Abstract
Mycobacterium tuberculosis (MTB) is a pathogenic bacterium, belonging to the family Mycobacteriaceae, that causes tuberculosis (TB). Toxoplasma gondii macrophage migration inhibitory factor (TgMIF), a protein homolog of macrophage migration inhibitory factor, has been explored for its potential to modulate immune responses during MTB infections. We observed that TgMIF that interacts with CD74, antizyme inhibitor 1 (AZIN1), and signal transducer and activator of transcription 1 (STAT1) modulates endocytosis, restoration of mitochondrial function, and macrophage polarization, respectively. These interactions promote therapeutic efficacy in mice infected with MTB, thereby presenting a potential route to host-directed therapy development. Furthermore, TgMIF, in combination with first-line TB drugs, significantly inhibited drug-resistant MTB strains, including multidrug-resistant TB. These results demonstrate that TgMIF is potentially a multifaceted therapeutic agent against TB, acting through immune modulation, enhancement of mitochondrial function, and dependent on STAT1 and AZIN1 pathways.
Toxoplasma gondii macrophage migration inhibitory factor has promising therapeutic potential against tuberculosis.
INTRODUCTION
Tuberculosis (TB) has been known since ancient times (1). In the second half of the 20th century, effective anti-TB drugs such as isoniazid (INH) and rifampicin (RIF) were implemented for treatment, and TB became a preventable and generally treatable disease (2). However, more than 10 million people continue to be infected with TB every year, and the emergence of drug-resistant TB strains poses significant treatment challenges (3). While the COVID-19 pandemic affected TB incidence, it is now back to pre-pandemic levels (4).
Mycobacterium tuberculosis (MTB) is a species of pathogenic bacteria in the family Mycobacteriaceae and the cause of TB (5). MTB typically affects the lungs, but it can spread to various organs from the respiratory tract. Alveolar macrophages are the first line of defense in activating immune mechanisms such as phagocytosis, expression of cytokines, autophagy, apoptosis, pyroptosis, and secretion of cytotoxic molecules [nitric oxide (NO) and reactive oxygen species (ROS)]. In addition, inducible nitric oxide synthase (iNOS), an enzyme that generates a cytotoxic molecule, contributes to defense against MTB (6–9). However, these innate defense mechanisms are extensively manipulated by various strategies of MTB, such as secreting virulence-related antigens, inducing mitochondrial dysfunction, reprogramming host metabolism, and interfering with macrophage polarization for successful intracellular infection (10–12). Treatment typically involves a combination of antibiotics known as anti-TB drugs. Commonly used medications include INH, RIF, ethambutol (EMB), and pyrazinamide (PZA). These drugs are often administered in combination, and the duration of treatment generally ranges from 6 to 9 months, varying based on the type of TB and the individuals (3, 13). Given that multidrug-resistant TB (MDR-TB) exhibits resistance to standard anti-TB drugs, understanding its interaction with macrophages becomes particularly crucial. Hence, manipulated macrophages are turned from foes into primary hosts for intracellular survival and replication of MTB (14). Therefore, understanding the mechanisms and restoring the function of macrophages altered by MTB infection might provide prospects for TB therapeutic strategies through the modulation of the immunobiological functions of macrophages.
Recently, biological products derived from microbiomes or parasites have been used to treat various diseases (15, 16), reduce adverse drug effects, and improve treatment outcomes. Toxoplasma gondii (T. gondii) is a protozoan parasite that causes toxoplasmosis, and approximately one-third of the human population worldwide is infected (17). Typically, T. gondii infections in individuals are asymptomatic but may lead to central nervous system disease and other severe conditions in immunocompromised patients, and infection during pregnancy can cause congenital infection (18, 19). Innate immunity is induced by T. gondii infection. However, T. gondii has evolved to manipulate host immune responses through the activity of several parasite-specific proteins (20, 21), such as dense granule proteins and rhoptry proteins (22–26).
Macrophage migration inhibitory factor (MIF), produced by the pituitary gland and various cell types, is a pleiotropic cytokine (27, 28). MIF initiates a signaling pathway via the MIF-CD74 axis and plays a crucial role in provoking an inflammatory response (27, 29). MIF is located on chromosome 22 (22q11.2) in the human genome, encodes a 114–amino acid nonglycosylated protein, and exhibits ~90% homology across mammals (30, 31). Furthermore, several parasites, such as Entamoeba histolytica, Plasmodium falciparum, Leishmania, Trichomonas vaginalis, and T. gondii have homologs to MIF, which promote manipulation of the host immune response (32). Although T. gondii MIF (TgMIF) has close structural, biochemical, and immunological characterization homology with mammalian MIFs (33), the mechanisms of TgMIF in regulating immune responses remain unclear.
This study aimed to investigate the interactions of TgMIF with binding proteins, CD74, antizyme inhibitor 1 (AZIN1), and signal transducer and activator of transcription 1 (STAT1), and explore their contributions to anti-MTB activity. This study focused on understanding the mechanisms by which TgMIF restores mitochondrial function and modulates macrophage polarization, potentially offering a treatment strategy for TB.
RESULTS
TgMIF modulates the inflammatory response via interaction with CD74, STAT1, and AZIN1
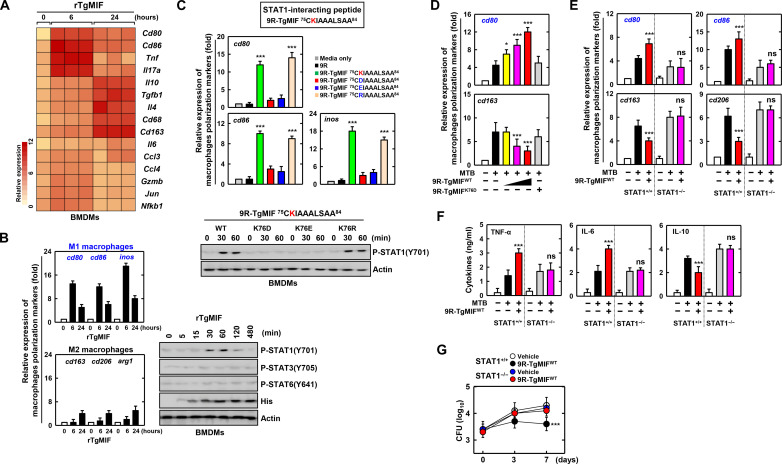
To investigate the functional restoration of MTB-infected macrophages by TgMIF, we established bacteria-synthesized His-tagged TgMIF proteins. Purified recombinant TgMIF (rTgMIF) (13 kDa) was validated using SDS–polyacrylamide gel electrophoresis and Western blotting (Fig. 1A and fig. S1A). Subsequently, we evaluated rTgMIF-induced cytotoxicity by confirming cell viability in murine bone marrow–derived macrophages (BMDMs). No significant difference was observed in cytotoxicity between rTgMIF and rVehicle in BMDMs (fig. S1B). To determine the effects of TgMIF on the expression of inflammatory cytokines, we first examined if TgMIF expression contributes to the effects of lipopolysaccharide (LPS). LPS treatment with proteinase K increased pro-inflammatory cytokines such as tumor necrosis factor–α (TNF-α), interleukin-6 (IL-6), and IL-12p40 (a subunit of the IL-12 family) (32, 33) as well as T helper 2 (TH2) cytokines such as IL-10, transforming growth factor–β1 (TGF-β1), and IL-4. However, rTgMIF that lost function due to proteinase K treatment or heating did not produce cytokines (fig. S1C). Furthermore, in TLR4 knockout mice, which play a crucial role in LPS recognition and signal initiation, TgMIF increased cytokines such as TNF-α, IL-6, IL-10, and TGF-β1 (fig. S1D). LPS measured in rTgMIF was less than 10 pg/mg of protein. Subsequently, it was demonstrated that rTgMIF without LPS provided reliable data. Consistent with previous reports, TgMIF caused elevated pro-inflammatory cytokines such as TNF-α, IL-6, and IL-12p40 (34, 35) and TH2 cytokines such as IL-10, TGF-β1, and IL-4 (Fig. 1B).
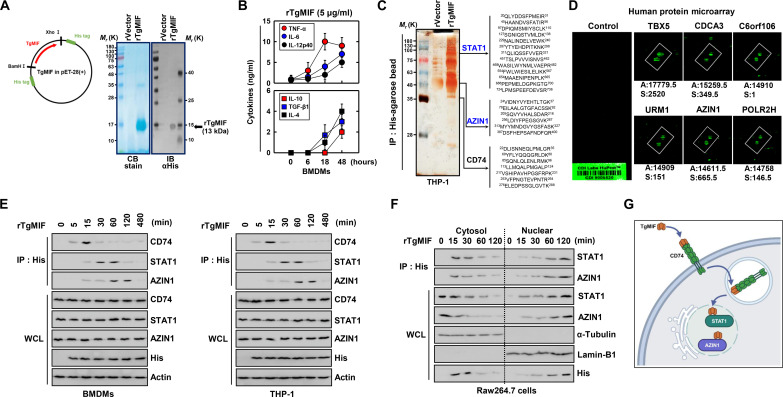
Fig. 1. TgMIF interacts with CD74, STAT1, and AZIN1.
(A) Bacterially purified 6xHis-TgMIF validation via Coomassie blue (CB) staining (left) or Western blotting with αHis (right). IB, immunoblotting. (B) Expression of pro-inflammatory cytokines (TNF-α, IL-6, and IL-12p40) and anti-inflammatory cytokines (IL-10, TGF-β1, and IL-4) in BMDMs treated with rTgMIF (5 μg/ml). The data shown are the means ± SD of five experiments. (C) Identification of STAT1, AZIN1, and CD74 using MS analysis in THP-1 cell lysates expressed with TgMIF or vector. (D) Binding partner identification using protein microarrays against rTgMIF. A, affinity; S, specificity. (E) Interaction with CD74, STAT1, and AZIN1 in BMDMs or THP-1 stimulated with rTgMIF (5 μg/ml) for the indicated times. WCL, whole cell lysate. (F) Western blot analysis of STAT1 and AZIN1 and their interactions in the nuclear and cytoplasmic fractions of Raw264.7 cells. (G) Schematic diagrams of the interaction of TgMIF and CD74, STAT1, and AZIN1. The data are representative of four independent experiments with similar results [(E) and (F)]. Full-length images of the blots presented in fig. S8.
To explore potential binding proteins and mechanisms involved in the inflammatory response to TgMIF, we performed coimmunoprecipitation coupled with mass spectrometry (co-IP/MS). STAT1, AZIN1, and CD74 were selected for further study based on robust signals intensities, abundant peptide counts, and consistent reproducibility and known relevance to immune response and inflammation (Fig. 1C) (36–38). In addition, a human proteome microarray identified 89 proteins that interacted with rTgMIF. In particular, six proteins (TBX5, CDCA3, C6orf106, URM1, POLR2H, and AZIN1) showed a high binding affinity for rTgMIF (Fig. 1D). We further investigated interactions between TgMIF and endogenous CD74, STAT1, or AZIN1. The results showed that significant interactions were observed between rTgMIF-stimulated BMDMs and THP-1 (human acute monocytic leukemia cell line) cells, with no binding observed with STAT3 and STAT6 (Fig. 1E and fig. S1C). The cytoplasmic and nuclear fractionation results localized the interaction of TgMIF with STAT1 and AZIN1 in the nucleus (Fig. 1F). In summary, these findings indicate that TgMIF interacts with CD74, STAT1, and AZIN1 in macrophages (Fig. 1G).
