Abstract
One of the characteristics of hepatitis C virus (HCV) is the high incidence of persistent infection. HCV core protein, in addition to forming the viral nucleocapsid, has multiple regulatory functions in host-cell transcription, apoptosis, cell transformation, and lipid metabolism and may play a role in suppressing host immune response. This protein is thought to be present in the bloodstream of the infected host as the nucleocapsid of infectious, enveloped virions. This study provides evidence that viral particles with the physicochemical, morphological, and antigenic properties of nonenveloped HCV nucleocapsids are present in the plasma of HCV-infected individuals. These particles have a buoyant density of 1.32 to 1.34 g/ml in CsCl, are heterogeneous in size (with predominance of particles 38 to 43 or 54 to 62 nm in diameter on electron microscopy), and express on their surface epitopes located in amino acids 24 to 68 of the core protein. Similar nucleocapsid-like particles are also produced in insect cells infected with recombinant baculovirus bearing cDNA for structural HCV proteins. HCV core particles isolated from plasma were used to generate anti-core monoclonal antibodies (MAbs). These MAbs stained HCV core in the cytoplasm of hepatocytes from experimentally infected chimpanzees in the acute phase of the infection. These chimpanzees had concomitantly HCV core antigen in serum. These findings suggest that overproduction of nonenveloped nucleocapsids and their release into the bloodstream are properties of HCV morphogenesis. The presence of circulating cores in serum and accumulation of the core protein in liver cells during the early phase of infection may contribute to the persistence of HCV and its many immunopathological effects in the infected host.
Hepatitis C virus (HCV) is an enveloped, positive-strand RNA virus with a genome of approximately 9,600 nucleotides, encoding a polyprotein precursor of about 3,000 amino acids (aa). This viral polyprotein is cleaved by the host-cell signal peptidase and a viral protease, which gives a series of proteins, including the capsid, two envelope proteins (E1 and E2), and seven nonstructural proteins (11, 15). The putative HCV virion consists of a viral envelope and an inner core. However, infectious HCV virions have not yet been isolated and characterized due to limiting viral yields in serum and in vitro propagation systems. HCV populations in serum are heterogeneous, because virions often bind immunoglobulins (5, 7, 13) and serum β-lipoproteins (28, 34, 41). It has been suggested that the serum of infected subjects contains defective particles (34) or partially enveloped nucleocapsids (7, 13, 14, 18), but viral populations of these types have not yet been isolated and characterized.
There are no diagnostic tests, based on immunological methods, able to detect HCV envelope proteins of circulating virions. This may be due to the low concentration of circulating virions, their association with lipoproteins and/or antibodies, or the inability of the available monoclonal antibodies (MAbs) to recognize HCV particles. HCV core protein is the only HCV antigen detected by immunological methods, after the treatment of serum samples by detergents or denaturing agents, which remove the envelope of the virion and expose its internal component (1, 19, 32, 38, 39). Such assays have proved useful for the detection of core antigen in the sera of HCV patients (19, 32, 40), especially in the antibody-negative early phase of HCV-related liver disease (33).
Core protein maps to the N-terminal 191 aa residues of the HCV polyprotein, and its primary function is formation of the viral nucleocapsid (38, 39). The sites of interaction with homologous and heterologous RNA have been mapped to the N-terminal region of the HCV core protein, whereas the main homotypic interaction domain maps to the tryptophan-rich sequence (aa 73 to 108) (30). The hydrophobic signal sequence for translocation of the E1 protein into the endoplasmic reticulum is cleaved at aa 172 by proteases associated with cell membranes. This processing results in the cleavage of the p23 core protein to give p21, which forms the viral nucleocapsid (36). It has been suggested, however, that the core protein is modified posttranslationally in infected hosts (38). Very little is known about the formation of infectious virions and the role of the core protein in this process, because expression of the structural region of the complete HCV genome in mammalian cells generates no virus particles, precluding analysis of virus assembly. To date, the infection of insect cells with a recombinant baculovirus is the only system from which virion-like particles have been successfully isolated and characterized (3, 4).
HCV core protein has many effects on host-cell signaling, including: modulation of host-cell gene expression (27, 37), apoptosis by interaction with the cytoplasmic tail of the lymphotoxin receptor (26) and with tumor necrosis factor receptor (44), transforming activity (16), and modulation of lipid metabolism (2, 35). It has also recently been suggested that HCV core protein suppresses the antiviral cytotoxic T-lymphocyte response by interacting with the C1q complement receptor, thereby playing a key role in the induction and maintenance of chronic HCV infection (20, 23).
In this report, virus particles displaying the properties of nonenveloped HCV nucleocapsids were isolated from the serum of HCV-infected patients and from a baculovirus expression system in vitro. The naturally occurring particles isolated from serum had properties similar to those of HCV nucleocapsids released from putative virions by detergent treatment. The presence of nonenveloped core particles in serum and accumulation of the core protein in liver cells may induce mechanisms leading to chronic disease and the multiple immunopathological effects of HCV infection.
MATERIALS AND METHODS
Serum and plasma samples.
Plasma and serum samples were obtained from volunteer blood donors with normal alanine transaminase (ALT) levels who tested positive for anti-HCV antibodies by MONOLISA anti-HCV PLUS (Bio-Rad, Marnes la Coquette, France) and RIBA (Ortho Diagnostics) and for HCV RNA by reverse transcription (RT)-PCR. Plasma samples were stored frozen at −80°C. Serum samples, obtained from chronic HCV carriers testing positive for HCV markers in routine assays as described above, were analyzed, without freezing, a few hours after blood sampling.
Serum samples from two chimpanzees (CH1537 and CH1572) experimentally infected with HCV were tested for the presence of circulating core antigen on days 0, 28, and 45 after inoculation for CH1537 and on days 0, 38, and 52 for CH1572. ALT levels increased above cutoff values on day 13 after inoculation in CH1537 and on day 7 in CH1572 and reached peak values on days 77 and 74 after inoculation for CH1537 and CH1572, respectively. Both chimpanzees tested positive for HCV RNA (5.8 log IU/ml in CH1537 and 5.7 log IU/ml in CH 1572 by Amplicor monitor; Roche). All serum samples from CH1537 and CH1572 were negative for anti-HCV core antibodies (RIBA HCV 3.0; Ortho Diagnostics). A sample from CH1572, obtained 45 days after inoculation, was positive for anti-NS4 (c100) and anti-NS3 (c33c) antibodies.
Recombinant core proteins.
Recombinant HCV core proteins produced in HepG2 cells were isolated as previously described (8, 11). Purified recombinant HCV core protein NC 360 (aa 1 to 120), produced in Escherichia coli, was kindly provided by J. F. Delagneau (Bio-Rad Labs, Marne la Coquette, France).
Monoclonal and polyclonal anti-HCV antibodies.
The anti-core VT MAb was from Valbiotech (Paris, France; immunogen not communicated). MAb ACAP27 was obtained by immunization of mice with a synthetic peptide corresponding to the amino acid sequence (aa 39 to 72) of the HCV core protein and was provided by J. F. Delagneau. MAb 1/1 was produced in this study by immunization of mice with nonenveloped nucleocapsids isolated from plasma of asymptomatic HCV carriers as described below. This antibody recognizes amino acid sequence (aa 45 to 68) of the core protein.
The anti-E1 (A4) and anti-E2 (A11) MAbs have been described elsewhere (8, 11). A globulin fraction (HCIG), prepared from the sera of HCV carriers strongly positive for anti-core, anti-NS3, and anti-NS4 antibodies in the Abbott HCV enzyme immunoassay, was kindly provided by Ali Fattom (Nabi, Rockville, Md.). Normal human globulins were obtained from Sigma.
Fractionation of HCV-positive plasma.
Plasma samples (RT-PCR titer, 105 to 107; genotype 1a or 1b) were thawed and clarified by centrifugation for 10 min at 20,000 × g. Virus particles were precipitated overnight at 4°C in 10% polyethylene glycol (PEG) 6000 supplemented with 0.4 M NaCl. The precipitate was collected by centrifugation for 1 h at 14,000 rpm in the SW41 rotor of a Beckman centrifuge. The pellet, containing all of the HCV RNA initially detected in plasma, was resuspended in 7.5 ml of a mixture containing 0.01 M Tris-HCl (pH 7.2), 0.15 M NaCl, and 10 mM EDTA. It was then subjected to centrifugation on a discontinuous CsCl density gradient (1.10 to 1.60 g/ml) in a Beckman SW41 rotor at 40,000 rpm for 48 h at 4°C. Fractions (0.7 ml) were collected from the bottom of the tube and were assayed for HCV RNA by RT-PCR and for the presence of HCV core antigen by enzyme-linked immunosorbent assay (ELISA). All reagents used in the purification procedure contained protease inhibitors: 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg of aprotinin per ml, and 10 mM EDTA.
ELISA for the detection of HCV core antigen.
Polyvinyl plates (Maxisorb, Nunc, Denmark) were coated with the VT or ACAP27 MAb, at a concentration of 5 μg/ml. They were then saturated with a mixture of 3% BSA and 0.05% Tween 20 in phosphate-buffered saline (PBS) and incubated with 100 μl of the sample to be tested for the presence of core antigen. The bound antigen was detected with the horseradish peroxidase (HRPO)-conjugated MAb ACAP27, with orthophenylenediamine as the substrate. A492 was determined with a Titertek Multiscan ELISA plate reader.
Preparation of MAbs against serum-derived core antigen.
Fractions obtained after centrifugation of HCV-positive plasma in the CsCl gradient and containing core particles were pooled, dialyzed against PBS, and concentrated with a Nanosep Centrifugal Concentrator 300K (Pall Filtron, St. Germain en Laye, France). Aliquots (50 μl) of the preparation were injected into the spleen of BALB/c mice. Three days after immunization, mouse spleen cells were fused with cells of the Sp2/0Ag myeloma cell line. Hybridoma supernatants were screened by ELISA with the purified recombinant core protein NC 360 (aa 1 to 120) and a positive hybridoma cloned by limiting dilution. The immunoglobulin (Ig) class of MAbs was determined by using anti-mouse IgG γ-chain (Amersham) and anti-mouse IgM μ-chain (Sigma) antibodies.
Epitope mapping of MAbs with synthetic core peptides.
The epitope specificity of anti-core MAbs (ACAP27, VT, and 1/1) and human anti-HCV globulins (HCIG) was determined with a panel of synthetic peptides corresponding to fragments of HCV core protein. Synthetic core peptides were kindly provided by A. Kolobov and J. F. Delagneau. ELISA was carried out as described above with core peptides at a concentration of 1 μg/ml to coat the plates. Bound antibodies were detected with HRPO-conjugated anti-mouse IgG (heavy plus light chains) F(ab′)2 fragments (Amersham), anti-mouse IgM (Sigma), or anti-human IgG (Dako).
Competitive inhibition assays.
Competitive inhibition assays were carried out to investigate the epitope specificity of anti-core MAbs and to analyze the capacity of human anti-core antibodies to inhibit the reactivity of MAbs with HCV core protein. For these assays, polyvinyl plates were coated with purified recombinant HCV core protein NC 360 (aa 1 to 120), at a concentration of 1 μg/ml. The plates were blocked and washed as described above. One hundred microliters of human anti-HCV globulins (HCIG), normal human globulins (as a control), or unlabeled MAbs, diluted in PBS, was added to the wells and incubated for 2 h at 37°C. The plates were washed; and peroxidase-conjugated anti-core MAb (ACAP27 or 1/1) was added. The plates were then incubated for 1 h at 37°C. The plates were developed and read as described above.
RT-PCR for determination of HCV RNA.
HCV RNA was determined by nested PCR, based on amplification of the cDNA from the core region of the viral genome. For RT-PCR, viral RNA was extracted by using the commercial RNable reagent (Eurobio, Les Ulis, France). RNA was reverse transcribed by using the primer 5′-CAT/CGTA/GAGGGTATCGATGAC-3′. The cDNA was amplified with this primer and a primer binding to the 5′ noncoding region: 5′-ACTGCCTGATAGGGTGCTTGCGAG-3′. Nested PCR was performed with the primers 5′-AGGTCTCGTAGACCGTGCATCATG-3′ and 5′-TTGCGG/TG/CACCTA/TCGCCGGGGGTC-3′.
Affinity-capture RT-PCR.
Affinity-capture-RT-PCR was carried out as described by Han et al. (12). PCR tubes were coated by incubation overnight with (per milliliter) 50 μg of anti-core MAb ACAP27 (IgG1), MAb 1/1 (IgM) or control MAbs unrelated to HCV of IgG1 and the IgM class or with 1% BSA in PBS. Serum samples (50 μl) from patients positive for HCV infection according to serological tests were then incubated in antibody-coated or control tubes. RNA was then extracted from the adsorbed material and used for RT-PCR, as described above.
Isolation of nucleocapsid-like particles from recombinant baculovirus-infected insect cells.
Nucleocapsid-like particles were isolated from Sf9 (Spodoptera frugiperda) cells infected with recombinant baculovirus, according to the procedure described by Baumert et al. (3, 4). A recombinant baculovirus containing a cDNA encoding the structural proteins of HCV was kindly provided by J. Liang, National Institutes of Health, Bethesda, Md. Insect cells were lysed as previously described (3, 4). Cell lysates were concentrated by precipitation overnight at 4°C in 4% PEG–0.4 M NaCl and were then layered onto a discontinuous 1.1- to 1.6-g/ml CsCl gradient and centrifuged for 24 h at 4°C at 41,000 rpm in the SW41 rotor of a Beckman ultracentrifuge. Fractions (0.5 ml) were tested for HCV antigens by ELISA. All reagents used for the purification of nucleocapsid-like particles contained protease inhibitor cocktail (Boehringer, Mannheim, Germany).
SPR analysis.
Surface plasmon resonance (SPR) analysis was carried out with a Biacore 2000 (Biacore AB, Uppsala, Sweden). All reagents, including the P20 surfactant, the amine-coupling kit containing N-hydroxy-succinimide and N-ethyl-N4-(3-diethylaminopropyl) carbodiimide, ethanolamine hydrochloride (EDC/NHS 1/1), and CM-5 sensor chips were obtained from Biacore. The running and dilution buffer (HBS-EP [pH 7.4]) consisted of 10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, and 0.005% P20 surfactant.
Recombinant core protein NC 360 (50 μg/ml in phosphate buffer at pH 7.2) was covalently coupled via primary amino groups to the CM-5 sensor chip by the amine-coupling procedure. The SPR signals for NC 360 protein were 4,500 resonance units (RU), where 1 RU corresponds to an immobilized protein concentration of 1 pg/mm2. Anticore MAbs (1 to 20 μg/ml) or human globulins (in concentrations of 20 to 200 μg/ml) or control antibodies unrelated to HCV (in corresponding concentrations) were injected in running buffer. Changes in surface concentration resulting from interaction of the antibody with surface-fixed antigen were detected as an optical phenomenon affecting the SPR signal, expressed in RU. Kinetic constants, association rate constants, and dissociation rate constants were calculated with BIAEVALUATION 3.1 software.
Affinity chromatography.
MAb ACAP27 was purified from ascitic fluid by chromatography, with a Hitrap protein A column (Amersham, Pharmacia Biotech). The MAb (5.3 mg) was bound to the Affi-Gel Hz (Bio-Rad) as recommended by the manufacturer. A serum sample (35 ml) from a chronic HCV carrier testing positive for HCV core antigen by ELISA was applied to the column, which was then incubated for 2 h at room temperature. The column was thoroughly washed with PBS, and bound antigen was eluted from the column with 0.1 M citric acid (pH 4.0). The pH of the eluate was immediately adjusted to neutral with 1 M Tris. The eluate was then tested by ELISA for core antigen, by PCR for HCV RNA, and by electron microscopy for virus particles.
Electron microscopy.
Formvar-coated microscope grids (200 or 300 mesh) were incubated with fractions of the gradient diluted 1:10 in PBS. Virus particles were stained with 1% uranyl acetate in distilled water, and the grids were observed in a Phillips CM-10 electron microscope. For solid-phase immunoelectron microscopy, formvar-coated grids were first incubated for 5 min with anticore (ACAP27 or 1/1), anti-E2 (A11), anti-E1 (A4), or control MAbs (unrelated to HCV) at a concentration of 5 μg/ml in Tris-HCl (pH.8.0). They were then washed with the same buffer. A drop of the preparation containing virus particles was then placed on the grid without drying and incubated for 10 min. Grids were then washed with PBS, stained with 1% uranyl acetate, and examined as described above.
Western immunoblotting.
Samples were solubilized by incubation in a mixture of Tris (pH 6.8), 2% sodium dodecyl sulfate (SDS), and 5% 2-mercaptoethanol for 2 min at 100°C. They were then subjected to electrophoresis in 12% polyacrylamide gels and electroblotted onto nitrocellulose membranes. Membrane strips were incubated overnight at 4°C with 5% skim milk powder and 0.1% Tween 20 in PBS and then washed and incubated for 1 h at 37°C with anti-core MAbs (ACAP27 or VT) or E1 (A4) and E2 (A11) diluted in 1% skim milk powder. HRPO-conjugated anti-mouse IgG (heavy plus light chains) F(ab′)2 fragments (Amersham) were also used. Blots were rinsed and developed with an enhanced chemiluminescence detection system (Amersham, Little Chalfont, United Kingdom).
Immunofluorescent staining of HCV-infected liver tissue.
The experimental protocols for HCV-infected chimpanzees, including details of animal care and housing, were approved by the Centers for Disease Control and Prevention Institutional Animal Care and Use Committee. Surgical liver biopsy specimens were obtained from two chimpanzees (CH1572 and CH1537) infected with HCV genotype 1a (CDC/Chiron US-1 strain) (6) 36 and 42 days after inoculation, respectively. In the specimens, 50 to 70% of hepatocytes contained HCV antigens, as assessed by staining with fluorescein isothiocyanate (FITC)-conjugated polyclonal IgG fractions from the sera of individuals with chronic HCV infection (21). Negative controls included biopsy specimens taken from CH1572 and CH1537 before inoculation with HCV, specimens from two uninfected, naive chimpanzees, and specimens from chimpanzees infected with either hepatitis A virus or hepatitis B virus.
Cryostat sections (5 to 8 μm thick) were fixed in anesthetic ether for 5 min, air dried, and incubated for 1 h at room temperature with MAb 1/1 preabsorbed onto a liver homogenate prepared from a naive chimpanzee. Sections were washed in PBS and incubated with FITC-conjugated goat F(ab′)2 fragment against mouse IgM (μ-chain) (Cappel, ICN Pharmaceuticals, Aurora, Ohio), at a concentration of 10 μg/ml, to detect bound MAb 1/1. The slides were examined with a Zeiss microscope equipped with an epifluorescence device and an HBO 100/W2 illuminator. Controls included stainings of cryostat sections of HCV-infected livers from CH1572 and CH1537 with the FITC-conjugated antimouse IgM antibody and stainings of these sections with PBS or an irrelevant mouse MAb of the IgM class in the place of primary antibody. Sections from HCV-infected CH1572 and CH1537 livers stained with MAb 1/1 were examined with a confocal laser microscope (Zeiss LSM 510).
RESULTS
ELISA for detection of the HCV core antigen.
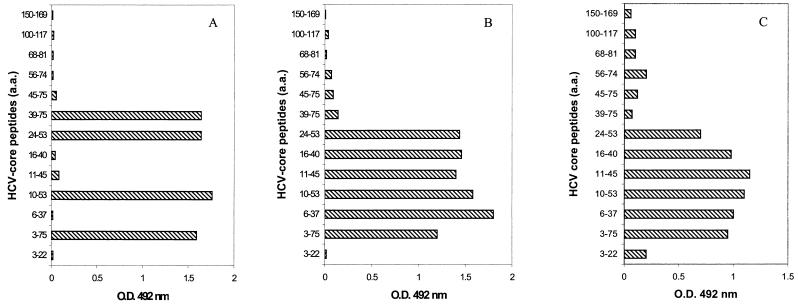
The specificity of MAbs used to detect HCV core antigen was ascertained by Western blotting with recombinant core protein produced in HepG2 cells. These MAbs reacted with protein bands corresponding to the two previously described forms (36) of the core protein: p23 and p21 (data not shown). The epitopes recognized by these MAbs were determined with a panel of synthetic peptides covering the HCV core protein: MAb VT reacted with aa 24 to 37, and MAb ACAP27 reacted with aa 40 to 53 (Fig. 1). These MAbs had extremely high affinity constants, as determined by SPR analysis (described below). The detection threshold of ELISA with either of these MAbs on the solid phase and peroxidase-conjugated MAb ACAP27 was about 1 ng, as determined with the recombinant NC 360 core protein (aa 1 to 110) as the reference antigen.
FIG. 1.
Epitope specificity of MAb ACAP27 (A) MAb VT (B) and human anti-HCV globulins (HCIG [C]) determined with a panel of synthetic core peptides.
Isolation of HCV core particles from plasma.
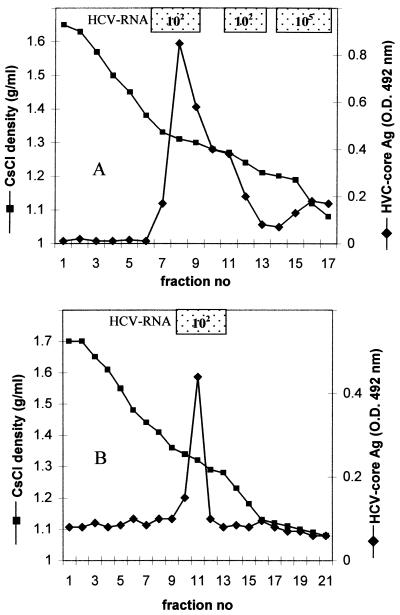
Analysis of the fractions collected after equilibrium centrifugation of HCV-positive plasma showed that the major peak of viral RNA (titer of 105, determined by RT-PCR) occurred at a density of 1.06 to 1.18 g/ml, corresponding to the putative β-lipoprotein-associated virions (41). Direct ELISA revealed the presence of HCV core antigen in a large peak at a density of 1.27 to 1.35 g/ml (Fig. 2A) in fractions containing 100 to 1,000 times less HCV RNA (titer of 102, as determined by RT-PCR) than fractions containing putative virions. Core antigen-positive fractions, subjected to a second centrifugation under the same conditions, banded at a density of 1.32 to 1.34 g/ml (not shown) Virus particles, heterogeneous in size, with predominant populations 38 to 43 and 54 to 62 nm in diameter were observed in these fractions by electron microscopy (Fig. 3A to C). These viral particles were bound to microscope grids by anticore MAbs (ACAP27 or 1/1), but not by anti-E1 and anti-E2 or control MAbs. The relative proportions of these particles differed between HCV preparations, and no particles in aggregates were observed.
FIG. 2.
(A) Isolation of naturally occurring HCV core particles from concentrated plasma by isopycnic centrifugation in a CsCl gradient. (B) Isolation of nucleocapsids from putative HCV virions by detergent treatment. A sample (1.5 ml) of the fraction of the gradient banding at a density of 1.10 g/ml (shown in panel A) and corresponding to the HCV-RNA peak was treated with 0.5% Tween 80 and centrifuged in a CsCl gradient as shown in panel A. Fractions (0.7 ml) were tested for HCV RNA by RT-PCR and for HCV core antigen by ELISA. O.D., optical density. Ag, antigen.
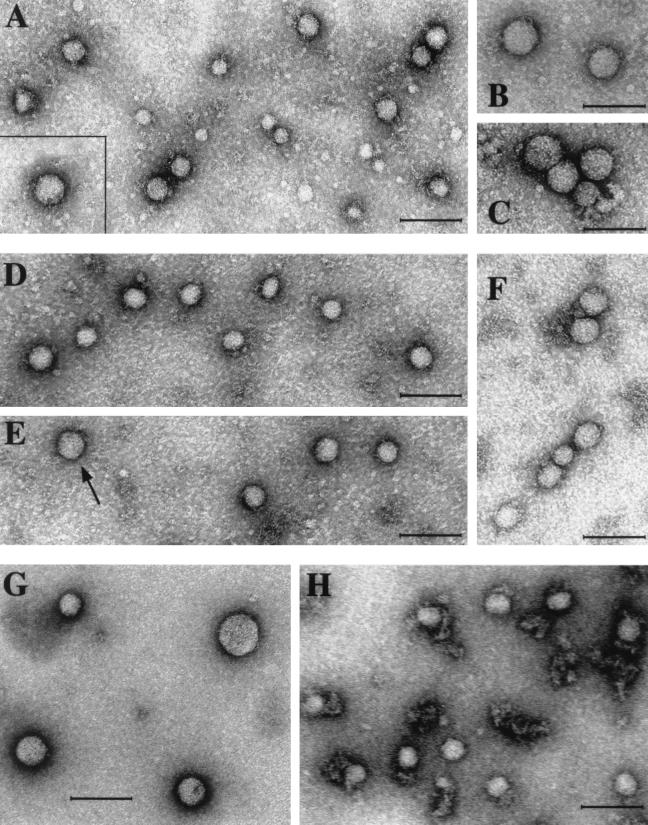
FIG. 3.
Analysis of HCV nucleocapsids by electron microscopy.(A) Direct staining of virus particles with 1% uranyl acetate. (B to H) Virus particles were adsorbed on anti-core MAb-coated microscope grids and were stained with 1% uranyl acetate. (A) Virus particles isolated from serum by affinity chromatography on anti-core MAb ACAP27 bound to the Affi-Gel Hz. (Insert) Larger particle 54 nm in diameter. (B and C) Virus particles 54 to 62 nm in diameter observed in core antigen-positive fractions from the CsCl gradient. In addition, 38- to 43-nm particles identical to those in panel A are shown. (D, E, and F) HCV nucleocapsids, isolated from the light fraction of the gradient (density, 1.10 g/ml) by treatment with Tween 80 (as shown in Fig. 2B). Most particles are 38 to 43 nm in diameter, but larger particles (shown in panel F and indicated by an arrow in panel D) were also observed. (G and H) Virus-like particles isolated from baculovirus-infected insect cells. (G) Free nucleocapsid-like particles banding in a CsCl gradient at a density of 1.35 to 1.36 g/ml in CsCl. (H) Membrane-bound nucleocapsid-like particles banding at a density of 1.25 g/ml in CsCl. Bars in all panels indicate 100 nm.
Isolation of HCV nucleocapsids from virions.
To compare the properties of HCV core particles naturally occurring in serum with those of HCV nucleocapsids isolated from putative HCV virions, an aliquot (1.5 ml) of a fraction corresponding to the HCV RNA peak (density of 1.10 g/ml) was treated with 0.5% Tween 80 and subjected to centrifugation in a CsCl gradient. HCV core antigen appeared at a density of 1.32 to 1.34 g/ml, accompanied by a shift of HCV RNA from the light region of the gradient (Fig. 2B). Virus particles, mostly 38 to 43 nm in diameter, but also larger particles 54 to 62 nm in diameter, were observed in these fractions by electron microscopy (Fig. 3D, E, and F). Both types of particles were bound to microscope grids by anti-core antibodies. This experiment showed that the HCV particles occurring naturally in the plasma of chronically infected patients and expressing core antigen at their surface had buoyant density and morphological and antigenic properties similar to those of HCV nucleocapsids released from virions by detergent treatment.
Production of new MAbs by immunization with core particles from serum.
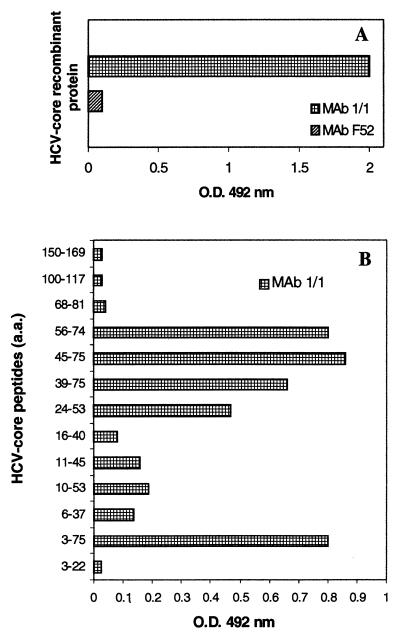
Naturally occurring core particles isolated from serum were used to induce anti-core MAbs. These new MAbs recognized epitopes mapping to aa 3 to 68 of the core protein. One of these MAbs (1/1), of the IgM class, reacted with the recombinant NC 360 core protein and recognized an epitope located between aa 45 and 68, as determined with a panel of synthetic core peptides (Fig. 4). This MAb was used for further studies.
FIG. 4.
Reactivity of MAb 1/1, generated by immunization with viral particles purified from plasma. (A) Reactivity with the recombinant core protein NC 360. O.D., optical density. (B) Reactivity with a panel of synthetic core peptides, in ELISA.
Circulating HCV core particles contain HCV RNA.
Further experiments were carried out to confirm that the core antigen-expressing particles were also present in native and unfractionated sera from HCV carriers and that the core particles circulating in serum contained HCV RNA. Fresh serum samples that had never been frozen (to exclude the possibility of virion degradation) were analyzed a few hours after blood sampling by affinity-capture RT-PCR. This method is based on the adsorption of viral particles by antibodies attached to PCR tubes. Viral RNA is then extracted and reverse transcribed, and the cDNA is amplified by PCR. The MAbs used to adsorb HCV core antigen-bearing particles were of the IgG (ACAP27) and IgM (1/1) classes to prevent possible false-positive reactions due to the presence of rheumatoid factor in the serum of most of the HCV carriers. These experiments (Fig. 5) demonstrated that the antigenic sites that reacted with anti-core antibodies were already detectable in freshly collected, unfractionated serum samples and were therefore not artifactually exposed by the fractionation procedure. They also demonstrate that at least a part of the HCV core antigen occurring naturally in serum was expressed on virus particles carrying HCV RNA.
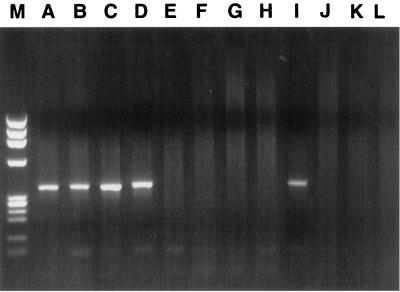
FIG. 5.
Affinity-capture RT-PCR performed on fresh unfractionated serum samples from HCV carriers. Lanes: M, molecular mass markers; A and B, PCR tubes coated with MAb 1/1, raised with serum core particles (IgM class); C and D, tubes coated with MAb ACAP27 (IgG class); E and F, tubes coated with irrelevant IgG control antibody; G and H, tubes coated with irrelevant IgM MAb; J and K, tubes coated with PBS or BSA; I, positive control for RT-PCR; L, negative control for RT-PCR.
Because the HCV core antigen was detected by direct ELISA in several unfractionated serum samples from HCV carriers, we subjected three such samples to affinity chromatography on Affi-Gel columns with the anti-core MAb ACAP27 bound to the solid support. HCV core antigen was eluted from the column, together with HCV RNA. Virus particles 38 to 43 nm in diameter and larger particles 54 to 62 nm in diameter, similar to those isolated from CsCl gradients, were observed in these preparations by electron microscopy (Fig. 3A).
Detection of HCV core antigen in serum in the presence of circulating anti-HCV antibodies.
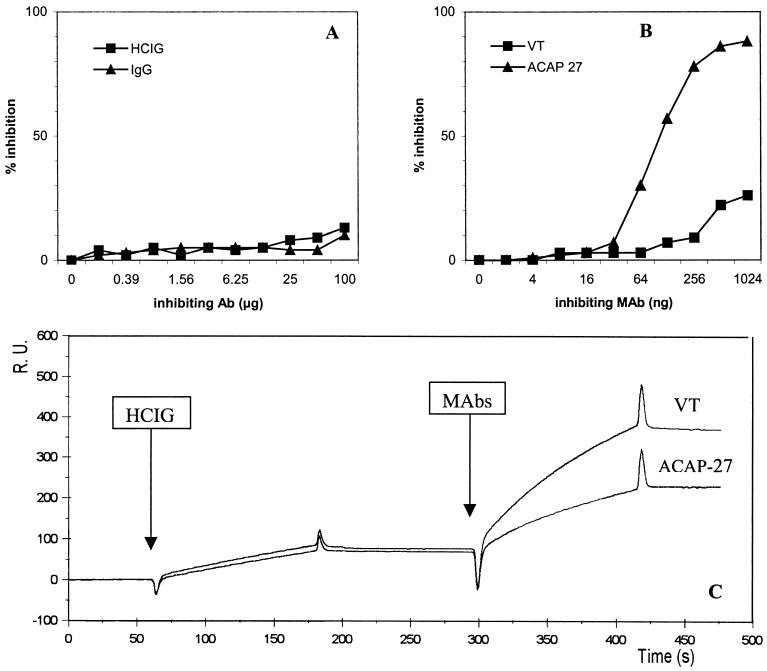
HCV core antigen was detected in several native and fractionated plasma and serum samples, despite the presence of circulating anti-HCV antibodies. We demonstrated, in competitive inhibition assays, that the reaction of mouse anti-core MAbs with the recombinant core protein was not inhibited by high concentrations (up to 200 μg/ml) of Igs isolated from the sera of HCV carriers (HCIG) (Fig. 6A). This preparation contained high levels of anti-core antibodies, as shown by routine assays and reactivity with synthetic core peptides (Fig. 1C). In contrast, homologous, unlabeled MAb ACAP27, used as a control, inhibited this system at nanogram concentrations (Fig. 6B).
FIG. 6.
Analysis of the reactivity of human and mouse anti-core antibodies with recombinant core protein. (A and B). Competitive binding assays with peroxidase-conjugated MAb ACAP27. Recombinant core protein (aa 1 to 110) was used as a solid-phase antigen. (A) A pool of globulins prepared from HCV-positive patients (HCIG), and normal human globulins (IgG) used as competitive antibodies. (B) Unlabeled MAb ACAP27 and MAb VT (recognizing different epitopes, not overlapping with that recognized by MAb ACAP27) used as competing antibodies. (C) Analysis of the reactivity of HCIG and the ACAP27 and VT MAbs with recombinant core protein NC 360 by SPR.
Further comparative analysis of the reactivity of anti-core antibodies of human and mouse origin was carried out by SPR (Biacore) (Fig. 6C). Recombinant core protein was immobilized on the sensor chip, and human anti-HCV globulins were then injected, followed by mouse MAbs. The binding of human antibodies (up 200 μg/ml) yielded only about 80 RU, whereas each of the ACAP27 and VT MAbs, injected sequentially (at a concentration of 20 μg/ml), showed strong binding (150 and 300 RU, respectively), despite the prior injection of HCIG (Fig. 6B). Complementary experiments performed with various concentrations of MAbs demonstrated an extremely high affinity for both MAbs (ACAP27 and VT), with an apparent dissociation constant Kaapp (5 × 10−11 M and 1.6 × 10−13 M, respectively), much higher than that for human anti-core antibodies (2 × 10−7 M). Overall, these data suggest that HCV core antigen could be detected in the serum of HCV-infected patients, due to the large difference in affinity between the mouse MAbs used in the detection assays and circulating human anti-core antibodies.
Nucleocapsid-like particles are produced in insect cells infected with recombinant baculovirus.
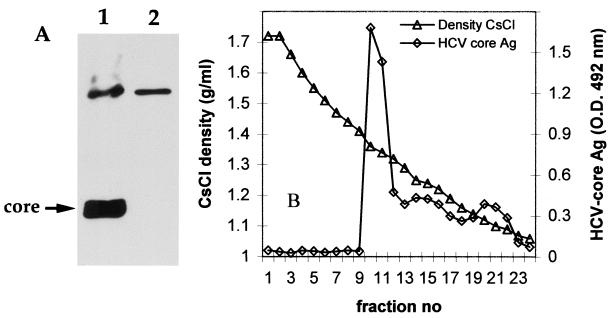
We investigated whether core particles similar to these isolated from human plasma were produced in insect cells infected in vitro with recombinant baculovirus. The HCV core protein was expressed in the infected Sf9 cells, as shown by Western blotting (Fig. 7A), but no protein bands corresponding to HCV envelope proteins were detected in these cell extracts with anti-E1 (A4) and E2 (A11) MAbs (data not shown). There was no evidence of secretion of HCV proteins to the cell supernatants by ELISA or Western blotting. The soluble fraction was therefore obtained after the lysis of infected cells as previously described (3, 4) and was subjected to isopycnic centrifugation in the CsCl gradient. ELISA detected a major core antigen peak at a density of 1.35 to 1.36 g/ml. (Fig. 7B). Nucleocapsid-like particles heterogeneous in size, from 33 to 62 nm in diameter, were observed in these fractions by electron microscopy and were bound to microscope grids coated with anti-core antibody (Fig. 3G). In a smaller peak of core antigen, at a density of 1.25 g/ml, virus particles 42 to 43 nm in diameter associated with fragments of membranes were observed by electron microscopy and were also bound to anti-core MAb-coated grids (Fig. 3H).
FIG. 7.
Isolation of virus-like particles from insect cells infected with recombinant baculovirus containing cDNA encoding the structural HCV proteins. (A) Western blot analysis of the production of core protein in Sf9 cells by using MAb ACAP27. Results for extracts from recombinant baculovirus-infected cells (lane 1) and extracts from uninfected cells (lane 2) are shown. (B) Purification of nucleocapsid-like particles from lysed cells on a CsCl gradient. HCV core antigen (Ag) was detected by ELISA, and virus particles from the core antigen peak were visualized by electron microscopy (Fig. 3G and H). O.D., optical density.
Localization of HCV core antigen in the liver of experimentally infected chimpanzees with MAb 1/1.
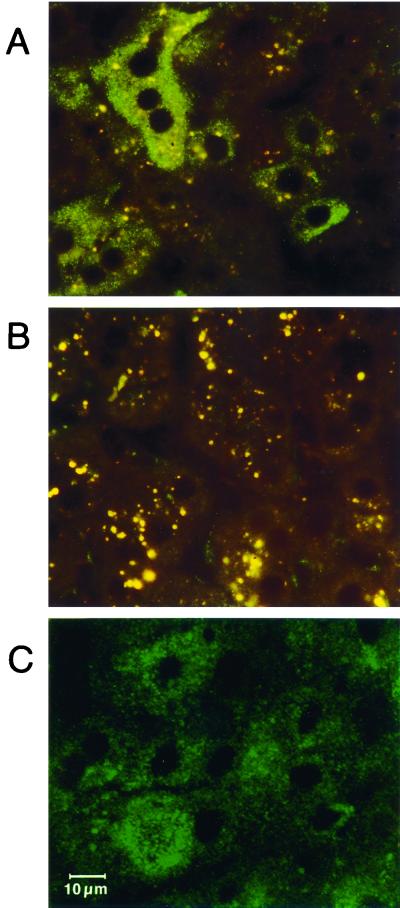
MAb 1/1, produced by immunization with serum core particles, reacted with the core antigen in the liver of chimpanzees experimentally infected with HCV. Immunostaining of liver tissue obtained during the early and viremic phase of the disease with MAb 1/1 resulted in granular fluorescence in the cytoplasm of hepatocytes. In CH1572, approximately 70% of liver cells contained small fluorescent granules, and 20% of hepatocytes showed much stronger granular and homogeneous fluorescence (Fig. 8A); in CH1537, the percentage of hepatocytes stained was similar, but the fluorescence was less intense. The selective cytoplasmic nature of the fluorescence was confirmed by observations with a confocal laser microscope (Fig. 8C). In liver specimens before inoculation, only a small number of powder-like granules of low-intensity fluorescence were identified in liver sinuses, sometimes in the hepatocytes in close vicinity (Fig. 8B). All other control specimens and immunochemical stainings were negative.
FIG. 8.
Immunofluorescence staining of a liver specimen from a chimpanzee (CH1572) experimentally inoculated with HCV. The experiment was performed with the anti-core MAb 1/1, raised by immunization with nonenveloped nucleocapsids purified from serum. (A) Indirect immunostaining with MAb1/1 followed by FITC-conjugated anti-mouse IgM, revealing a granular, fairly homogeneous pattern in the cytoplasm of hepatocytes. A group of four stained hepatocytes with one nucleus and two positive hepatocytes with two or more nuclei, displaying apple-green fluorescence, are shown. (B) A negative control, CH1572, before inoculation, immunostained with MAb 1/1, displaying rare weakly labeled sinusoidal granules. Both panels A and, especially, B, show coarse granular, orange-yellow autofluorescence of lipofuchsin. (C) Analysis of a liver section stained with MAb 1/1, examined by confocal microscopy. Shown are hepatocytes with granular or more homogeneous deposits binding MAb1/1 (indirect immunostaining), as in panel A, limited to the cytoplasm. The scale marker in panel C applies to all panels.
Detection of circulating HCV core antigen in serum of HCV-infected chimpanzees.
Selected serum samples from CH1537 and CH1572 with HCV core in hepatocytes identified by immunohistochemistry were tested for the presence of circulating HCV core antigen by ELISA in the early phase of the infection. HCV core could be detected directly in serum of both chimpanzees: 38 days after inoculation in CH1537 and 28 days after inoculation in CH1572. ELISA optical density readings were 4 to 5 standard deviations above the mean values of serum samples from the same chimpanzees before inoculation (negative controls). Both chimpanzees tested negative for circulating HCV core antigen on days 52 and 45, respectively. Serum samples positive for core antigen contained HCV RNA, but were negative for anti-core antibodies and were obtained substantially before a major peak of ALT: 39 days for CH1537 and 46 days for CH1572.
DISCUSSION
In this study, we show that virus particles that express on their surface core antigen occur naturally in the serum of HCV-infected individuals. These virus particles display physicochemical properties, antigenic reactivity, and morphology similar to those of HCV nucleocapsids isolated by the treatment of putative HCV virions with detergent. The buoyant density of these virus particles (1.32 to 1.34 g/ml in CsCl) is that expected for nonenveloped, RNA-containing nucleocapsids. Indeed, by using affinity RT-PCR, we confirmed that the HCV core antigen in serum was associated with HCV RNA and was therefore located on HCV RNA-bearing particles. Naturally occurring core particles were heterogeneous in size, with the predominant population 38 to 43 nm in diameter. Larger particles, 54 to 62 nm in diameter, were also consistently observed in core antigen preparations by electron microscopy and were also bound to the microscope grids by anti-core antibodies. Similar virus particles, mostly 37 to 43 nm in diameter, but also some larger 54- to 62-nm particles, were observed by electron microscopy in preparations of viral nucleocapsids isolated by detergent treatment of putative HCV virions.
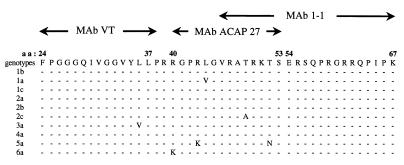
The principles of assays used in this study for detection of HCV core are different from those of all other assays published before (1, 19, 32, 33, 38–40) or recently commercialized (Ortho Diagnostics), which use detergents or denaturing agents for pretreatment of serum samples or in a “sample diluent.” These assays mainly quantify the core protein from denatured HCV virions. In our study, HCV core antigen was detected directly by ELISA and by affinity RT-PCR in several fresh, unfractionated and untreated serum and plasma samples. We were also able to isolate core particles directly from plasma by affinity chromatography with anti-core antibodies. Thus, core epitopes were not artifactually exposed by the fractionation procedure, but were instead naturally present on circulating HCV particles. At least three epitopes were found to map to the sequence between aa 24 and 68 of the serum core particle, and this sequence seems to be well conserved in different HCV genotypes (Fig. 9).
FIG. 9.
Epitope mapping of the core antigen on circulating virus particles, obtained with anti-core MAbs, and consensus within the sequence between aa 24 and 67 of the HCV core protein of various genotypes. MAbs VT and ACAP27 were used in ELISA for detection of serum core particles, and MAb 1/1 was generated by immunization with HCV particles purified from plasma.
Circulating core particles reacted with MAbs despite the presence of human anti-HCV antibody in the samples analyzed. Indeed, human anti-core antibodies from the sera of HCV-infected individuals did not compete with mouse MAbs for these antigenic sites, probably due to the large differences in affinity demonstrated by SPR analysis. Therefore, HCV core antigen can be detected directly with immunological assays involving high-affinity MAbs, not only in the initial infection phase (window period) (33), but also, as shown here, during chronic disease, even if anti-HCV antibodies are present in the serum. The detection of circulating, envelope-free HCV nucleocapsids in serum has potential diagnostic applications. (A patent is pending.)
Because the overproduction and release of nucleocapsids may be a feature of HCV morphogenesis, we investigated whether nucleocapsid-like HCV particles were also produced in insect cells infected with recombinant baculovirus containing cDNA encoding the HCV core and envelope proteins. Indeed, a population of subviral particles that was reactive with anti-core MAbs was isolated from baculovirus-infected insect cells. In accordance with previous observations (3, 4), these particles were not secreted into cell culture supernatant, and a mild detergent treatment (the same as previously used by these authors to isolate enveloped virus-like particles) was required to isolate core-like particles from infected cells. These nucleocapsid-like particles banded at a density of 1.35 to 1.36 g/ml in the CsCl gradient and were very heterogeneous in size (30 to 68 nm). The density of these particles suggested that they contained RNA, consistent with observations that the formation of nucleocapsid-like particles in vitro requires interaction of the core protein with RNA for encapsidation (22). Another population of core particles, isolated in this study from baculovirus-infected insect cells, banded at a density of 1.25 g/ml, was more homogeneous in size (42 to 43 nm) and cosedimented with membrane fragments. These two populations of nucleocapsid-like particles may correspond to the two subpopulations of nucleocapsids reported for duck hepatitis B virus: cytosolic core particles secreted from cells in a nonenveloped form and membrane-bound core particles secreted from infected cells as enveloped virions (22). Although subviral nucleocapsid-like particles have not yet been isolated from baculovirus-infected insect cells, Baumert et al. (4) reported that some of the virus-like particles produced in insect cells reacted with anti-core antibodies and stimulated anti-core antibody responses.
In this study, we generated new MAbs by immunization of mice with HCV core particles naturally occurring in serum. One of these MAbs, used for the immunostaining of liver tissue from experimentally infected chimpanzees, enabled us to demonstrate the presence of HCV core antigen in the cytoplasm of hepatocytes at the acute and viremic phase of the disease. In previous studies, polyclonal sera from HCV-infected patients containing antibodies against several structural and nonstructural recombinant HCV proteins have been used for immunostainings of HCV antigens in liver (21), but the reactivity of these probes with HCV core in chimpanzee liver could not be evidenced by absorption studies. