Abstract
To better understand the mechanisms responsible for the observed effects of deletions in the promoter region of the latency-associated transcript (LAT) gene in impairing herpes simplex virus (HSV) reactivation, we generated mice transgenic for a 5.5-kb HSV type 2 (HSV-2) genomic fragment spanning the major LAT, along with the LAT promoter and flanking regions, in the C57BL/6 background. The mice expressed abundant 2.2-kb major LATs in trigeminal ganglia (TG) and other tissues. The effects of the transgene on HSV-2 infection, latency, and reactivation were assessed. When infected with wild-type (WT) HSV-2 or its LAT promoter deletion (LAT−) mutant, primary lung fibroblast lines established from normal C57BL/6 and transgenic mice supported virus growth equally well. The replication of these viruses in the mouse eye and their spread to TG and brains were similar. The quantities of latent viral DNA in TG of transgenic and normal mice, as determined by real-time PCR, were comparable. UV light-induced reactivation of the LAT− mutant from transgenic mice (0 to 7%) was no more frequent than that from normal mice (0 to 14%), while WT virus was reactivated from 13 to 54% of normal mice and 22 to 54% of transgenic mice. The cumulative data indicate that, when expressed transgenically, the HSV-2 major LAT cannot influence HSV-2 infection or latency and cannot complement the defect in reactivation of the LAT− mutant. These results imply that the phenotype of reduced reactivation associated with the LAT− mutant is related to a function encoded in the LAT promoter but not to the major LAT itself.
Herpes simplex virus (HSV) type 2 (HSV-2) infects 23% of American adults, many of whom experience recurrent genital lesions and symptoms. The pathogenesis of the infection involves the replication of virus in the genital mucosa, axonal transport to neuronal nuclei within sacral dorsal root ganglia, and the establishment there of lifelong latency that is punctuated by episodic reactivation.
The nature and regulation of HSV-2 latency and the factors that induce reactivation have been the subjects of extended study. Of the more than 80 gene products expressed by the 150-kb HSV-2 genome during acute infection, only one family of transcripts is readily detected in latency. The gene encoding these latency-associated transcripts (LATs) (5, 22) is about 9 kb long, extending antisense to and overlapping the ICP0 and ICP34.5 open reading frames (ORFs) (Fig. 1). Its counterpart in HSV-1 (6, 7, 12, 18, 21) has been studied more extensively.
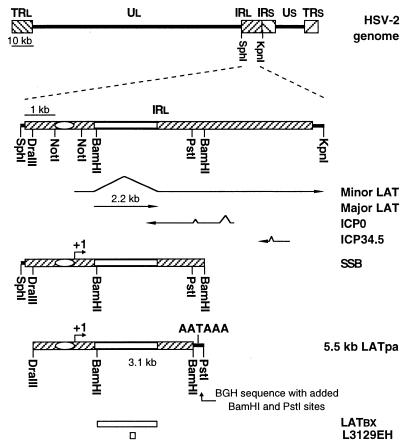
FIG. 1.
Organization of the HSV-2 genome and the relevant segments examined in this study. The top line represents the HSV-2 genome, with internal and terminal repeats (IRL, IRS, TRL, and TRS) flanking the unique long and short segments (Ul and Us, respectively). The second line highlights the IRL and the restriction enzyme recognition sites relevant to this study. The locations of LATs and the genes that they overlap are depicted below the IRL. The SphI-SalI-BamHI fragment from which the transgene construct was generated is shown as segment SSB. The oval and +1 represent the LAT promoter and the LAT transcription initiation site, respectively. The open bar represents the coding region for the major LAT. The recombinant transgene, LATpa, is a 5.5-kb DNA fragment consisting of the viral DraIII-PstI fragment and the BGH polyadenylation signal with added BamHI and PstI sites. Cloned DNA fragments LATBX and L3129EH served as templates for in vitro transcription of RNA probes for in situ hybridization.
The dominant species of HSV RNA that accumulates in latently infected neurons, the major LAT, is a stable intron spliced from the primary (minor) LAT (9, 29). Promoter activity driving HSV-2 LAT expression resides within a 624-bp segment spanning a TATA box, a cyclic AMP-responsive element, a novel GC-rich element, and a neuron-responsive domain (26). An HSV-2 mutant with a deletion of this promoter region (termed LAT− HSV-2) expresses no detectable major LAT during latency and, importantly, exhibits a reduced rate of spontaneous genital reactivation in guinea pigs (13, 27).
The reason for the impaired reactivation of LAT− HSV-2 and similar HSV-1 mutants is not known. There are no convincing data to indicate that the LATs encode a protein that regulates reactivation. Because the 3′ end of the major LAT overlaps the 3′ end of ICP0 and the larger, primary LAT extends through both ICP0 and ICP34.5, it has been postulated that LATs regulate HSV reactivation through an antisense mechanism (6, 21). Whatever the mechanism is, extensive studies of HSV-1 deletion mutants have localized such effects on reactivation to the region encompassing the 5′ portion of the major LAT and its upstream sequences, including its promoter region (1, 4, 10, 13, 24, 25).
Studies involving HSV-1 mutants in which polyadenylation signals were added at various positions within the LAT coding region implied that the sequences responsible for the effect of LAT deletions on reactivation reside within the 5′ end of the major LAT or upstream of that position (1). Mutant viruses with deletions between the LAT promoter and the major LAT were reactivated normally, implying a function critical for viral reactivation within the LAT, its promoter, or their upstream sequences (28).
