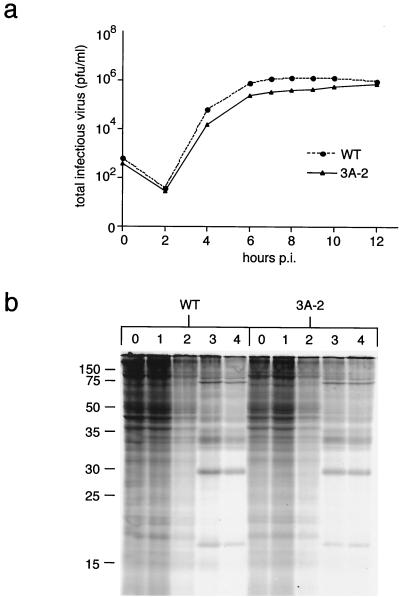
An alternative hypothesis to the observed differences in secretion of beta interferon, IL-6, and IL-8 between 3A-2- and wild-type-infected cells is that infection with 3A-2 mutant virus causes a more potent induction of beta interferon, IL-6, and IL-8 mRNA transcription or accelerates the synthesis and secretion pathways of these cytokines by some other mechanism. Such a scenario could be envisaged if, for example, 3A-2 mutant virus infection proceeded very slowly, delaying the inhibition of host translation; increased amounts of cytokines would be synthesized early in infection with 3A-2 mutant virus. However, as shown in Fig. 2, the yields of wild-type and 3A-2 mutant virus in a single-cycle infection in MG63 cells were not substantially different (Fig. 3a). Furthermore, MG63 cells infected with both viruses showed very similar time courses of inhibition of host protein synthesis, as shown by the gradual reduction in the amount of background labeling with [35S]methionine during the course of single-cycle infections (Fig. 3b). Therefore, the increased amounts of cytokines beta interferon, IL-6, and IL-8 in the medium of 3A-2 mutant-infected cells is not likely to be due to increased synthesis of these cytokines.
FIG. 3.
Effect of 3A-2 mutation on viral growth and inhibition of host protein synthesis. (a) Growth curves of wild-type (WT) and 3A-2 mutant poliovirus in MG63 cells at 37°C. MG63 cells were infected with wild-type or 3A-2 mutant virus at 0.1 PFU/cell. Cells were harvested at the times indicated postinfection (p.i.), lysates were prepared, and virus yield was determined by plaque assay of the lysates. (b) Total proteins synthesized after infection of MG63 cells at 20 PFU/cell with wild-type and 3A-2 mutant poliovirus are shown. At the indicated times postinfection (hours), cells were labeled for 15 min with [35S]methionine/cysteine, lysates were prepared, and proteins were displayed on an SDS–14% PAGE gel. Sizes are shown on the left (in kilodaltons).
Wild-type and 3A-2 mutant poliovirus infections allow comparable amounts of beta interferon mRNA synthesis.
The synthesis of beta interferon, IL-6, and IL-8 mRNAs and protein involves complex autocrine loops. For example, the transcription of beta interferon mRNA is induced by infection with both RNA and DNA viruses. This induction is thought to be mediated, at least in part, by the accumulation of intracellular double-stranded RNA. However, beta interferon protein, once it is translated and secreted from the infected cell, can bind to the alpha/beta interferon receptors on the cell in which it was made as well as neighboring uninfected cells, inducing transcriptional induction of at least 50 different mRNAs, including beta interferon mRNA (13, 21, 53, 57).