TgMIF binding mapping: CD74, STAT1, and AZIN
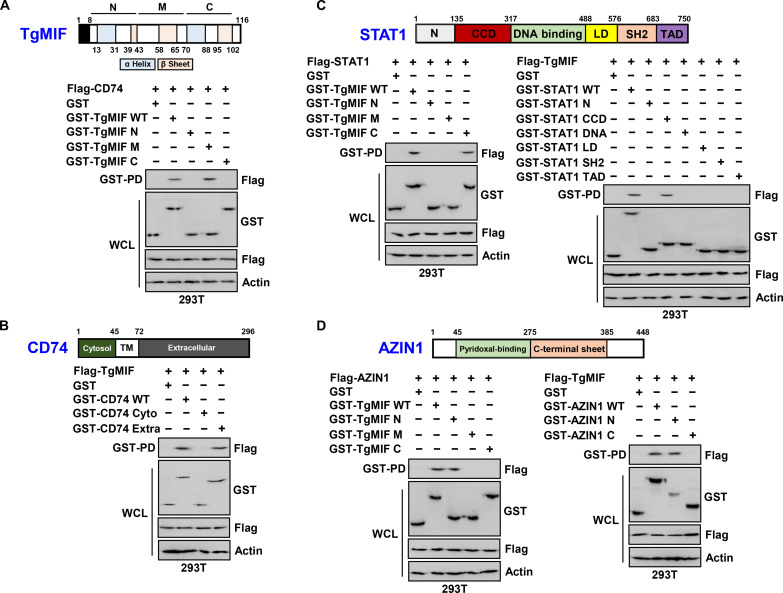
To elucidate the regions of TgMIF responsible for interactions with CD74, AZIN1, and STAT1, we engineered variants of the glutathione S-transferase (GST)–TgMIF fusion protein comprising N-terminal (amino acid 13–43), middle (amino acid 44–69), and/or C-terminal domains (amino acid 70–102). In 293T cells, the GST-TgMIF wild-type (WT) and GST-TgMIF middle region fusion proteins successfully pulled down Flag-CD74, indicating a specific interaction between the middle region of TgMIF and CD74 (Fig. 2A). Further mapping of the TgMIF-binding domain on CD74 revealed that TgMIF binds to the extracellular domain (amino acid 72–296) of CD74, as demonstrated by GST pulldown assays using truncated mutants of GST-CD74 (Fig. 2B). In addition, we investigated the interaction domains of TgMIF, STAT1, and AZIN1. The GST-TgMIF C-terminal and N-terminal domains successfully pulled down Flag-STAT1 and Flag-AZIN1, respectively, indicating the presence of distinct interaction sites (Fig. 2, C and D). Subsequent mapping studies using truncated mutants of GST-STAT1 and Flag-TgMIF in 293T cells revealed that the coiled-coil domain (CCD) (amino acid 135–317) of STAT1 was crucial for its interaction with TgMIF (Fig. 2C). Furthermore, mapping studies demonstrated that the AZIN1 N terminus (amino acid 45–275) was required for the interaction with TgMIF (Fig. 2D).
Fig. 2. Mapping TgMIF binding sites.
(A) Exploration of the CD74 binding site of TgMIF using GST-vector or GST-TgMIF and its truncated constructs. (B) Identification of the TgMIF binding site of CD74 using GST-vector or GST-CD74 and its truncated constructs. (C) Exploration of TgMIF binding sites on STAT1 or STAT1 on TgMIF using GST-vector, GST-TgMIF, or GST-STAT1 and its truncated constructs. (D) Identification of TgMIF binding sites on AZIN1 or AZIN1 on TgMIF using GST-vector, GST-TgMIF, or GST-AZIN1 and its truncated constructs. The data are representative of four independent experiments with similar results [(A) to (D)]. Full-length images of the blots presented in fig. S8.
In summary, our findings highlight that TgMIF has N-terminal (amino acid 13–43), middle (amino acid 44–69), and C-terminal (amino acid 70–102) domains that interact with AZIN1, CD74, and STAT1, respectively.
TgMIF-induced inflammatory responses through CD74-mediated endocytosis
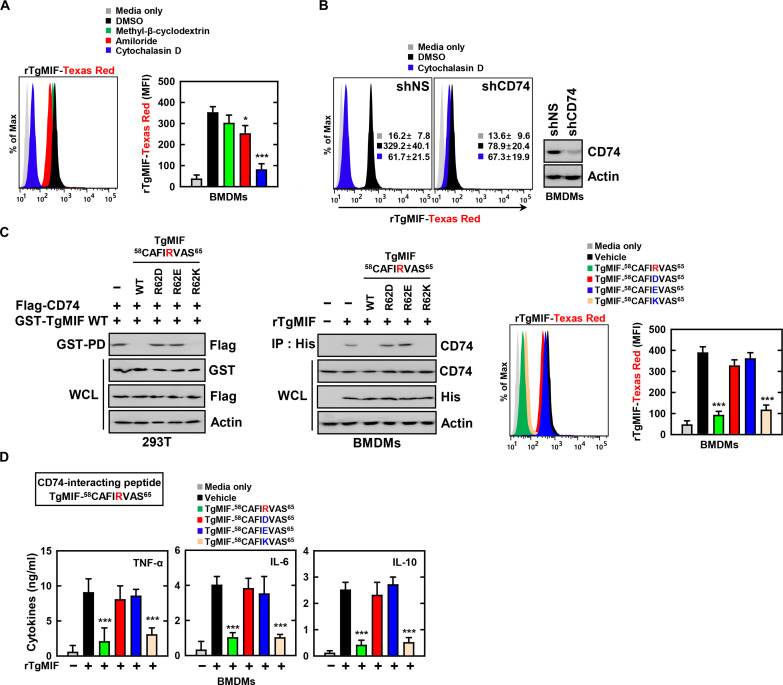
To determine the endocytosis pathway of TgMIF, we applied three different endocytosis inhibitors: methyl-β-cyclodextrin (a caveolin-mediated endocytosis inhibitor), amiloride (a macropinocytosis inhibitor), and cytochalasin D (a phagocytosis/macropinocytosis inhibitor). Cytochalasin D treatment significantly reduced TgMIF expression compared to that of the untreated control BMDMs (Fig. 3A).
Fig. 3. Endocytosis pathway of TgMIF and its interaction with CD74.
(A) Effect of endocytosis inhibitors (10 mM methyl-β-cyclodextrin, cholesterol chelator; 1 μM amiloride, inhibits Na+/H+ exchange; 10 μM cytochalasin D, inhibitor of actin polymerization) on TgMIF expression in BMDMs. MFI, mean fluorescence intensity; DMSO, dimethyl sulfoxide. (B) Inhibition of TgMIF expression by shCD74 treatment in BMDMs. (C) Binding specificity of the TgMIF peptide (1 μM) with CD74. (D) Inflammatory responses of TgMIF peptide (1 μM) in BMDMs. The data are representative of four independent experiments with similar results [(B) and (C)]. Statistical significance was determined by the Student’s t test with Bonferroni adjustment (*P < 0.05; ***P < 0.001). Full-length images of the blots presented in fig. S8.
Next, we investigated the role of CD74 in TgMIF endocytosis by evaluating TgMIF expression in BMDMs treated with short hairpin RNA oligos of CD74 (shCD74). Similar to cytochalasin D treatment, shCD74 treatment significantly inhibited TgMIF expression (Fig. 3B). Our findings suggest that the middle domain of TgMIF is crucial for its interaction with CD74 (Fig. 2, A and B). Subsequently, we explored the binding specificity of TgMIF-58-65 to CD74 in 293T cells. To further refine our understanding, we designed specific amino acid fragments (TgMIF-58-65, TgMIF-15-27, and TgMIF-75-84) representing the minimal peptide sequences that interact with CD74, AZIN1, and STAT1, respectively, using the PROFbval method. As shown in Fig. 3C, TgMIF-58-65 WT competitively inhibited the interaction between TgMIF and CD74. Arginine (R) and lysine (K) are positively charged amino acids due to their side chains containing positively charged groups, while aspartic acid (D) and glutamic acid (E) are negatively charged amino acids due to their side chains containing negatively charged groups. These electrostatic properties of the side chains critically influence the protein structure and function by affecting protein folding, stability, and interactions with other molecules (39). The R62 amino acid with charged side chains in TgMIF-58-65 was identified as essential for its interaction with CD74. R62D and R62E are amino acid substitutions where the original arginine (R) at position 62 is replaced by aspartic acid (D) and glutamic acid (E), respectively. TgMIF-58-65, R62D, and R62E did not interfere with the interaction between TgMIF and CD74. In contrast, TgMIF-58-65 R62K competitively inhibited the interaction between TgMIF and CD74 (Fig. 3C). Furthermore, TgMIF-58-65 R62K demonstrated the interaction with endogenous CD74 by competitively inhibiting the binding of rTgMIF to the BMDMs (Fig. 3C). Measurement of CD74 protein expression indicated that TgMIF-58-65 R62K was crucial for the binding of TgMIF to CD74 (Fig. 3C).
Last, we assessed the effect of the CD74-interacting peptide of TgMIF on inflammatory responses. While rTgMIF increased inflammatory cytokines such as TNF-α, IL-6, and IL-10, treatment with TgMIF-58-65 R62K significantly inhibited cytokine production in BMDMs (Fig. 3D). In summary, these results demonstrate the essential role of R62 amino acids with charged side chains in the TgMIF-58-65 fragment in the interaction with CD74 and indicate that CD74 is indispensable for TgMIF-induced inflammatory responses.