During the past decade, members of our laboratory have been investigating the HSV-2 LAT gene and HSV-2 latency and reactivation. To further clarify the function of the HSV-2 major LAT and its flanking sequences, transgenic mice were generated in which the HSV-2 major LAT was expressed under the control of its native promoter. LAT accumulated in the nuclei of neurons of trigeminal ganglia (TG) in these mice. We studied the effects of this LAT transgene on HSV-2 replication in primary cultured transgenic mouse cells and during acute ocular infection of mice, on the establishment and maintenance of virus latency in TG, and on rates of UV-induced virus reactivation in vivo. Specific experiments addressed the capacity of the LAT transgene to complement the deficient reactivation from latency of an LAT promoter deletion mutant, LAT− HSV-2, that expresses no detectable major LAT and is reactivated poorly.
MATERIALS AND METHODS
Viruses.
The LAT− mutant was derived from HSV-2 strain 333 by targeted deletion of the 624-bp NotI-NotI fragment (Fig. 1) from the virus genome as described previously (13, 27). The wild-type (WT) virus used here is the clonal rescued virus, ΔR, of the LAT− mutant obtained through site-specific recombination that restored the NotI-NotI fragment. These viruses were propagated and titrated in Vero cells by plaque assays (16).
HSV-2 one-step growth curve for primary fibroblasts.
Lungs were dissected from 7-week-old transgenic mice and their transgene-negative littermates. The resultant primary mouse lung fibroblast cells were expanded in FGM2 medium supplemented with the FGM2 Bullet kit (BioWhittaker, Walkersville, Md.) and used to seed six-well plates at a density of 106 cells/well. Two days later, the cultures were infected with the LAT− mutant or WT virus at a multiplicity of infection of 0.5 in 0.5 ml of the growth medium, and one-step growth curves from 0 to 24 h postinfection (p.i.) were determined as described previously (13).
Plasmids.
Plasmid pSSB contains the SphI-SalI-BamHI fragment (Fig. 1) from the IRL region of HSV-2 strain 333 inserted into the SphI-BamHI region of vector pCAT.Basic (Promega, Madison, Wis.). To create the desired transgene, a DNA segment containing the polyadenylation signal of the bovine growth hormone (BGH) gene was inserted into the PstI site downstream of the major LAT, and the resulting plasmid was named pSSBLATpa. To generate plasmid pLATBX, a 1,793-bp BamHI-XhoI fragment of the major LAT was inserted into vector pGEM4Z (Promega) between its BamHI and KpnI sites. Plasmid pG.L3129EH contains a 187-bp PCR product amplified from the HSV-2 major LAT (Fig. 1) inserted into pGEM3zf(+) (Promega) between its EcoRI and HindIII sites. To generate plasmid p2gDs, a 1.22-kb PCR product amplified from the HSV-2 strain S glycoprotein D (gD) gene was inserted into the KpnI and EcoRI sites of vector pcDNA3 (Invitrogen, Carlsbad, Calif.). All constructs were verified by DNA sequencing.
Creation of LATpa transgenic mice.
The 5,511-bp transgene DNA, LATpa DNA (Fig. 1), included a 5,277-bp genomic DNA fragment of HSV-2 strain 333 consisting of the sequences 1,167 bp upstream and 4,114 bp downstream of the LAT transcription initiation site (26). This viral DNA encompasses the LAT promoter and some of its upstream region, the entire 2.2-kb major LAT, and 1,170 bp further downstream of the major LAT splicing adapter (nucleotides 118,332 to 123,608 of the comparable strain HG52 sequence). The poly(A) signal of BGH (Fig. 1) was added to the 3′ end of the viral fragment to complete the transcription unit. The transgene fragment was cleaved from plasmid pSSBLATpa with restriction enzymes DraIII and PstI, resolved on an agarose gel, and recovered using a QIAquick gel extraction kit (QIAGEN, Valencia, Calif.). This fragment was used to construct transgenic mice in a C57BL/6 background (NCI-Frederick, Frederick, Md.) at the National Institute of Allergy and Infectious Diseases (NIAID) Transgenic Mice Facility in accordance with protocols approved by the Frederick Cancer Research and Development Center and NIAID Animal Care and Use Committees. Briefly, 3- to 4-week-old females were induced to superovulation with 5 IU of pregnant mares' serum gonadotropin followed 48 h later by 5 IU of human chorionic gonadotropin and then mated with C57BL/6 males. Fertilized oocytes were harvested on the next day, and pronuclei were injected with LATpa DNA at 5 ng/μl. Embryos that divided into two cells overnight were transferred to the oviducts of pseudopregnant B6D2F1 recipients. Tail DNA was obtained from pups at weaning and prepared using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.).
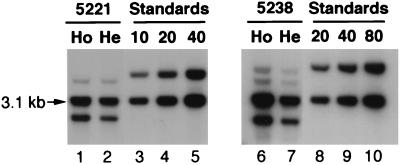
To identify transgenic mice, 4 μg of mouse tail DNA was digested with BamHI and subjected to Southern analysis with 32P-labeled LATpa DNA as the probe. The transgene copy number per cell was estimated by comparing the density of the primary 3.1-kb transgene band from mouse tail DNA to that of reference standards comprised of 4 μg of normal mouse tail DNA alone or spiked with 4 to 80 copies of plasmid pSSBLATpa/cell (Fig. 2). The expression of the transgene in the heterozygotes was determined by Northern blot assays.
FIG. 2.
