Abstract
Human immunodeficiency virus (HIV) type 1 encodes an essential protein, Rev, which functions to transport unspliced and singly spliced viral transcripts from the nucleus to the cytoplasm to allow expression of the viral structural proteins. It has previously been reported that Sam68 synergistically stimulates Rev activity (T. Reddy et al., Nat. Med. 5:635–642, 1999). Here we report that the Sam68-like mammalian proteins SLM1 and SLM2 also stimulate Rev activity. Their stimulation ability cannot be attributed to a shuttling property, since Sam68, SLM1, and SLM2 do not display significant shuttling activity alone or in the presence of Rev. In addition, Sam68, SLM1, and SLM2 do not affect the equilibrium between unspliced and completely spliced HIV RNA. The C-terminally truncated Sam68 mutant (Sam68ΔC) previously observed to inhibit the Sam68-mediated stimulation of Rev activity (Reddy et al., 1999) also inhibits SLM1- and SLM2-mediated stimulation of Rev activity. This suggests that the mechanism by which Sam68, SLM1, and SLM2 stimulate Rev activity may be common. Sam68ΔC does not inhibit Rev activity by inhibiting Rev from shuttling between the nucleus and cytoplasm. Inhibition by Sam68ΔC is a consequence of its mislocalization to the cytoplasm, as evidenced by the fact that addition of an exogenous nuclear localization signal to Sam68ΔC restores nuclear localization and stimulation of Rev activity. We demonstrate that Sam68ΔC causes perinuclear accumulation of unspliced HIV env RNA and propose that Sam68ΔC inhibits Rev activity by sequestering Rev-responsive RNA away from the translation apparatus.
Expression of the full complement of human immunodeficiency virus type 1 (HIV-1) proteins requires that incompletely spliced viral transcripts be transported from the nucleus to the cytoplasm for translation into the structural proteins of the virus (10, 12, 19, 28). Nuclear export of the incompletely spliced transcripts is achieved through the action of an essential HIV accessory protein, Rev (11, 43). Expressed from fully spliced viral RNA, Rev localizes to the nucleus through the interaction of its nuclear localization signal (NLS) (5, 21, 27, 35) with the import receptor importin β (49). Once in the nucleus, Rev interacts with the incompletely spliced viral transcripts. This interaction is mediated by a 240-nucleotide sequence, termed the Rev responsive element (RRE), present within the terminal intron of the incompletely spliced and unspliced viral transcripts (3, 18, 20, 28, 40, 56). Nuclear export of the Rev-RNA complex is then mediated by interaction of the Rev nuclear export signal (NES) (13, 31) with the nuclear export receptor CRM1 (14, 45). Translation of the unspliced and singly spliced viral transcripts results in production of the structural proteins, and some accessory proteins of the virus (6). Rev thus acts as a regulator of the switch from early to late viral gene expression (9, 25).
It has recently been reported that Sam68, the 68-kDa Src-associated substrate during mitosis (15, 47), is a functional homologue of Rev and a synergistic activator of Rev activity (36). Furthermore, it has been demonstrated that a C-terminal deletion of Sam68 resulted in a transdominant-negative mutant (Sam68ΔC) that can inhibit Rev activity and HIV replication (36). Sam68 associates with various SH2 and SH3 domain-containing signaling molecules and is a substrate for various cellular tyrosine kinases (7, 32, 48, 53). Sam68 also contains a KH domain, an RNA binding motif originally defined in hnRNP K (16, 42). It has been shown to bind single-stranded RNA, homopolymeric RNA, and single-stranded and double-stranded DNA in vitro (47, 51, 54), but its specific RNA target is as yet unknown. Sam68 is also capable of self-association in an RNA and KH domain-dependent manner (1). The KH motif is embedded in a larger motif, the GSG domain, which is found in a growing family of proteins, including two Sam68-like mammalian proteins, SLM1 and SLM2 (8). Like Sam68, both SLM1 and SLM2 contain proline-rich motifs, arginine-glycine repeats, and a C-terminal tyrosine-rich region. Moreover, they both also have RNA binding properties and can self-associate as well as heterodimerize with Sam68 (8).
Here we report that the Sam68-like proteins SLM1 and SLM2 can functionally replace Rev under some circumstances, that they stimulate Rev activity, and that stimulation by both is also inhibited by Sam68ΔC. To explore the mechanisms of Sam68- and SLM-mediated stimulation and Sam68ΔC-mediated inhibition of Rev activity, we have examined their abilities to shuttle, to transport Rev-responsive RNA, and to affect HIV RNA metabolism. We have determined that Sam68, SLM1, and SLM2 do not display significant shuttling activity and that Sam68ΔC does not impede the shuttling behavior of Rev. Furthermore, stimulation of Rev activity by Sam68, SLM1, and SLM2 does not work by increasing the abundance of target RRE-RNA available for Rev to export. We demonstrate that inhibition of Rev activity by Sam68ΔC is a consequence of its mislocalization to the cytoplasm, as addition of an NLS to Sam68ΔC restores nuclear localization of the fusion protein and stimulation of Rev activity. Finally, we demonstrate here that Sam68ΔC inhibits Rev function by localizing the unspliced Rev-responsive RNA to perinuclear bundles, hindering the RRE transcript from being translated.
MATERIALS AND METHODS
Expression constructs.
The following plasmids have been described previously: BlSVhygro, SVH6Rev (34), SVH6M10 (33), pDM128 (21), BlCMVCATpA, pgTat (28), BlenvHindIII (41), pgGP160 puro (46), and pcDNA3.1 (Invitrogen). pcDNA3.1-based Myc-tagged Sam68, Sam68ΔC, Sam68 FmR:184I→N, Sam68ΔL1, Sam68ΔL4, Sam68 Gld:178G→D, SLM1, and SLM2 have also been described previously (2, 8). FmRΔC, ΔL1ΔC, ΔL4ΔC, and GldΔC were all created by XhoI digestion of the respective parent plasmid to remove the region encoding the C terminus of Sam68 and subsequent ligation of the remaining backbone.
Myc-tagged NLSSam68ΔC was constructed by amplification of the Sam68ΔC reading frame with primers 5′ Sam PstI (5′-AAC TGC AGC CCA GCG CCG GGA CGA TCCT-3′) and 3′ pcDNA3 (5′-CGG GAT CCT AGA AGG CAC AGT CGA GG-3′) using Sam68ΔC as the template. The amplicon was digested with PstI/BamHI and used to replace the Rev gene in the B1-SVNLSRev construct (44). The resulting construct was then excised with EcoRI/NotI and cloned into B1-CMVmycpA to add the Myc tag to the amino terminus of the protein, generating B1-CMV mycNLS Sam68ΔC pA. To provide probe for the RNase protection assays, the 336-bp HindIII/BamHI fragment of pgGP160 puro was cloned into Bluescript-SK (Stratagene).
pSPVA and 5′VA probe plasmids have been described previously (55) and were generously provided by B. Blencowe with the approval of D. L. Bentley. Gag-RRE has been described previously (50) and was generously provided by S. Venkatesan.