TgMIF modulates mitochondrial dynamics and metabolism through AZIN1
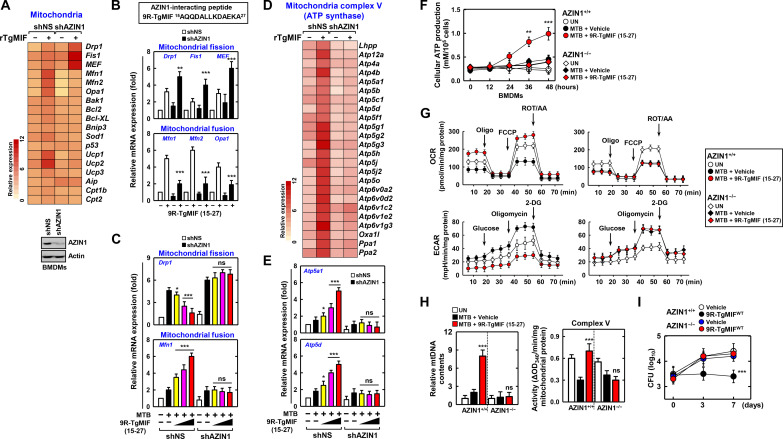
To profile the genes and mechanisms affected by AZIN1, which interacts with TgMIF, we conducted differential gene expression (DGE) analysis of BMDMs treated with rTgMIF and either non-silencing shRNA (shNS), transfected with non-silencing control, or shAZIN1, based on RNA sequencing (RNA-seq) data. RNA-seq results revealed altered DGE patterns related to mitochondria when comparing shAZIN1 to shNS (fig. S2A). Specifically, within the subset of genes with altered expression, members of the mitochondrial carrier family (SLC25) facilitate the transport of a variety of compounds across the inner membrane of mitochondria. Previous studies reported that SLC25A46, a member of the SLC25 family, is implicated in mitochondrial fusion and fission (40, 41). Furthermore, TOMM40 and TOMM22, components of the translocase of the outer mitochondrial membrane (TOM) complex, were reported to be involved in mitochondrial fusion and fission (42). These findings collectively suggest that the mitochondrial genes are prominent players in the cellular response to AZIN1 modulation. The expression of genes involved in rTgMIF-induced mitochondrial fission (DRP1, FIS1, and MEF) and fusion (MFN1, MFN2, and OPA1) was regulated by AZIN1 (Fig. 4A). Drp1, Fis1, and MEF, which were up-regulated by rTgMIF, exhibited a marked increase following shAZIN1 treatment compared with shNS. Conversely, Mfn1, Mfn2, and Opa1, which were elevated by TgMIF, decreased in macrophages treated with shAZIN1 (fig. S2B). Furthermore, even in the absence of AZIN1, the TgMIF variant 9R-TgMIF (15–27) significantly increased mitochondrial fission gene expression, whereas mitochondrial fusion gene expression increased by 9R-TgMIF (15–27) was significantly decreased by AZIN1 (Fig. 4B).
Fig. 4. TgMIF modulates mitochondrial dynamics and metabolism through AZIN1.
(A to C) AZIN1-dependent regulation of mitochondrial fission and fusion gene expression by TgMIF (1, 5, and 10 μM). (D and E) AZIN1-dependent modulation of ATP synthase–related gene expression by TgMIF. (F) Cellular ATP production in MTB-infected BMDMs is dependent on TgMIF and AZIN1. (G) Restoration of mitochondrial respiration and ECARs by MTB infection with TgMIF and AZIN1. Oligo, oligomycin; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; 2-DG, 2-deoxyglucose. (H) Measurement of mtDNA content and mitochondria complex V activity in MTB-infected macrophages. (I) Number of CFU of MTB. Statistical significance was determined by the Student’s t test with Bonferroni adjustment (*P < 0.05; **P < 0.01; ***P < 0.001). ns, not significant. Full-length images of the blots presented in fig. S8.
To assess the restorative effect of AZIN1-dependent TgMIF on MTB-induced mitochondrial dysfunction, we measured Drp1 and Mfn1 expression. Drp1 expression significantly increased following MTB infection compared to non-infected macrophage, and treatment with 9R-TgMIF (15–27) inhibited Drp1 expression depending on AZIN1 expression. Conversely, 9R-TgMIF (15–27) significantly increased Mfn1 expression in MTB-infected macrophages with AZIN1 (Fig. 4C). Western blotting demonstrated that TgMIF suppressed phospho-Drp1 (P-Drp1) and Drp1 expression, which was increased by MTB infection, depending on AZIN1 expression (fig. S2C). MFN1 and MFN2 expression were increased by TgMIF, along with AZIN1 expression in MTB-infected macrophages (fig. S2D). Furthermore, the elevated mitochondrial mass implicated in fusion was increased by 9R-TgMIF (15–27) and decreased by AZIN1 in MTB-infected macrophages (fig. S2E).
RNA-seq results revealed the involvement of TgMIF in the mitochondrial complex, particularly the fifth mitochondrial complex [adenosine 5′-triphosphate (ATP) synthase] (Fig. 4D and fig. S3A). For instance, Atp5a1 and Atp5d, mitochondrial ATP synthase–associated genes, were up-regulated by 9R-TgMIF (15–27) treatment but suppressed by AZIN1 knockdown (Fig. 4E and fig. S3B). Up-regulated Atp5A1 protein expression by MTB infection and TgMIF treatment was suppressed by AZIN1 knockdown (fig. S3C). In addition, 9R-TgMIF (15–27) treatment increased cellular ATP production in MTB-infected BMDMs compared to MTB-infected BMDMs expressing shAZIN1 (Fig. 4F).
Subsequently, we measured oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) to investigate the mitochondrial function and metabolic characteristics of the cells. Substrate oxidation comparisons in BMDMs indicated a significant increase in basal respiration, ATP-linked response, and maximal respiration, which were reduced by MTB infection, and this increase was manifested through 9R-TgMIF (15–27). However, in AZIN1-deficient mouse macrophages, no increase in basal respiration, ATP-linked response, or maximum respiration was observed following TgMIF treatment (Fig. 4G). The ECAR results revealed that MTB infection induced a significant mitochondrial response in BMDMs, which was subsequently reduced by 9R-TgMIF (15–27) treatment. However, no such reduction was observed in AZIN1-deficient mouse macrophages (Fig. 4G).
ROS, which originate from molecular oxygen and are generated through redox reactions or electronic excitation (43), can induce mitochondrial DNA (mtDNA) mutations and damage the mitochondrial respiratory chain, decreased with TgMIF treatment depending on the expression of AZIN1 in MTB-infected macrophages (fig. S3D). Furthermore, TgMIF increased mtDNA content and mitochondrial complex V activity in MTB-infected macrophages but decreased them in shAZIN1-treated macrophages (Fig. 4H). No changes were observed in the activities of complexes I, II-III, or IV upon treatment with TgMIF and AZIN1 (fig. S3E). We measured nicotinamide adenine dinucleotide (NAD+) and lactate levels to assess if TgMIF promoted the recovery of mitochondrial dysfunction. TgMIF increased NAD+ levels and decreased intracellular lactate levels compared to the vehicle in MTB-infected macrophages (fig. S3F). In addition, functional restoration of mitochondria by AZIN1-dependent TgMIF resulted in decreased MTB colony-forming units (CFU) (Fig. 4I).
In summary, these results demonstrate that TgMIF plays a crucial role in the increase in mitochondrial function against MTB infection by regulating mitochondrial fission and fusion, ATP synthesis, and metabolism in an AZIN1-dependent manner.
TgMIF-induced STAT1-dependent macrophage polarization enhances host defense
We identified candidate genes showing time-dependent differential expression patterns in response to TgMIF (Fig. 5A and fig. S4A). M1 macrophage markers, such as cd80, cd86, and iNOS, transiently increased after 6 hours of TgMIF treatment, whereas M2 macrophage markers, including cd163, cd206, and Arg1, showed no significant changes. In addition, TgMIF induced a temporary increase in STAT1 phosphorylation at 30 and 60 min (Fig. 5B). Using TgMIF mutants, we highlighted the significance of the K76 amino acid with charged side chains in TgMIF-75-84 for the interaction with STAT1. TgMIF-75-84 K76D and K76E failed to increase cd80, cd86, and iNOS expression, whereas TgMIF-75-84 K76 and TgMIF-75-84 K76R significantly increased their expression. Furthermore, TgMIF-75-84 WT and TgMIF-75-84 K76R transiently increased p-STAT1 expression at 30 and 60 min (Fig. 5C).
Fig. 5. TgMIF-induced STAT1-dependent macrophage polarization enhances host defense.
(A) Differential expression of macrophage polarization markers in TgMIF-treated BMDMs. (B) Expression of phosphorylation of STAT1, STAT3, and STAT6 in BMDMs treated with rTgMIF (5 μg/mL) for the indicated times. (C) Expression of M1 macrophage polarization markers and phosphorylation of STAT1 by amino acid variants of the TgMIF peptide. (D) The expression of macrophage polarization markers changes depending on various concentrations of the TgMIF (1, 5, and 10 μM) or 9R-TgMIF K76D (10 μM). (E) STAT1-dependent M1 and M2 macrophage polarization regulation by TgMIF. (F) Inflammatory cytokine production in MTB-infected BMDMs is dependent on TgMIF and STAT1. (G) Number of CFU of MTB. Statistical significance was determined by the Student’s t test with Bonferroni adjustment (*P < 0.05; ***P < 0.001). Full-length images of the blots presented in fig. S8.
To investigate if macrophage polarization markers are regulated by TgMIF in MTB-infected macrophages, we observed a dose-dependent increase in cd80, cd86, and iNOS expression following treatment with TgMIF WT compared to TgMIF K76D. cd206 is commonly expressed on tissue-resident macrophages that exhibit an M2-polarized phenotype, often associated with a reparative function (44, 45). However, in most tissues, both resident and newly recruited macrophages predominantly adopt an M1 polarization profile (46). Moreover, while M0 and M2 macrophages exhibit elevated cd206 expression, M1 macrophages typically show lower levels of cd206 expression (47). Therefore, we confirmed the expression of M2 macrophage markers such as cd163 and Arg1 in addition to cd206 (48, 49). As a result, cd163, cd206, and Arg1 expression decreased in cells treated with TgMIF WT, whereas TgMIF K76D showed no change (Fig. 5D and fig. S4B). In addition, pro-inflammatory cytokines TNF-α and IL-6 increased, and the anti-inflammatory cytokine IL-10 decreased with TgMIF WT (fig. S4D).
Subsequently, we explored the STAT1 dependency of macrophage polarization by TgMIF in MTB-infected macrophages. The increase in M1 macrophage markers (cd80, cd86, and iNOS) induced by TgMIF was inhibited by STAT1 blockade, and the decrease in M2 macrophage markers (cd163, cd206, and Arg1) was blocked by STAT1 inhibition (Fig. 5E and fig. S4C). Furthermore, the influence of STAT1 on cytokine production induced by TgMIF in MTB-infected macrophages revealed a reduction in pro-inflammatory cytokines (TNF-α and IL-6) and an increase in anti-inflammatory IL-10 in the absence of STAT1 (Fig. 5F).
The STAT1-dependent TgMIF-induced polarization of M1 macrophages inhibited MTB growth in a dose-dependent manner (Fig. 5G and fig. S4E). In summary, these results highlight the role of STAT1 in TgMIF-induced macrophage polarization and its subsequent impact on MTB growth.
TgMIF rescues TB lung injury and mortality via STAT1 and AZIN1
To evaluate the therapeutic effects of TgMIF against lung injury in MTB-induced mice, we designed a conjugated TgMIF peptide and its mutants, TgMIF MT1 (CD74-binding mutant) and TgMIF MT2 (STAT1-binding mutant). In the CD74-binding mutant (TgMIF MT1), an arginine (R) at position 62 is replaced by aspartic acid (D). In the STAT1-binding mutant (TgMIF MT2), a lysine (K) at position 75 is replaced by aspartic acid (D). These specific substitutions were engineered to investigate the alterations in TgMIF’s binding properties with CD74 and STAT1, respectively (fig. S5A). Stability, Cytochromes P450 (CYP) enzyme activity, and Pharmacokinetic (PK) parameters were measured to assess the bioavailability, metabolic rate, and adverse reactions in response to the peptide (fig. S5B).