Southern analysis of mouse tail DNA. Four micrograms of mouse tail DNA was digested with BamHI and subjected to Southern hybridization with the [32P]dCTP-labeled LATpa fragment. Also analyzed were copy number standards consisting of 4 μg of normal mouse tail DNA and pSSBLATpa DNA equivalent to 10, 20, 40, and 80 copies per cell, respectively. The copy numbers of the transgene in transgenic mouse cells were estimated by comparing the density of the 3.1-kb band to that of the standards. Shown are data obtained with one heterozygous mouse and one homozygous mouse of transgenic lines 5221 and 5238. Lanes 1 and 2, transgenic line 5221 homozygote (Ho) and heterozygote (He), estimated to have about 30 and 15 copies per cell, respectively; lanes 3, 4, and 5, standards equivalent to 10, 20, and 40 copies per cell, respectively; lanes 6 and 7, transgenic line 5238 homozygote (Ho) and heterozygote (He), with estimated copy numbers of 80 and 40 per cell, respectively; lanes 8, 9, and 10, standards equivalent to 20, 40, and 80 copies per cell, respectively.
In situ hybridization.
The distribution of LAT in transgenic mouse tissues was revealed by in situ hybridization. Mice were perfused with 4% formaldehyde–phosphate-buffered saline, and tissues were dissected and fixed in 4% formaldehyde–phosphate-buffered saline overnight and embedded in paraffin. Tissue sections were hybridized as described previously (6) with [35S]UTP-labeled LAT-specific single-stranded RNA probe LATBX or L3129, transcribed in vitro from plasmid pLATBX or pG.L3129EH, respectively (Fig. 1).
RNA purification.
To quantify the LAT expressed in mice, individual tissues were dissected from transgenic or normal mice at the age of 7 to 8 weeks. Total RNA was purified with an RNeasy mini kit (QIAGEN). To determine the level of LAT expression in latently infected mice, each TG was dissected at 65 or 69 days p.i. RNA was purified from the TG by the same method as that used above but with an additional DNase digestion step recommended for samples to be analyzed by reverse transcription (RT)-PCR. To confirm the expression of LAT in infected primary cultured fibroblasts, cells were harvested at various times from 0 to 24 h postinoculation, and RNA was isolated with the RNeasy mini kit.
DNA purification.
TG were dissected from uninfected or latently infected mice at day 65 or 69 p.i. and incubated in 500 μl of the lysis buffer from the Puregene DNA isolation kit with 0.6 mg of proteinase K/ml at 56°C overnight. DNA was then isolated according to the manufacturer's instructions.
Northern analysis.
The expression of the 2.2-kb LAT in mouse tissues or in primary fibroblasts was discerned by Northern blot assays using [32P]dCTP-labeled LATpa (Fig. 1) as a probe. Briefly, 0.32 μg of RNA from infected fibroblasts or 1 μg of tissue RNA was heated with RNA denaturing buffer (5prime-3prime, Inc., Boulder, Colo.) at 65°C for 15 min and resolved on an 0.8% agarose–1.2 M formaldehyde–1× 3-morpholinepropanesulfonic acid (MOPS) gel. RNAs were transferred to Nytran membranes (Schleicher & Schuell, Keene, N.H.) and processed by following the manufacturer's instructions.
Acute virus infection in mice.
Eight-week-old transgenic mice (line 5238) or normal control mice were divided into two groups each, of 15 mice/group, with a balanced distribution by gender. All mice were studied at the Animal Care and Use Committee-approved NIAID Animal Care Facility under an approved protocol. Mice were infected ocularly with 104 PFU of WT or LAT− mutant virus as described previously (16). On days 3, 5, and 7 p.i., 5 mice from each group were euthanatized. Tissues were dissected and homogenized separately in 1 ml (eyes and TG) or 5 ml (brain) of MEM-199 medium (Quality Biological, Gaithersburg, Md.) using a Tissumite (Tekmar-Dohrmann, Cincinnati, Ohio). After centrifugation at 1,800 × g and 4°C for 5 min, the supernatants were titrated by plaque assays.
UV-induced in vivo reactivation of latent virus.
Transgenic and normal mice were inoculated by corneal scarification with 104 PFU of WT or LAT− mutant virus and injected with 0.5 ml of 6% human immunoglobulin intraperitoneally at day 0 or 1 to limit mortality. To assess the effect of the transgene on virus reactivation from latently infected TG, mice were anesthetized 40 or 50 days p.i., and both eyes were exposed to UV irradiation at a wavelength of 302 nm (720 mJ/eye) (14). Two days later, the mice were euthanatized, and their TG were dissected and homogenized in 0.5 ml of culture medium. Following centrifugation, the supernatants were used to inoculate Vero cell monolayers. The cultures were observed for cytopathic effects daily for 15 days.
Quantitative DNA PCR.
The viral genome copy numbers in latently infected TG were determined by quantitative DNA PCR using a Perkin-Elmer TaqMan PCR system. The primers and probe were based on the HSV-2 gD sequence: forward primer gDTQ3, 5′ TCAGCGAGGATAACCTGGGA 3′; reverse primer gDTQ4, 5′ GGGAGAGCGTACTTGCAGGA 3′; and probe gDP4, 5′ FAM-CCAGTCGTTTATCTTCACTAGCCGCAGGTA-TAMARA 3′. Each 25-μl PCR mixture contained 100 ng of ganglionic DNA, 1× Universal TaqMan PCR Master Mix, 250 nM each the forward and reverse primers, and 200 nM probe. In each PCR experiment, a standard panel was run consisting of serial fourfold dilutions of plasmid p2gDs from 80 to 0.020 fg/reaction. All samples were run in triplicate. The reactions were carried out using an ABI PRISM 7700 sequence detector with optimized cycle conditions: 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 20 s, and 60°C for 1 min. By comparison of the threshold cycle (Ct) of each sample to the Cts of the standards, the copy number for each experimental reaction was estimated. TG DNA samples from three to seven mice per group were assessed.
Quantitative RT-PCR.
