Abstract
Varicella-zoster virus (VZV) open reading frame 63 (ORF63), located between nucleotides 110581 and 111417 in the internal repeat region, encodes a nuclear phosphoprotein which is homologous to herpes simplex virus type 1 (HSV-1) ICP22 and is duplicated in the terminal repeat region as ORF70 (nucleotides 118480 to 119316). We evaluated the role of ORFs 63 and 70 in VZV replication, using recombinant VZV cosmids and PCR-based mutagenesis to make single and dual deletions of these ORFs. VZV was recovered within 8 to 10 days when cosmids with single deletions were transfected into melanoma cells along with the three intact VZV cosmids. In contrast, VZV was not detected in transfections carried out with a dual deletion cosmid. Infectious virus was recovered when ORF63 was cloned into a nonnative AvrII site in this cosmid, confirming that failure to generate virus was due to the dual ORF63/70 deletion and that replication required at least one gene copy. This requirement may be related to our observation that ORF63 interacts directly with ORF62, the major immediate-early transactivating protein of VZV. ORF64 is located within the inverted repeat region between nucleotides 111565 and 112107; it has some homology to the HSV-1 Us10 gene and is duplicated as ORF69 (nucleotides 117790 to 118332). ORF64 and ORF69 were deleted individually or simultaneously using the VZV cosmid system. Single deletions of ORF64 or ORF69 yielded viral plaques with the same kinetics and morphology as viruses generated with the parental cosmids. The dual deletion of ORF64 and ORF69 was associated with an abnormal plaque phenotype characterized by very large, multinucleated syncytia. Finally, all of the deletion mutants that yielded recombinants retained infectivity for human T cells in vitro and replicated efficiently in human skin in the SCIDhu mouse model of VZV pathogenesis.
Varicella-zoster virus (VZV) is a ubiquitous human herpesvirus that causes varicella during primary infection of susceptible individuals (2). VZV is a lymphotropic virus, with the capacity to infect CD4 and CD8 T cells, permitting its spread to mucocutaneous sites and producing the vesicular rash commonly referred to as chicken pox. VZV is a member of the alphaherpesvirus group and also exhibits the neurotropism characteristic of these viruses; it establishes latency in sensory nerve ganglia, and its reactivation results in herpes zoster, a localized dermatomal exanthem.
The VZV genome is a double-stranded DNA molecule with open reading frames (ORFs) that are known or predicted to encode at least 69 distinct gene products. The genome consists of two main coding regions, the unique long (UL) and unique short (US) segments, each of which is flanked by internal repeat (IR) and terminal repeat (TR) sequences. Functions have been assigned to only about half of the VZV gene products, and many of these are presumed because of their partial sequence homologies with herpes simplex virus type 1 (HSV-1), which is the prototype of the alphaherpesviruses. Whereas generating mutant VZV strains by homologous recombination is complicated by the extreme cell-associated nature of VZV replication, mutational analysis of VZV genes can now be accomplished efficiently with cosmid vectors (5, 25). In our system, four cosmids that contain overlapping fragments of the complete VZV genome, derived from the Oka vaccine strain (V-Oka), are used to make selected deletions of particular ORFs. The initial objective in generating VZV recombinants with deletions or disrupted expression of specific genes is to determine whether the gene is required for replication in vitro and, if not, whether its absence is associated with an altered phenotype in standard tissue culture cells, in specialized cells in culture, and finally, in differentiated cells in the SCIDhu model in vivo. Defining VZV genes required for infection of T cells and skin is of particular interest because VZV pathogenesis is characterized by tropism for these cells, especially during primary infection (2).
The purpose of these experiments was to examine whether genes that are duplicated in the VZV repeat regions are required for viral replication. VZV differs from HSV-1 in having three sets of diploid genes encoded within the repeats flanking the US segment: ORF62/71, ORF63/70, and ORF64/69. Since ORF62 protein is the major transactivating protein in VZV, its functions are predicted to be essential. Therefore, the other two pairs of genes, ORF63/70 and ORF64/60, were the targets of these mutational analyses. The ORF63 gene is encoded by nucleotides (nt) 110581 to 111417 and is duplicated between nt 118480 and 119316; its HSV homologue is ICP22 (21). The gene encodes a protein of 278 amino acids with a predicted molecular mass of 30.5 kDa but produces an extensively modified 47-kDa protein within infected cells. Its function was first characterized in transient expression experiments which showed downregulation of the ORF62 promoter and upregulation of the thymidine kinase promoter (16), but later reports indicate that it has little effect on immediate-early or early promoters (22). ORF63 is of particular interest in VZV pathogenesis because it has been reported to be transcribed and translated during latency in rat and human sensory ganglia (10, 18, 23, 24). ORF64 follows ORF63 in the genome; it is located between nt 111565 and 112107 and duplicated between nt 117790 and 118332. The role of this diploid gene in viral replication is not known. Its HSV-1 homologue is Us10, which occurs as a single ORF within the US region.
In these experiments, deleting both copies of the ORF63/70 gene pair showed that at least one copy of ORF63 was required for VZV replication; restoring ORF63 by expression from a nonnative site resulted in recovery of infectivity. Along with analyses showing that ORF63 binds to ORF62, these observations suggest that interactions between these regulatory proteins may be essential for VZV replication. A single copy of ORF63 was sufficient for infection of primary human T cells. Although both ORF64 and ORF69 could be deleted without blocking infectivity in vitro, deletion of both ORFs caused an altered phenotype, with an abnormal, large syncytium plaque morphology. Infectivity for human T cells was preserved, suggesting that the ORF64/69 gene pair may not be required for VZV lymphotropism. All of the deletion mutants retained virulence for cutaneous cells in the SCIDhu model.
MATERIALS AND METHODS
Cosmids and plasmids for mutational analysis.
Four overlapping fragments of genomic DNA from V-Oka in SuperCos 1 cosmid vectors (Stratagene, La Jolla, Calif.) were kindly provided by George Kemble, Aviron, Inc., Mountain View, Calif. (17). ORF64/69 and ORF63/70 are in the repeat regions in cosmid pvSpe21ΔAvr. In the pvSpe21ΔAvr cosmid, the inverted repeats and US region are in the inverted orientation relative to the UL region, such that the ORFs in the inverted repeats and the US region are in the opposite orientation, i.e., ORF71 is adjacent to ORF61 and ORF62 is at the end of the genome. Deletion of an AvrII site at SuperCos 1 nt 3359 produced a unique AvrII site at VZV nt 117038 (Fig. 1).
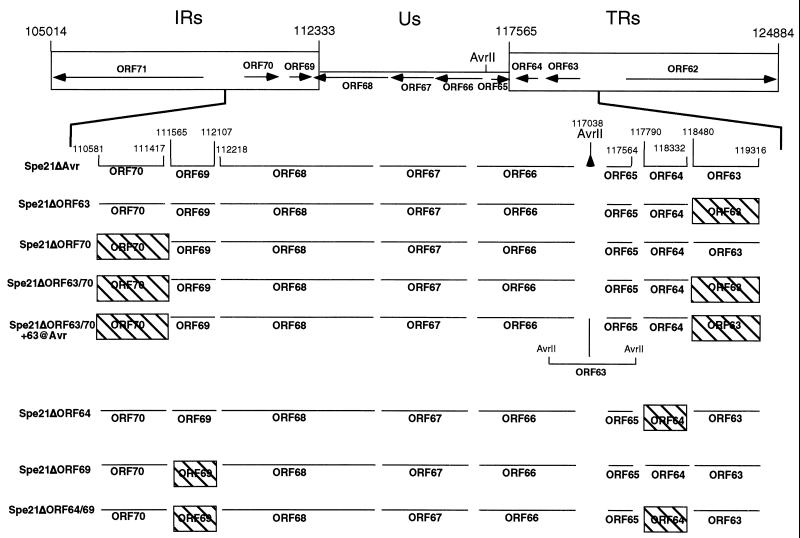
FIG. 1.
Schema of cosmid mutations. The upper section depicts the VZV IR-US-TR region of the genome containing the coding regions of ORFs 62 to 71, and the unique AvrII site in the US region is indicated. The lower section is an expanded view of the region between ORFs 70 and 63. The nucleotide numbers of the start and stop sites of the ORFs that were deleted (ORFs 63, 64, 69 and 70), as well as the nucleotide numbers of the relevant adjacent ORFs, are given. The designations of the various cosmids generated are shown on the left. Hatched boxes, deleted ORFs.
In order to delete the desired ORFs, regions of the pvSpe21ΔAvr cosmid were subcloned into plasmid vectors. The pvSpe21ΔAvr cosmid was digested with PstI and AvrII to obtain a 15.4-kb fragment (nt 101623 to 117039) that contained ORF61 and ORFs 71 to 66. This fragment was subcloned into pLitmus28 (New England Biolabs, Beverly, Mass.) to make plasmid pLitmusVZV. A second plasmid vector was constructed by digesting pvSpe21ΔAvr with AvrII and EcoRI to generate a 7.8-kb fragment (nt 117039 to 124884) which contains ORFs 65 to 62. This fragment was cloned into pLitmus28 to generate pLitmus VZV.2. A shuttle vector, pvSpe21ΔTRs, was generated by digesting pvSpe21 with AvrII and religating the 27-kb DNA fragment. This cloning step removed a portion of the US region, all of the inverted repeat, and a portion of the SuperCos 1 vector not required for replication.
Deletion of ORF63/70.
Two sets of PCR primers were designed as follows: primer 1, 5′-GTTTGGTCTTACGAATCCTCGG-3′ (5′ end anneals at nt 107843 in the IRs and at nt 122045 in the TRs); primer 2,5′-TGCAAATCTAGACCTTGGGG-3′ (5′ end anneals at nt 110590 in the IRs and at nt 119307 in the TRs); primer 3, 5′-CCATGGCGTCTAGACTTTATAA-3′ (5′ end anneals at nt 111405 in the IRs and at nt 118485 in the TRs); and primer 4, 5′-CTACGGATACGGAAGAAGAG-3′ (5′ end anneals at nt 112306 in the IRs and at nt 117591 in the TRs). Primers 1 and 4 anneal upstream and downstream of SphI restriction sites that are found on either side of the ORF63/70 coding regions. Primers 2 and 3 anneal over the ORF63/ORF70 start and stop codons and change the nucleotides indicated in boldface to create an XbaI site (TCTAGA). Using pLitmusVZV or pLitmus VZV.2 as templates, PCRs were carried out using the Elongase enzyme mix (GIBCO/BRL, Gaithersburg, Md.) with primers 1 and 2 and with primers 3 and 4. The 2,728- and 892-nt PCR products were isolated and digested with SphI and XbaI, and the resulting 2,127- and 464-nt products were reisolated. The isolated PCR products were sequenced to verify that no additional mutations were introduced during the PCRs. pLitmusVZV and pLitmusVZV.2 were digested with SphI, yielding a 14.8- and a 7.2-kb fragment. A triple ligation was set up using the PCR products and the SphI-digested pLitmusVZV or pLitmusVZV.2 to create plasmid clones with deletions of ORF63 (pLitmusVZVΔORF63) or ORF70 (pLitmusVZVΔORF70). Clones were screened to verify the presence of the XbaI site in place of the ORF63/ORF70 coding regions and to ensure the correct orientation of the SphI fragments. To transfer the region of DNA with ORF63 deleted from pLitmusVZVΔORF63 to the pvSpe21ΔAvr cosmid, the SrfI/AvrII fragment from the shuttle cosmid vector pvSpe21 TRs was replaced with the SrfI/AvrII fragment, with ORF63 deleted, from pLitmusVZVΔORF63 to generate pvSpe21ΔTRsΔORF63. The NheI/AvrII fragment from pvSpe21ΔTRsΔORF63 was then isolated and used to replace the wild-type NheI/AvrII fragment contained in pvSpe21ΔAvr to generate pvSpe21ΔORF63. To transfer the region of DNA with ORF70 deleted from pLitmusVZVΔORF70 to pvSpe21ΔAvr, the 7.0-kb AvrII/AscI fragment from pLitmusVZVΔORF70 was isolated and used to replace the wild-type 7.8-kb AvrII/AscI fragment from pvSpe21ΔAvr, creating pvSpe21ΔORF70. A cosmid clone with both gene copies deleted was constructed by isolating the 7.0-kb AvrII/AscI fragment from pvSpe21ΔORF70 and using it to replace the wild-type 7.8-kb AvrII/AscI fragment from pvSpe21ΔORF63, yielding pvSpe21ΔORF63/70.