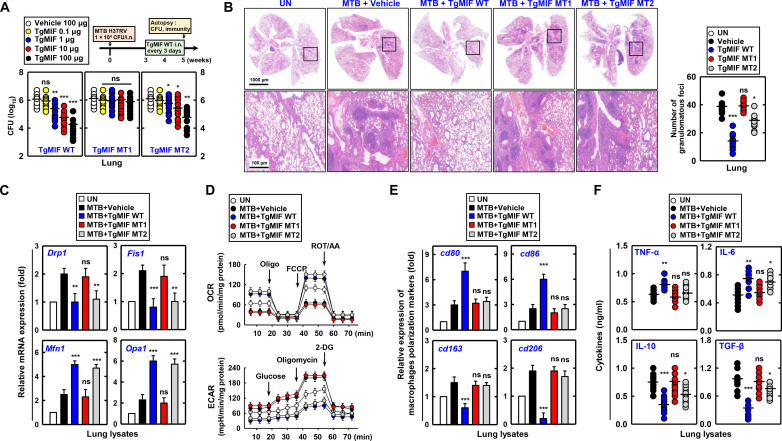
In the timeline designed for the TB mouse model, TgMIF WT and TgMIF MT2 post-treatment resulted in a dose-dependent decrease in MTB CFU (Fig. 6A). However, pretreatment and cotreatment with TgMIF did not completely control MTB, and a reduction was observed only at high concentrations (fig. S5C). Furthermore, reduced infiltration of immune cells and diminished lung damage were observed in MTB H37Rv strain-infected mice. However, these effects were not observed in the TgMIF MT1–treated mice (Fig. 6B). Further investigation of the effect of TgMIF on mitochondrial function in MTB-infected mice revealed that TgMIF WT and TgMIF MT2 treatment suppressed Drp1 and Fis1 expression and elevated Mfn1 and Opa1 expression in the lungs. In contrast, TgMIF MT1 did not regulate the expression of mitochondrial fission or fusion genes (Fig. 6C). Moreover, TgMIF enhanced mitochondrial function, as demonstrated by the restoration of mitochondrial respiration, suppression of ECARs, increased ATP production and ATP synthase activity, and reduction in mitochondrial ROS levels (Fig. 6D and fig. S5D).
Fig. 6. Therapeutic effects of TgMIF on lung injury and immune response in MTB-induced mice.
(A and B) Therapeutic effects of TgMIF peptide and its mutants, TgMIF MT1 (CD74-binding mutant) and TgMIF MT2 (STAT1-binding mutant), against lung injury in MTB-induced mice (n = 25). i.n., intranasally. UN, untreated. (C) Effect of TgMIF peptide on the restoration of mitochondrial function in MTB-infected mouse lung lysates. (D) Modulation of mitochondrial function-alterations in mitochondrial respiration and ECARs by TgMIF treatment. (E) Alteration in macrophage polarization marker expression in MTB-infected mice treated with TgMIF peptide. (F) Inflammatory cytokine production in the lungs of MTB-infected mice following TgMIF treatment. Statistical significance was determined by the Student’s t test with Bonferroni adjustment (*P < 0.05; **P < 0.01; ***P < 0.001).
In BMDMs, the interaction between TgMIF and STAT1 induces M1 macrophage polarization (Fig. 5). Consistent with these findings, in the lungs of MTB-infected mice, TgMIF increased the expression of P-STAT1, CD80, and CD86. However, TgMIF MT1 and MT2 showed no changes in CD163 or CD206 expression (figs. S5E and S6E).
To elucidate the precise localization of TgMIF in the organs, the biodistribution of TgMIF was assessed at each time point using in vivo imaging system (IVIS) imaging. As a result, high-contrast images of MTB-infected mice were captured 1 to 6 hours after the administration of TgMIF-Cy5.5 via intranasal delivery, revealing distribution primarily in the lungs (fig. S5F).
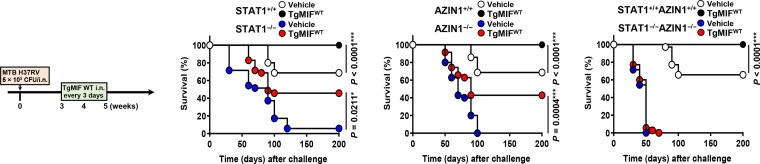
Alterations in immune cell population and inflammatory cytokine production have been observed in the lungs of MTB-infected mice. Innate immune cells increased in number, whereas CD4 T helper cells, CD8 cytotoxic T cells, and FOXP3+ regulatory T cells showed no change (fig. S6A). TgMIF increased the expression of TNFα and IL-6 in the lungs while decreasing IL-10 and TGF-β, with no observed change in interferon-γ (IFN-γ) (Fig. 6F and fig. S6B). We investigated the post-treatment efficacy of WT TgMIF, dependent on STAT1, AZIN1, or both STAT1 and AZIN1, against mortality induced by high-dose MTB H37Rv strain in mice.
In STAT1+/+ mice, the TgMIF WT–treated group exhibited a notably higher survival rate compared to the vehicle-treated group. In STAT1−/− mice, both vehicle-treated and TgMIF WT–treated groups showed reduced survival, but TgMIF WT improved survival even in the absence of STAT1. Similarly, in AZIN1+/+ mice, the TgMIF WT–treated group experienced a significantly higher survival rate compared to the vehicle-treated group. In AZIN1−/− mice, both vehicle-treated and TgMIF WT–treated groups showed diminished survival, but TgMIF WT enhanced survival even in the absence of AZIN1. In STAT1+/+AZIN1+/+ mice displayed similar survival rates for TgMIF treatment. However, in STAT1−/−AZIN1−/− mice, both vehicle-treated and TgMIF WT–treated groups showed a marked reduction in survival. These findings collectively suggest that both STAT1 and AZIN1 are critical for the protective efficacy of TgMIF in MTB-infected mice. While TgMIF WT can interact with either STAT1 or AZIN1 to exert its protective effects, the presence of at least one functional protein (either STAT1 or AZIN1) is essential for maintaining significant survival rates. Complete deficiency in STAT1 and AZIN1 abolishes the therapeutic effect of TgMIF WT and significantly reduces survival (Fig. 7).
Fig. 7. TgMIF treatment affects the TB survival rate after MTB infection.
The efficacy of TgMIF on MTB-induced mortality is dependent on STAT1 and AZIN1 (n = 35). Significant differences in comparison relative to the control mice are indicated (log-rank test).
In summary, TgMIF exhibited therapeutic efficacy against MTB-induced lung injury and mortality through diverse mechanisms, including immune modulation, enhancement of mitochondrial function, and dependence on the STAT1 and AZIN1 pathways.
TgMIF synergizes with TB drugs against drug-resistant strains
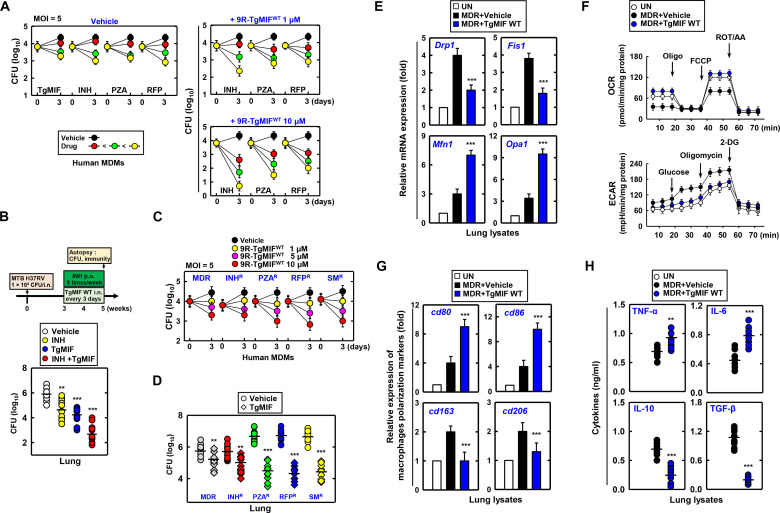
To enhance the bactericidal action against MTB in human monocyte-derived macrophages (MDMs), we investigated the efficacy of TgMIF in combination with the first-line TB treatment drugs INH, PZA, and RFP. We observed dose-dependent inhibition of MTB growth with TgMIF, INH, PZA, and RFP in human MDMs. Furthermore, the CFU inhibition efficiency was enhanced by combining TgMIF with INH, PZA, and RFP, and particularly significant additive effects were observed when combined with INH (Fig. 8A). This additive effect was extended to in vivo studies in which the combination of INH and TgMIF inhibited MTB growth in the lungs of MTB-infected mice (Fig. 8B).
Fig. 8. Therapeutic synergy of TgMIF with TB drugs against drug-resistant TB strains.
(A) Effect of the combination of TgMIF with INH (0.05, 0.1, and 0.5 μg/ml), PZA (5, 10, and 20 μg/ml), and RFP (0.025, 0.1, and 0.5 μg/ml) on MTB growth inhibition in human MDMs. Low (red circle), medium (green circle), and high (yellow circle) concentration of drugs. MOI, multiplicity of infection. (B) Inhibition of MTB growth in the lungs of MTB-infected mice by the combination of INH (15 mg/kg) and TgMIF (10 μg) (n = 35). p.o., per os. (C and D) Reduction in bacterial colony counts by TgMIF in drug-resistant TB strains. (E and F) Restoration of mitochondrial function in lungs infected with MDR-TB following TgMIF treatment. (G) Regulation of macrophage polarization markers by TgMIF in MDR-infected lungs. (H) Induction of inflammatory cytokine production by TgMIF in MTB-infected macrophages. Statistical significance was determined by the Student’s t test with Bonferroni adjustment (**P < 0.01; ***P < 0.001).
We evaluated the therapeutic efficacy of TgMIF against TB strains resistant to INH, PZA, RFP, streptomycin (SM), and MDR-TB. TgMIF significantly decreased the bacterial colony counts in human MDMs and lungs across all drug-resistant TB strains (Fig. 8, C and D). These effects were accompanied by reduced immune cell infiltration and lung damage (fig. S7).
In lungs infected with MDR-TB, TgMIF treatment exerted a regulatory effect on mitochondrial function. TgMIF inhibited Drp1 and Fis1 expression while elevating Mfn1 and Opa1 expression in lung lysates obtained from MDR-infected mice (Fig. 8E). Furthermore, TgMIF increased the OCR, which was decreased by MDR infection, and decreased the ECAR, which was enhanced by MDR infection (Fig. 8F).
In MDR infections, TgMIF demonstrates a regulatory effect on macrophage polarization markers. cd80 and cd86 were increased, whereas cd163 and cd206 were reduced by TgMIF in the lungs of MDR-infected mice (Fig. 8G). In addition, TgMIF induced the production of inflammatory cytokines in MTB-infected macrophages, with significantly increased TNF-α and IL-6 and marked decreases in IL-10 and TGF-β in lung lysates obtained from MDR-infected mice (Fig. 8H).