LAT RNA in latently infected ganglia was quantitated with a Perkin-Elmer One-Step RT-PCR system. The sequence of forward primer TQ201 was 5′ GTCAACACGGACACACTCTTTT 3′, and the sequence of reverse primer TQ202 was 5′ CGAGGCCTGTTGGTCTTTATC 3′. The sequence of probe TQP201 was 5′ FAM-CACCCACCAAGACAGGGAGCCA-TAMARA 3′. All sequences were located within the HSV-2 major LAT. Each 25-μl reaction mixture contained 500 pg of total ganglionic RNA, 1,600 nM each forward and reverse primers, 200 nM probe, One-Step RT-PCR Master Mix, and 1× RT with RNasin (omitted in RT-negative reactions). RNA samples from four or five mice per group were assessed. For each sample, both RT-positive and RT-negative reactions were tested in triplicate. Tenfold serial dilutions of RNA from transgenic mouse liver were included as reference standards. The reactions were carried out using the ABI PRISM 7700 sequence detector with optimized cycle conditions: 48°C for 30 min, 95°C for 10 min, 40 cycles at 95°C for 20 s, and 60°C for 1 min. By comparison of the Ct of each sample to the Cts of the standards, the relative amounts of LAT in the samples were estimated.
Statistical analyses.
Student's t test was used to compare the means between any two groups; Fisher's exact test was used for analyzing the percentages of UV-induced reactivation in vivo. All computations were carried out using JMP version 3.0 software (SAS Institute, Cary, N.C.).
RESULTS
Establishment of mouse lines transgenic for the HSV-2 major LAT.
The LATpa transgene spans the upstream LAT promoter regulatory region, the major LAT coding sequence, about 1 kb downstream of the 3′ end of the major LAT, and the BGH polyadenylation signal. All viable progeny arising from C57BL/6 mouse oocytes microinjected with the LATpa transgene (Fig. 1) were screened by Southern hybridization. Six independent founders were identified with transgene copy numbers ranging from 4 to 50 per cell (Fig. 2). Four transgenic mouse lines were established by breeding the founders with normal mice. Their heterozygous offspring were further bred to obtain additional heterozygotes as well as homozygotes. Lines 5238 and 5221, with 40 and 15 transgenes per cell, respectively, were bred to homozygosity (Fig. 2). Heterozygotes of line 5238 were used predominantly in the experiments described in this report. There was no excess fetal loss or abnormal development of any transgenic mice at both the gross and the histologic levels.
Expression of the transgene in mice.
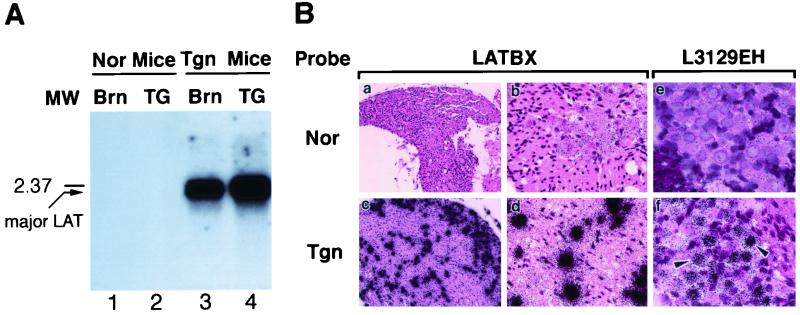
Large amounts of the 2.2-kb HSV-2 major LAT were detected in transgenic mouse TG, brains (Fig. 3A), and other tissues (data not shown). The cellular and subcellular localizations of the major LAT in the TG were determined by in situ hybridization. Intense LAT-specific signals were identified over neurons (Fig. 3B, images a to d). The signal was so strong that the subcellular localization could not be well distinguished. By using a shorter probe (L3129EH), the LAT signal was reduced and could be localized over neuronal nuclei (Fig. 3B, image f).
FIG. 3.
Expression of the transgene in mouse tissues. (A) Northern analysis of RNA from transgenic line 5238 and normal mice. One microgram of total RNA from brains (Brn) or TG of normal (Nor) mice (lanes 1 and 2) or transgenic (Tgn) mice (lanes 3 and 4) was denatured and resolved on a 1.2 M formaldehyde denaturing gel. The Northern transfer membrane was hybridized with a [32P]dCTP-labeled LATpa probe. The arrow indicates the intense signal of the 2.2-kb major LAT in the transgenic mouse tissues, migrating just ahead of a 2.37-kb size marker. (B) Cellular localization of the LAT in transgenic mouse TG. Ganglionic tissue sections prepared from 7-week-old transgenic (Tgn) or normal (Nor) mice were hybridized with [35S]UTP-labeled single-stranded RNA probe LATBX (images a to d). The LAT signal was intense in the transgenic mouse tissues (magnifications, ×100 in image c and ×400 in image d) but was not detected in the normal mouse tissues (images a and b). Cells with intense LAT signals appear to be neurons. Sections in images e and f were hybridized with a shorter probe, L3129EH, and exposed more briefly so that the signal was reduced and the intracellular localization of LAT could be better observed. The major LAT signal was evident over the nuclei of neurons (arrowheads) of transgenic mice (image f; magnification, ×400). No signal was detected from normal mouse tissues (image e).
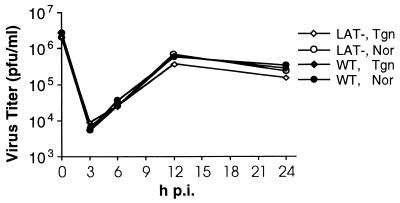
Kinetics of HSV-2 replication in primary fibroblasts from transgenic mice are normal.
Primary cultures of lung fibroblasts from transgenic or normal mice were infected with LAT− or WT HSV-2, harvested at multiple time points, and titrated on Vero cell monolayers. Cells from normal and transgenic mice supported the growth of WT and LAT− viruses equally well (Fig. 4). By Northern hybridization, we confirmed that the transgene was expressed strongly in fibroblasts from transgenic mice during the infection (data not shown).
FIG. 4.
HSV-2 replication in mouse lung fibroblasts from transgenic and normal mice. Primary lung fibroblasts from 7-week-old transgenic (Tgn) or normal (Nor) mice were infected with LAT− mutant or WT virus at a multiplicity of infection of 0.5 to 1. At 0, 3, 6, 12, and 24 h p.i., cells were harvested and cell-free viruses were prepared and then titrated on Vero cell monolayers.
HSV-2 replicates and spreads normally in LAT transgenic mice.
Having generated transgenic mice, we determined whether the accumulation of LAT would affect the replication and spread of exogenous HSV-2 following ocular infection. Transgenic or normal mice were infected with WT or LAT− HSV-2. Anticipating that these viruses may differ in their capacities to become established or reactivated from latency and may be differentially susceptible to the effects of the LAT transgene, we first assessed their behaviors during acute infection.
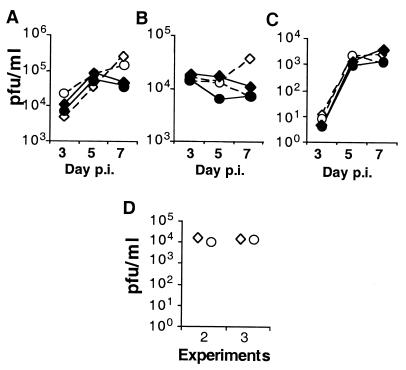
Viral titers in eyes, TG, and brains at days 3, 5, and 7 p.i. were determined. The titers of WT and LAT− viruses in infected eyes from each group was similar on days 3 and 5 p.i., but the LAT− mutant achieved slightly higher titers in both normal and transgenic mice at day 7 (Fig. 5A). The titers of the LAT− mutant in ganglia of infected transgenic mice were also higher than those in normal mice at day 7 (P = 0.08) (Fig. 5B). However, these small differences were not reproducible in two further experiments (Fig. 5D). The virus titers in brains from normal and transgenic mice were similar at each time point p.i. and followed the expected time course of spread from the periphery to the brain (Fig. 5C). We conclude that the LAT− and WT viruses replicate similarly in both normal and LAT transgenic mice. The transgene has no effect on the acute-phase replication and spread of HSV-2 in mice.
FIG. 5.
Effect of the LAT transgene on HSV-2 replication during acute ocular infection. Mice were infected corneally with 104 PFU of the LAT− mutant or WT HSV-2 strain 333. On days 3, 5, and 7 p.i., eyes (A), TG (B), and brains (C) of five mice from each group were dissected and homogenized. The virus titers were determined by plaque assays, and the group means are depicted. Symbols: ◊, LAT− mutant in transgenic mice; ○, LAT− mutant in normal mice; ⧫, WT virus in transgenic mice; ●, WT virus in normal mice. (D) Results from second and third experiments. Virus titers for TG harvested from transgenic mice (◊) or normal mice (○) 7 days after infection with the LAT− mutant.
The LAT transgene has no effect on the latent HSV-2 load.
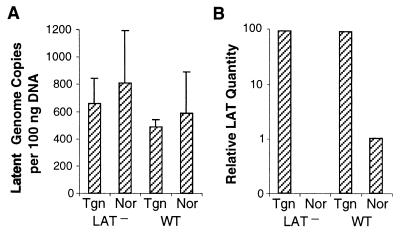
Prior experiments with the guinea pig genital herpes model indicated that the quantity of latent HSV-2 genomes, the latent viral load, correlates with rates of spontaneous reactivation (15). To determine whether the accumulation of LAT in transgenic mice influences the latent load of exogenous virus administered through the ocular route, ganglia were harvested 60 or 65 days p.i. and analyzed by quantitative real-time PCR. Two independent experiments were conducted using transgenic mouse line 5238 (Fig. 6A). The viral genome copy numbers in ganglia latently infected with the LAT− mutant were similar for transgenic and normal mice. The latent loads of WT virus were also comparable in transgenic and normal mice. The cumulative data reveal no gross defect in the establishment or maintenance of latency for the LAT− mutant and, moreover, that the LAT transgene does not influence latent HSV-2 DNA loads.
FIG. 6.
HSV-2 genome loads and quantity of LAT in latently infected TG. Transgenic (Tgn) mice were infected with 104 PFU of LAT− mutant or WT HSV-2. As controls, normal (Nor) mice were infected similarly with LAT− mutant or WT HSV-2. At day 69 p.i., TG were dissected and DNA or RNA was isolated. (A) Viral copy numbers in total DNA were determined by real-time PCR with primers and a probe specific for the HSV-2 gD gene. The means for three to five mice from each group were plotted. The P values (Student's t test) for transgenic versus normal mice infected with LAT− mutant virus or WT virus were 0.75 and 0.79, respectively. (B) The abundance of the LAT was estimated by real-time RT-PCR with primers and a probe specific for the HSV-2 LAT. The mean quantity of LAT in ganglia of four or five mice from each group is shown here in logarithmic scale. The amount of LAT in transgenic mice infected with LAT− or WT virus was 89- or 88-fold that in normal mice infected with WT virus.
The major LAT is expressed in latently infected transgenic mice.