Deletion of ORF64/69.
Two sets of PCR primers were designed as follows: primer 1, 5′-TGCCGCCTCGTCCACAAAGT-3′ (5′ end anneals at nt 105870 in the IRs and at nt 124008 in the TRs); primer 2, 5′-TCCGCAGAGATCTAGACCAC-3′ (5′ end anneals at nt 111571 in the IRs and at nt 118318 in the TRs); primer 3, 5′-GAGAGATCTAGACACCCCAT-3′ (5′ end anneals at nt 112096 in the IRs and at nt 117794 in the TRs); primer 4, 5′-CATTATCTCCGCCCTCTTAT-3′ (5′ end anneals at nt 116979 in the IRs); and primer 5, 5′-GTTCCGACCCTGCCGCTTAC-3′ (anneals within the pLitmusVZV.2 vector). Primers 1, 4, and 5 anneal upstream and downstream of BamHI restriction sites that are found on either side of the ORF64/69 coding regions. Primers 2 and 3 anneal over the ORF64/ORF69 start and stop codons and change the nucleotides indicated in boldface to create an XbaI site (TCTAGA). PCR products were generated, isolated, digested with the appropriate enzymes, and reisolated. The isolated PCR products were sequenced to verify that no additional mutations were introduced during the PCRs. Triple ligations were performed using digested pLitmusVZV and pLitmusVZV.2 to obtain pLitmusVZVΔORF64 and pLitmusVZVΔORF69. Clones were screened to verify the presence of the XbaI site in place of the ORF64/ORF69 coding regions, and to ensure correct orientation. Cosmids pvSpe21ΔORF64, pvSpe21ΔORF69, and pvSpe21ΔORF64/69 were generated as described above.
Insertion of ORF63 at a nonnative site in the genome.
Two PCR primers, primer 1 (5′-GCAACCCAATCCTAGGTCTC-3′ [5′ end anneals at nt 110406]) and primer 2 (5′-GGGTGCTCACCTAGGGATCC-3′ [5′ end anneals at nt 111542]), were designed to introduce AvrII sites (CCTAGG) at both ends of the PCR product (nucleotide changes are indicated by boldface). The ORF63 sequence, including the putative promoter region and downstream elements, was amplified, and the 1.1-kb PCR product was isolated, digested with AvrII, and reisolated. Cosmid pvSpe21ΔORF63/70 was digested at the unique AvrII site located between ORF65 and ORF66 (Fig. 1). The linearized cosmid and the 1.1-kb PCR product were ligated to produce pvSpe21ΔORF63/70+63@Avr.
Transfection and virus isolation.
Before use in cotransfections, the intact and mutated pvSpe21 cosmids and three VZV cosmids, designated pvFsp4, pvSpe5, and pvPme19, that span the complete VZV genome were electroporated into Top 10F′ competent cells (Invitrogen Inc., Carlsbad, Calif.), grown in Luria broth containing kanamycin and ampicillin, and purified using a plasmid maxi prep kit (Qiagen, Inc., Chatsworth, Calif.). Cosmids were digested with AscI and mixed in water to a final concentration of 100 ng of pvFsp4, pvSpe5, or pvPme19/μl and 50 ng of pvSpe21/μl (25). Transfections were carried out with human melanoma cells using 20 or 30 μl of the cosmid mix in 31.5 μl of 2 M CaCl2 in water and HEPES-buffered saline. Human melanoma cells were grown in tissue culture medium (Dulbecco's modified Eagle's medium; GIBCO/BRL, Grand Island, N.Y.) supplemented with heat-inactivated fetal calf serum.
After transfection, the melanoma cells were kept at 37°C for 3 to 4 days, trypsinized, and transferred to a 75-cm2 flask; plaques appeared 5 to 10 days after transfection with intact cosmids. Cells transfected with mutant cosmids were passed at a 1:3 ratio every 3 to 4 days. Infectious virus was propagated in melanoma cells, and titrations were carried out as previously described (5).
PCR analysis of viral DNA.
VZV cosmid DNA was purified using Qiagen columns, and recombinant virus DNA was recovered from infected cells using DNazol (GIBCO/BRL, Inc., Grand Island, N.Y.). PCR was performed using Elongase enzyme mix (GIBCO BRL, Grand Island, N.Y.). The primers used to assess deletions of ORF63 and ORF64 were 5′-CACCGTTCGCACTTTCTTTC-3′ (primer 1) and 5′-TTTACCTCGCCACATTTAGC-3′ (primer 2). To assess deletions of ORF70 and ORF69, primer 1 was used together with 5′-CCACACAAACATCACCTG-3′ (primer 3). To analyze the region containing the unique AvrII site, primer 3 was used together with 5′-TTACCACCGCTTCCATCA-3′ (primer 4).
DNA sequence analysis of viral DNA amplified from infected cells.
Following PCR, using viral infected cell DNA as a template, DNA was isolated using the Qiagen Gel Extraction kit, or PCR products were cloned into the pCR-TOPO cloning vector (Invitrogen). Sequencing reactions were primed by using either the M13 fwd and rev primers contained in the pCR-TOPO vector or custom primers. To sequence the ORF63 region, primer 5′-ACCCAAGTAGCCTTATTC-3′ was used. This primer anneals within the repeat region and was also used to sequence across the ORF70 region. To sequence across the ORF64 region, primer 5′-CAGTACGCTTTTATC-3′ was used. Sequence analysis was carried out at the Stanford University Protein and Nucleic Acid (PAN) Facility.
Confocal microscopy for VZV protein expression.
At increasing intervals after infection, VZV-infected monolayers were fixed with 2% paraformaldehyde containing 0.05% Triton X-100 and washed extensively with phosphate-buffered saline (PBS) (0.01 M; pH 7.4). Antibodies included monoclonal antibody (MAb) 5C6 (anti-IE62), MAb 711 (anti-gE), and MAb 258 (anti-gH). After fixation, primary murine MAbs to viral proteins were added, and the plates were incubated overnight on a rotor at 4°C and then washed extensively. A goat anti-mouse antibody conjugated with Texas Red or Oregon Green was added for 1h; cellular nuclei were stained with Toto-3 dye. Monolayers were washed again, Fluorguard was added and coverslips were placed, and specimens were examined using a Bio-Rad 1024 laser scanning confocal microscope located in the Central Microscopy Research Facility of the University of Iowa. All fluoroprobes were purchased from Molecular Probes, Inc., Eugene, Oreg.
Preparation of infected melanoma nuclear extracts.
Melanoma cells were grown to 80% confluency and then infected with 50 μl of a frozen stock of VZV-infected melanoma cells. The infection was allowed to proceed for approximately 2 days. The cells were then washed with 10 ml of sterile PBS. Next 2 ml of trypsin-EDTA was added to the cells for 2 min at 37°C. Ten milliliters of fresh minimal essential medium (containing fetal bovine serum [FBS] and gentamicin) was added to the cells, and the cells were then collected by low-speed centrifugation. The cells were washed with 10 ml of PBS and repelleted. The pellet volume was then determined, and an equivalent volume of PBS was used to resuspend the pellet, followed by low-speed centrifugation for 5 min at room temperature. Next, a 1× volume of buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM dithiothreitol [DTT]) was used to resuspend the pellet. The resuspension was placed on ice for 15 min. The cells were broken by passage through a 25-gauge 5/8 needle and repelleted. Next, 2/3 volume of buffer C (20 mM HEPES [pH 7.9], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA [pH 8.0], 0.5 phenylmethylsulfonyl fluoride, and 0.5 mM DTT) was used to resuspend the pellet. This resuspension was stirred on ice for 30 min and then centrifuged at 23,000 × g for 10 min at 4°C. The supernatant was dialyzed in buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 0.1 M KCl, 0.2 mM EDTA [pH 8.0], 0.5 mM phenylmethylsulfonyl fluoride, and 0.5 mM DTT) for 2 h at 4°C. After dialysis the extract was centrifuged at 23,000 × g for 10 min at 4°C, and the supernatant (VZV-infected melanoma nuclear extract) was used in the coimmunoprecipitation experiments.
Coimmunoprecipitation of the ORF63 protein and IE62 using an anti-IE62 MAb.
Suspensions of 100 μl of protein G-Sepharose (Amersham, Uppsala, Sweden) were blocked with a 4% milk-PBS solution for 1 h at 4°C. VZV-infected melanoma nuclear extracts (500 μg) were added to the protein G-Sepharose in combination with an anti-IE62 MAb (25 μl). The reaction mixture was incubated at 4°C for 6 h. After the incubation the reaction products were spun down and washed three times with a PBS–0.1% Tween 80 solution. The beads were then resuspended in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis SDS-PAGE loading buffer and boiled for 10 min. The final volume was 100 μl. Bound proteins were separated on an SDS–10% PAGE gel and transferred to nitrocellulose membranes. To detect IE62, a polyclonal αIE62 antibody was used as the primary antibody, and to detect ORF63, a polyclonal anti-ORF63 antibody was used as the primary antibody, in the Western blots. Reactive bands were visualized using goat anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase (Chemicon, Temecula, Calif.) in conjunction with Supersignal West Pico Chemiluminescence substrate (Pierce, Rockford, Ill.).
Assays for IE62 and ORF63 protein interactions.
Experiments to assess binding of the ORF63 protein to IE62 protein were done using ORF63 expressed as a fusion protein with maltose binding protein (MBP) using the pMalCT plasmid as described by Stevenson et al. (48). Synthesis of the ORF63 protein was demonstrated using an antiserum to the ORF63 protein kindly provided by P. R. Kinchington (University of Pittsburgh). The fusion protein was purified by amylose affinity chromatography. The IE62 protein was generated by expression of ORF62 in baculovirus and purified by using Q Sepharose and Sp Sepharose, as described by Spengler et al. (45). Generation of C-terminal deletions of IE62 expressed as glutathione S-transferase (GST) fusion proteins and purification of these fusion proteins were performed as previously described by our laboratories (45).