Cell lines, transfections, heterokaryon assays, and chloramphenicol acetyltransferase (CAT) assays.
HeLa and 293 cells were maintained in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% fetal bovine serum (FBS), 50 μg of gentamicin sulfate/ml, and 2.5 μg of amphotericin B/ml. For transient expression studies, vectors were introduced by calcium phosphate transfection (24). Two days posttransfection, cells were harvested.
Transfections to analyze the effect of Sam68, SLM1, SLM2, or Sam68-based mutants on Rev function were carried out in triplicate, per condition, as follows (amounts indicated are for 105 293 cells): 0.125 μg of pDM128, 0.025 μg of BISVhygro or B1SVH6Rev, 0.5 μg of pcDNA3.1 or the myc-tagged Sam68/SLM-based vectors as described in Results. Transfections to analyze the effect of Sam68ΔC on Sam68-, SLM1-, or SLM2-mediated stimulation of Rev function were carried out in triplicate, per condition, as follows (amounts indicated are for 105 293 cells): 0.125 μg of pDM128, 0.025 μg of B1SVhygro or B1SVH6Rev, 0.1 μg of Sam68, SLM1, or SLM2, and 1 μg of Sam68ΔC as described in Results. DNA was equalized to 1.25 μg per transfection with pcDNA3.1. CAT assays were performed as previously described (17). Conversions were normalized by protein concentration determined by Bradford assay (Bio-Rad).
Transfections for heterokaryon assays were carried out on 105 HeLa cells as follows: 2 μg of BISVH6M10, BISVH6Rev, myc-Sam68, myc-Sam68ΔC, myc-SLM1, or myc-SLM2. At 24 h after transfection, the cells were split and plated together as follows: M10 plus myc-Sam68, M10 plus myc-Sam68ΔC, M10 plus myc-SLM1, M10 plus myc-SLM2, Rev plus myc-Sam68, Rev plus myc-Sam68ΔC, Rev plus myc-SLM1, or Rev plus myc-SLM2. At 24 h after coplating, fusion was carried out as previously described (44).
Transfections for in situ hybridization were performed on 3 × 105 HeLa cells as follows: 1.25 μg of pgTat or pgGP160puro, 0.25 μg of B1SVhygro or B1SVH6Rev, and 5 μg of pcDNA3.1 or the myc-tagged Sam68, Sam68ΔC, SLM1, SLM2, or NLSSam68ΔC vector as described in Results. DNA was equalized to 6.5 μg per transfection.
Transfections for harvesting RNA for RNase protection analyses were performed on 107 293 cells as follows: 5 μg of pgGP160 puro, 1 μg of pSPVA, 1 μg of B1SVhygro or B1SVH6Rev, and 20 μg of pcDNA3.1 or myc-tagged Sam68, SLM1, or SLM2 vector as described in Results. pSPVA is a polIII reporter that was used as an internal control for monitoring transfection efficiency. DNA was equalized to 27 μg per transfection.
Transfections for Western blotting of myc-tagged proteins and Rev were carried out as follows on 106 293 cells: 0.5 μg of pDM128, 0.1 μg of B1SVhygro or B1SVH6Rev, and 2 μg of pcDNA3.1 or myc-tagged Sam68-based vectors as described in Results. DNA was equalized to 2.6 μg per transfection. Transfections for enzyme-linked immunosorbent assay (ELISA) analysis of p24 expression were carried out as follows on 105 293 cells: 1 μg of Gag-RRE, 0.1 μg of B1SVhygro or BISVH6Rev, 1 μg of pSVexTat, and 2 μg of myc-tagged Sam/SLM vectors as described in Results.
Immunofluorescence and in situ hybridization.
Cells grown on coverslips were processed for immunofluorescence as previously described (35). A rabbit polyclonal antibody was used to detect Rev. A monoclonal antibody to the Myc epitope (Invitrogen) was used to detect myc-tagged proteins. Fluorescein-labeled anti-rabbit and Texas Red-labeled anti-mouse antibodies (Jackson ImmunoResearch) were used to detect the polyclonal and monoclonal antibodies, respectively. Immunofluorescence was observed using a Leica DMR microscope at a magnification of either ×400 or ×630.
In situ hybridization was performed as previously described (41). A digoxigenin-labeled Env-HindIII probe, antisense to HIV-1 env mRNA, was used to probe unspliced HIV RNA. The probes were synthesized using the digoxigenin RNA labeling kit (Roche) to transcribe XhoI-digested BlenvHindIII. Cotransfected myc-tagged proteins were detected with the monoclonal anti-myc antibody described above, while Rev was detected with the polyclonal anti-Rev antibody described above. A fluorescein isothiocyanate (FITC)-conjugated sheep anti-digoxigenin antibody (Boehringer Mannheim), a Texas Red-labeled anti-mouse antibody, and a Texas Red-labeled anti-rabbit antibody were used to detect the antisense probes, myc-tagged proteins, and Rev, respectively.
Western blotting and ELISAs.
Total cell lysates for probing Sam/SLM, Rev, and gp160/120 abundance were prepared by washing cells with phosphate-buffered saline (PBS) and harvesting in radioimmunoprecipitation assay (RIPA) buffer (4). Total lysates were fractionated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels with 5% (anti-gp 120), 10% (anti-myc) or 15% (anti-Rev blots) polyacrylamide, transferred to polyvinylidene difluoride (PVDF) membranes (Schleicher and Schuell) by wet immunotransfer, and processed for Western blotting. Blots were probed with an antibody to gp120 (generously provided by D. Branch), the myc-epitope tag, or Rev. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (Jackson ImmunoResearch, Bio/Can Scientific) and the ECL kit (Amersham) were used to detect bound antibodies. Chemiluminescence was detected by exposure to X-Omat AR film (Kodak). ELISAs for p24 were performed according to the manufacturer's instructions (Beckman Coulter).
RNase protection assays.