To rule out the possibility that the lack of effect of the transgene on latent virus load was due to a lack of transgene expression during latent virus infection, total RNA was extracted from latently infected ganglia (69 days p.i.). By quantitative real-time RT-PCR, LAT RNA was undetectable in ganglia of normal mice infected with LAT− virus, as expected. Small quantities of LAT were detected in normal mice infected with WT HSV-2. In contrast, LAT levels in transgenic mice infected with LAT− or WT virus were 89- or 88-fold, respectively, that in normal mice infected with WT virus (Fig. 6B).
UV-induced reactivation of the LAT− mutant virus.
It has been reported that the LAT− virus is reactivated suboptimally in guinea pigs (13). Were it to behave similarly in mice, we could use the mouse ocular model to determine whether the accumulation of LAT in transgenic mouse neurons complements this reactivation defect. Three independent experiments with two transgenic mouse lines were conducted to assess the capability of the transgene to affect the reactivation of LAT− and WT HSV-2 from latently infected ganglia.
Transgenic or normal mice were infected via the ocular route with LAT− or WT HSV-2 and housed for 40 to 50 days. The eyes of latently infected mice were exposed to UV irradiation. In experiment 1, no reactivation of the LAT− mutant was detected in either transgenic or normal mice. In contrast, the WT virus was reactivated from 54% of ganglia in both transgenic and normal mice. Subsequent confirmatory experiments, experiment 2 with the same transgenic line and experiment 3 with a second transgenic line, showed similar, although variable, results (Table 1). The cumulative data verify that the LAT− virus is reactivated very poorly in normal mice, as in guinea pigs. In addition, the LAT transgene does not have a notable effect on the reactivation of WT virus. More importantly, the LAT transgene does not complement UV-induced reactivation of the LAT− mutant from mouse TG.
TABLE 1.
UV-induced reactivation of HSV-2 from transgenic mice
| Expt | No.a (%) of cultures in which reactivation was detected with the following virus and mice:
|
|||
|---|---|---|---|---|
| LAT− mutant
|
WT
|
|||
| Transgenic | Normal | Transgenic | Normal | |
| 1 | 0/16 (0)b | 0/8 (0)b | 7/13 (54)c | 7/13 (54)c |
| 2 | 0/18 (0)d | 2/14 (14)d | ND | 1/8 (13) |
| 3 | 1/14 (7)e | 0/16 (0)e | 4/18 (22) | ND |
| Total | 1/48 (2.1) | 2/38 (5.3) | 11/31 (35.5) | 8/21 (38.1) |
Number showing reactivation/number tested. ND, not determined.
P = 1.00.
P = 1.00.
P = 0.18.
P = 0.47.
DISCUSSION
That HSV-1, HSV-2, and other related neurotropic alphaherpesviruses contain genes that are abundantly expressed during latency argues that the products that they encode evolved and were retained for some biological purpose. In the over 13 years since the HSV LAT was first described, no studies have yielded compelling answers as to its role. Many studies have shown that mutants in which the LAT promoter or the LAT 5′ coding regions were altered or deleted are impaired in establishment or reactivation from latency (1, 4, 10, 13, 24, 25). Prior to this study, it remained uncertain, however, whether the factor responsible for these various effects is the major LAT itself, upstream or downstream transcripts colinear with it, or some other factors embodied in the LAT promoter region. Moreover, the studies have not indicated the mechanism by which the LAT may act, whether through antisense effects on downstream viral regulatory genes, through a still-undetected protein product, or through the mere presence of the sequence itself in a critical genome region. The creation of mice transgenic for the HSV-2 major LAT afforded a novel opportunity to address these issues.
The transgene (Fig. 1) was engineered to encode the major LAT under the control of its native promoter. All upstream sequence elements reported to confer high-level expression in neurons (26) were included, as were the GC-rich domains believed to harbor secondary promoters (3) and enhancers (28) for the major LAT. This design was chosen so that transcriptional regulation of the transgene would resemble that which occurs within the viral genome. The transgene extended 1,170 bp downstream of the 3′ end of the major LAT and terminated in an added poly(A) site to accommodate the splicing sites for the major LAT, so that the entire major LAT can be spliced from within the body of the primary LAT transcript, as it is in the context of virus infection and latency. The primary transcript from the transgene is expected to overlap 1,673 bases of the ICP0 transcript, so that any elements within it that may participate in antisense counterregulation of ICP0 expression will be expressed. Analyses of cells and tissues derived from multiple transgenic founders and their progeny confirmed the expression of the desired 2.2-kb major LAT, with particularly high levels in neurons (Fig. 3). Although sequencing of the LAT generated in transgenic mice was not done to prove absolute identity with authentic viral LAT, we can conclude that the length and cellular localization of the transgenic LAT are the same as those of the major LAT made from virus.
The analysis of transgene expression in different murine tissues verified the previous in vitro finding (26) that the native LAT promoter is responsive to neuronal factors. An interesting observation, however, is that the promoter is not strictly neuron specific. Extensive in situ and Northern hybridization experiments revealed high levels of LAT accumulating in the liver and other tissues (data not shown).
The HSV-2 LAT region was studied here with an ocular model system. Although it could be argued that a genital infection model would be more relevant to HSV-2 biology, with its greater ease of manipulation, as detailed below, the ocular model proved sufficient for our purposes here. First, we showed that following ocular inoculation, HSV-2 strain 333 is well able to infect the mouse eye, spread to and establish latency in TG, and be reactivated efficiently in vivo after UV irradiation. Second, and more importantly, the failure of an LAT− virus to be reactivated effectively was recapitulated in the mouse ocular model (Table 1), allowing us to clearly discern whether the transgene possesses any complementary functions.
The present data strongly indicate that the major LAT region, when expressed in a transgenic mouse model, does not alter the course or severity of acute infection, nor does it modulate establishment, maintenance, or reactivation from latency.