Immunoprecipitation of proteins from VZV-infected cell lysates was performed as previously described (45). Briefly, VZV-infected melanoma cells were harvested at 80% cytopathic effect (CPE) and lysed in PBS by passage through a 25-gauge needle. Proteins were precipitated by addition of a MAb to IE62 (H6), followed by addition of protein G-Sepharose beads to enhance precipitation. Precipitates were washed with PBS containing 0.1% Tween 80. Pellets were boiled in sample buffer, and the proteins present were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The presence of IE62 and the ORF63 proteins was detected using polyclonal antibodies against these proteins.
An enzyme-linked immunosorbent assay (ELISA) method was used to detect binding of baculovirus-expressed IE62 protein and GST fusion proteins containing C-terminal truncations of IE62 to an MBP-ORF63 fusion protein. The construction, expression, and purification of the GST-IE62 truncations have been described previously (45). For the ELISAs, 500 ng of target protein (MBP-ORF63, MBP, or bovine serum albumin [BSA]) was adsorbed onto 96-well plates at 4°C overnight. The plates were blocked with 2% BSA and then washed and probed with intact IE62 or the GST-IE62 truncations. The plates were developed and read as described by Spengler et al. (45).
Infection of T cells.
Primary T cells were isolated from human tonsils obtained from the Department of Pathology, Stanford University Medical Center. The tissue was disassociated, resuspended in prewarmed RPMI medium plus 10% FBS, and incubated at 37°C for 30 min to remove adherent cells. The nonadherent cells were loaded on an affinity T-cell column (Pierce, Inc.) to enrich for T cells, yielding more than 90% purity. The MRC-5 cell monolayer was inoculated with virus-infected cells showing a CPE of 3 to 4+ (ca. 75 to 90% of the cells exhibiting altered morphology) at a ratio of 1 infected cell to 10 uninfected cells. After 24 h, when a CPE of 1 to 2+ (ca. 25 to 50% of the cells exhibiting altered morphology) was observed, 5 × 106 to 6 × 106 T cells were added to the infected monolayer and incubated in RPMI medium plus 10% FBS supplemented with 4 U of interleukin-2, 5 μM β-mercaptoethanol, and 10 μg of gentamicin at 37°C. T cells were harvested 48 h after infection. To identify VZV-infected tonsillar T cells, MAbs were used that were specific for human CD4 (clone S3.5, conjugated with phycoerythrin [PE]), human CD8 (clone 3B5, conjugated with fluorescein isothiocyanate [FITC]), and VZV-immune or non-VZV-immune polyclonal human serum (IgG purified) along with PE- or FITC-conjugated goat anti-human IgG (CalTag Laboratories, South San Francisco, Calif.).
For fluorescence-activated cell sorter (FACS) analysis, aliquots of VZV-infected tonsillar T cells (approximately 106 cells) were washed and resuspended in 100 μl of FACS staining buffer (PBS with 1% fetal calf serum–0.2% sodium azide). The cells were incubated with human anti-VZV immune serum on ice for 30 min, washed in staining buffer, and incubated with PE- or FITC-labeled goat anti-human IgG (Caltag, Inc., Burlingame, Calif.) and PE-labeled goat anti-mouse IgG (Jackson Laboratories, West Grove, Pa.) on ice for another 30 min to detect VZV antigens. The cells were then washed and incubated with peridinine chlorophyll protein (PerCp)–anti-CD4 (Becton Dickinson, Inc., San Jose, Calif.) on ice for 30 min, after which they were washed, resuspended in staining buffer, and analyzed on a FACSCalibur (Becton Dickinson, Inc.). As negative controls, cells were incubated with the appropriate isotype control antibodies.
Infection of SCIDhu skin implants.
Skin implants were prepared and inoculated as described by Moffat et al. (31). Briefly, 8 weeks after implantation, mice were anesthetized and bilateral skin implants were exposed for inoculation with the test virus grown in MRC-5 cells; 50 μl of the inoculum was injected per implant using a 27-gauge needle. Control implants were inoculated with an equal number of uninfected MRC-5 cells. At 21 and 28 days postinoculation, the implants were dissected from the murine skin and divided; one half was fixed in 4% paraformaldehyde, and the other half was minced for viral titration and then stored in PBS (140 mM NaCl, 2.7 mM KCl, 15 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.6]) at −20°C. Viral titers were measured by infectious focus assay (31). Immunohistochemical staining was done with a polyclonal human anti-VZV serum, a secondary biotinylated goat anti-human antibody, and a streptavidin-alkaline phosphatase conjugate (Fast Red substrate mix [2% dimethylformamide, 0.1% Fast Red, 0.02% naphthol AS-MX phosphate, 100 mM Tris {pH 8.2}]) (5). The slides were rinsed, counterstained with hematoxylin, and examined by light microscopy.
RESULTS
Effects of ORF63, ORF70, and ORF63/70 deletions on VZV replication.
Following the deletion of ORF63, ORF70, or ORF63/70 from the pvSpe21ΔAvr cosmid, transfections were done using the cosmid clones shown in Fig. 1 in order to assess the effects of these changes on VZV replication. Transfection of the three intact cosmids, pvFsp4, pvSpe5, and pvPme19, and intact pvSpe21ΔAvr yielded 3+ CPE in melanoma cells within 8 to 10 days (Table 1). Parallel transfections, in which pvSpe21ΔORF63 or pvSpe21ΔORF70 was substituted for pvSpe21ΔAvr, showed equivalent CPE at 8 to 10 10 days. These viruses were designated rOkaΔORF63 and rOkaΔORF70. The plaque morphology was indistinguishable from that observed with rOka generated with intact pvSpe21. These experiments were repeated three times using at least two independently derived clones of each of the mutated cosmids; the results were the same in all experiments. In contrast, transfection of intact cosmids with pvSpe21ΔORF63/70 yielded no infectious virus in a series of three separate experiments. Cells were split every 4 to 5 days and held for 28 to 30 days to ensure that a virus with a slow-growth phenotype was not missed. These observations suggest that one copy of ORF63 or ORF70 is required for VZV replication in culture.
TABLE 1.
Infectivities of cosmids with deletions of ORF63, ORF70, or ORF63/70
| Cosmid cotransfected with pvFsp4, pvSpe5, and pvPme19 | CPEa | Days post-transfection | Designation |
|---|---|---|---|
| pvSpe21ΔAvr | +++ | 8–10 | rOka |
| pvSpe21ΔORF63 | +++ | 8–10 | rOkaΔORF63 |
| pvSpe21ΔORF70 | +++ | 8–10 | rOkaΔORF70 |
| pvSpe21ΔORF63/70 | − | 28–30b | NAc |
| pvSpe21ΔORF63/70+63@Avr | +++ | 8–10 days | rOka/ORF63rev |
Symbols indicate proportions of cells exhibiting altered morphology: +, 25 to 50%; ++, 50 to 75%; +++, 75 to 90%; −, no CPE detected.
Cells were split every 4 to 5 days to ensure that a slow-growth phenotype would not be missed.
NA, not applicable.
Construction and characterization of the repaired virus, rOka/ORF63rev.
In order to confirm that the failure to recover virus from transfections done using pvSpe21ΔORF63/70 was due to the absence of the diploid gene, the pvSpe21ΔORF63/70 cosmid was further modified to insert the ORF63 sequence into the noncoding AvrII site between ORFs 65 and 66, which is located just before the US TR segment (Fig. 1). The requirement for at least one copy of the ORF63/70 gene for VZV replication was demonstrated by the recovery of infectious virus, designated rOka/ORF63rev, in transfections done with pvSpe21ΔORF63/70+63@AvrII and the three cosmids pvFsp4, pvSpe5, and pvPme19 in two independent experiments (Table 1). The growth kinetics and plaque morphology of the repaired virus were in distinguishable from those of rOka, rOkaΔORF63, and rOkaΔORF70 viruses.
PCR analysis of rOkaΔORF63, rOkaΔORF70, and the repaired virus, rOka/ORF63rev.
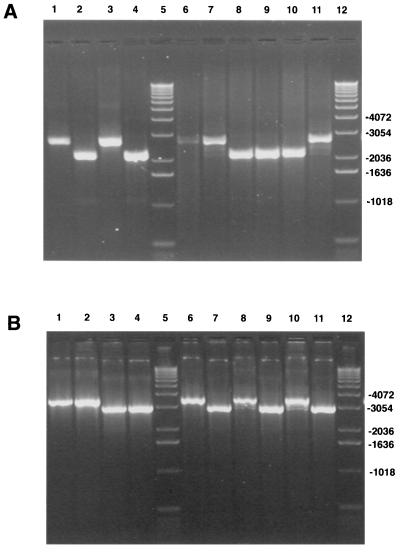
PCR analysis was done on DNA isolated from cells infected with rOka, rOkaΔORF63, rOkaΔORF70, and rOka/ORF63rev in order to confirm that the recombinant viruses produced from transfections using the ORF63 and ORF70 deletion cosmids, as well as the repaired cosmid, had the expected genetic changes. The cosmids used to generate the viruses, pvSpe21ΔAvr, pvSpe21ΔORF63, pvSpe21ΔORF70, and pvSpe21ΔORF63/70+63@Avr, as well as the double deletion cosmid, pvSpe21ΔORF63/70, were also tested by PCR. PCRs were carried out using primers designed to amplify specifically either ORF63, ORF70, or the AvrII site. As shown in Fig. 2, PCR of the cosmid DNAs yielded bands of the expected sizes, which were 2,672 nt for intact pvSpe21ΔAvr and pvSpe21ΔORF70 versus 1,837 nt for pvSpe21ΔORF63 and pvSpe21ΔORF63/70, using the ORF63 primers (upper panel, lanes 2 to 6). Using the ORF70 PCR primers, a 3,528-nt product was generated from the pvSpe21ΔAvr and pvSpe21ΔORF63 cosmids compared with a 2,692-nt product from the cosmids containing ORF70 deletions. The repaired cosmid yielded a 3,809-nt product because the AvrII site is located between the ORF70 primers (Fig. 2, lower panel, lanes 2 to 6). A product of 1,123 nt was obtained from all cosmids, except for the repaired cosmid, pSpe21ΔORF63/70+63@ Avr, from which a 2,240-nt product was obtained using the AvrII site primers (data not shown). All band sizes were as predicted for the intact, deleted, and repaired cosmids.
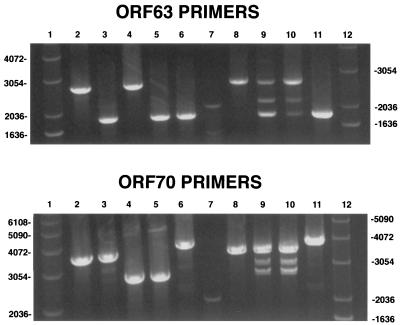
FIG. 2.
PCR analysis of rOkaΔORF63, rOkaΔORF70, and rOka/ORF63rev. PCR analysis was done with cosmid DNA (lanes 2 to 6) or DNA isolated from infected cells (lanes 8 to 11) as described in Materials and Methods. (Upper panel) Results obtained using the ORF63 primers; (lower panel) PCR products obtained using the ORF70 primers. Lanes 1 and 12, 1-kb DNA ladder; lanes 2, pvSpe21ΔAvr; lanes 3, pvSpe21ΔORF63; lanes 4, pvSpe21ΔORF70; lanes 5, pvSpe21ΔORF63/70; lanes 6, pvSpe21ΔORF63/70+63@Avr; lanes 7, 100-bp DNA ladder; lanes 8, rOka; lanes 9, rOkaΔORF63; lanes 10, rOkaΔORF70; lanes 11, rOka/ORF63rev.
In contrast to the analysis of cosmids, PCRs done using DNA from cells infected with the mutant viruses yielded some unexpected results. As shown in Fig. 2, PCR of the repaired virus, rOka/ORF63rev, and of rOka yielded a single product of the expected size using either the ORF63 or ORF70 primers (lanes 8 and 11). However, three discrete products were observed with each set of primers when DNA from cells infected with either rOkaΔORF63 or rOkaΔORF70 was used as the PCR template (Fig. 2, lanes 9 to 10). The slowest-migrating product was of the size expected from intact viral DNA, while the fastest-migrating product was as expected after deletion of ORF63 or ORF70 from the viral genome. The appearance of the third product, of intermediate size, was not predicted. Use of the ORF63 primers was expected to generate a single product of 1,837 nt from the rOkaΔORF63 virus and a single product of 2,672 nt from the rOkaΔORF70 virus; similar results were predicted with the ORF70 primers. The same results were obtained when this experiment was repeated using infected-cell DNA produced by viruses made in three independent transfections with pvSpe21ΔORF63 and two transfections with pvSpe21ΔORF70 (data not shown). In order to verify further that these observations were not due to an artifact of the PCRs or the primers used, two new sets of primers were designed and the PCRs were repeated. Again, a single product was obtained from the cosmid DNA with ORF63 or ORF70 deleted and from DNA from cells infected with repaired rOka/ORF63rev or rOka, while three products were generated using DNA from cells infected with rOkaΔORF63 and rOkaΔORF70, as observed using the first sets of PCR primers (data not shown).
Sequence analysis of PCR products from rOkaΔORF63 and rOkaΔORF70 mutants.
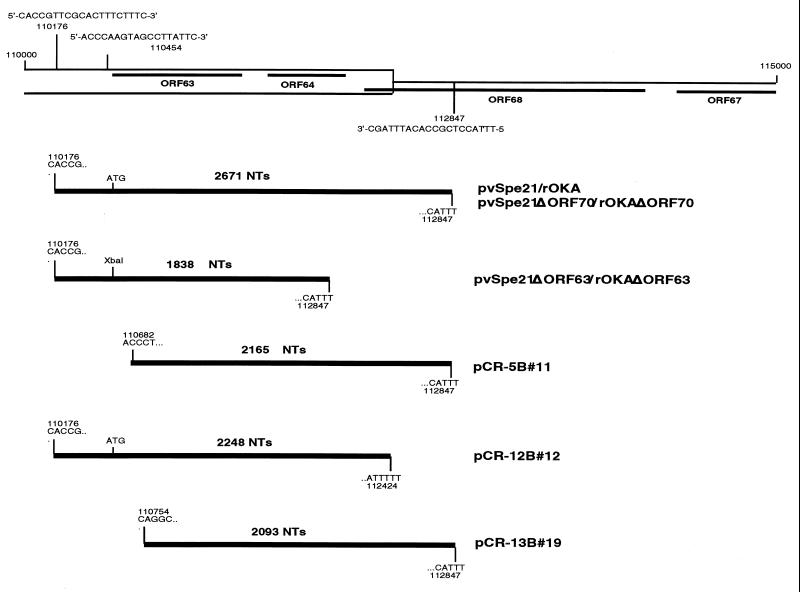
To characterize the DNA products amplified by the ORF63 and ORF70 primers, and to analyze the middle band amplified from the rOkaΔORF63 and rOkaΔORF70 viruses, the products of the individual PCRs were resolved on agarose gels, isolated, and cloned into pCR-TOPO cloning vector (Invitrogen). Positive clones were subjected to DNA sequence analysis using either priming sites found on the plasmid vector or custom-designed sequencing primers. As predicted, slow- and fast-migrating PCR products were observed when the ORF63 or ORF70 PCR primers, corresponding to the full-length and deleted regions of ORF63 or ORF70, were used (Fig. 3). The full-length products contained the ORF63 or ORF70 start codon at the expected location, while the deletion product contained the XbaI restriction enzyme site inserted in place of ORF63 or ORF70. Three clones containing the intermediate DNA product amplified with the ORF63 primers were isolated; these clones were designated pCR-5B#11, pCR-12B#12, and pCR-13B#19. Sequence analyses of the intermediate fragments generated when the ORF63 primers were used to amplify infected-cell DNA yielded unexpected results. Instead of showing the deletion of an internal region of about 400 nt, each fragment was missing nucleotides from either the beginning or the end of the expected DNA sequence. One clone containing the intermediate DNA product amplified using the ORF70 primers, pCR-29B#13, was isolated. Similar results were obtained by sequence analysis of the intermediate product made with the ORF70 primer pair; the 2,962-nt fragment was missing about 500 nt from the 3′ end of the insert (data not shown). At this time we do not know whether these aberrant PCR products arise from partially deleted VZV genomes or from fragments of viral DNA not incorporated into genomes, since the PCRs were done using total cellular DNA harvested from infected cells.
FIG. 3.
Sequence analysis of ORF63 deletion viruses. The upper section is a diagram of a portion of the IRs-US; the locations and sequences of the primers used for PCR (positions 110176 and 112847) and DNA sequence analysis (position 110454) are indicated. The lower section comprises diagrams of the PCR products described in Fig. 2 that were cloned into pCR2.1. The nucleotide number and sequence of the beginning and end of each PCR product are noted.
In vitro characterization of ORF63 interactions with IE62.
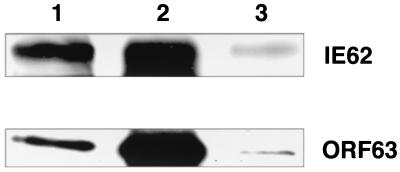
Since the gene deletion experiments indicated that one copy of the ORF63/70 gene pair was required for VZV replication, we hypothesized that the protein encoded by this gene may interact with the ORF62 gene product either directly or indirectly and as a result may modify the transactivating properties of IE62 at critical stages of viral infection. Interaction between these two regulatory proteins was demonstrated by coimmunoprecipitation of the ORF63 protein from VZV-infected cells using an anti-IE62 MAb (H6) and detection of the ORF63 protein by Western blotting using polyclonal anti-ORF63 antiserum (Fig. 4). Under these experimental conditions, a significant fraction of the ORF63 protein present in the nuclear extracts was found to be associated with the fraction of IE62 protein precipitated with the anti-IE62 MAb.
FIG. 4.
The ORF63 protein interacts with IE62 in situ. IE62 was precipitated from infected melanoma cell extracts using the anti-IE62 MAb H6 and protein G-Sepharose and was visualized as described in Materials and Methods. (Upper panel) Results from a gel probed with a rabbit polyclonal IgG specific for IE62; (lower panel) results from an identical gel probed with a polyclonal anti-ORF63 IgG. Lanes 1, 15 μl of the final sample after resuspension in SDS-PAGE buffer; lanes 2, positive control containing 2 μg of nuclear extract; lanes 3, negative control using sample generated in the presence of protein G alone.
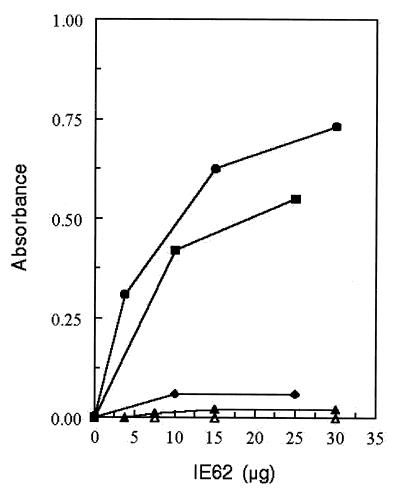
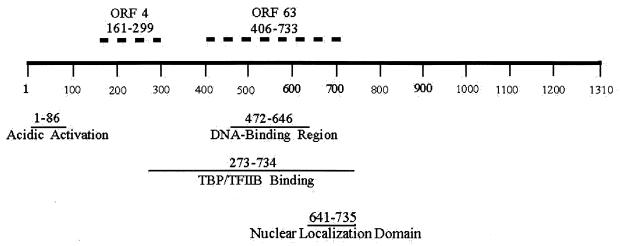
A direct interaction between the ORF62 and ORF63 gene products was documented by ELISA experiments using purified recombinant proteins (Fig. 5). Microtiter plates were coated with MBP-63 protein or MBP or BSA as a control; IE62 protein produced in baculovirus was added, and binding was assessed using the anti-IE62 MAb. The binding site for ORF63 protein within the IE62 primary structure was localized to amino acids 406 to 733 using a set of C-terminal truncations of IE62 fused to GST. In this case binding of the IE62 fragments was detected using an anti-GST antibody. As shown in Fig. 5, a fragment of IE62 encompassing amino acids 1 to 733 showed binding to MBP-ORF63 similar to that observed with full-length IE62. In contrast, titration of a fragment of IE62 encompassing amino acids 1 to 406 resulted in a signal which was just above background level (Fig. 5). Thus the region of IE62 which interacts with the ORF63 protein lies between amino acids 406 and 733. These residues overlap the DNA-binding domain of IE62 (amino acids 472 to 646) and partially overlap sequences recently shown to bind the cellular transcription factors TPB and TFIIB (15) (Fig. 6).
FIG. 5.
ORF63 binds IE62 in vitro. Affinity-purified MBP-ORF63 was adsorbed to microtiter wells and reacted with increasing amounts of recombinant IE62 purified from baculovirus or with GST fusion proteins containing C-terminal deletions of IE62. Bound IE62 or IE62 fragments were detected by ELISA using either an anti-IE62 MAb or an anti-GST polyclonal antibody. Solid circles, full-length IE62; solid squares, GST-IE62 (amino acids 1 to 733); solid diamonds, GST-IE62 (amino acids 1 to 406). Solid and open triangles represent controls in which either MBP or BSA, respectively, was adsorbed to plates and probed with intact IE62. Control experiments using MBP and the IE62 fusion proteins also showed no detectable interaction (data not shown).
FIG. 6.
Mapping of protein interaction sites in the primary structure of IE62. The location of the region of IE62 required for ORF63 identified in this study is shown along with that for ORF4, which has been mapped in our laboratories using the same strategy. Also indicated are the locations of functional domains and other protein interaction sites mapped by our laboratories and others.
Effects of ORF64, ORF69, and ORF64/69 deletions on VZV replication.