Nuclear and cytoplasmic fractions of RNA were harvested from 293 cells 48 h after transfection as previously described (22, 30) with the following modifications. Cells were lifted by 2 mM EDTA treatment, and EDTA was added to the hypotonic lysis buffer to a final concentration of 10 mM in order to remove any remaining polysomal RNA from the nuclear fraction (52). The template for the gp160 probe synthesis, Bl-env H/B (containing the HindIII/BamHI fragment of Hxb2 env), was linearized with XhoI (New England Biolabs) and transcribed with T3 RNA polymerase (MBI Fermentas) in the presence of [32P]UTP (Amersham). A riboprobe against control VA RNA was synthesized as previously described (30). To gel purify the probes, they were run on 4% polyacrylamide–8 M urea gels, cut out, and eluted into 0.5 M ammonium acetate–0.1% SDS–1 mM EDTA. Ten micrograms of RNA and ∼100,000 cpm of each probe were hybridized at 49°C overnight. The unhybridized RNA was subsequently digested with RNase T1 (Sigma) and RNase A (Boehringer Mannheim) as previously described (30). To visualize the protected RNA, the resultant RNA was run out on 8% polyacrylamide–8 M urea gels. The gels were dried, exposed to phosphor screens, and scanned using a PhosphorImager (both from Molecular Dynamics). Bands were quantitated using ImageQuant.
RESULTS
The Sam68-like mammalian proteins SLM1 and SLM2 synergistically activate Rev-dependent gene expression.
It has previously been reported that Sam68 can synergistically activate Rev-dependent gene expression as well as inducing viral structural protein expression in the absence of Rev (36). In those studies, deletions of the N-terminal (Δ42–329), C-terminal (Δ330–443 and Δ410–443), and KH domains all failed to enhance RRE-mediated transactivation (36). Since SLM1 and SLM2 have high sequence identity with Sam68 in their GSG regions (72 and 69%, respectively), which include the KH domain (8), we wanted to determine whether they, too, could replace and/or stimulate Rev activity. The ability of exogenously expressed Sam68, SLM1, and SLM2 to replace and/or enhance Rev function was assessed and compared using an RRE-regulated CAT reporter gene, pDM128. Transport to the cytoplasm and expression of the intronic CAT gene is Rev and RRE dependent, as previously described (21). In the absence of Rev, Sam68, SLM1, and SLM2 were found to induce expression of CAT from pDM128 (Fig. 1A). The NES mutant of Rev, M10, and Sam68ΔC were found to have no effect. To test the effect of these proteins on Rev function, 293 cells were transiently transfected with pDM128, Rev, and Sam68, SLM1, or SLM2. As previously reported, Sam68 stimulates Rev activity in this functional assay when coexpressed (Fig. 1B). SLM1 and SLM2 also stimulate Rev activity and do so to an extent similar to that with Sam68, causing a 6- to 10-fold increase in CAT activity compared to cells transfected with Rev alone (Fig. 1B). Stimulation of Rev transactivation by Sam68, SLM1, and SLM2 is dose dependent (6- to 10-fold induction with 20:1 overexpression of the Sams/SLMs in Fig. 1B relative to a 2- to 4-fold induction with 4:1 overexpression in Fig. 1C). The effects observed are not due to a general effect on gene expression, as neither SLM1 nor SLM2 significantly enhances CAT activity from the Rev-independent reporter (Bl-CMVCATpA) (Fig. 1B).
FIG. 1.
SLM1 and SLM2 synergize with Rev in RRE-directed reporter gene expression. (A) SLM1 and SLM2 are able to stimulate HIV-1 gene expression in the absence of Rev. Approximately 105 COS-7 cells were transfected with 1.0 μg of the RRE-CAT reporter plasmid (pDM128) in the presence or absence of 1 μg of expression vector expressing Rev, M10, Sam68, Sam68ΔC, SLM1, or SLM2. At 48 h after transfection, cells were harvested and CAT assays were performed. (B) SLM1 and SLM2, like Sam68, stimulate Rev-dependent gene expression. Approximately 105 293 cells were transfected with 0.125 μg of the RRE-CAT reporter plasmid (pDM128) or the Bl-CMVCATpA reporter in the presence or absence of 0.025 μg of Rev expression plasmid and indicated expression vectors (0.5 μg). At 48 h after transfection, cells were harvested and CAT assays were performed. (C) Sam68ΔC inhibits Rev activity as well as SLM1, SLM2, and Sam68 stimulation of Rev-dependent gene expression. Approximately 105 293 cells were transfected with 0.125 μg of pDM128, 0.025 μg of Rev, 0.1 μg of Sam68, SLM1, or SLM2, and 1 μg of Sam68ΔC.
It was also previously demonstrated that the C-terminally truncated Sam68 mutants not only are unable to stimulate Rev-mediated gene expression but also act as transdominant-negative inhibitors of Sam68-stimulated Rev activity (36). We therefore wished to assess whether SLM1- and SLM2-mediated stimulation of Rev activity are also inhibited by Sam68ΔC (Δ347–443). Indeed, coexpression of Sam68ΔC does inhibit the synergistic activation of Rev-dependent gene expression by SLM1 and SLM2 (Fig. 1C), suggesting that they share, with Sam68, a common mechanism for stimulation of Rev activity. Inhibition of Rev activity by the transdominant mutant was complete, resulting in background activity akin to that observed when Rev is absent.
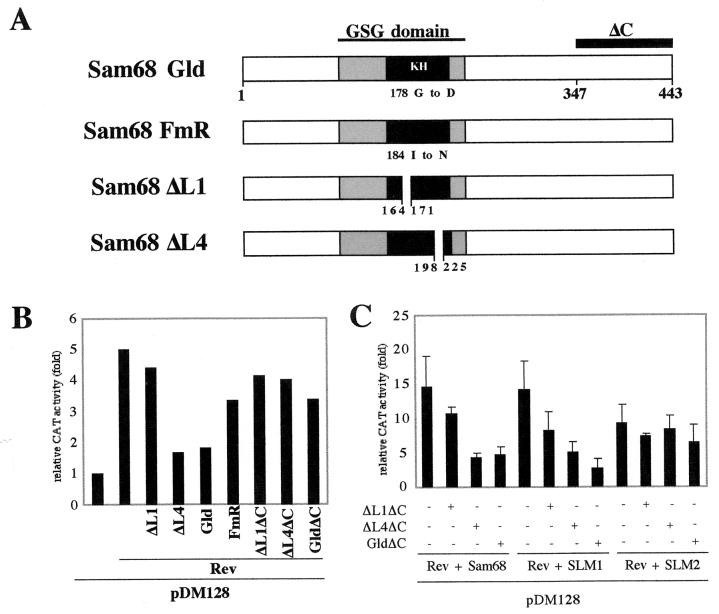
In an effort to identify more precisely which regions of Sam68 and Sam68ΔC are responsible for their stimulatory and inhibitory effects on Rev activity, respectively, RNA binding mutants (Sam68 FmR:184I→N and Sam68 Gld:178G→D) and multimerization-defective mutants (Sam68ΔL1 [amino acids {aa} 164 to 171 deleted] and Sam68ΔL4 [Δ aa 198–225]) (2, 8) were tested for their effects on Rev activity in 293 cells (Fig. 2A). In the wild-type Sam68 context, these mutants had little ability to stimulate Rev activation of pDM128 expression (Fig. 2B), with both ΔL4 and Gld inhibiting Rev-dependent gene expression to a significant extent. In the Sam68ΔC context, ΔL1ΔC, ΔL4ΔC, and GldΔC had slight to intermediate abilities to inhibit Rev-dependent gene expression or stimulation of Rev activity by Sam68, SLM1, or SLM2 (Fig. 2B and C). FmRΔC behaves as a general activator of gene expression, as it stimulated CAT activity from Bl-CMVCATpA (Fig. 1B). Western blotting revealed that none of the mutants had any effect on Rev expression levels, nor were their expression levels correlated with their inabilities to stimulate or inhibit Rev activity (data not shown). These results suggest that the RNA binding and RNA-dependent multimerization domains of Sam68 are required for its ability to stimulate Rev-dependent gene expression and, in the context of Sam68ΔC, for its ability to fully inhibit Rev-dependent gene expression.
FIG. 2.
Requirement for Sam68 multimerization and RNA binding for function. (A) Schematic of Sam68 mutants tested. (B) Mutants of Sam68 and Sam68ΔC have slight to intermediate abilities to stimulate or inhibit Rev-dependent gene expression, respectively. Transfections were performed as described in the legend to Fig. 1B for mutants of Sam68. (C) Mutants of Sam68ΔC have little to intermediate abilities to inhibit stimulation of Rev activity by Sam68, SLM1 or SLM2. Transfections were performed as described in the legend to Fig. 1C for mutants of Sam68ΔC. Each bar represents fold induction of CAT activity over pDM128 in the absence of Rev and is the average of at least three separate experiments performed in triplicate.
As verification that the effects of Sam68, SLM1, SLM2, and Sam68ΔC on Rev function were not limited to the reporter assay used in the above experiments, assays were repeated using either p24 or gp120 levels as readouts. As shown in Fig. 3, Sam68, SLM1, and SLM2 coexpression resulted in significant increases in p24 (Fig. 3A) and gp120 (Fig. 3B) production over that seen with Rev alone. In addition, Sam68ΔC coexpression resulted in complete suppression of Rev-dependent gene expression.
FIG. 3.
Sam68, SLM1, and SLM2 stimulation and Sam68ΔC inhibition of Rev-induced p24 and gp120 expression. 293 cells were transfected with Gag-RRE (A) or pgGP160 puro (B) in the absence or presence of Rev and Sam68, Sam68ΔC, SLM1, or SLM2 as indicated. At 48 h after transfection, cell were harvested and equal amounts of supernatants or lysates were analyzed by ELISA for p24 (A) or by Western blotting for gp120 (B), respectively, as described in Materials and Methods.
Addition of an NLS to Sam68ΔC causes its nuclear relocalization and loss of inhibitory effect on Rev activity.
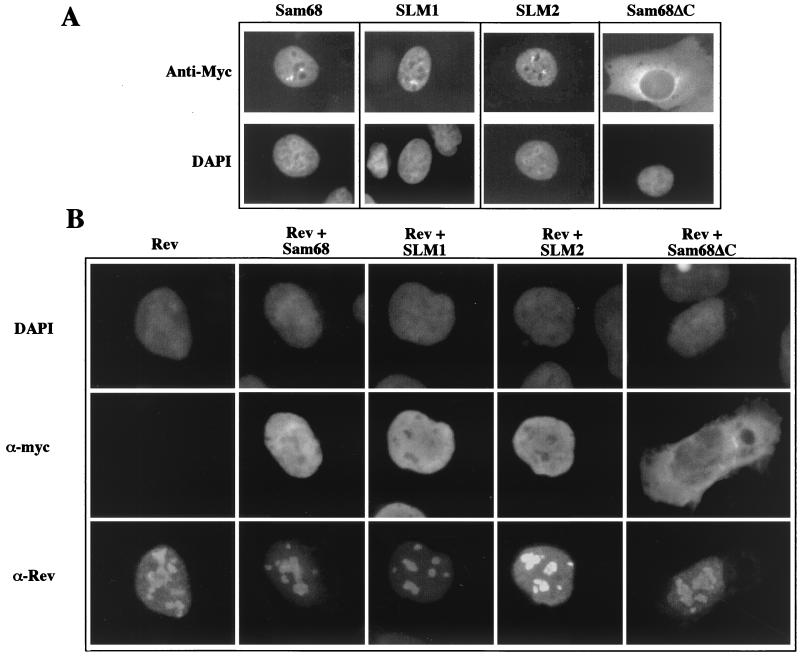
Sam68, SLM1, and SLM2 localize in the nucleoplasm of cells, while Sam68ΔC is in the cytoplasm (Fig. 4A) (2, 36). To assess whether these factors may elicit an effect through changes in Rev subcellular localization, cotransfected cells were examined for the localization of the proteins. In all instances, no effect on Rev subcellular distribution was found (Fig. 4B), the protein displaying accumulation in the nucleolus upon coexpression of all the factors tested. Our finding that coexpression of Sam68ΔC has no effect on Rev distribution differs from that previously reported (36), and we are currently unable to explain the discrepancy in the observations.
FIG. 4.
Effect of Sam68, SLM1, SLM2, or Sam68ΔC expression on Rev subcellular distribution. Cells were transfected with vectors encoding myc-tagged Sam68, SLM1, SLM2, or Sam68ΔC alone (A) or with a vector expressing Rev (B). Forty-eight hours posttransfection, cells were fixed and processed for immunofluorescent localization of the proteins. Shown are representative samples of the distribution patterns observed following staining of cells with anti-myc antibody (α-myc), anti-Rev antibody (α-Rev), and DAPI.
It was previously reported that a single point mutation, R439→A, in the NLS of Sam68 results in both its cytoplasmic accumulation and loss of its stimulatory effect on Rev activity (37). These observations led us to investigate whether relocalization of Sam68ΔC to the nucleus results in a loss of its inhibitory activity. Therefore, the effect of the addition of an exogenous NLS to Sam68ΔC on the ability of Sam68ΔC to inhibit Rev activity was assessed. The simian virus 40 (SV40) large T antigen NLS (SVNLS) was fused to the N terminus of Sam68ΔC, creating NLSSam68ΔC (Fig. 5A), and the effect of its expression on Rev activity was determined. Addition of the SVNLS to Sam68ΔC results in nuclear, nonnucleolar localization of the fusion protein (Fig. 5B) akin to the subcellular localization observed with Sam68, SLM1, and SLM2. Addition of the NLS also correlates with an alteration in the activity of Sam68ΔC, as NLSSam68ΔC does not inhibit Rev activity but rather, like Sam68, stimulates Rev activity ∼7-fold (Fig. 5C). Moreover, NLSSam68ΔC is no longer able to inhibit Sam68-, SLM1-, or SLM2-mediated stimulation of Rev activity (data not shown). These observations suggest that the inhibitory effect of Sam68ΔC is due to its localization to the cytoplasm.
FIG. 5.
Rescue of stimulatory activity upon addition of a heterologous NLS to Sam68ΔC. (A) Schematic of NLSSam68ΔC. (B) Subcellular localization of NLSSam68ΔC. The subcellular localization of the myc-tagged NLSSam68ΔC was determined by immunostaining with an anti-myc antibody followed by a secondary anti-mouse antibody conjugated to Texas Red (upper panel). Nuclei were DAPI stained (lower panel). (C) Effect of NLSSam68ΔC on Rev activity. Transfections were performed as described in the legend to Fig. 1B. Each bar represents fold CAT activity compared with pDM128 in the absence of Rev and is the average of at least three separate experiments performed in triplicate.
Sam68ΔC does not inhibit Rev shuttling.
The previous observation that Sam68 can replace Rev and interact with RRE-RNA (36) led us to examine whether Sam68, SLM1, and SLM2 can shuttle between the nucleus and cytoplasm as one means by which they could exert an effect on Rev-dependent RNA expression. Heterokaryon assays were thus performed, using HeLa cells transfected with M10 and coplated with HeLa cells transfected with Sam68, SLM1, or SLM2. Heterokaryons were subsequently formed by PEG-induced fusion, and protein localization was determined 3 h postfusion. M10 localizes to the nucleus and nucleolus and harbors a mutation in its NES, rendering it export deficient. Since M10 is export incompetent, it cannot shuttle and does not equilibrate into the Sam68-, SLM1-, or SLM2-expressing nuclei (Fig. 6A). If Sam68, SLM1, or SLM2 can shuttle between the nucleus and cytoplasm, then, in a heterokaryon, they would be expected to equilibrate into the M10-expressing nucleus. None of the factors were observed to equilibrate into M10-positive nuclei (Fig. 6A), suggesting that Sam68, SLM1, and SLM2 do not shuttle between the nucleus and cytoplasm of cells to any significant extent within the time period (3 h) of this assay.
FIG. 6.
Analysis of Sam68, SLM1, and SLM2 shuttling. Heterokaryon assays were performed using HeLa cells transfected with M10 (A) or Rev (B) fused to HeLa cells transfected with the myc-tagged proteins. Cells were immunostained with an anti-Rev antibody followed by a secondary anti-rabbit antibody conjugated to FITC (left panels). The myc-tagged Sam68, SLM1, SLM2, and Sam68ΔC were immunostained with an anti-myc antibody followed by a secondary anti-mouse antibody conjugated to Texas Red (middle panels). Nuclei were DAPI stained (right panels). (A) HeLa cells transfected with M10 were fused to other HeLa cells transfected with myc-tagged Sam68, SLM1, or SLM2, as indicated. Thin arrows, M10 nuclei; thick arrows, Sam68 or SLM nuclei. (B) HeLa cells transfected with wild-type Rev were fused to other HeLa cells transfected with myc-tagged Sam68, SLM1, SLM2, or Sam68ΔC, as indicated.
An interaction between Rev and Sam68 was also previously observed in vitro (36), raising the possibility that Sam68, SLM1, and SLM2 could stimulate, and Sam68ΔC could inhibit, Rev activity by affecting Rev transport across the nuclear membrane. To ascertain whether Sam68, SLM1, or SLM2 may be coshuttling with Rev to affect its activity, Sam68-, SLM1-, SLM2-, and Sam68ΔC-expressing cells were subsequently fused with HeLa cells expressing wild-type Rev. As shown in Fig. 6B, Rev equilibrates into Sam68, SLM1-, and SLM2-positive nuclei in the heterokaryons, while Sam68, SLM1, and SLM2 fail to equilibrate into the Rev-positive nuclei of the same heterokaryons. This suggests that none of these factors synergistically activates Rev function by coshuttling with Rev. Furthermore, Sam68ΔC does not impede the equilibration of Rev into all nuclei of the heterokaryons (Fig. 6B), suggesting that Sam68ΔC does not inhibit Rev function by inhibiting its nuclear export or subsequent nuclear reimport.
Sam68, SLM1, and SLM2 do not affect RRE-RNA stability or splicing.
We next wanted to investigate whether Sam68 and its related proteins stimulate Rev activity by increasing the stability of the unspliced RNA and thus the pool of RNA available for Rev to transport. To test this, the effect of the factors on the abundance of unspliced and spliced env RNA was examined in RNase protection assays (RPAs). Total RNA was harvested from cells transfected with pgGP160 puro, Rev, and/or the Sam or SLM vectors. Unspliced and spliced env RNAs were differentiated by the probe schematized in Fig. 7A. Coexpression of Sam68, Sam68ΔC, or SLM1 had no significant effect on the abundance of unspliced env RNA in the absence or presence of Rev (Fig. 7B). (SLM2 also had no effect [data not shown].) To address whether the difference between the effects of Sam68 and Sam68ΔC is exerted at the level of unspliced env RNA export, RNA was extracted from nuclear and cytoplasmic fractions, and levels of unspliced env RNA were assayed. As shown in Fig. 7C, analysis of the accumulation of unspliced viral RNA in the presence of either Sam68 or Sam68ΔC revealed comparable levels upon cotransfection of Rev.
FIG. 7.
Effects of Sam68, SLM1, and Sam68ΔC on HIV-1 env RNA abundance, splicing, and transport. (A) Schematic of the gp160 probe (∼400 bases) against env RNA. Unspliced gp160 RNA is detected as a protected ∼360-base fragment, while spliced RNA is detected as a protected ∼98-base fragment. (B) Effects of Sam68, Sam68ΔC, and SLM1 on viral RNA abundance. RNA was harvested from 293 cells and 10 μg of RNA was hybridized to a labeled gp160 probe. Shown is total RNA harvested from cells expressing env RNA in the absence (−) or presence (+) of Rev, Sam68, Sam68ΔC, or SLM1, as indicated. The control for transfection efficiency and sample loading is the VA transcript, detected as a protected fragment of 70 bases. (C) Effects of Sam68 and Sam68ΔC on viral RNA transport. Transfections were performed as outlined for panel B, cells were harvested, and nuclear and cytoplasmic fractions were prepared as described in Materials and Methods.
Sam68ΔC recruits unspliced HIV-1 env RNA to perinuclear bundles.
Given that the Sam and SLM proteins affect Rev activity but do not themselves shuttle, the Sam68 and SLM proteins may be directly affecting the localization of the unspliced env mRNA. To examine this possibility, in situ analysis of unspliced Rev-dependent RNA was performed. Subcellular localization of unspliced RRE-RNA was detected by probing a region of the Tat/Rev intron present within the Env gene of the reporter construct pgTat (identical results were obtained using the pgGP160 puro expression vector). Unspliced RNA is trapped in the nucleus in the absence of Rev (Fig. 8A, top panel) and is only detected in the cytoplasm upon coexpression of Rev (Fig. 8B, top panel). Unlike Rev, Sam68, SLM1, and SLM2 are unable to induce cytoplasmic accumulation of unspliced RNA on their own (Fig. 8A). Coexpression of Sam68, SLM-1, or SLM-2 with Rev results in cytoplasmic accumulation of unspliced viral RNA to an extent comparable to that seen upon expression of Rev alone (Fig. 8B).
FIG. 8.
Effects of Sam68, SLM1, and SLM2 on HIV-1 RNA subcellular distribution. At 48 h after transfection, cells were fixed and hybridized to an antisense digoxigenin (DIG)-labeled RNA probe corresponding to intronic sequence. Hybridized probe was then detected with an anti-DIG antibody conjugated to FITC (left panels). Cotransfected myc-tagged proteins were immunostained with an anti-myc antibody that was subsequently detected with a secondary antibody conjugated to Texas Red (middle panels). Nuclei were DAPI stained (right panels). (A) Localization of unspliced env RNA in the absence of Rev expression; (B) localization of unspliced env RNA in the presence of Rev expression.
Expression of Sam68ΔC has no effect on the subcellular localization of unspliced RNA in the absence of Rev (Fig. 9A). However, coexpression of Rev and Sam68ΔC results in a marked change in the localization of the unspliced RNA and Sam68ΔC (Fig. 9B). The unspliced RNA is still exported from the nucleus by Rev but does not permeate the cytoplasm either diffusely or in a punctate manner, as normally observed (Fig. 8B). Instead, bundles of RNA are detected at the nuclear membrane and the periphery of the nucleus, colocalizing with similar concentrated bundles of Sam68ΔC (Fig. 9C). Sam68ΔC also often displays a less-concentrated diffuse cytoplasmic stain. These observations suggest that Sam68ΔC may inhibit Rev activity by sequestering the RRE-RNA RNPs at the nuclear periphery, preventing their interaction with components of the translational apparatus.
FIG. 9.
Subcellular distribution of unspliced HIV-1 env RNA (FITC, green) upon Sam68ΔC (Texas Red, red) coexpression. (A) In the absence of Rev, the unspliced RNA is detected in the nucleus. (B) Upon Rev and Sam68ΔC coexpression, the unspliced RNA accumulates in perinuclear bundles that colocalize with Sam68ΔC. (C) A z-series of images reveals that unspliced env RNA and Sam68ΔC colocalize (yellow staining) to the nuclear periphery.
DISCUSSION
The previously reported observation that Sam68 stimulates Rev activity (36) prompted us to examine whether the Sam68-like mammalian proteins SLM1 and SLM2 could similarly stimulate Rev activity. Indeed, in the CAT-based reporter (Fig. 1), gp120, and Gag-based reporter assays (Fig. 3), SLM1 and SLM2 were found to stimulate Rev activity to a similar extent as Sam68. Deletion of the C-terminal region of Sam68, which harbors tyrosine phosphorylation sites and an NLS, results in a dominant-negative inhibitor of Sam68-stimulated Rev activity, Sam68ΔC (36). Here we demonstrate that Sam68ΔC also inhibits stimulation of Rev activity by SLM1 and SLM2 (Fig. 1C). This suggests that the mechanism by which Sam68, SLM1, and SLM2 stimulate Rev activity may be shared.
SLM1 and SLM2 share the basic organization of Sam68, including a GSG domain in which is embedded a KH domain with high sequence homology (∼70%) to that of Sam68 (8). The KH domain is a highly conserved domain found in several RNA binding proteins (16). Point mutations in the KH domain of Sam68, which abolish the RNA-dependent multimerization or RNA-binding properties of Sam68 (2), also reduce the ability of Sam68 to stimulate Rev activity and that of Sam68ΔC to inhibit Rev activity (Fig. 2). These results suggest that the RNA-binding properties are critical for the ability of Sam68 to affect Rev activity. SLM1 and SLM2 can heterodimerize with Sam68 (8), again suggesting that they may all affect Rev activity by a common mechanism. The specific RNA targets for Sam68, SLM1, and SLM2 have not yet been determined. However, a SELEX experiment with Sam68 determined that Sam68 strongly interacts with AU-rich sequences (26), several of which can be found throughout the unspliced HIV-1 env RNA sequence. SLM1 and Sam68 have similar homopolymeric RNA-binding properties, while those of SLM2 are different (8). Recent experiments have demonstrated that Sam68 can increase gene expression in multiple contexts, including indirect binding of Rev to the target mRNA mediated by fusion to the MS2 binding domain, transactivation by human T-lymphotropic virus type 1 Rex/equine infectious anemiavirus Rev (39), and constitutive export mediated by the constitutive transport element (38). In light of the significant sequence variation among these elements, it is possible that the Sam/SLMs may function either through indirect contact with the RNA or through direct binding at sites in the RNA other than the RRE.
Sam68, SLM1, and SLM2 display nuclear, nonnucleolar localization, while Sam68ΔC is diffusely cytoplasmic (Fig. 4A). Coexpression with Rev was not observed to effect changes in the subcellular distribution of any of the proteins (Fig. 4B). The NLS of Sam68 is located in its C terminus (23, 37), and a single point mutation (P439→R) has been previously demonstrated to result in cytoplasmic accumulation of Sam68-P439R and failure to stimulate Rev-dependent gene expression (37). Furthermore, Sam68-P439R was demonstrated to act as an inhibitor of Rev activity, although less efficiently than Sam68ΔC (37). This suggests that the inhibitory activity of Sam68ΔC is a consequence of its cytoplasmic localization. Alternatively, the mutation might affect some other, as yet unidentified function of this region. In order to determine whether nuclear localization could cause Sam68ΔC to stimulate Rev activity, the SV40 large T antigen NLS was fused to the amino terminus of Sam68ΔC. This results in nuclear localization of NLSSam68ΔC (Fig. 5B) and correlative ability to stimulate Rev-dependent gene expression (Fig. 5C). Consequently, the C terminus of Sam68 is likely a domain for regulation of its activity (7, 51) but is not required for stimulation of Rev-dependent gene expression, as its deletion in the context of a fusion to an NLS has activity comparable to that of Sam68 (Fig. 5C). Given that the RNA binding domain in the KH region of Sam68 is required for its ability to stimulate Rev-dependent gene expression and that the amino terminus (aa 1 to 96) can be removed with no effect on Rev activity (36), only the GSG domain and nuclear localization appear to be required for its effect on Rev function.
Given that Sam68, SLM1, and SLM2 share the ability to bind RNA and that Sam68 has previously been reported to interact with RRE-RNA (36), we wished to determine whether the abilities of these factors to stimulate Rev activity could reflect an ability to shuttle between the nucleus and cytoplasm. It has previously been reported that Sam68, unlike hnRNP A1, hnRNP K, and Rev, does not accumulate in the cytoplasm upon inhibition of transcription (29). Here we demonstrate through heterokaryon assays that Sam68, SLM1, and SLM2 do not shuttle to any significant extent under the conditions and time frame (3 h) tested. The previously reported observation that Sam68 can bind Rev in vitro (36) led us to examine whether Sam68, SLM1, and SLM2 could shuttle through an interaction with Rev. In this report, we demonstrate that Sam68, SLM1, and SLM2 do not colocalize with Rev to any significant extent (Fig. 4B), nor do they not coshuttle with Rev (Fig. 6B). Thus, any interaction between Sam68 and Rev is transient and likely restricted to the nucleus. Furthermore, the ability of Sam68ΔC to inhibit Rev activity cannot be attributed to an ability to inhibit Rev from shuttling, as Rev was able to equilibrate into all nuclei of heterokaryons formed between cells expressing Sam68ΔC and cells expressing Rev (Fig. 6B).
Since Sam68, SLM1, and SLM2 did not demonstrate an ability to shuttle in the heterokaryon assays, we examined whether their stimulatory effects on Rev activity could be due to a direct effect on transport of the unspliced env RNA. However, no difference in the localization of unspliced env RNA was observed when Sam68, SLM1, or SLM2 was coexpressed (Fig. 8). Furthermore, RNase protection analysis revealed that Sam68, Sam68ΔC, SLM1, and SLM2 do not affect the abundance of unspliced to spliced RNA (Fig. 7B). Therefore, their effects cannot be attributed to stabilization of unspliced Rev substrate RNA or alteration in splicing. The mechanism by which Sam68, SLM1, and SLM2 stimulate Rev activity thus remains to be determined.
In situ analysis with Sam68ΔC reveals that this mutant blocks the Rev-mediated transport of unspliced viral RNA throughout the cytoplasm (Fig. 8B and 9). Upon expression of Sam68ΔC and Rev, perinuclear bundles are formed that contain both unspliced env RNA and Sam68ΔC. This observation suggests that the mechanism by which Sam68ΔC inhibits Rev-dependent gene expression is trapping of the Rev-dependent RNA away from the translation machinery and in close juxtaposition to the nuclear periphery. This hypothesis can account for the observation that Sam68ΔC inhibits the stimulatory abilities of Sam68, SLM1, and SLM2 (Fig. 1C) and loses such inhibitory activity when redirected to the nucleus by addition of a heterologous NLS (Fig. 5). The cytoplasmic Sam68ΔC, unable to localize to the nucleus, acts downstream of all three stimulators of Rev activity, whose stimulatory effects are conducted in the nucleus. In light of these observations, one potential model for Sam68, SLM1, and SLM2 stimulation of Rev activity is that they act in the nucleus to facilitate intranuclear transport of viral RNA in complex with Rev to the nucleoplasmic face of the nuclear pore complex. Sam68ΔC, present within the cytoplasm, would appear to bind to the RNA emerging from the nucleus and inhibit its movement away from the nuclear periphery. Accumulation of Sam68ΔC at the nuclear periphery is dependent on Rev-mediated transport of viral RNA to the cytoplasm, indicating that the protein's activity is triggered by its interaction with the emerging RNA. Further study of these proteins may provide insight into the mechanisms controlling RNA movement to and from the nuclear pore.
ACKNOWLEDGMENTS
This work was supported by grants from the Ontario HIV Treatment Network (OHTN) and the Medical Research Council (MRC) of Canada to A.W.C., as well as grants from the Cancer Research Society Inc. and the MRC (MT-13377) to S.R. A.W.C. is supported by a scientist award from the OHTN. S.R. is a scholar of the MRC. V.B.S. is supported by an OHTN studentship.
We thank B. Blencowe for use of the confocal microscope and D. Branch for his assistance.
REFERENCES
- 1.Chen T, Boisvert F M, Bazett-Jones D P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk-1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cochrane A W, Chen C-H, Rosen C. Specific interaction of the HIV Rev transactivator protein with a structured region in the env mRNA. Proc Natl Acad Sci USA. 1990;87:1198–1201. doi: 10.1073/pnas.87.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cochrane A W, Golub E, Volsky D, Ruben S, Rosen C A. Functional significance of phosphorylation to the human immunodeficiency virus Rev protein. J Virol. 1989;63:4438–4440. doi: 10.1128/jvi.63.10.4438-4440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cochrane A W, Perkins A, Rosen C A. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen B. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 7.Derry J, Richard S, Valderrama Carvajal H, Ye X, Vasioukhin V, Cochrane A, Chen T, Tyner A. Sik/BRK phosphorylates Sam68 in the nucleus and negatively regulates its RNA binding ability. Mol Cell Biol. 2000;20:6114–6126. doi: 10.1128/mcb.20.16.6114-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Fruscio M, Chen T, Richard S. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc Natl Acad Sci USA. 1999;96:2710–2715. doi: 10.1073/pnas.96.6.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan L, Oakes J, Ferraro A, Bagasra O, Pomerantz R. Tat and Rev differentially affect restricted replication of human immunodeficiency virus type 1 in various cells. Virology. 1994;199:474–478. doi: 10.1006/viro.1994.1148. [DOI] [PubMed] [Google Scholar]
- 10.Emerman M, Vazeux R, Peden K. The Rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell. 1989;57:1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg M B, Jarrett R F, Aldovini A, Gallo R C, Wong-Staal F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986;46:807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- 12.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. The Rev protein of HIV-1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornerod M, Ohno M, Yoshida M, Mattaj I. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 15.Fumagalli S, Totty N, Hsuan J, Courtneidge S. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 16.Gibson T J, Thompson J D, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins. FEBS Lett. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- 17.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadzopoulou-Cladaras M, Felber B K, Cladaras C, Athanassopoulos A, Tse A, Pavlakis G N. The Rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammarskjöld M L, Heimer J, Hammarskjöld B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heaphy S, Dingwall C, Ernberg I, Gait M J, Green S M, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element. Cell. 1990;60:685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- 21.Hope T J, Huang X, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping of cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S, Spector D L. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishidate T, Yoshihara S, Kawasaki Y, Roy B C, Toyoshima K, Akiyama T. Identification of a novel nuclear localization signal in Sam68. FEBS Lett. 1997;409:237–241. doi: 10.1016/s0014-5793(97)00455-9. [DOI] [PubMed] [Google Scholar]
- 24.Kriegler M. Gene transfer and expression: a laboratory manual. 1st ed. New York, N.Y: Stockton Press; 1990. [Google Scholar]
- 25.Laughlin M, Pomerantz R. Retroviral latency. R. G. Austin, Tex: Landes Company; 1994. [Google Scholar]
- 26.Lin Q, Taylor S, Shalloway D. Specificity and determinants of Sam68 RNA binding: implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 27.Malim M H, Bohnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 28.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 29.McBride A E, Taylor S J, Shalloway D, Kirkegaard K. KH domain integrity is required for wild-type localization of Sam68. Exp Cell Res. 1998;241:84–95. doi: 10.1006/excr.1998.4047. [DOI] [PubMed] [Google Scholar]
- 30.McCracken S, Rosonina E, Fong N, Sikes M, Beyer A, O'Hare K, Shuman S, Bentley D L. Role of RNA polymerase II carboxy-terminal domain in coordinating transcription with RNA processing. Cold Spring Harbor Symp Quant Biol. 1998;63:301–309. doi: 10.1101/sqb.1998.63.301. [DOI] [PubMed] [Google Scholar]
- 31.Meyer B, Meinkoth J, Malim M. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumeister E N, Zhu Y, Richard S, Terhorst C, Chan A C, Shaw A S. Binding of ZAP-70 to phosphorylated T-cell receptor zeta and eta enhances its autophosphorylation and generates specific binding sites for SH2 domain-containing proteins. Mol Cell Biol. 1995;15:3171–3178. doi: 10.1128/mcb.15.6.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen H, Beidas S, Dillon P, Rosen C A, Cochrane A W. Mutational analysis of the HIV-1 Rev protein and its target sequence, the Rev responsive element. J Acquir Immune Defic Syndr. 1991;4:558–567. [PubMed] [Google Scholar]
- 34.Olsen H S, Cochrane A W, Dillon P J, Nalin C M, Rosen C A. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent upon multimer formation mediated through a basic stretch of amino acids. Genes Dev. 1990;4:1357–1364. doi: 10.1101/gad.4.8.1357. [DOI] [PubMed] [Google Scholar]
- 35.Perkins A, Cochrane A W, Ruben S M, Rosen C A. Structural and functional characterization of the human immunodeficiency virus Rev protein. J Acquir Immune Defic Syndr. 1989;2:256–263. [PubMed] [Google Scholar]
- 36.Reddy T, Xu W, Mau J, Goodwin C, Suhasini M, Tang H, Frimpong K, Rose D, Wong-Staal F. Inhibition of HIV replication by dominant-negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat Med. 1999;5:635–642. doi: 10.1038/9479. [DOI] [PubMed] [Google Scholar]
- 37.Reddy T R. A single point mutation in the nuclear localization domain of Sam68 blocks the Rev/RRE-mediated transactivation. Oncogene. 2000;19:3110–3114. doi: 10.1038/sj.onc.1203637. [DOI] [PubMed] [Google Scholar]
- 38.Reddy T R, Tang H, Xu W, Wong-Staal F. Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA. Oncogene. 2000;19:3570–3575. doi: 10.1038/sj.onc.1203676. [DOI] [PubMed] [Google Scholar]
- 39.Reddy T R, Xu W, Wong-Staal F. General effect of Sam68 on Rev/Rex regulated expression of complex retroviruses. Oncogene. 2000;19:4071–4074. doi: 10.1038/sj.onc.1203749. [DOI] [PubMed] [Google Scholar]
- 40.Rosen C A, Terwilliger E, Dayton A, Sodroski J G, Haseltine W A. Intragenic cis-acting Art gene-responsive sequences of the human immunodeficiency virus. Proc Natl Acad Sci USA. 1988;85:2071–2075. doi: 10.1073/pnas.85.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seguin B, Staffa A, Cochrane A. Control of HIV-1 RNA metabolism: the role of splice sites and intron sequences in unspliced viral RNA subcellular distribution. J Virol. 1998;72:9503–9513. doi: 10.1128/jvi.72.12.9503-9513.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siomi H, Matunis M J, Michael W M, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sodroski J, Goh W C, Rosen C, Dayton A, Terwilliger E, Haseltine W A. A second post-transcriptional transactivator gene required for the HTLV-III replication. Nature. 1986;321:412–417. doi: 10.1038/321412a0. [DOI] [PubMed] [Google Scholar]
- 44.Soros V, Cochrane A. Alterations in HIV-1 Rev transport in response to cell stress. Virology. 2001;280:199–210. doi: 10.1006/viro.2000.0752. [DOI] [PubMed] [Google Scholar]
- 45.Stade K, Ford C, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 46.Swenarchuk L, Harakidas P, Cochrane A. Regulated expression of HIV-1 Rev function in mammalian cell lines. Can J Microbiol. 1999;45:480–490. [PubMed] [Google Scholar]
- 47.Taylor S, Shalloway D. An RNA-binding protein associated with src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 48.Taylor S J, Anafi M, Pawson T, Shalloway D. Functional interaction between c-src and its mitotic target. J Biol Chem. 1995;270:10120–10124. doi: 10.1074/jbc.270.17.10120. [DOI] [PubMed] [Google Scholar]
- 49.Truant R, Cullen B. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venkatesan S, Gerstberger S, Park H, Holland S, Nam Y. Human immunodeficiency virus type 1 Rev activation can be achieved without Rev-responsive element RNA if Rev is directed to the target as a Rev/MS2 fusion protein which tethers the MS2 operator RNA. J Virol. 1992;66:7469–7480. doi: 10.1128/jvi.66.12.7469-7480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L L, Richard S, Shaw A S. p62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem. 1995;270:2010–2013. doi: 10.1074/jbc.270.5.2010. [DOI] [PubMed] [Google Scholar]
- 52.Weil D, Boutain S, Audibert A, Dautry F. Mature mRNAs accumulated in the nucleus are neither the molecules in transit to the cytoplasm nor constitute a stockpile for gene expression. RNA. 2000;6:962–975. doi: 10.1017/s1355838200000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weng Z, Thomas S M, Rickles R, Taylor J, Brauer A W, Seidel-Dugan C, Michael W M, Dreyfuss G, Brugge J. Identification of Src, Fyn, and Lyn SH3-binding proteins: implications for a function of SH3 domains. Mol Cell Biol. 1994;14:4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong G, Muller O, Clark R, Conroy R, Moran M F, Polakis P, McCormick F. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell. 1992;69:551–558. doi: 10.1016/0092-8674(92)90455-l. [DOI] [PubMed] [Google Scholar]
- 55.Yankulov K, Blau J, Purton S, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 56.Zapp M, Green M. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]