In summary, when combined with anti-TB drugs, TgMIF exhibits significant efficacy against MTB and drug-resistant TB strains. This combination therapy resulted in reduced bacterial colony count, immune modulation, mitigation of lung damage, and restoration of mitochondrial function. These findings highlight the potential of TgMIF as a valuable component in the treatment of drug-resistant TB.
DISCUSSION
This study aimed to investigate the role of TgMIF in modulating the immune response during MTB infection. We explored the interactions of TgMIF with binding proteins (CD74, AZIN1, and STAT1) and their contributions to anti-MTB activity. In this study, we revealed the potential therapeutic effects of TgMIF on TB through the restoration of mitochondrial function and modulation of macrophage polarization.
The approval of 17 new chemical entities for clinical trials, either independently or in combination with nine existing anti-TB drugs, marks a significant advancement compared to previous years (50–52). However, MTB has developed resistance to drugs currently used in clinical practice. The drug resistance of MTB, particularly MDR-TB, which is difficult to treat due to resistance to the anti-TB drugs INH and RIF, has presented a significant health and economic burden worldwide (53). Therefore, novel treatments that are more effective and less toxic, such as natural products (probiotics and polyphenols) and biological products (antimicrobial peptides), have been tested to overcome antibiotic resistance against MTB (54).
T. gondii is an intracellular parasite that infects various organs and tissues during acute infection and particularly targets the brain in chronic infection (55, 56). One of the most notable immunological characteristics of T. gondii infection is the robust and continuous cell-mediated immune response, which plays a crucial role in protecting the host against the rapid proliferation of tachyzoites and consequent pathology (57). This immunoregulatory mechanism observed during T. gondii infection has prompted numerous investigations into its potential applications for treating other immune-related diseases. Consequently, proteins derived from T. gondii have emerged as promising candidates for therapeutic intervention under challenging and incurable conditions (22–25, 58–60).
MIF, involved in both innate and adaptive immune responses, exhibits pro-inflammatory activity that plays a crucial role in biological processes, including the activation of extracellular signal–regulated kinase (ERK) pathways triggered by the MIF-CD74 interaction (61, 62). In addition, the ERK signaling pathway is promoted by MIF and TgMIF (33). Several studies have suggested that TgMIF may induce ERK signaling through the interaction with the MIF receptor CD74 (33, 63). Our results demonstrated that TgMIF binds with CD74 and results in enhanced cytokine production, including TNF-α, IL-6, and IL-10. Therefore, TgMIF shares structural similarities with MIF, enabling it to exert similar biological functions, particularly modulating immune responses the through activation of specific signaling pathways.
Previous studies reported that MIF is implicated in the innate immune response against MTB in humans, triggering the release of early pro-inflammatory cytokines such as TNF-α and IL-1β (64, 65). In addition, MIF-deficient mice infected with MTB exhibited enhanced lung pathology and suppressed the production of innate cytokines, including TNF-α, IL-12, and IL-10 (66). Moreover, MTB exploits host mitochondria and regulates host immune signaling, activities that are crucial for successful infection. MTB infection induces distinct alterations in mitochondrial dynamics, including fission and fusion, which are essential for maintaining mitochondrial function and ATP production. These alterations in mitochondria play a critical role in the immunometabolism of host immune cells, including dendritic cells, T cells, and macrophages (67). A prior study indicated that the expression of mitochondrial fission proteins such as Drp1 and p-MFF was elevated in MTB-infected macrophages (68). Another study demonstrated that MTB infection induces mitochondrial fusion by enhancing the expression of Mfn1, resulting in increased ATP production (69). This is consistent with previous findings that MTB enhances the expression of fission and fusion proteins. However, depending on AZIN1 expression, TgMIF significantly suppresses fission protein Drp1 and enhances fusion protein Mfn1. Furthermore, previous studies have reported a decrease in mitochondrial ATP production in MTB-infected BMDMs (68, 70). Consistent with these findings, our results reveal an increase in ATP production in an AZIN1-dependent manner following TgMIF treatment, despite the reduction in ATP production caused by MTB. Overall, TgMIF, through its interaction with AZIN1, modulates mitochondrial dynamics and ATP production in MTB-infected cells, potentially influencing the host immune response against MTB infection.
This study reveals that TgMIF interacts with STAT1 in macrophages, with specific binding domains identified for this interaction. The specificity of TgMIF interaction with STAT1, in contrast to STAT3 or STAT6, may be attributed to the unique structural and functional characteristics of each STAT protein. STAT1, STAT3, and STAT6 all belong to the STAT family and have distinct functions in cellular signaling pathways. STAT1, STAT3, and STAT6 have CCDs that are reported to be important for protein-protein interaction and play a crucial role in mediating the formation of STAT dimers (71). A potential reason for the specific interaction between TgMIF and STAT1 could be the differences in the amino acid sequences or structural features of the CCD within the STAT proteins (72–74). Macrophage polarization and functions are regulated through the activation of several implicated pathways; the equilibrium between STAT1 and STAT3/STAT6 activation plays an important role. In addition, STAT1 activation promotes M1 macrophage polarization, leading to cytotoxic and pro-inflammatory functions. In contrast, activation of STAT3 and STAT6 through IL-4/IL-13 and IL-10 signaling promotes M2 macrophage polarization, which is associated with active tolerance and tissue repair (75). Our results demonstrated an increase in ATP production, which was reduced by MTB, in an AZIN1-dependent manner with TgMIF treatment. TgMIF interacted with STAT1 and increased the expression of M1 macrophage markers cd80, cd86, and iNOS, elevated pro-inflammatory cytokines TNF-α and IL-6, and decreased the anti-inflammatory cytokine IL-10. These findings suggest that TgMIF modulates macrophage polarization and function through STAT1 activation, potentially influencing the host immune response against pathogens such as MTB.
In addition, in vitro results indicate that CD74 is crucial for the endocytosis and inflammatory response induced by TgMIF. Inhibition of CD74 significantly reduced TgMIF expression, and its interaction with CD74 was essential for inflammatory cytokine production. These findings are consistent with the in vivo results, where TgMIF MT1, which lacked CD74 binding ability, did not exhibit similar protective effects. Specifically, TgMIF MT1 without CD74 binding did not show effectiveness in reducing MTB CFU, relieving lung damage, or regulating mitochondrial function. In conclusion, the in vivo results with TgMIF MT1 confirm the importance of CD74, which aligns with the in vitro findings.
Mutational and genetic drivers, in addition to metabolic processes, play significant roles in the evolution of persistent drug-resistant MTB cells. These factors collectively contribute to the development and maintenance of drug resistance in MTB (50). Our results show the therapeutic effect of TgMIF in drug-resistant MTB-infected mouse lungs by improving mitochondrial dysfunction and inducing M1 macrophage polarization in both first-line and MDR MTB-induced MDMs. On the basis of our results, TgMIF shows promise as a therapeutic agent against drug-resistant MTB strains by targeting mitochondrial dysfunction and promoting an immune response that favors M1 macrophage polarization.
We made significant contributions by elucidating the role of TgMIF in modulating immune responses during MTB infection. However, several aspects that require improvement should be considered in future research. First, we investigated the molecular mechanisms activated by the TgMIF-CD74, TgMIF-AZIN1, and TgMIF-STAT1 interactions. Understanding the detailed molecular mechanisms involved may provide insight into the regulation of immune responses during MTB infection. We explored the role of AZIN1 in mitochondrial dynamics and its interaction with TgMIF. Further investigation into the functions of AZIN1 and its role in mitochondrial fission and fusion may reveal additional aspects of its involvement in host defense against MTB. Second, we investigated the potential development of resistance to TgMIF, either by MTB or by the host. Understanding resistance mechanisms is crucial for predicting the long-term effectiveness of TgMIF-based therapeutic strategies.
In summary, this study suggests that TgMIF has promising therapeutic potential against MTB infection by influencing immune responses, restoring mitochondrial function, and enhancing the effects of conventional TB drugs. These findings provide directions for further research and the potential development of TgMIF-based therapeutic strategies for TB, especially in the context of drug-resistant strains.
MATERIALS AND METHODS
Mice and cell culture
WT C57BL/6 mice were purchased from Samtako Bio (Gyeonggi-do, Korea). AZIN1−/− (C57BL/6Smoc-Azin1em1Smoc) mice were generated by the Shanghai Model Organisms Center Inc., and STAT1−/− [B6.129S(Cg)-Stat1tm1Dlv/J] mice were purchased from the Jackson Laboratory (Bar Harbor, ME, United States). STAT1−/− mice were crossed with AZIN1−/− mice, and the offspring were intercrossed to generate AZIN1−/− STAT1−/− mice, all on a C57BL6/J background. Primary BMDMs were isolated from WT, AZIN1−/−, or STAT1−/− mice and cultured in Dulbecco’s modified essential medium (DMEM) for 3 to 5 days in the presence of macrophage colony-stimulating factor (R&D Systems, 416-ML), as described previously (76). The mouse experimental design and protocols used in this study were approved at the Institute of Microbiology, Chinese Academy of Sciences (IMCAS) by the Research Ethics Committee (permit SQIMCAS2020013) and the Institutional Animal Care and Use Committee of Hanyang University (protocol 2020-075). Human embryonic kidney (HEK) 293T [ATCC-11268, American Type Culture Collection (ATCC)] and Raw264.7 (ATCC TIB-71) cells were cultured in DMEM (Gibco, Waltham, MA, United States) containing 10% fetal bovine serum (FBS) (Gibco), sodium pyruvate, nonessential amino acids, penicillin G (100 IU/ml), and SM (100 μg/ml). Human monocytic THP-1 (ATCC TIB-202) cells were grown in RPMI 1640/GlutaMAX supplemented with 10% FBS and treated with 20 nM phorbol 12-myristate 13-acetate (Sigma-Aldrich) for 24 hours to induce differentiation into macrophage-like cells, followed by washing three times with phosphate-buffered saline (PBS).
Reagents and antibodies
Methyl-β-cyclodextrin (C4555), amiloride hydrochloride (PHR1839), and cytochalasin D (C8273) were purchased from Sigma-Aldrich.
Antibodies specific for CD74 (AB245693) and AZIN1 (PA5-97491) were purchased from Abcam (Cambridge, United Kingdom) and Thermo Fisher Scientific (Waltham, MA, United States), respectively. STAT1 (9172), STAT3 (9139), STAT6 (9362), P-Drp1 (S637) (4867), Drp1 (8570), ATP5A1 (18023), P-STAT1(Y701) (9167), P-STAT3(Y705) (9145), and P-STAT6(Y641) (9361) were purchased from Cell Signaling Technology (Danvers, MA, United States). Antibodies against His (H-3), FLAG (D-8), GST (B-14), actin (I-19), lamin B1 (B-10), tubulin (5F131), MFN1 (D-10), and MFN2 (F-5) were purchased from Santa Cruz Biotechnology (Dallas, TX, United States).