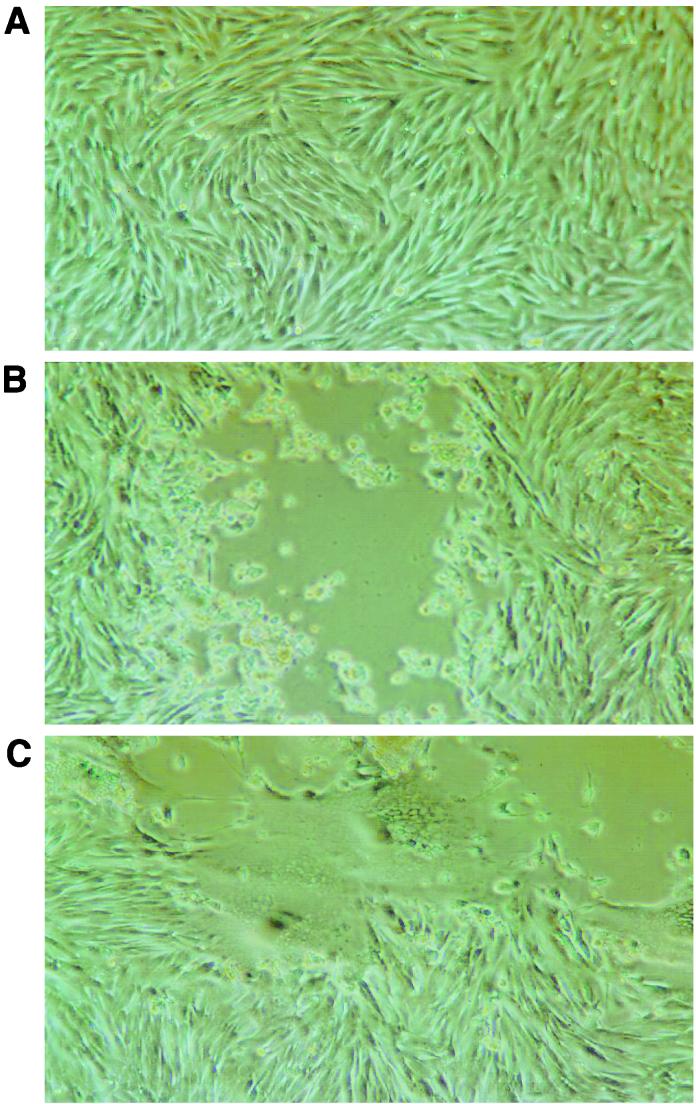
Following the removal of ORF64, ORF69, or ORF64/69 from the pvSpe21ΔAvr cosmid (Fig. 1), transfections were done using the mutant cosmid and the three intact cosmids in order to assess the effects of these deletions on VZV replication. Melanoma cells transfected with the four intact cosmids showed 3+ CPE within 8 to 10 days (Table 2). Parallel transfections in which pvSpe21ΔORF64 or pvSpe21ΔORF69 was substituted for pvSpe21ΔAvr showed equivalent CPE at 8 to 10 days and yielded viruses designated rOkaΔORF64 and rOkaΔORF69. The plaque morphology was indistinguishable from that observed when rOKA was generated with intact pvSpe21ΔAvr (Fig. 7B); these observations were reproducible when repeated three times using at least two independent clones of each mutated cosmid. Transfections of intact cosmids with pvSpe21ΔORF64/69 also yielded infectious virus within 8 to 10 days; the virus generated from these transfections was designated rOkaΔORF64/69. However, the pattern of replication of all rOKAΔORF64/69 mutants was highly unusual in melanoma cells; monolayers exhibited extensive fusion and a very enlarge plaque morphology, characterized by abnormal aggregates of multinucleated cells (Fig. 7C). This phenotype was observed with viruses generated from three separate mutant cosmid clones used to make recombinants in at least two separate transfections.
TABLE 2.
Infectivities of cosmids with deletions of ORF64, ORF69, or ORF64/69
| Cosmid cotransfected with pvFsp4, pvSpe5, and pvPme19 | CPEa | Virus designation | Phenotype |
|---|---|---|---|
| pvSpe21 | +++ | rOka | Normal |
| pvSpe21ΔORF64 | +++ | rOkaΔORF64 | Normal |
| pvSpe21ΔORF69 | +++ | rOkaΔORF69 | Normal |
| pvSpe21ΔORF64/69 | +++ | rOkaΔORF64/69 | Abnormal fusion |
At 8 to 10 days posttransfection. Symbols indicate proportions of cells exhibiting altered morphology: +, 25 to 50%; ++, 50 to 75%; +++, 75 to 90%.
FIG. 7.
Plaque morphology of rOkaΔORF64 and rOkaΔORF64/69. Melanoma cells that were either mock transfected (A), transfected with the three wild-type cosmids and pvSpe21ΔORF64 (B), or transfected with the three wild-type cosmids and pvSpe21ΔORF64/69 (C) were photographed at 9 days posttransfection. Panels B and C represent a single plaque taken from a flask containing numerous plaques with a similar phenotype.
PCR analysis of rOkaΔORF64, rOkaΔORF69, and rOkaΔORF64/69 mutant viruses.
To confirm that recombinant viruses produced from transfections using the pvSpe21ΔORF64, pvSpe21ΔORF69, and pvSpe21ΔORF64/69 cosmids had the expected mutations, PCR analysis was done on DNA isolated from cells infected with rOka, rOkaΔORF64, rOkaΔORF69, and rOkaΔORF64/69. The cosmids used to generate these viruses were included in parallel reactions. The preparations of cosmid DNA yielded bands of the expected sizes using the ORF64 primers; these were 2,672 nt for pvSpe21ΔAvr and pvSpe21ΔORF69 and 2,130 nt for pvSpe21ΔORF64 and pvSpe21ΔORF64/69 (Fig. 8A, lanes 1 to 4). The ORF69 primers generated the expected 3528 nt product from the pvSpe21 and pvSpe21ΔORF64 cosmids and the expected 2986 nt product from cosmids with the ORF69 deletions (Fig. 8B, lanes 1 to 4). PCRs done with DNA from cells infected with rOka, rOkaΔORF64, rOkaΔORF69, or rOkaΔORF64/69 yielded products of the expected sizes, as observed for the cosmid DNAs (Fig. 8, lanes 6 to 11). The PCR products were isolated and cloned into pCR-TOPO cloning vectors for sequence analysis. As expected, the larger PCR products, which were 2,672 nt with the ORF64 primers and 3,528 nt with the ORF69 primers, were determined to be wild-type sequences. The smaller PCR products, which were 2,130 nt with the ORF64 primers and 2,986 nt using the ORF69 primers, had either ORF64 or ORF69 deleted and contained the XbaI site in place of the original ORF (data not shown). Although the size difference between the full-length and deleted PCR products was smaller than that observed for the rOkaΔORF63 and rOkaΔORF70 viruses (Fig. 2), three products were also generated in PCRs using DNA from cells infected with rOkaΔORF64 and rOkaΔORF69. These results provided further evidence that the appearance of three PCR products in cells infected with the single deletion mutants was not due to a PCR artifact. We speculate that they may be generated by inversion events that occur during the replication cycle. In the case of wild-type virus such an event is undetectable, since both repeats are identical. However, in our deletion mutants, one repeat is shorter than the other, so that when the inversion occurs, it is detectable as a faster-migrating PCR product.
FIG. 8.
PCR analysis of rOkaΔORF64, rOkaΔORF69, and rOkaΔORF64/69. PCR analysis was done using cosmid DNA (lanes 1 to 4) or DNA isolated from infected cells (lanes 6 to 11). Results obtained with the ORF64 primers (A) and with the ORF69 primers (B) are shown. Lanes 1, pvSpe21ΔAvr; lanes 2, pvSpe21ΔORF64; lanes 3, pvSpe21ΔORF69; lanes 4, pvSpe21ΔORF64/69; lanes 5 and 12, 1-kb DNA ladder; lanes 6, rOka; lanes 7 and 11, rOkaΔORF69; lanes 8 and 10, rOkaΔORF64; lanes 9, rOkaΔORF64/69.
Titration of wild-type and mutant viruses.
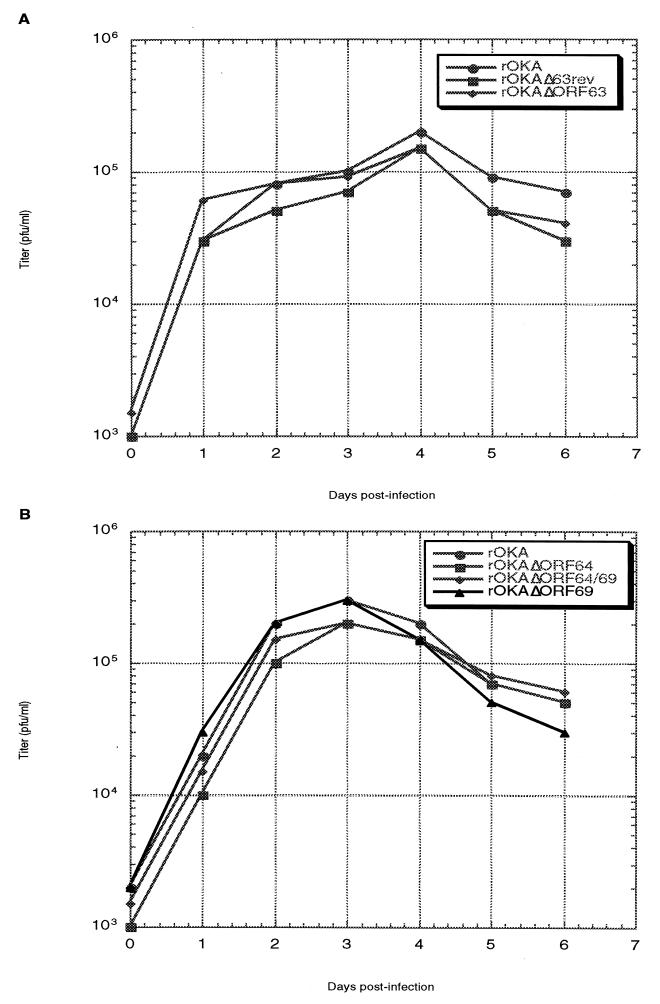
Infectious virus recovered following transfection of melanoma cells was titered on Vero cell monolayers. As shown in Fig. 9A, no significant differences in viral titers were observed among the wild-type virus, the ORF63 deletion virus, and the ORF63 restored virus at any of the six time points assayed. The results of titrations of the ORF64/69 deletion viruses were similar (Fig. 9B). These results demonstrate that removing one copy of the ORF63/70 pair, or one or both copies of the ORF64/69 pair, does not affect viral replication in vitro.
FIG. 9.
Replication kinetics of ORF63 and ORF64/69 deletion mutants. Virus-infected melanoma cells were seeded onto fresh monolayers of melanoma cells. At days 1 through 6 postinfection, the infected monolayer was harvested, and the infected cells were serially diluted and used to infect monolayers of Vero cells in duplicate. At 7 to 8 days postinfection, the Vero cell monolayers were stained with crystal violet, and the number of plaques were counted.
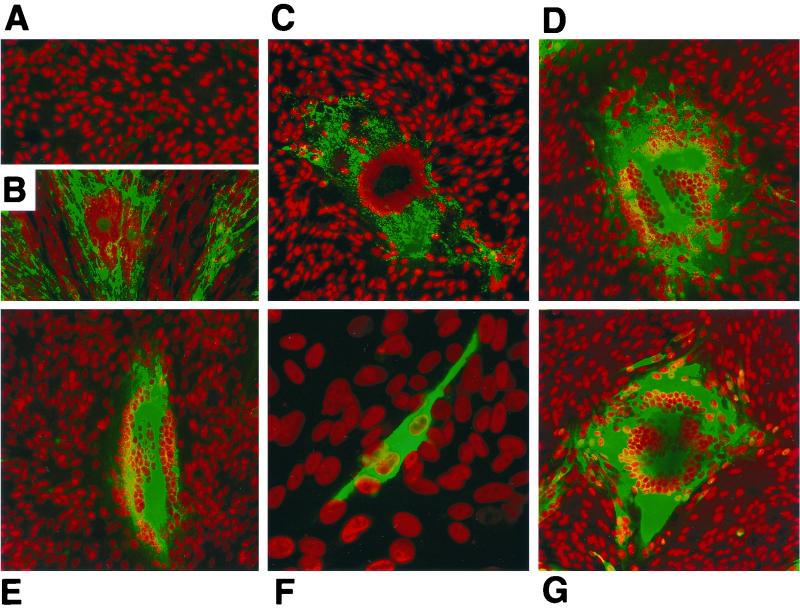
Analysis of the abnormal phenotype of the rOkaΔORF64/69 mutant.
In order to further characterize the unusual plaque morphology of the rOkaΔORF64/69 mutant, confocal microscopy was done using antibodies to gE, gH, and IE62 to stain melanoma cells infected with this mutant, rOkaΔORF64, or rOka (Fig. 10). Cells infected with the rOkaΔORF64/69 virus and stained using a MAb to gE (Fig. 10D), gH (Fig. 10E), or IE62 (Fig. 10G) displayed the unusual plaque phenotype seen by light microscopy. Cells infected with rOkaΔORF64/69 exhibited extensive fusion, and the monolayer showed many abnormal aggregates of multinucleated cells. In contrast, cells infected with rOkaΔORF64 and stained with a MAb to gE (Fig. 10C) or IE62 (Fig. 10F) exhibited the usual plaque morphology observed in melanoma cells infected with rOKA (Fig. 10B) or wild-type VZV. In addition, inspection of the confocal micrographs stained with gE reveals an increase in the amounts of gE expressed in rOkaΔORF64/69-infected cells compared to those in rOka- or rOkaΔORF64-infected cells.
FIG. 10.
Confocal microscopy analysis of ORF64/69 deletion virus-infected cells. Melanoma cells that were either mock infected (A), or with either rOka (B), rOkaΔORF64 (C and F), or rOkaΔORF64/69 (D, E, and G) were reacted with a MAb against IE62 (F and G), gE (B through D), or gH (E). This was followed by an anti-mouse antibody conjugated with Oregon Green. Nuclei were stained with Toto-3 dye and appear red in the image. Cells were examined with a Bio-Rad 1024 laser scanning confocal microscope. Magnifications, ×18.6 (A through E and G) and ×55.8 (F).
Infectivities of rOkaΔ63, rOkaΔORF64, and rOkaΔORF64/69 for human T cells.
In order to determine whether the removal of one or both copies of ORF64 altered infectivity for differentiated human T cells, tonsillar T cells were added to monolayers of human embryo lung cells that had been infected with rOkaΔORF63, rOkaΔORF64, rOkaΔORF64/69, or rOka. The percent infected T cells was determined by FACS analysis (data not shown). The percent VZV-infected T cells was 8.8% for rOkaΔORF63 versus 9.6% for rOka. In experiments with the ORF64 and ORF69 mutants, the percentage was 18.3% for rOkaΔORF64 versus 28% for rOka, and 18.2% for rOkaΔORF64/69 versus 28% for rOka (data not shown). These differences are not considered to be significant. These results suggest that deletion of ORF63 and ORF64/69 does not alter VZV infectivity for human T cells.
Infectivities of rOkaΔORF63, rOkaΔORF63rev, rOkaΔORF64, and rOkaΔORF64/69 in SCIDhu skin implants.
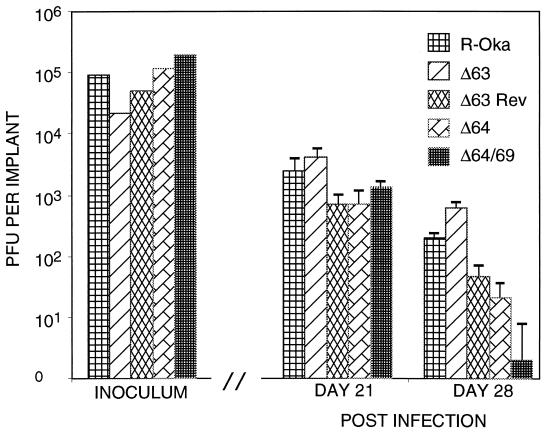
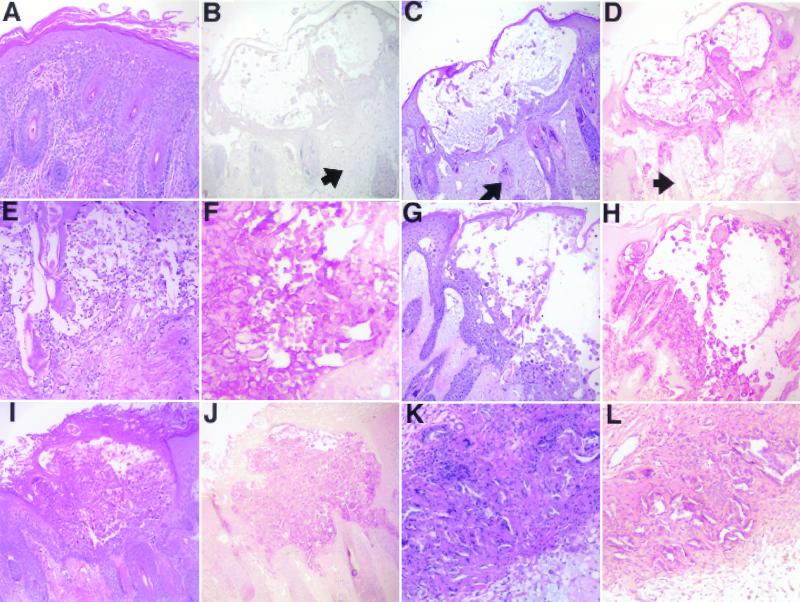
As shown in Fig. 11, none of the deletion mutants had impaired replication in SCIDhu skin implants compared with that of rOka. The inoculum titers were equivalent for all viruses, and each virus was tested in triplicate. As expected, infectious virus titers decreased by day 28, because the implant tissue was depleted of intact cells by viral replication. There was no difference in infectivity among rOka, rOkaΔ63, and rOkaΔ63rev (Fig. 12D, F, and H), indicating again that the deletion of both copies of ORF63/70 did not introduce other mutations that might have altered replication capacity. The dual deletion mutant, rOkaΔORF64/69, remained fully infectious compared with rOka and rOkaΔORF64 (Fig. 12D, J, and L). These results demonstrate that only a single copy of the ORF63/70 gene pair is required, and that the gene products of ORF64/69 are not required, for viral spread in skin implants.
FIG. 11.
Replication of mutants in SCIDhu skin implants. Skin implants in SCIDhu mice were inoculated with rOka, rOkaΔORF63, rOkaΔORF63rev, rOKAΔORF64, or rOKAΔORF64/69. Viral titers were measured by infectious focus assay (31). Each bar represents the mean titer of infectious virus recovered from infection of three animals at 21 or 28 days after inoculation. Error bars, standard errors. The titers of the inocula are shown in the left section of the graph.
FIG. 12.
Infectivity of rOka, rOkaΔORF63, rOkaΔORF63rev, rOKAΔORF64, or rOKAΔORF64/69 in SCIDhu skin implants. Skin implants that were either mock infected (A) or infected with either rOka (B, C, and D), rOkaΔORF63 (E and F), rOkaΔORF63rev (G and H), rOKAΔORF64 (I and J), or rOKAΔORF64/69 (K and L) were fixed in formalin, paraffin embedded, and cut into 3-μm-thick sections. Panels A, C, E, G, I, and K were stained with hematoxylin and eosin. Panels D, F, H, J, and L were stained with a polyclonal human anti-VZV serum. Panel B shows the absence of staining with a nonimmune serum. Arrows in Panels B through D indicate counterstained areas that had no infected cells.
DISCUSSION
This analysis of the contributions of the diploid VZV genes, ORF63/70 and ORF64/69, to VZV replication suggested that the gene product of ORF63/70 was necessary for VZV replication, whereas replication can occur in the absence of ORF64/69, even though plaque morphology is extremely aberrant. The presence of one copy of the ORF63/70 or the ORF64/69 gene pair was sufficient for normal patterns of VZV replication and plaque formation in cell culture. Infectivity was restored by insertion of ORF63 into a nonnative site, indicating that the lack of viral replication was due to the simultaneous deletion of ORF63/70, rather than to a disruption of promoter sequences or other regulatory regions affecting adjacent genes in the repeat segment or to random mutations elsewhere in the VZV genome.
This evidence that ORF63 is indispensable for lytic infection, considered along with the observations that ORF63 transcripts are predominant in latently infected ganglia and that ORF63 protein is also made during latency, suggest that the ORF63 protein plays a critical role in all stages of VZV pathogenesis (7, 23, 42). Relatively little is known about the functions of the ORF63 gene product. It is designated ORF63 protein because it is expressed at immediate-early times in VZV-infected cells and it is present in virions, as a component of the tegument (19). It is a phosphoprotein that localizes predominantly to the nuclei of infected and transfected cells, and it has a nuclear localization signal between amino acids 210 and 278 (9, 19, 48). Unlike its HSV homologue, ICP22, ORF63 does not have spliced transcripts, but its transcription is complex; it has multiple initiation and polyadenylation sites (19, 20, 22). It is phosphorylated by cellular casein kinase II, and recent evidence from our laboratories indicates that it is heavily phosphorylated by recombinant VZV ORF47 kinase (T. K. Kenyon, J. Lynch, W. Ruyechan, and C. Grose, unpublished data). ORF47 phosphorylation apparently can be substituted for by cellular kinases, based on studies with ORF47 deletion mutants (14, 15, 47). When it is detected in latently infected neurons, ORF63 protein is localized primarily to the cytoplasm, suggesting that its transfer into the nucleus is a characteristic of lytic rather than latent infection (9, 23). In skin biopsy specimens analyzed by immunohistochemical or in situ hybridization methods, early lesions showed ORF63 protein in keratinocytes and in dermal nerves and perineural type I dendrocytes (1). The VZV ORF63 protein is also a major target of VZV-specific CD4 and CD8 memory T cells, indicating that it is processed by antigen-presenting cells during primary infection (1).
The VZV ORF63/70 homologue in HSV is ICP22. This protein is phosphorylated by the HSV UL13 and US3 viral kinases (38, 39) and is nucleotidylated by casein kinase II (28, 29). In contrast to our findings with the ORF63/70 protein, ICP22 has been shown to be dispensable for growth in tissue culture. The efficiency of replication of ICP22 mutants is cell type dependent (34, 37, 44); the levels of ICP0 and a subset of gamma genes are diminished and ICP22 mutants are highly attenuated in experimental animal models (34, 39, 44). Studies examining RNA polymerase II in HSV-infected cells indicate that ICP22 is required for the production of an altered phosphorylation state of that enzyme (40, 41). None of these functions of ICP22 appears to be absolutely required for replication of the virus.