Accumulated LAT did not inhibit or enhance HSV replication, as was first revealed by a single-step growth study conducted with primary cells derived from the transgenic mice (Fig. 4). This finding contrasts with that of Mador et al., using stably transformed neuronal cells (17), and that of Thomas et al., who showed that expression of a putative ORF in the HSV-1 major LAT increased HSV-1 replication (23). These differences could be related to the use of different promoters and cell types (17), differences in the splicing of the recombinant major LAT (23), or differences, should any exist, between HSV-1 and HSV-2.
The studies that we conducted with the transgenic and control mice showed no consistent attenuation of the course, severity, or spread of virus from eyes to ganglia to brain (Fig. 5). This result is similar to experience with this LAT− mutant in the guinea pig vaginal model (13, 27).
The effects of the transgene on virus latency and reactivation were assessed in several ways. First, we determined the latent viral load by real-time PCR. The numbers of latent genome copies were comparable in transgenic and normal mice (Fig. 6A), although we cannot comment here on the relative proportions of neurons infected in each setting. In addition, we showed that the latent load established in mice infected with the LAT− HSV-2 mutant was similar to that in mice infected with WT HSV-2, in agreement with some but not all recent reports with similar HSV-1 mutants (19, 20, 24, 28).
The present data are in accord with prior reports that LAT− HSV-2 establishes and maintains latency normally in the guinea pig genital model (13, 27). Moreover, we found that the rates of virus reactivation from TG of normal mice latently infected with WT or LAT− mutant virus were identical ex vivo following explantation (data not shown). These studies verified that both LAT− mutant and WT HSV-2 are competent to establish and maintain latency in mouse TG. However, the LAT− mutant virus was not able to reactivate as efficiently as the WT virus following exposure of mouse eyes to UV light (Table 1). Thus, impaired reactivation of this LAT− HSV-2 mutant has been verified now with both mouse and guinea pig models (13, 27). More importantly, no complementation of this deficiency could be detected in transgenic mice that harbored prominent amounts of the major LAT in their neurons in TG.
The cumulative data indicate that the major LAT, when expressed from a transgene, cannot function in trans. We cannot envision a mechanism by which a transcript would fail to function in trans in an antisense capacity, regardless of whether it is derived from the viral or the host genome. Thus, the data strongly argue against an antisense mechanism as underlying an effect of the major LAT. Any protein encoded from the transgene would, of necessity, also function in trans. Thus, any protein encoded from the major LAT or from sequences proximate to it cannot modulate viral reactivation. The present data do not exclude some cis-acting function of the LAT promoter and major LAT region. Thus, the complicated GC-rich structure of this region may be crucial for regulating the conformation of the latent viral genome episome and the accessibility of the viral genome to transcription factors during reactivation. It is also possible that a combination of active transcription in the LAT region itself and another unknown function is required for reactivation. These latter hypotheses are consistent with the similarity of the GC content and in both strong promoter activity during latency HSV-1 and HSV-2 LAT regions, despite the regional low degree of sequence homology. Finally, the data do not exclude the possibility of trans-acting transcripts or peptide products encoded by further upstream sequences that extend into or overlap the LAT promoter region, as suggested in a recent report by Inman et al. (11), or sequences further downstream within the domain of the primary (minor) LAT, such as sequences antisense to ICP34.5 or ICP4 (2).
Thirteen years of study of the major LAT have failed to reveal a mechanism by which these transcripts may influence HSV reactivation. Here, we found that when expressed from a transgene, the major LAT is unable to complement the defective reactivation of an HSV-2 mutant with a deletion of the LAT promoter. This result agrees with the observation made for HSV-1 LAT by Fareed and Spivack (8), in which latency and reactivation were not affected by serial interruptions of all predicted ORFs in the major LAT. We conclude that the LAT region influences reactivation, but not through actions of the major LAT itself. Now, the focus of investigations should be broadened to more distant sequences flanking the major LAT, sequences whose properties and activities have not been studied in nearly as much detail.
ACKNOWLEDGMENTS
We thank Judith Hewitt at the Transgenic/Knockout Mice Facility of NIAID for helping with microinjection, obtaining tail DNA, and maintaining the breeding of the transgenic colonies. We appreciate the generous help with statistical analyses provided by Claire Hallahan and the continuing advice and encouragement given by Jeffrey Cohen.
REFERENCES
- 1.Bloom D C, Hill J M, Devi-Rao G, Wagner E K, Feldman L T, Stevens J G. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol. 1996;70:2449–2459. doi: 10.1128/jvi.70.4.2449-2459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S H, Kramer M F, Schaffer P A, Coen D M. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1997;71:5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Schmidt M C, Goins W F, Glorioso J C. Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections. J Virol. 1995;69:7899–7908. doi: 10.1128/jvi.69.12.7899-7908.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook S D, Paveloff M J, Doucet J J, Cottingham A J, Sedarati F, Hill J M. Ocular herpes simplex virus reactivation in mice latently infected with latency-associated transcript mutants. Investig Ophthalmol Vis Sci. 1991;32:1558–1561. [PubMed] [Google Scholar]
- 5.Croen K D, Ostrove J M, Dragovic L, Straus S E. Characterization of herpes simplex virus type 2 latency-associated transcription in human sacral ganglia and in cell culture. J Infect Dis. 1991;163:23–28. doi: 10.1093/infdis/163.1.23. [DOI] [PubMed] [Google Scholar]
- 6.Croen K D, Ostrove J M, Dragovic L J, Smialek J E, Straus S E. Latent herpes simplex virus in human trigeminal ganglia. Detection of an immediate early gene “anti-sense”transcript by in situ hybridization. N Engl J Med. 1987;317:1427–1432. doi: 10.1056/NEJM198712033172302. [DOI] [PubMed] [Google Scholar]
- 7.Deatly A M, Spivack J G, Lavi E, Fraser N W. RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia of latently infected mice. Proc Natl Acad Sci USA. 1987;84:3204–3208. doi: 10.1073/pnas.84.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fareed M U, Spivack J G. Two open reading frames (ORF1 and ORF2) within the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1 are not essential for reactivation from latency. J Virol. 1994;68:8071–8081. doi: 10.1128/jvi.68.12.8071-8081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell M J, Dobson A T, Feldman L T. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci USA. 1991;88:790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill J M, Maggioncalda J B, Garza H H, Jr, Su Y H, Fraser N W, Block T M. In vivo epinephrine reactivation of ocular herpes simplex virus type 1 in the rabbit is correlated to a 370-base-pair region located between the promoter and the 5′ end of the 2.0-kilobase latency-associated transcript. J Virol. 1996;70:7270–7274. doi: 10.1128/jvi.70.10.7270-7274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inman M, Perng G C, Henderson G, Ghiasi H, Nesburn A B, Wechsler S L, Jones C. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J Virol. 2001;75:3636–3646. doi: 10.1128/JVI.75.8.3636-3646.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krause P R, Croen K D, Straus S E, Ostrove J M. Detection and preliminary characterization of herpes simplex virus type 1 transcripts in latently infected human trigeminal ganglia. J Virol. 1988;62:4819–4823. doi: 10.1128/jvi.62.12.4819-4823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krause P R, Stanberry L R, Bourne N, Connelly B, Kurawadwala J F, Patel A, Straus S E. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J Exp Med. 1995;181:297–306. doi: 10.1084/jem.181.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBlanc R A, Pesnicak L, Godleski M, Straus S E. Treatment of HSV-1 infection with immunoglobulin or acyclovir: comparison of their effects on viral spread, latency, and reactivation. Virology. 1999;262:230–236. doi: 10.1006/viro.1999.9891. [DOI] [PubMed] [Google Scholar]
- 15.Lekstrom-Himes J A, Pesnicak L, Straus S E. The quantity of latent viral DNA correlates with the relative rates at which herpes simplex virus types 1 and 2 cause recurrent genital herpes outbreaks. J Virol. 1998;72:2760–2764. doi: 10.1128/jvi.72.4.2760-2764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lekstrom-Himes J A, Wang K, Pesnicak L, Krause P R, Straus S E. The comparative biology of latent herpes simplex virus type 1 and type 2 infections: latency-associated transcript promoter activity and expression in vitro and in infected mice. J Neurovirol. 1998;4:27–37. doi: 10.3109/13550289809113479. [DOI] [PubMed] [Google Scholar]
- 17.Mador N, Goldenberg D, Cohen O, Panet A, Steiner I. Herpes simplex virus type 1 latency-associated transcripts suppress viral replication and reduce immediate-early gene mRNA levels in a neuronal cell line. J Virol. 1998;72:5067–5075. doi: 10.1128/jvi.72.6.5067-5075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rock D L, Nesburn A B, Ghiasi H, Ong J, Lewis T L, Lokensgard J R, Wechsler S L. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987;61:3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawtell N M, Thompson R L. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol. 1992;66:2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sedarati F, Izumi K M, Wagner E K, Stevens J G. Herpes simlex virus type 1 latency-associated transcription plays no role in establishment or maintenance of a latent infection in murine secsory neurons. J Virol. 1989;63:4455–4458. doi: 10.1128/jvi.63.10.4455-4458.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 22.Tenser R B, Edris W A, Hay K A, de Galan B E. Expression of herpes simplex virus type 2 latency-associated transcript in neurons and nonneurons. J Virol. 1991;65:2745–2750. doi: 10.1128/jvi.65.5.2745-2750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas S K, Gough G, Latchman D S, Coffin R S. Herpes simplex virus latency-associated transcript encodes a protein which greatly enhances virus growth, can compensate for deficiencies in immediate-early gene expression, and is likely to function during reactivation from virus latency. J Virol. 1999;73:6618–6625. doi: 10.1128/jvi.73.8.6618-6625.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson R L, Sawtell N M. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J Virol. 1997;71:5432–5440. doi: 10.1128/jvi.71.7.5432-5440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trousdale M D, Steiner I, Spivack J G, Deshmane S L, Brown S M, MacLean A R, Subak-Sharpe J H, Fraser N W. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J Virol. 1991;65:6989–6993. doi: 10.1128/jvi.65.12.6989-6993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Krause P R, Straus S E. Analysis of the promoter and cis-acting elements regulating expression of herpes simplex virus type 2 latency-associated transcripts. J Virol. 1995;69:2873–2880. doi: 10.1128/jvi.69.5.2873-2880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Pesnicak L, Straus S E. Mutations in the 5′ end of the herpes simplex virus type 2 latency-associated transcript (LAT) promoter affect LAT expression in vivo but not the rate of spontaneous reactivation of genital herpes. J Virol. 1997;71:7903–7910. doi: 10.1128/jvi.71.10.7903-7910.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshikawa T, Stanberry L R, Bourne N, Krause P R. Downstream regulatory elements increase acute and latent herpes simplex virus type 2 latency-associated transcript expression but do not influence recurrence phenotype or establishment of latency. J Virol. 1996;70:1535–1541. doi: 10.1128/jvi.70.3.1535-1541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zabolotny J M, Krummenacher C, Fraser N W. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J Virol. 1997;71:4199–4208. doi: 10.1128/jvi.71.6.4199-4208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]