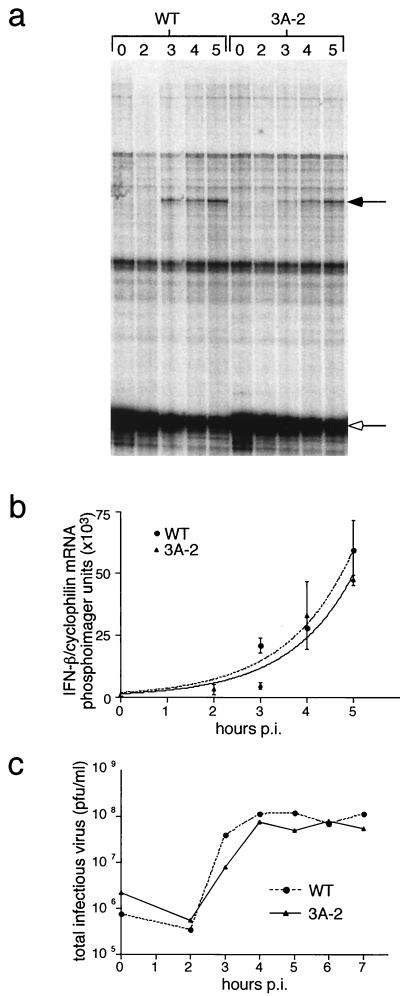
To obtain an accurate assessment of the amount of beta interferon mRNA made in the absence of an autocrine regulatory loop, we monitored beta interferon mRNA accumulation in HEC-1B cells, known to lack a functional alpha/beta interferon receptor (1, 22, 64). When HEC-1B cells were infected with wild-type or 3A-2 mutant poliovirus, no accumulation of beta interferon mRNA could be observed by RNase protection (data not shown). However, when wild-type- and 3A-2-infected cells were treated with double-stranded RNA 1.5 h after infection, the synthesis of beta interferon mRNA could be detected after 3 h of infection, and it continued to increase in abundance throughout both infections (Fig. 4a). When normalized to the amounts of cyclophilin mRNA, which is constitutively expressed, no differences were detected in the amounts of beta interferon mRNA that accumulated in HEC-1B cells infected with wild-type and 3A-2 mutant virus (Fig. 4b). Therefore, there is no difference between wild-type- and 3A-2-infected cells in their ability to support the transcriptional induction of beta interferon mRNA synthesis.
FIG. 4.
Beta interferon mRNA and secreted protein levels from HEC-1B cells, which lack a functional alpha/beta interferon receptor, infected with wild-type (WT) and 3A-2 mutant poliovirus and treated with double-stranded RNA. (a) RNase protection assay. HEC-1B cells were infected with wild-type poliovirus or 3A-2 mutant poliovirus at 20 PFU/cell. After 1.5 h of incubation at 37°C, cells were treated with 175 μg of poly(I):poly(C) and 800 μg of DEAE-dextran per ml in serum-free medium. RNA was collected at the indicated times (hours) postinfection (p.i.), and the RNase protection assay was performed as described in Materials and Methods. The solid arrow denotes beta interferon mRNA, and the open arrow identifies cyclophilin mRNA. (b) PhosphoImager quantitative analysis from the gel in panel a along with replicate experiments analyzed on the same gel. Standard error of replicated experiments is shown. (c) Single-cycle growth curve for HEC-1B cells infected at 20 PFU/cell with either wild-type (WT) or 3A-2 mutant virus.
Cells infected with wild-type and 3A-2 mutant virus display similar ultrastructural changes.
Wild-type 3A protein, expressed in isolation, inhibits ER-to-Golgi traffic (16) and causes the swelling of the ER membranes in COS-1 cells (15). We were concerned that the differential effects of the wild-type 3A and mutant 3A-2 proteins on ER biochemistry could result in alterations in the ability of these virus-infected cells to form the membranous vesicles on which poliovirus RNA replication occurs (5, 7, 12, 54, 63). We and others had previously speculated that the inhibition of ER-to-Golgi traffic by viral 3A protein was involved mechanistically in the formation of these virus-induced vesicles during infection (15–17, 55, 65). Furthermore, recent data from this laboratory have shown that a combination of viral proteins 3A and 2BC was required to mimic the ultrastructure of poliovirus-infected cells, arguing that 3A may play a direct role in vesicle formation (60).
To test the effect of the 3A-2 mutant allele on cellular membrane rearrangements during viral infection, the ultrastructure of cells infected with wild-type and 3A-2 mutant poliovirus was examined. High-pressure cryofixation followed by freeze-substitution was chosen as a preparative technique for its ability to preserve transient and unstable membrane morphologies (11, 25, 41). Previous studies demonstrated that this method preserved the complex membrane morphology of vesicles that accumulate in the centrosomal region of both COS and HeLa cells during poliovirus infection (15, 54, 60). COS-1 cells were found to have superior high-resolution cytoplasmic structures in comparison with other cell lines that did not freeze or stain well. Therefore, they were chosen instead of MG63 cells for the study of membrane changes after wild-type and 3A-2 infection.