One explanation for the critical role of ORF63/70 in VZV replication may be its interaction with the ORF62 gene product. The VZV IE62 protein is encoded by the diploid gene pair ORF62/71, adjacent to ORF63/70. The IE62 protein is the major viral regulatory protein of VZV and the most abundant component of the virion tegument (19, 35). Possible interaction between the IE62 and ORF63 proteins was first reported using transient expression assays and suggested that the ORF63 protein had the capacity to downregulate the ORF62 promoter. However, a subsequent analysis indicated that ORF63 protein had minimal enhancing or suppressing effects on the ORF62 promoter, or on promoters of other immediate-early or early genes. In contrast, recent data from our laboratories show that the ORF63 protein enhances the transcription of the VZV gI promoter activated by the IE62 protein in transient transfections both in a continuous T-cell line and in melanoma cells (J. Lynch, K. Matteson, J. Hay, and W. Ruyechan, unpublished data). Thus, the effect of the ORF63 protein may be promoter dependent; understanding this aspect of ORF63 protein function will necessitate examination of an extensive panel of VZV promoters representing all three kinetic classes of viral genes. The experiments presented here show that ORF63 protein binds to IE62 protein during lytic infection of tissue culture cells. This protein-protein interaction was confirmed using purified recombinant IE62 and ORF63 proteins in vitro, and ORF63 protein was shown to bind to a region of the IE62 protein encompassing amino acids 406 to 733. This region of the IE62 protein has been shown to be involved in two other important macromolecular interactions. Amino acids 472 to 646 contain the DNA-binding domain of IE62 (50, 52), and amino acids 273 to 734 have recently been shown to interact with mammalian TATA-binding protein (TBP) and the general transcription factor TFIIB (36). Thus, ORF63 protein could, upon binding, alter the affinity or specificity of the IE62 protein for a given promoter region. Similarly, binding of ORF63 protein could alter the interaction of IE62 protein with TBP and/or TFIIB, resulting in a change in the efficiency of the interaction of the basal cellular transcription machinery at IE62-activated promoters.
It is important to note that the ORF63 protein binding region within the IE62 amino acid sequence is distinct from the regions of IE62 protein that we found were required for interaction with the ORF4 and ORF9 products (45; M. Spengler, W. Ruyechan, and J. Hay, unpublished data). This difference suggests that the activity of the IE62 protein can be influenced at specific sites by several distinct binding partners, both viral and cellular. This possibility is currently being investigated in our laboratories. Finally, the in vitro experiments demonstrating an interaction between IE62 and ORF63 proteins were performed with purified recombinant proteins, demonstrating that a direct physical interaction does not require any additional viral or cellular factors. We do not yet know if the phosphorylation state of either IE62 or ORF63 proteins significantly affects this interaction, as has been shown to be the case with interaction between IE62 and IE4 proteins (46). Since these data were obtained with baculovirus and bacterially expressed proteins, phosphorylation by either of the viral kinases is not a requirement for interaction.
With respect to the ORF64 and ORF69 gene pair, these experiments demonstrated that this diploid gene was dispensable for VZV replication in tissue culture. However, deletion of both copies of VZV ORF64/69 was associated with extensive syncytial changes and formation of large polykaryocytes, most of which contained more than 50 nuclei. We interpret these results as indicating that the phenotype is attributable to the double deletion of ORF64/69, and not to other, unidentified mutations, because viruses that had a single deletion of each gene had the phenotype of viruses derived from nonmutated cosmids, and the abnormal phenotype was observed with multiple mutants derived from independently generated ORF64/69 deletion cosmids. Although HSV-1 mutants designed to remove only the HSV-1 homologue, Us10, have not been described, early experiments demonstrated that a cluster of genes in the US component of the HSV-1 genome, including Us10, was dispensable for growth in tissue culture cells. The absence of Us10, as part of a segment of the genome missing from another HSV-1 isolate, also had no effect on replication or plaque morphology (51), and Us10 was not required for mouse neurovirulence (33, 53). Of note, HSV-1 Us10 is present in only one copy, and it is outside the repeats flanking the US segment. VZV ORF64 is one of the genes that seem to have moved from the US into the repeats during the evolution of the alphaherpesviruses (8, 9, 27). Simian varicella virus (SVV) has a gene designated RS3, which has a predicted 56% amino acid identity to VZV ORF64 (13); gene 66 of equine herpesvirus 1 (EHV-1) is another homologue (49). Overall, the homologies among ORF64, SVV RS3, and Us10 are limited, but Us10 is twice as large as ORF64 and RS3, and these two genes are more closely related to each other than to Us10 or EHV-1 gene 66. The functions of this gene family have not been defined. ORF64, like the related genes encoded by other alphaherpesviruses, has an atypical zinc finger motif characteristic of regulatory proteins that bind to specific DNA or RNA sequences. There is some evidence that Us10 may be a virion-associated protein (3).
The formation of the large polykaryocytes by the rOkaΔORF64/69 mutant is of interest because it indicates that fusion of VZV-infected cells may be modulated by expression of genes that have not been previously associated with fusion. The VZV gH-gL heterodimer is known to mediate fusion in transient expression assays; gL is required as a chaperone for gH transport to the cell membrane (11, 12, 26). In the absence of gL, the gH glycoprotein does not fully mature, nor does it act as a potent fusogen. However, when gL is lacking in the VZV transfection system, gE can provide compensation and allow gH to traffic to the plasma membrane. Surprisingly, when both gB and gE were inserted into the same vaccinia virus genome and coexpressed, abundant fusion and syncytium formation ensued, to an extent that was similar to that with gH and gL coexpression (L. Maresova, T. Pasieka, and C. Grose, submitted for publication). The hypothesis which best explains the unanticipated role of gE in facilitating gB-mediated fusion is based on the history of herpesvirus genomic evolution described by McGeoch and Davison (27). Their analysis suggests that the VZV US segment is the most recent addition to the alphaherpesvirus genome. Since the VZV US segment contains the fewest glycoproteins, it is considered to be more primitive than the HSV US segment, which contains the important gD fusogen. In this study, confocal microscopy experiments suggested that gE expression was more abundant in cells infected with the rOkaΔORF64/69 mutant than in those infected with the recombinant vOka strain or ORF64 or ORF69 single-deletion mutants. The VZV US segment encodes only two glycoprotein genes, gE and gI, and has no gD or gG genes. In the absence of gD, VZV gE may have assumed a fusion-regulatory role. If so, the ORF64/69 gene product, located in the repeats flanking the US region, may downmodulate gE gene expression. Without the ORF64/69 protein, the expression of gE appears to be increased, with a resulting enhancement of cell-cell fusion.
We noted that the replication of recombinants generated from single deletions of ORF63 or ORF70 and from single deletions of ORF64 or ORF69 in cell culture was associated with the production of DNA that represented a partial sequence of the gene. The input cosmids in the transfection system contain the S region in a single orientation. However, DNA isolated from recombinant virus or from infected cells yields restriction enzyme patterns consistent with inversion of the S segment (17). Davison and Scott have suggested that inversion occurs via recombination events during the early rounds of replication in which the viral DNA is being replicated via theta intermediates (9). The identification of truncated sequences of the deleted ORFs raises the possibility that specific sites and/or regions of specific length are required for recombination leading to inversion. The deletion of sequences within the inverted repeats could lead to abortive events, with truncation of sequences at or near the site of recombination. The R4 repeat, one of the five repeat elements made up of variable numbers of a 27-bp repeat found within the VZV genome, occurs in an intragenic region adjacent to the two copies of the viral origin of DNA replication within the IR-TR repeat element (4, 9) and just upstream of the ORF63/70 and ORF64/69 genes. This sequence is likely to be single-stranded during the earliest events of replication, based on its proximity to the viral DNA replication origins, and could provide the site for a specific crossover event during this phase of replication. Since total infected-cell DNA was used for PCR in this study, it is not possible to state whether the unexpected products represent viral genomic DNA. However, the availability of the single-deletion mutants will allow an investigation of their source, and if they are genomic, potential mechanisms for recombination and inversion of the S segment of VZV DNA can be explored.
Because of the extreme cell-associated nature of VZV replication, the cosmid mutagenesis approach has been an essential tool for defining VZV ORFs that are dispensable for replication (3, 9). The genes that can be deleted without blocking infectivity for cell culture include ORF1 (membrane protein), ORF8 (dUTPase), ORF9A, ORF10 (transactivator/tegument), ORF13 (thymidylate synthetase), ORF14 (gC), ORF19 (ribonucleotide reductase), ORF32, ORF36 (thymidine kinase), ORF47 (protein kinase), ORF57, ORF59 (uracil-DNA glycosylase), ORF61 (transactivator/transrepressor), ORF66 (protein kinase), ORF67 (gI) (5, 25) and, most recently, ORF S/L (17). Our experiments demonstrate that the diploid gene ORF64/69 can be added to this list of dispensable ORFs. However, removal of both copies alters VZV plaque morphology, as was observed after deletion of gI or ORFS/L (6, 17, 25). Previously, gK, encoded by ORF5, was the only VZV protein that had been proved to be essential for replication in cell culture by demonstrating restoration of infectivity by insertion of the gene into a nonnative site (30). These experiments demonstrate that one copy of ORF63/70 is also required for replication.
The SCIDhu mouse appears to be an authentic model for VZV cell-to-cell spread and fusion formation in human epidermal implants (31). The model makes it possible to define VZV cell spread phenotypes, including limited cell spread (VZV V-Oka), normal cell spread (VZV parent Oka strain and other low-passage-number clinical isolates), and accelerated cell spread (VZV-MSP) (31, 32, 43). Many VZV genes are dispensable in cell culture, but experiments in the SCIDhu mouse model using an ORF47 deletion mutant showed that such genes may be essential for VZV replication in differentiated human cells which become critical targets of infection during VZV pathogenesis in the human host (31). The fact that ORF64/69 was not required for cell spread in the SCIDhu mouse model further suggests that the ORF64/69 gene product is a fusion modulatory factor and not an essential virion structural protein. In our experiments, only one copy of ORF63/70 was sufficient to preserve infectivity for primary human T cells in culture and for skin implants in the SCIDhu model. ORF63 protein was detected in skin lesions, and it is notable for its expression in latently infected ganglia (23). Since the final stage of VZV pathogenesis is infection of neurons and satellite cells within dorsal root ganglia, our hypothesis is that the diploid ORF63/70, and perhaps also the ORF64/69 genes, may be required for VZV neurotropism and the establishment and maintenance of latency.