Plasmid construction
FLAG-tagged CD74, STAT1, and AZIN1 plasmids were purchased from Addgene (Cambridge, United Kingdom). Plasmids encoding different regions of TgMIF (8-116, 13-43, 44-69, and 70-102) were generated via polymerase chain reaction (PCR) amplification from full-length TgMIF cDNA and subcloned into a pEBG derivative encoding an N-terminal GST epitope tag between the Bam HI and Not I sites. All transient constructs in mammalian cells were derived from the pEBG-GST mammalian fusion vector and the pEF-IRES-Puro expression vector. All the constructs were sequenced using an ABI PRISM 377 automatic DNA sequencer (Foster City, CA, United States) to verify 100% correspondence with the original sequence.
Identification of rTgMIF-binding proteins through HuProt microarrays
Human protein microarrays (CDI Labs, United States) containing over 20,000 full-length recombinant human proteins were used to identify rTgMIF-binding proteins. Briefly, the protein microarrays were incubated with a blocking buffer (2% bovine serum albumin in PBS with 0.1% Tween 20) for 2 hours, and 3 μg of biotinylated proteins was treated onto the arrays for 8 hours at 4°C. Subsequently, the arrays were incubated with 1 μg of streptavidin-conjugated Alexa Fluor 532 for 1 hour at 4°C. Microarray results were obtained using a GenePix 4100A microarray laser scanner (Molecular Devices, United States).
GST pulldown, immunoblot, and immunoprecipitation analysis
THP-1, 293T, and BMDMs were treated as indicated and processed for analysis using GST pulldown, Western blotting, and co-IP, as previously described (76, 77). For GST pulldown, 293T cells were harvested and lysed in NP-40 buffer supplemented with a complete protease inhibitor cocktail (Roche, Basel, Switzerland). After centrifugation, the supernatants were precleared with protein A/G beads at 4°C for 2 hours. Precleared lysates were mixed with a 50% slurry of glutathione-conjugated Sepharose beads (Amersham Biosciences, Amersham, United Kingdom), and the binding reaction was incubated for 4 hours at 4°C. The precipitate was washed extensively with a lysis buffer. Proteins bound to the glutathione beads were eluted with an SDS loading buffer by boiling for 5 min. For immunoprecipitation, the cells were harvested and lysed in the NP-40 buffer supplemented with a complete protease inhibitor cocktail (Roche, Basel, Switzerland). After preclearing with protein A/G agarose beads for 1 hour at 4°C, whole-cell lysates were used for immunoprecipitation with the indicated antibodies. Generally, 1 to 4 μg of a commercial antibody was added to 1 ml of cell lysates and incubated at 4°C for 8 to 12 hours. After the addition of protein A/G agarose beads for 6 hours, immunoprecipitates were washed extensively with a lysis buffer and eluted with the SDS loading buffer by boiling for 5 min. For immunoblotting, polypeptides were resolved by SDS–polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Bio-Rad, Hercules, CA, United States). Immunodetection was performed by specific antibodies. Antibody binding was visualized via chemiluminescence (ECL, Millipore, Burlington, MA, United States) and detected using a Vilber chemiluminescence analyzer (Fusion SL 3, Vilber Lourmat).
Enzyme-linked immunosorbent assay
Cell culture supernatants and mouse sera were analyzed for cytokine content using the BD OptEIA ELISA set (BD Pharmingen, Franklin Lakes, NJ, United States) for the detection of TNF-α, IL-6, IL-10, TGF-β, and IFN-γ. All assays were performed according to the manufacturer’s instructions.
PCR array
PCR array was carried out with a StepOnePlus real-time PCR system (ABI) using mouse RT2 Profiler PCR Array kits (Qiagen, Hilden, Germany, PAMM-087Z; PAMM-008Z; PAMM-097Z) or the mouse AccuPower qPCR Array System: Immune qPCR Panel Kit (Bioneer, S-6042-PM2, Daejeon, Korea) with RT2 SYBR Green ROX qPCR Master Mix (Qiagen, 330502). The PCR array kit contained five reference genes, RPLP0, HPRT1, ACTB, B2M, and GAPDH, whose data were analyzed using NormFinder algorithms. On the basis of the result, it was determined that the optimum number of control genes was one, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) ranked first for normalization based on our samples and genes of interest. Subsequently, it was used for quantitative PCR (qPCR) analysis. The expression of 84 genes contributing to female infertility relative to reference GAPDH levels was estimated using the 2−ΔΔCt formula. Each experiment was performed in triplicate.
Peptides
9R-conjugated TgMIF peptides were commercially synthesized and purified in acetate salt form by Peptron (Seoul, Korea) to avoid abnormal cell responses. The amino acid sequences of the peptides used in this study are shown in Figs. 3 to 5 and fig. S5. The endotoxin content, as measured by the Limulus amebocyte lysate assay (BioWhittaker, Walkersville, MD, United States), was less than 3 to 5 pg/ml at the concentrations of peptides used in the experiments.
Measurement of ATP production
ATP levels were measured using the luciferin/luciferase method with an ATP Bioluminescence Assay Kit (PerkinElmer, Shelton, CT, United States, 6016943) according to the manufacturer’s instructions. Briefly, ~2 × 105 BMDMs were infected with MTB and washed with PBS. The ATP release buffer was then added to the cell culture. Next, luciferin and luciferase were mixed (50 μl, final volume) in a separate vial, to which 50 μl of lysate (liberated ATP) from the cell cultures was added. Luminescence was analyzed after a 30-s delay using a FluoroScan luminometer (Labsystems, Helsinki, Finland). An ATP standard curve prepared at the same time according to the manufacturer’s instructions was used to calculate the concentration of ATP in each sample.
mtDNA quantification
To quantify the mtDNA copy number, we measured the mitochondrial (mt) to nuclear (n) DNA ratio, as described previously (78). Pyruvate kinase (Pklr) was used as a marker for nDNA, and the reduced form of NAD+ (NADH) dehydrogenase subunit 1 (mt-Nd1) was used as a marker for mtDNA. Real-time PCR reactions were performed according to the manufacturer’s instructions (QuantiFast SYBR Green PCR Master Mix, Qiagen, 204052), and thermal cycling was performed in a QuantStudio 3 (ABI). The mtDNA content was normalized to the nucleic acid content. The primer pairs used for PCR are as follows: mND1 (197 bp), sense: ggcccattcgcgttattctt, antisense: tcgtaacggaagcgtggata; mPKLR (191 bp), sense: atctacattgacgacgggct, antisense: acattatgctccaccccgaa.
Mitochondrial membrane potential measurements
The mitochondrial membrane potential (ΔΨm) of intact cells was measured as described previously (78), with modifications. The cells were then washed with PBS and trypsinized. The protein concentration of the cells was adjusted to 0.2 mg/ml in DMEM without phenol red (Life Technologies, Invitrogen), FBS, or antibiotics. Tetramethylrhodamine ethyl ester (TMRE) (200 nM; Molecular Probes/Invitrogen, T669) was added to the cell suspension. Cells were incubated at 37°C for 30 min in the dark. ΔΨm was measured using flow cytometry, and the data were analyzed using the FlowJo software. TMRE fluorescence was measured using an FL2 channel (582 nm).
OXPHOS complex activity assay
The activity of oxidative phosphorylation (OXPHOS) complexes was determined using MitoTox Complex I, II-III, IV, and V OXPHOS Activity Microplate Assay kits from Abcam (ab109903, ab109904, ab109905, ab109907, respectively, Cambridge, MA), following the manufacturer’s instructions. The activity of complexes was measured by monitoring the change in absorbance at 340 nm over a period of 1 hour at 30°C. Oligomycin (O4876, Sigma-Aldrich) was used as a positive control.
Metabolic assays
The lactate levels in the medium were determined using the Lactate Assay Kit (Sigma-Aldrich, MAK064) according to the manufacturer’s instructions. Briefly, cells were infected with MTB in the presence or absence of 9R-TgMIF, and the supernatants were then collected and stored at −80°C to inactivate lactate dehydrogenase. The reaction mix was added to the samples, which were then analyzed using a microplate reader [optical density at 570 nm (OD570)]. The NAD+/NADH ratio was measured in whole-cell lysates using the NAD+/NADH Quantification Colorimetric Kit (Sigma-Aldrich, MAK037) according to the manufacturer’s instructions. For real-time analysis of the ECAR and OCR, BMDMs were analyzed using an XF-24 Extracellular Flux Analyzer (Seahorse Bioscience, Santa Clara, CA, United States). In brief, BMDMs were plated in XF-24 cell culture microplates (2 × 105 cells per well in 200 μl) and then infected with MTB in the presence or absence of 9R-TgMIF. At the indicated time points, the medium was removed and the cells were washed and analyzed in an XF Running Buffer [unbuffered RPMI, 10 mM glucose, 10% fetal calf serum, 100 U/ml penicillin/SM, 2 mM l-glutamine, and granulocyte-macrophage colony-stimulating factor (20 ng/ml)] according to the manufacturer’s instructions to analyze the real-time values of the OCR and ECAR. Where indicated, the ECAR and/or OCR were analyzed in response to oligomycin (2 μg/ml), 0.3 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone, 2 μM rotenone, 1 mM antimycin, 10 mM glucose, and 100 mM 2-deoxyglucose (all from Sigma-Aldrich).
Preparation of mycobacterial strains
The H37Rv strain of M. tuberculosis subsp. tuberculosis (ATCC 27294) was obtained from the ATCC (Rockville, MD, United States). MDR-TB (KMRC 00116-00250), INH-resistant TB (KMRC 00120-00137), PZA-resistant TB (KMRC 00130-00064), RIF-resistant TB (KMRC 00121-00341), and SM-resistant TB (KMRC 00122-00123) were purchased from the Korean Mycobacterium Resource Center (KMRC) in the Korean Institute of Tuberculosis (Osong, South Korea). Mycobacteria used in this study were prepared as described previously (77). Briefly, they were cultured in the Middlebrook 7H9 medium (Difco, Sparks, MD, United States, 271310) supplemented with 10% oleic albumin dextrose catalase (OADC; BD Biosciences, San Diego, CA, 212240), 0.5% glycerol, and 0.05% Tween 80 (7H9-OADC) on a rotary shaking incubator (140 rpm) at 37°C to an OD600 of 0.4 to 0.6. All bacteria-related procedures were reviewed and approved by the Institutional Biosafety Committee of Hanyang University (HY-IBC-2022-03).
MTB infection in vitro and in vivo
For the in vitro experiments, cells were infected with MTB for 2 to 4 hours. Then, cells were washed with PBS to remove extracellular bacteria, supplied with a fresh medium, and incubated at 37°C for indicated time points. For in vivo experiments, female specific pathogen–free C57BL/6 mice were 6 to 8 weeks old and age-matched and sex-matched in each experiment. No additional randomization or blinding was performed to allocate the experimental groups. Mice were intranasally injected with MTB (1 × 104 or 5 × 105 CFU per mouse). After 5 weeks, the mice were euthanized, and the lungs, spleens, and livers were harvested. The mice were maintained in biosafety level 3 laboratory facilities. All animal experiments were approved by the Biomedical Research Ethics Committee of the Institute of Microbiology (Chinese Academy of Sciences, Beijing, China).
CFU assay
To analyze bacterial survival in murine macrophages, MTB-infected cells (multiplicity of infection: 1 or 10) were incubated for 4 hours and washed with Dulbecco's Phosphate-Buffered Saline (DPBS) to remove extracellular bacteria. The infected cells were incubated in a fresh medium for the indicated periods. Thereafter, cells were lysed in sterile distilled water for 40 min, and intracellular bacteria were collected. Cell lysates were diluted with DPBS and spotted onto Middlebrook 7H10 agar containing 10% OADC. Colonies were counted to assess the intracellular bacterial viability after 2 to 3 weeks.
Histology
For immunohistochemistry of tissue sections, mouse lungs were fixed in 10% formalin and embedded in paraffin. Paraffin sections (4 μm) were cut and stained with hematoxylin and eosin. Histopathological scores were established based on the numbers and distribution of inflammatory cells and the severity of inflammation within the tissues (79, 80). A board-certified pathologist (M.-K. Kim, Kim Min-Kyung Pathology Clinic, Seoul) independently scored each organ section without prior knowledge of the treatment groups. Histological scores ranging from 0 to 4 were ascribed to each specimen.
Miscellaneous procedures
Details of recombinant protein production, protein purification, MS, flow cytometry, and in vivo imaging are provided in the Supplementary Materials.
Statistical analysis
All data were analyzed using the Student’s t test with Bonferroni adjustment or analysis of variance (ANOVA) for multiple comparisons, and the data are represented as the means ± SD. Statistical analyses were conducted using the SPSS (version 12.0) statistical software (SPSS, Chicago, IL, United States). Differences were considered statistically significant at P < 0.05. For survival, data were graphed and analyzed using the product limit method of Kaplan-Meier, using the log-rank (Mantel-Cox) test for comparisons with GraphPad Prism (version 5.0, La Jolla, CA, United States).
Acknowledgments
We would like to thank all members of the Infection Biology Lab for critically reading and discussing the manuscript.
Funding: This work was supported by a National Research Foundation of Korea grant funded by the Korean government (MSIP) (grant nos. 2019R1I1A2A01064237 and 2021R1A4A5032463) and a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health and Welfare, Republic of Korea (HI22C0884). C.Y. was supported by a research fund from Hanyang University (HY-2024-0554).
Author contributions: C.Y., H.K.K., Y.S.H., W.J.G., S.-J.M., and E.C. performed molecular and animal experiments and analyzed the data. J.-M.Y. contributed to the conception and critically reviewed the manuscript. C.-S.Y. designed and conceptualized the study, supervised the experimental work, analyzed the data, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Experimental Procedures
REFERENCES AND NOTES
- 1.Barberis I., Bragazzi N. L., Galluzzo L., Martini M., The history of tuberculosis: From the first historical records to the isolation of Koch's bacillus. J. Prev. Med. Hyg. 58, E9–E12 (2017). [PMC free article] [PubMed] [Google Scholar]
- 2.Quenelle D. C., Winchester G. A., Staas J. K., Barrow E. L., Barrow W. W., Treatment of tuberculosis using a combination of sustained-release rifampin-loaded microspheres and oral dosing with isoniazid. Antimicrob. Agents Chemother. 45, 1637–1644 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R., Dwivedi S. P., Gaharwar U. S., Meena R., Rajamani P., Prasad T., Recent updates on drug resistance in Mycobacterium tuberculosis. J. Appl. Microbiol. 128, 1547–1567 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Schildknecht K. R., Pratt R. H., Feng P. I., Price S. F., Self J. L., Tuberculosis—United States, 2022. MMWR Morb. Mortal. Wkly Rep. 72, 297–303 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delogu G., Sali M., Fadda G., The biology of Mycobacterium tuberculosis infection. Mediterr. J. Hematol. Infect. Dis. 5, e2013070 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bo H., Moure U. A. E., Yang Y., Pan J., Li L., Wang M., Ke X., Cui H., Mycobacterium tuberculosis-macrophage interaction: Molecular updates. Front. Cell. Infect. Microbiol. 13, 1062963 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautam K., Negi S., Saini V., Targeting endogenous gaseous signaling molecules as novel host-directed therapies against tuberculosis infection. Free Radic. Res. 55, 655–670 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Lee K. I., Choi S., Choi H. G., Kebede S. G., Dang T. B., Back Y. W., Park H. S., Kim H. J., Recombinant Rv3261 protein of Mycobacterium tuberculosis induces apoptosis through a mitochondrion-dependent pathway in macrophages and inhibits intracellular bacterial growth. Cell. Immunol. 354, 104145 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Maphasa R. E., Meyer M., Dube A., The macrophage response to Mycobacterium tuberculosis and opportunities for autophagy inducing nanomedicines for tuberculosis therapy. Front. Cell. Infect. Microbiol. 10, 618414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bar-Oz M., Meir M., Barkan D., Virulence-associated secretion in Mycobacterium abscessus. Front. Immunol. 13, 938895 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J. H., Shim D., Kim K. E. S., Lee W., Shin S. J., Understanding metabolic regulation between host and pathogens: New opportunities for the development of improved therapeutic strategies against Mycobacterium tuberculosis infection. Front. Cell. Infect. Microbiol. 11, 635335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramon-Luing L. A., Palacios Y., Ruiz A., Téllez-Navarrete N. A., Chavez-Galan L., Virulence factors of Mycobacterium tuberculosis as modulators of cell death mechanisms. Pathogens 12, 839 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dartois V. A., Rubin E. J., Anti-tuberculosis treatment strategies and drug development: Challenges and priorities. Nat. Rev. Microbiol. 20, 685–701 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guirado E., Schlesinger L. S., Kaplan G., Macrophages in tuberculosis: Friend or foe. Semin. Immunopathol. 35, 563–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khanna S., Sims M., Louie T. J., Fischer M., LaPlante K., Allegretti J., Hasson B. R., Fonte A. T., McChalicher C., Ege D. S., Bryant J. A., Straub T. J., Ford C. B., Henn M. R., Wang E. E. L., von Moltke L., Wilcox M. H., SER-109: An oral investigational microbiome therapeutic for patients with recurrent Clostridioides difficile infection (rCDI). Antibiotics 11, 1234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Summers R. W., Elliott D. E., Urban J. F. Jr., Thompson R., Weinstock J. V., Trichuris suis therapy in Crohn's disease. Gut 54, 87–90 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djurković-Djaković O., Dupouy-Camet J., Van der Giessen J., Dubey J. P., Toxoplasmosis: Overview from a One Health perspective. Food Waterborne Parasitol. 15, e00054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abo-Al-Ela H. G., Toxoplasmosis and psychiatric and neurological disorders: A step toward understanding parasite pathogenesis. ACS Chem. Nerosci. 11, 2393–2406 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Daher D., Shaghlil A., Sobh E., Hamie M., Hassan M. E., Moumneh M. B., Itani S., El Hajj R., Tawk L., El Sabban M., El Hajj H., Comprehensive overview of Toxoplasma gondii-induced and associated diseases. Pathogens 10, 1351 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lima T. S., Lodoen M. B., Mechanisms of human innate immune evasion by Toxoplasma gondii. Front. Cell. Infect. Microbiol. 9, 103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasai M., Yamamoto M., Innate, adaptive, and cell-autonomous immunity against Toxoplasma gondii infection. Exp. Mol. Med. 51, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J. S., Lee D., Kim D., Mun S. J., Cho E., Son W., Yang C. S., Toxoplasma gondii GRA8-derived peptide immunotherapy improves tumor targeting of colorectal cancer. Oncotarget 11, 62–73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J. S., Mun S. J., Cho E., Kim D., Son W., Jeon H. I., Kim H. K., Jang K., Yang C. S., Toxoplasma gondii GRA9 regulates the activation of NLRP3 inflammasome to exert anti-septic effects in mice. Int. J. Mol. Sci. 21, 8437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y. R., Kim J. S., Yun J. S., Kim S., Kim S. Y., Jang K., Yang C. S., Toxoplasma gondii GRA8 induces ATP5A1-SIRT3-mediated mitochondrial metabolic resuscitation: A potential therapy for sepsis. Exp. Mol. Med. 50, e464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koh H. J., Kim Y. R., Kim J. S., Yun J. S., Jang K., Yang C. S., Toxoplasma gondii GRA7-targeted ASC and PLD1 promote antibacterial host defense via PKCα. PLOS Pathog. 13, e1006126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomita T., Guevara R. B., Shah L. M., Afrifa A. Y., Weiss L. M., Secreted effectors modulating immune responses to Toxoplasma gondii. Life 11, 988 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris J., VanPatten S., Deen N. S., Al-Abed Y., Morand E. F., Rediscovering MIF: New tricks for an old cytokine. Trends Immunol. 40, 447–462 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Tierney T., Patel R., Stead C. A., Leng L., Bucala R., Buckingham J. C., Macrophage migration inhibitory factor is released from pituitary folliculo-stellate-like cells by endotoxin and dexamethasone and attenuates the steroid-induced inhibition of interleukin 6 release. Endocrinology 146, 35–43 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Gil-Yarom N., Radomir L., Sever L., Kramer M. P., Lewinsky H., Bornstein C., Blecher-Gonen R., Barnett-Itzhaki Z., Mirkin V., Friedlander G., Shvidel L., Herishanu Y., Lolis E. J., Becker-Herman S., Amit I., Shachar I., CD74 is a novel transcription regulator. Proc. Natl. Acad. Sci. U.S.A. 114, 562–567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budarf M., McDonald T., Sellinger B., Kozak C., Graham C., Wistow G., Localization of the human gene for macrophage migration inhibitory factor (MIF) to chromosome 22q11.2. Genomics 39, 235–236 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Calandra T., Roger T., Macrophage migration inhibitory factor: A regulator of innate immunity. Nat. Rev. Immunol. 3, 791–800 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh S., Jiang N., Farr L., Ngobeni R., Moonah S., Parasite-produced MIF cytokine: Role in immune evasion, invasion, and pathogenesis. Front. Immunol. 10, 1995 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sommerville C., Richardson J. M., Williams R. A., Mottram J. C., Roberts C. W., Alexander J., Henriquez F. L., Biochemical and immunological characterization of Toxoplasma gondii macrophage migration inhibitory factor. J. Biol. Chem. 288, 12733–12741 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., Wang J., Zhou N., Fang Q., Cai H., Du Z., An R., Liu D., Chen X., Wang X., Li F., Yan Q., Chen L., Du J., Protozoan-derived cytokine-transgenic macrophages reverse hepatic fibrosis. Adv. Sci. 11, e2308750 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou N., Chen Y., Wang J., An R., Cai H., Liu S., Yao L., Tang Y., Chen L., Du J., TgMIF promotes hepatocyte pyroptosis and recruitment of proinflammatory macrophages during severe liver injury in acute toxoplasmosis. J. Infect. Dis. 227, 1417–1427 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Noe J. T., Mitchell R. A., MIF-dependent control of tumor immunity. Front. Immunol. 11, 609948 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lian J., Liang Y., Zhang H., Lan M., Ye Z., Lin B., Qiu X., Zeng J., The role of polyamine metabolism in remodeling immune responses and blocking therapy within the tumor immune microenvironment. Front. Immunol. 13, 912279 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Jiang T., Li M. Q., Zheng X. L., Zhao G. J., Transcriptional regulation of macrophages polarization by MicroRNAs. Front. Immunol. 9, 1175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou H. X., Pang X., Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 118, 1691–1741 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuettpelz J., Janer A., Antonicka H., Shoubridge E. A., The role of the mitochondrial outer membrane protein SLC25A46 in mitochondrial fission and fusion. Life Sci. Alliance 6, e202301914 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruprecht J. J., Kunji E. R. S., The SLC25 mitochondrial carrier family: Structure and mechanism. Trends Biochem. Sci. 45, 244–258 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.N. V. Yang, S. Rogers, R. Guerra, D. J. Pagliarini, E. Theusch, R. M. Krauss, TOMM40 and TOMM22 of the Translocase Outer Mitochondrial Membrane Complex rescue statin-impaired mitochondrial dynamics, morphology, and mitophagy in skeletal myotubes. bioRxiv 2023.06.24.546411 [Preprint] (2023). 10.1101/2023.06.24.546411. [DOI]
- 43.Sies H., Jones D. P., Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 21, 363–383 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Novak M. L., Koh T. J., Macrophage phenotypes during tissue repair. J. Leukoc. Biol. 93, 875–881 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Chaffee T. S., LaRue R. S., Huggins D. N., Witschen P. M., Ibrahim A. M., Nelson A. C., Machado H. L., Schwertfeger K. L., Tissue-resident macrophages promote extracellular matrix homeostasis in the mammary gland stroma of nulliparous mice. eLife 9, e57438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Funes S. C., Rios M., Escobar-Vera J., Kalergis A. M., Implications of macrophage polarization in autoimmunity. Immunology 154, 186–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao X., van den Hil F. E., Mummery C. L., Orlova V. V., Generation and functional characterization of monocytes and macrophages derived from human induced pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 52, e108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han S., Bao X., Zou Y., Wang L., Li Y., Yang L., Liao A., Zhang X., Jiang X., Liang D., Dai Y., Zheng Q. C., Yu Z., Guo J., d-lactate modulates M2 tumor-associated macrophages and remodels immunosuppressive tumor microenvironment for hepatocellular carcinoma. Sci. Adv. 9, eadg2697 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinhäuser I., Pereira-Martins D. A., Almeida L. Y., Hilberink J. R., Silveira D. R. A., Quek L., Ortiz C., Araujo C. L., Bianco T. M., Lucena-Araujo A., Mota J. M., Hogeling S. M., Sternadt D., Visser N., Diepstra A., Ammatuna E., Huls G., Rego E. M., Schuringa J. J., M2 macrophages drive leukemic transformation by imposing resistance to phagocytosis and improving mitochondrial metabolism. Sci. Adv. 9, eadf8522 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boshoff H. I. M., Warner D. F., Gold B., Editorial: Drug-resistant Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 13, 1215294 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edwards B. D., Field S. K., The struggle to end a millennia-long pandemic: Novel candidate and repurposed drugs for the treatment of tuberculosis. Drugs 82, 1695–1715 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandes G. F. S., Thompson A. M., Castagnolo D., Denny W. A., Dos Santos J. L., Tuberculosis drug discovery: Challenges and new horizons. J. Med. Chem. 65, 7489–7531 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Song W. M., Li Y. F., Ma X. B., Liu J. Y., Tao N. N., Liu Y., Zhang Q. Y., Xu T. T., Li S. J., Yu C. B., Gao L., Cui L. L., Li H. C., Primary drug resistance of Mycobacterium tuberculosis in Shandong, China, 2004–2018. Respir. Res. 20, 223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arrigoni R., Ballini A., Topi S., Bottalico L., Jirillo E., Santacroce L., Antibiotic resistance to Mycobacterium tuberculosis and potential use of natural and biological products as alternative anti-mycobacterial agents. Antibiotics 11, 1431 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang Y. S., Shin J. H., Yang J. P., Jung B. K., Lee S. H., Shin E. H., Characteristics of infection immunity regulated by Toxoplasma gondii to maintain chronic infection in the brain. Front. Immunol. 9, 158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin E. H., Chun Y. S., Kim W. H., Kim J. L., Pyo K. H., Chai J. Y., Immune responses of mice intraduodenally infected with Toxoplasma gondii KI-1 tachyzoites. Korean J. Parasitol. 49, 115–123 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim W. H., Shin E. H., Kim J. L., Yu S. Y., Jung B. K., Chai J. Y., Suppression of CD4 T-Cells in the spleen of mice infected with Toxoplasma gondii KI-1 tachyzoites. Korean J. Parasitol. 48, 325–329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S. G., Seo S. H., Shin J. H., Yang J. P., Lee S. H., Shin E. H., Increase in the nuclear localization of PTEN by the Toxoplasma GRA16 protein and subsequent induction of p53-dependent apoptosis and anticancer effect. J. Cell. Mol. Med. 23, 3234–3245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seo S. H., Kim S. G., Shin J. H., Ham D. W., Shin E. H., Toxoplasma GRA16 inhibits NF-κB activation through PP2A-B55 upregulation in non-small-cell lung carcinoma cells. Int. J. Mol. Sci. 21, 6642 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seo S. H., Shin J. H., Ham D. W., Shin E. H., PTEN/AKT signaling pathway related to hTERT downregulation and telomere shortening induced in Toxoplasma GRA16-expressing colorectal cancer cells. Biomed. Pharmacother. 153, 113366 (2022). [DOI] [PubMed] [Google Scholar]
- 61.Farr L., Ghosh S., Moonah S., Role of MIF cytokine/CD74 receptor pathway in protecting against injury and promoting repair. Front. Immunol. 11, 1273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitchell R. A., Metz C. N., Peng T., Bucala R., Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic phospholipase A2 activation by macrophage migration inhibitory factor (MIF). Regulatory role in cell proliferation and glucocorticoid action. J. Biol. Chem. 274, 18100–18106 (1999). [DOI] [PubMed] [Google Scholar]
- 63.Gomes A. O., Barbosa B. F., Franco P. S., Ribeiro M., Silva R. J., Gois P. S. G., Almeida K. C., Angeloni M. B., Castro A. S., Guirelli P. M., Cândido J. V., Chica J. E. L., Silva N. M., Mineo T. W. P., Mineo J. R., Ferro E. A. V., Macrophage migration inhibitory factor (MIF) prevents maternal death, but contributes to poor fetal outcome during congenital toxoplasmosis. Front. Microbiol. 9, 906 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oddo M., Calandra T., Bucala R., Meylan P. R., Macrophage migration inhibitory factor reduces the growth of virulent Mycobacterium tuberculosis in human macrophages. Infect. Immun. 73, 3783–3786 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orme I. M., Cooper A. M., Cytokine/chemokine cascades in immunity to tuberculosis. Immunol. Today 20, 307–312 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Das R., Koo M. S., Kim B. H., Jacob S. T., Subbian S., Yao J., Leng L., Levy R., Murchison C., Burman W. J., Moore C. C., Scheld W. M., David J. R., Kaplan G., MacMicking J. D., Bucala R., Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, E2997–E3006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mohareer K., Banerjee S., Mycobacterial infection alters host mitochondrial activity. Int. Rev. Cell Mol. Biol. 377, 87–119 (2023). [DOI] [PubMed] [Google Scholar]
- 68.Lee J., Choi J. A., Cho S. N., Son S. H., Song C. H., Mitofusin 2-deficiency suppresses Mycobacterium tuberculosis survival in macrophages. Cells 8, 1355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ning Y., Cai Y., Dai Y., Li F., Mo S., Werz O., Chen X., Mitochondrial fusion mediated by mitofusin 1 regulates macrophage mycobactericidal activity by enhancing autophagy. Infect. Immun. 89, e0030621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cumming B. M., Addicott K. W., Adamson J. H., Steyn A. J., Mycobacterium tuberculosis induces decelerated bioenergetic metabolism in human macrophages. eLife 7, e39169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mengie Ayele T., Tilahun Muche Z., Behaile Teklemariam A., Bogale Kassie A., Chekol Abebe E., Role of JAK2/STAT3 signaling pathway in the tumorigenesis, chemotherapy resistance, and treatment of solid tumors: A systemic review. J. Inflamm. Res. 15, 1349–1364 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Godava M., Vrtel R., Vodicka R., STAT6—Polymorphisms, haplotypes and epistasis in relation to atopy and asthma. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech Repub. 157, 172–180 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Hüntelmann B., Staab J., Herrmann-Lingen C., Meyer T., A conserved motif in the linker domain of STAT1 transcription factor is required for both recognition and release from high-affinity DNA-binding sites. PLOS ONE 9, e97633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manoharan S., Balakrishnan A., Hemamalini V., Perumal E., Screening of potent STAT3-SH2 domain inhibitors from JAK/STAT compound library through molecular dynamics simulation. Mol. Divers. 27, 1297–1308 (2023). [DOI] [PubMed] [Google Scholar]
- 75.Parisi L., Gini E., Baci D., Tremolati M., Fanuli M., Bassani B., Farronato G., Bruno A., Mortara L., Macrophage polarization in chronic inflammatory diseases: Killers or builders? J. Immunol. Res. 2018, 8917804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim J. S., Kim H. K., Lee J., Jang S., Cho E., Mun S. J., Yoon S., Yang C. S., Inhibition of CD82 improves colitis by increasing NLRP3 deubiquitination by BRCC3. Cell. Mol. Immunol. 20, 189–200 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koh H. J., Kim Y. R., Kim J. S., Yun J. S., Kim S., Kim S. Y., Jang K., Yang C. S., CD82 hypomethylation is essential for tuberculosis pathogenesis via regulation of RUNX1-Rab5/22. Exp. Mol. Med. 50, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang C. S., Kim J. J., Lee H. M., Jin H. S., Lee S. H., Park J. H., Kim S. J., Kim J. M., Han Y. M., Lee M. S., Kweon G. R., Shong M., Jo E. K., The AMPK-PPARGC1A pathway is required for antimicrobial host defense through activation of autophagy. Autophagy 10, 785–802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J. K., Kim Y. S., Lee H. M., Jin H. S., Neupane C., Kim S., Lee S. H., Min J. J., Sasai M., Jeong J. H., Choe S. K., Kim J. M., Yamamoto M., Choy H. E., Park J. B., Jo E. K., GABAergic signaling linked to autophagy enhances host protection against intracellular bacterial infections. Nat. Commun. 9, 4184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buchweitz J. P., Karmaus P. W., Harkema J. R., Williams K. J., Kaminski N. E., Modulation of airway responses to influenza A/PR/8/34 by Delta9-tetrahydrocannabinol in C57BL/6 mice. J. Pharmacol. Exp. Ther. 323, 675–683 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Experimental Procedures