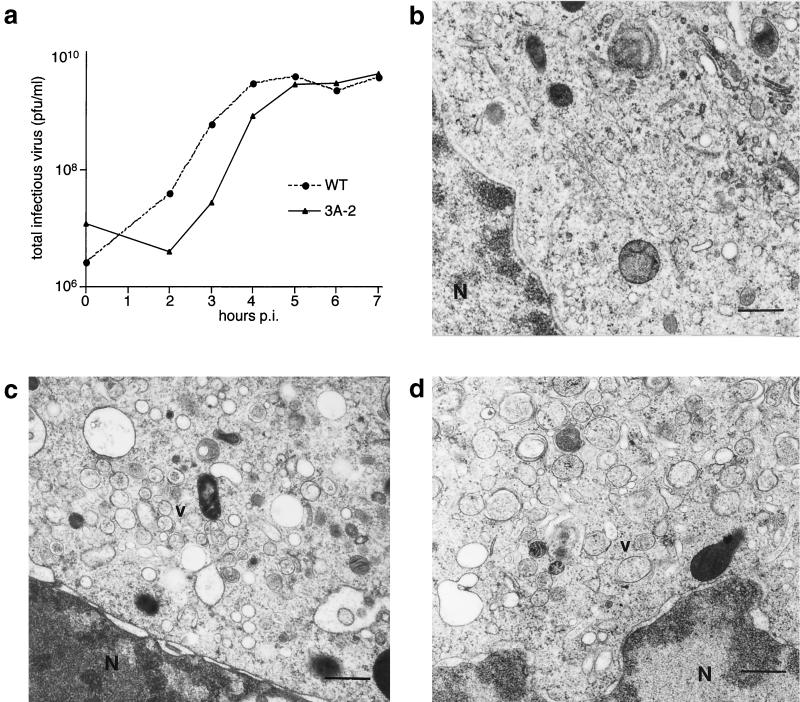
In COS-1 cells, although some differences in the growth of 3A-2 virus were observed, similar yields of virus were obtained at later time points (Fig. 5a). At these comparable time points, cells infected with wild-type and 3A-2 polioviruses showed similar ultrastructural changes. In both cases, the centrosomal region of the cytoplasm, where Golgi stacks are normally found in uninfected cells, was occupied by a cluster of vesicles, many limited by double or multiple membranes (Fig. 5b to d). Therefore, the 3A-2 mutation did not inhibit the membrane rearrangements induced by poliovirus infection.
FIG. 5.
Ultrastructure of COS-1 cells infected with wild-type and 3A-2 mutant poliovirus. The yields of intracellular virus as a function of time in these COS-1 cells are shown for both wild-type (WT) virus and 3A-2 mutant virus infected at 20 PFU/cell (a). Electron microscopy of uninfected cells (b), cells infected with wild-type poliovirus (c), and 3A-2 mutant poliovirus (d) for 4.5 h at 37°C. Bars, 500 nm. N, nucleus; v, poliovirus-induced vesicles.
DISCUSSION
Poliovirus 3A protein is known to have numerous functions in the viral replicative cycle, but the relationship between these functions is not known. Mutations in the 3A coding region, including the 3A-2 mutation at the restrictive growth temperature, are known to cause defects in viral RNA synthesis (3, 23). The larger polypeptide 3AB, which contains the 22-amino-acid protein primer for viral RNA synthesis fused to its carboxyl terminus, binds to 3D, the poliovirus RNA-dependent RNA polymerase (27, 66) and, when purified in the presence of detergent, stimulates polymerase activity (33, 45, 48, 51, 66). Recently, we have shown that viral proteins 2BC and 3A, expressed together, can mimic the ultrastructure and membrane rearrangements of poliovirus-infected cells (60), suggesting a role for 3A in vesicle formation during infection.
When expressed in isolation, viral 3A protein localizes to the ER, where it causes a three- to fivefold reduction in the rate of ER-to-Golgi traffic. In the presence of 3A, ER membranes assume a distended configuration, and protein cargo otherwise destined for secretion accumulates in these swollen cisternae. This dramatic decrease in transport rate is not seen in cells that express 3A-2, a mutant allele (15). The reduced ability of the 3A-2 mutant protein to inhibit ER-to-Golgi traffic does not correlate with reduced yield of 3A-2 mutant virus in single-cycle infections, with reduced binding of 3AB proteins that contain the 3A-2 mutation to poliovirus 3D polymerase in the two-hybrid system (data not shown), or with altered cellular ultrastructure during viral infection (Fig. 5). Therefore, we conclude that the ability of poliovirus 3A protein to inhibit ER-to-Golgi traffic is not likely to be absolutely required for viral replication in tissue culture.
The reduced ability of the 3A-2 mutant protein to block secretion does correlate, however, with an increase in the amounts of secreted cytokine from infected cells. Specifically, threefold greater amounts of beta interferon, IL-6, and IL-8 are secreted from MG63 cells infected with 3A-2 mutant virus than from those infected with wild-type virus (Fig. 2). This difference in the extracellular abundance of three different cytokines cannot be attributed to changes in the ability of the 3A-2 mutant virus to replicate in MG63 cells (Fig. 3a), to inhibit host translation (Fig. 3b), or to support the transcriptional induction of beta interferon mRNA (Fig. 4). Therefore, we conclude that the increased amount of cytokines beta interferon, IL-6, and IL-8 secreted from 3A-2 mutant poliovirus-infected cells is the direct result of the inability of the 3A-2 mutant protein to inhibit ER-to-Golgi traffic as effectively as the wild-type 3A protein.
Many picornaviruses are known to be poor inducers of interferon. Of five types of viruses tested for their ability to induce alpha a beta interferons in cultures of human leukocytes after multiple infectious cycles, rhinovirus, the only picornavirus tested, was the least effective (46). Nonetheless, alpha interferon has been observed in nasal secretions of humans infected with rhinovirus (35), and all strains of poliovirus tested thus far cause the induction of small amounts of alpha/beta interferons in human leukocytes in tissue culture (47). For Theiler's virus, secretion of alpha/beta interferons significantly affects the outcome of disease. Mice that lack the alpha/beta interferon receptor die of severe encephalitis within 2 weeks of infection with Theiler's virus, whereas their heterozygous littermates resolve the initial encephalitis and become chronically infected (20). For mengovirus and encephalomyocarditis virus, it has been suggested that the attenuation of virulence caused by decreasing the length of poly(C) tracts in the 5′ noncoding regions correlates with reduced induction of the interferon-induced kinase protein kinase R (38). Although the amount of potent antiviral cytokines such as alpha/beta interferons may be low during some picornaviral infections, they can exert profound effects on the course of infection.
In its natural host organism, humans, poliovirus causes either subclinical enteric infection or, in the fewer than 1% of infected individuals in whom the virus spreads to the nervous system, paralytic poliomyelitis (50). After being ingested, the virus can be found in tonsillar tissue and the Peyer's patches of the intestinal ileum. The role of an inflammatory response has not been documented during this early phase of virus production at these initial infected sites.
Poliomyelitis is characterized by the virus-induced destruction of motor neurons, mostly within the anterior horn of the spinal cord, but also in the brain stem and the motor cortex (50). At the sites of cell necrosis, some authors have reported an interstitial inflammatory response involving polymorphonuclear leukocytes, some mononuclear leukocytes, and an occasional eosinophil (26, 30, 42). However, symptoms of encephalitis only rarely have been found to accompany poliomyelitis (42); the inflammatory response has long been thought to be secondary to the virus-induced killing of motor neurons. As stated by Bodian, “in many areas of severe nerve cell destruction the inflammatory response is relatively quite mild” (9). In another study, lymphocytes were found to be more numerous around tissue infected with less virulent strains of virus and at later stages of infection (30). Other than these few pieces of data that are relatively dated, very little about the inflammatory response to poliovirus has been reported.
Here, we suggest that the inhibition of protein secretion by 3A is not required for viral RNA replication itself, but serves as a virulence factor, reducing the amounts of antiviral and proinflammatory cytokines secreted from infected cells. It is possible that this is related to the observed phenotype of an I46T mutation in 3A, which leads to a defect in cell lysis (34). We have recently shown that the inhibition of ER-to-Golgi traffic by 3A also serves to reduce the rate of transport of MHC class I molecules to the surface of poliovirus-infected cells and therefore the amount of MHC class I-dependent activation of CD8+ T cells (14). Therefore, the ability of poliovirus 3A protein to inhibit ER-to-Golgi traffic through its effects on cytokine secretion, antigen presentation in the context of MHC class I, and potentially other secreted or membrane-associated proteins, is likely to affect viral pathogenesis within the host by increasing virus yield, reducing virus-associated inflammation, or both. The effect of inhibiting ER-to-Golgi traffic by the wild-type function of 3A in poliovirus pathogenesis in a transgenic mouse model is under investigation.
ACKNOWLEDGMENTS
We thank Peter Sarnow, Edward S. Mocarski, John Lyle, and Sunny Choe for insightful comments on the manuscript and Tom Maniatis and Nancy C. Reich for generous provision of reagents.
This work was supported by NIH grants AI-25166 and AI-07328 and by the Hutchison Program for Translational Medicine, Stanford University.
REFERENCES
- 1.Bazzigher L, Pavlovic J, Haller O, Staeheli P. Mx genes show weaker primary response to virus than other interferon-regulated genes. Virology. 1992;186:154–160. doi: 10.1016/0042-6822(92)90069-2. [DOI] [PubMed] [Google Scholar]
- 2.Belsham G J, Jackson R J. Translation initiation on picornavirus RNA. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 869–900. [Google Scholar]
- 3.Bernstein H D, Baltimore D. Poliovirus mutant that contains a cold-sensitive defect in viral RNA synthesis. J Virol. 1988;62:2922–2928. doi: 10.1128/jvi.62.8.2922-2928.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein H D, Sarnow P, Baltimore D. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J Virol. 1986;60:1040–1049. doi: 10.1128/jvi.60.3.1040-1049.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160:220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- 6.Bienz K, Egger D, Pfister T. Characteristics of the poliovirus replication complex. Arch Virol Suppl. 1994;9:147–157. doi: 10.1007/978-3-7091-9326-6_15. [DOI] [PubMed] [Google Scholar]
- 7.Bienz K, Egger D, Troxler M, Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J Virol. 1990;64:1156–1163. doi: 10.1128/jvi.64.3.1156-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billiau A, Edy V G, Heremans H, Van Damme J, Desmyter J, Georgiades J A, De Somer P. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–15. doi: 10.1128/aac.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodian D. Histopathologic basis of clinical findings in poliomyelitis. Am J Med. 1949;6:563–578. doi: 10.1016/0002-9343(49)90130-8. [DOI] [PubMed] [Google Scholar]
- 10.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 11.Dahl R, Staehelin L A. High-pressure freezing for the preservation of biological structure: theory and practice. J Electron Microsc Tech. 1989;13:165–174. doi: 10.1002/jemt.1060130305. [DOI] [PubMed] [Google Scholar]
- 12.Dales S, Eggers H J, Tamm I, Palade G E. Electron microscopic study of the formation of poliovirus. Virology. 1965;26:379–389. doi: 10.1016/0042-6822(65)90001-2. [DOI] [PubMed] [Google Scholar]
- 13.David M. Transcription factors in interferon signaling. Pharm Ther. 1995;65:149–161. doi: 10.1016/0163-7258(94)00050-d. [DOI] [PubMed] [Google Scholar]
- 14.Deitz S B, Dodd D A, Cooper S, Parham P, Kirkegaard K. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc Natl Acad Sci USA. 2000;97:13790–13795. doi: 10.1073/pnas.250483097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doedens J R, Giddings T H, Jr, Kirkegaard K. Inhibition of ER-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol. 1997;71:9054–9064. doi: 10.1128/jvi.71.12.9054-9064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doedens J R, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egger D, Teterina N, Ehrenfeld E, Bienz K. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J Virol. 2000;74:6570–6580. doi: 10.1128/jvi.74.14.6570-6580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 549–574. [Google Scholar]
- 19.Enoch T, Zinn K, Maniatis T. Activation of the human beta-interferon gene requires an interferon-inducible factor. Mol Cell Biol. 1986;6:801–810. doi: 10.1128/mcb.6.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiette L, Aubert C, Muller U, Huang S, Aguet M, Brahic M, Bureau J F. Theiler's virus infection of 129Sv mice that lack the interferon alpha/beta or interferon gamma receptors. J Exp Med. 1995;181:2069–2076. doi: 10.1084/jem.181.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsberg K, Paulsson Y, Westermark B. Effect on platelet-derived growth factor-induced mitogenesis of double-stranded RNA: evidence for an autocrine growth inhibition mediated by interferon-beta. J Cell Physiol. 1988;136:266–272. doi: 10.1002/jcp.1041360208. [DOI] [PubMed] [Google Scholar]
- 22.Fuse A, Ashino-Fuse H, Kuwata T. Binding of 125I-labeled human interferon to cell lines with low sensitivity to interferon. Gann. 1984;75:379–384. [PubMed] [Google Scholar]
- 23.Giachetti C, Hwang S S, Semler B L. cis-Acting lesions targeted to the hydrophobic domain of a poliovirus membrane protein involved in RNA replication. J Virol. 1992;66:6045–6057. doi: 10.1128/jvi.66.10.6045-6057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giachetti C, Semler B L. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J Virol. 1991;65:2647–2654. doi: 10.1128/jvi.65.5.2647-2654.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilkey J C, Staehelin L A. Advances in ultrarapid freezing for the preservation of cellular ultrastructure. J Electron Microsc Tech. 1986;3:177–210. [Google Scholar]
- 26.Hashimoto I, Hagiwara A, Komatsu T. Ultrastructural studies on the pathogenesis of poliomyelitis in monkeys infected with poliovirus. Acta Neuropathol. 1984;64:53–60. doi: 10.1007/BF00695606. [DOI] [PubMed] [Google Scholar]
- 27.Hope D A, Diamond S E, Kirkegaard K. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J Virol. 1997;71:9490–9498. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horn F, Henze C, Heidrich K. Interleukin-6 signal transduction and lymphocyte function. Immunobiology. 2000;202:151–167. doi: 10.1016/S0171-2985(00)80061-3. [DOI] [PubMed] [Google Scholar]
- 29.Hovi T, Pitkaranta A, Macadam A, Minor P, Almond J. Covariance of lowered capacity to induce interferon in human leukocytes and temperature sensitivity of type 3 poliovirus. J Interferon Res. 1991;11:105–110. [PubMed] [Google Scholar]
- 30.Hurst E W. The histology of experimental poliomyelitis. J Pathol. 1929;32:457–477. [Google Scholar]
- 31.Johannes G, Carter M S, Eisen M B, Brown P O, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lama J, Paul A V, Harris K S, Wimmer E. Properties of purified recombinant poliovirus protein 3aB as substrate for viral proteinases and as cofactor for RNA polymerase 3Dpol. J Biol Chem. 1994;269:66–70. [PubMed] [Google Scholar]
- 34.Lama J, Sanz M A, Carrasco L. Genetic analysis of poliovirus protein 3A: characterization of a non cytopathic mutant virus defective in killing Vero cells. J Gen Virol. 1998;79:1911–1921. doi: 10.1099/0022-1317-79-8-1911. [DOI] [PubMed] [Google Scholar]
- 35.Levandowski R A, Horohov D W. Rhinovirus induces natural killer-like cytotoxic cells and interferon alpha in mononuclear leukocytes. J Med Virol. 1991;35:116–120. doi: 10.1002/jmv.1890350208. [DOI] [PubMed] [Google Scholar]
- 36.Li J-P, Baltimore D. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J Virol. 1988;62:4016–4021. doi: 10.1128/jvi.62.11.4016-4021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maniatis T, Falvo J V, Kim T H, Kim T K, Lin C H, Parekh B S, Wathelet M G. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 38.Martin L R, Neal Z C, McBride M S, Palmenberg A C. Mengovirus and encephalomyocarditis virus poly(C) tract lengths can affect virus growth in murine cell culture. J Virol. 2000;74:3074–3081. doi: 10.1128/jvi.74.7.3074-3081.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maynell L A, Kirkegaard K, Klymkowsky M W. Inhibition of poliovirus replication by brefeldin A. J Virol. 1992;66:1985–1994. doi: 10.1128/jvi.66.4.1985-1994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller M D, Krangel M S. Biology and biochemistry of the chemokines: a family of chemotactic and inflammatory cytokines. Crit Rev Immunol. 1992;12:17–46. [PubMed] [Google Scholar]
- 41.Moor H. Theory and practice of high pressure freezing. In: Steinbrecht R A, Zierold K, editors. Cryotechniques in biological electron microscopy. Berlin, Germany: Springer-Verlag; 1987. pp. 175–191. [Google Scholar]
- 42.Neal J B. Symptomatology. In: Milbank J, editor. Poliomyelitis: International Committee for the Study of Infantile Paralysis. Baltimore, Md: The Williams and Wilkins Company; 1932. pp. 162–215. [Google Scholar]
- 43.Novak J E, Jarvis T C, Kirkegaard K. RNA: transcription, transfection and quantitation. In: Mahy B W J, Kangro H O, editors. Virology methods manual. London, England: Academic Press; 1996. pp. 165–190. [Google Scholar]
- 44.Oppenheim J J, Feldmann M, editors. Cytokine reference. Vol. 1. San Diego, Calif: Academic Press; 2001. [Google Scholar]
- 45.Paul A V, Cao X, Harris K S, Lama J, Wimmer E. Studies with poliovirus polymerase 3Dpol. J Biol Chem. 1994;269:29173–29181. [PubMed] [Google Scholar]
- 46.Pitkaranta A, Hovi T. Induction of interferon in human leukocyte cultures by natural pathogenic respiratory viruses. J Interferon Res. 1993;13:423–426. doi: 10.1089/jir.1993.13.423. [DOI] [PubMed] [Google Scholar]
- 47.Pitkaranta A, Linnavuori K, Hovi T. Virus-induced interferon production in human leukocytes: a low responder to one virus and be a high responder to another virus. J Interferon Res. 1991;11:17–23. doi: 10.1089/jir.1991.11.17. [DOI] [PubMed] [Google Scholar]
- 48.Plotch S J, Palant O. Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase 3Dpol. J Virol. 1995;69:7169–7179. doi: 10.1128/jvi.69.11.7169-7179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Racaniello V R, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- 50.Racaniello V R, Ren R. Poliovirus biology and pathogenesis. Curr Top Microbiol Immunol. 1996;206:305–325. doi: 10.1007/978-3-642-85208-4_15. [DOI] [PubMed] [Google Scholar]
- 51.Richards O C, Ehrenfeld E. Effects of poliovirus 3AB protein on 3D polymerase-catalyzed reaction. J Biol Chem. 1998;273:12832–12840. doi: 10.1074/jbc.273.21.12832. [DOI] [PubMed] [Google Scholar]
- 52.Roebuck K A. Regulation of interleukin-8 gene expression. J Interferon Cytokine Res. 1999;19:429–438. doi: 10.1089/107999099313866. [DOI] [PubMed] [Google Scholar]
- 53.Samuel C E. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 54.Schlegel A, Giddings T H, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlegel A, Kirkegaard K. Cell biology of enterovirus infection. In: Rotbart H A, editor. Human enterovirus infections. Washington, D.C.: American Society for Microbiology; 1995. pp. 135–154. [Google Scholar]
- 56.Schmid M, Wimmer E. IRES-controlled protein synthesis and genome replication of poliovirus. Arch Virol Suppl. 1994;9:279–289. doi: 10.1007/978-3-7091-9326-6_28. [DOI] [PubMed] [Google Scholar]
- 57.Sen G C, Lengyel P. The interferon system. A bird's eye view of its biochemistry. J Biol Chem. 1992;267:5017–5020. [PubMed] [Google Scholar]
- 58.Shuai K. Interferon-activated signal transduction to the nucleus. Curr Opin Cell Biol. 1994;6:253–259. doi: 10.1016/0955-0674(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 59.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 60.Suhy D A, Giddings T H, Jr, Kirkegaard K. Remodeling the ER by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thanos D, Maniatis T. Identification of the rel family members required for virus induction of the human beta interferon gene. Mol Cell Biol. 1995;15:152–164. doi: 10.1128/mcb.15.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Towner J, Semler B L. Determinants of membrane association on poliovirus protein 3AB. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- 63.Troxler M, Egger D, Pfister T, Bienz K. Intracellular localization of poliovirus RNA by in situ hybridization at the ultrastructural level using single-stranded riboprobes. Virology. 1992;191:687–697. doi: 10.1016/0042-6822(92)90244-j. [DOI] [PubMed] [Google Scholar]
- 64.Wathelet M G, Clauss I M, Content J, Huez G A. Regulation of two interferon-inducible human genes by interferon, poly(rI).poly(rC) and viruses. Eur J Biochem. 1988;174:323–329. doi: 10.1111/j.1432-1033.1988.tb14101.x. [DOI] [PubMed] [Google Scholar]
- 65.Wimmer E, Hellen C U T, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 66.Xiang W, Cuconati A, Paul A V, Cao X, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA. 1995;1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 67.Yalamanchili P, Weidman K, Dasgupta A. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology. 1997;239:176–185. doi: 10.1006/viro.1997.8862. [DOI] [PubMed] [Google Scholar]
- 68.Zinn K, DiMaio D, Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983;34:865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]