ACKNOWLEDGMENTS
This work was supported by grants AI36884 and AI18449 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Annunziato P W, Lungu O, Panagiotidis C, Zhang J H, Silvers D N, Gershon A A, Silverstein S J. Varicella-zoster virus proteins in skin lesions: implications for a novel role of ORF29p in chickenpox. J Virol. 2000;74:2005–2010. doi: 10.1128/jvi.74.4.2005-2010.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvin A M. Varicella-zoster virus, P. 2547–2586. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 3.Beers D R, Henkel J S, Schaefer D C, Rose J W, Stroop W G. Neuropathology of herpes simplex virus encephalitis in a rat seizure model. J Neuropathol Exp Neurol. 1993;52:241–252. doi: 10.1097/00005072-199305000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Casey T A, Ruyechan W T, Flora M N, Reinhold W, Straus S E, Hay J. Fine mapping and sequencing of a variable segment in the inverted repeat region of varicella-zoster virus DNA. J Virol. 1985;54:639–642. doi: 10.1128/jvi.54.2.639-642.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen J I, Seidel K E. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc Natl Acad Sci USA. 1993;90:7376–7380. doi: 10.1073/pnas.90.15.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen J L, Nguyen H. Varicella-zoster virus glycoprotein I is essential for growth of virus in Vero cells. J Virol. 1997;71:6913–6920. doi: 10.1128/jvi.71.9.6913-6920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohrs R J, Randall J, Smith J, Gilden D H, Dabrowski C, van Der Keyl H, Tal-Singer R. Analysis of individual human trigeminal ganglia for latent herpes simplex virus type 1 and varicella-zoster virus nucleic acids using real-time PCR. J Virol. 2000;74:11464–11471. doi: 10.1128/jvi.74.24.11464-11471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison A J. Structure of the genome termini of varicella-zoster virus. J Gen Virol. 1984;65:1969–1977. doi: 10.1099/0022-1317-65-11-1969. [DOI] [PubMed] [Google Scholar]
- 9.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 10.Debrus S, Sadzot-Delvaux C, Nikkels A F, Piette J, Rentier B. Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J Virol. 1995;69:3240–3245. doi: 10.1128/jvi.69.5.3240-3245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duus K M, Grose C. Multiple regulatory effects of varicella-zoster virus (VZV) gL on trafficking patterns and fusogenic properties of VZV gH. J Virol. 1996;70:8961–8971. doi: 10.1128/jvi.70.12.8961-8971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duus K M, Hatfield C, Grose C. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology. 1995;210:429–440. doi: 10.1006/viro.1995.1359. [DOI] [PubMed] [Google Scholar]
- 13.Gray W L, Gusick N J, Ek-Kommonen C, Kempson S E, Fletcher T M. The inverted repeat regions of the simian varicella virus and varicella-zoster virus genomes have a similar genetic organization. Virus Res. 1995;39:181–193. doi: 10.1016/0168-1702(95)00091-7. [DOI] [PubMed] [Google Scholar]
- 14.Heineman T C, Cohen J I. The varicella-zoster virus (VZV) open reading frame 47 (ORF47) protein kinase is dispensable for viral replication and is not required for phosphorylation of ORF63 protein, the VZV homolog of herpes simplex virus ICP22. J Virol. 1995;69:7367–7370. doi: 10.1128/jvi.69.11.7367-7370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heineman T C, Seidel K, Cohen J I. The varicella-zoster virus ORF66 protein induces kinase activity and is dispensable for viral replication. J Virol. 1996;70:7312–7317. doi: 10.1128/jvi.70.10.7312-7317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackers P, Defechereux P, Baudoux L, Lambert C, Massaer M, Merville L M, Rentier B, Piette J. Characterization of regulatory functions of the varicella-zoster virus gene 63-encoded protein. J Virol. 1992;66:3899–3903. doi: 10.1128/jvi.66.6.3899-3903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemble G W, Annunziato P, Lungu O, Winter R E, Cha T A, Silverstein S J, Spaete R R. Open reading frame S/L of varicella-zoster virus encodes a cytoplasmic protein expressed in infected cells. J Virol. 2000;74:11311–11321. doi: 10.1128/jvi.74.23.11311-11321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy P G, Grinfeld E, Bell J E. Varicella-zoster virus gene expression in latently infected and explanted human ganglia. J Virol. 2000;74:11893–11898. doi: 10.1128/jvi.74.24.11893-11898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinchington P R, Bookey D, Turse S E. The transcriptional regulatory proteins encoded by varicella-zoster virus open reading frames (ORFs) 4 and 63, but not ORF 61, are associated with purified virus particles. J Virol. 1995;69:4274–4282. doi: 10.1128/jvi.69.7.4274-4282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinchington P R, Fite K, Turse S E. Nuclear accumulation of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, is inhibited by phosphorylation mediated by the VZV open reading frame 66 protein kinase. J Virol. 2000;74:2265–2277. doi: 10.1128/jvi.74.5.2265-2277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinchington P R, Cohen J I. Viral proteins. In: Arvin A M, Gershon A A, editors. Varicella zoster virus: basic virology and clinical management. Cambridge, United Kingdom: Cambridge University Press; 2000. pp. 74–104. [Google Scholar]
- 22.Kost R G, Kupinsky H, Straus S E. Varicella-zoster virus gene 63: transcript mapping and regulatory activity. Virology. 1995;209:218–224. doi: 10.1006/viro.1995.1246. [DOI] [PubMed] [Google Scholar]
- 23.Lungu O, Panagiotidis C, Annunziato P, Gershon A, Silverstein S. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc Natl Acad Sci USA. 1998;95:780–785. doi: 10.1073/pnas.95.12.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahalingam R, Wellish M, Cohrs R, Debrus S, Piette J, Rentier B, Gilden D H. Expression of protein encoded by varicella-zoster virus open reading frame 63 in latently infected human ganglionic neurons. Proc Natl Acad Sci USA. 1996;93:2122–2124. doi: 10.1073/pnas.93.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallory S, Sommer M, Arvin A M. Mutational analysis of the role of glycoprotein I in varicella-zoster virus replication and its effects on glycoprotein conformation and trafficking. J Virol. 1997;71:8279–8288. doi: 10.1128/jvi.71.11.8279-8288.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maresova L, Kutinova L, Ludvikova V, Zak R, Mares M, Nemeckova S. Characterization of interaction of gH and gL glycoproteins of varicella-zoster virus: their processing and trafficking. J Gen Virol. 2000;81:1545–1552. doi: 10.1099/0022-1317-81-6-1545. [DOI] [PubMed] [Google Scholar]
- 27.McGeoch D J, Davison A J. The molecular evolutionary history of the herpesviruses. In: Domingo E, Webster R, Holland J, editors. Origin and evolution of viruses. London, United Kingdom: Academic Press; 1999. pp. 441–465. [Google Scholar]
- 28.Mitchell C, Blaho J A, McCormick A L, Roizman B. The nucleotidylylation of herpes simplex virus 1 regulatory protein α22 by human casein kinase II. J Biol Chem. 1997;272:25394–25400. doi: 10.1074/jbc.272.40.25394. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell C, Blaho J A, Roizman B. Casein kinase II specifically nucleotidylylates in vitro the amino acid sequence of the protein encoded by the α22 gene of herpes simplex virus 1. Proc Natl Acad Sci USA. 1994;91:11864–11868. doi: 10.1073/pnas.91.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo C, Suen J, Sommer M, Arvin A M. Characterization of varicella-zoster virus glycoprotein K (open reading frame 5) and its role in virus growth. J Virol. 1999;73:4197–4207. doi: 10.1128/jvi.73.5.4197-4207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moffat J F, Stein M D, Kaneshima H, Arvin A M. Tropism of varicella-zoster virus for human CD4+ and CD8+ T lymphocytes and epidermal cells in SCID-hu mice. J Virol. 1995;69:5236–5242. doi: 10.1128/jvi.69.9.5236-5242.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffat J F, Zerboni L, Sommer M H, Heineman T C, Cohen J I, Kaneshima H, Arvin A M. The ORF47 and ORF66 putative protein kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-hu mouse. Proc Natl Acad Sci USA. 1998;95:11969–11974. doi: 10.1073/pnas.95.20.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishiyama Y, Kurachi R, Daikoku T, Umene K. The US 9, 10, 11, and 12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology. 1993;194:419–423. doi: 10.1006/viro.1993.1279. [DOI] [PubMed] [Google Scholar]
- 34.Ogle W O, Roizman B. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J Virol. 1999;73:4305–4315. doi: 10.1128/jvi.73.5.4305-4315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perera L P, Mosca J D, Ruyechan W T, Hayward G S, Straus S E, Hay J. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J Virol. 1993;67:4474–4483. doi: 10.1128/jvi.67.8.4474-4483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perera L P. The TATA motif specifies the differential activation of minimal promoters by varicella-zoster virus immediate-early regulatory protein IE62. J Biol Chem. 2000;275:487–496. doi: 10.1074/jbc.275.1.487. [DOI] [PubMed] [Google Scholar]
- 37.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 38.Purves F C, Ogle W O, Roizman B. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purves F C, Roizman B. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein α22. Proc Natl Acad Sci USA. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice S A, Lam V. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J Virol. 1994;68:823–833. doi: 10.1128/jvi.68.2.823-833.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice S A, Long M C, Lam V, Schaffer P A, Spencer C A. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J Virol. 1995;69:5550–5559. doi: 10.1128/jvi.69.9.5550-5559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadzot-Delvaux C, Debrus S, Nikkels A, Piette J, Rentier B. Varicella-zoster virus latency in the adult rat is a useful model for human latent infection. Neurology. 1995;45:S18–S20. doi: 10.1212/wnl.45.12_suppl_8.s18. [DOI] [PubMed] [Google Scholar]
- 43.Santos R A, Hatfield C C, Cole N L, Padilla J A, Moffat J F, Arvin A M, Ruyechan W T, Hay J, Grose C. Varicella-zoster virus gE escape mutant VZV-MSP exhibits an accelerated cell-to-cell spread phenotype in both infected cell cultures and SCID-hu mice. Virology. 2000;275:306–317. doi: 10.1006/viro.2000.0507. [DOI] [PubMed] [Google Scholar]
- 44.Sears A M, Halliburton I W, Meigner B, Silver S, Roizman B. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spengler M L, Ruyechan W T, Hay J. Physical interaction between two varicella-zoster virus gene regulatory proteins, IE4 and IE62. Virology. 2000;272:375–381. doi: 10.1006/viro.2000.0389. [DOI] [PubMed] [Google Scholar]
- 46.Spengler M L, Ruyechan W T, Hay J. Interactions among structural proteins of varicella-zoster virus. Arch Virol Suppl. 2001;17S:71–79. doi: 10.1007/978-3-7091-6259-0_8. [DOI] [PubMed] [Google Scholar]
- 47.Stevenson D, Colman K L, Davison A J. Characterization of the putative protein kinases specified by varicella-zoster virus genes 47 and 66. J Gen Virol. 1994;75:317–326. doi: 10.1099/0022-1317-75-2-317. [DOI] [PubMed] [Google Scholar]
- 48.Stevenson D, Xue M, Hay J, Ruyechan W T. Phosphorylation and nuclear localization of the varicella-zoster virus gene 63 protein. J Virol. 1996;70:658–662. doi: 10.1128/jvi.70.1.658-662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Telford E A, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 50.Tyler J K, Everett R D. The DNA binding domains of the varicella-zoster virus gene 62 and herpes simplex virus type 1 ICP4 transactivator proteins heterodimerize and bind to DNA. Nucleic Acids Res. 1994;22:711–721. doi: 10.1093/nar/22.5.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Umene K. Conversion of a fraction of the unique sequence to part of the inverted repeats in the S component of the herpes simplex virus type 1 genome. J Gen Virol. 1986;67:1035–1048. doi: 10.1099/0022-1317-67-6-1035. [DOI] [PubMed] [Google Scholar]
- 52.Wu C L, Wilcox K W. The conserved DNA-binding domains encoded by the herpes simplex virus type 1 ICP4, pseudorabies virus IE180, and varicella-zoster virus ORF62 genes recognize similar sites in the corresponding promoters. J Virol. 1991;65:1149–1159. doi: 10.1128/jvi.65.3.1149-1159.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamada H, Daikoku T, Yamashita Y, Jiang Y M, Tsurumi T, Nishiyama Y. The product of the US10 gene of herpes simplex virus type 1 is a capsid/tegument-associated phosphoprotein which copurifies with the nuclear matrix. J Gen Virol. 1997;78:2923–2931. doi: 10.1099/0022-1317-78-11-2923. [DOI] [PubMed] [Google Scholar]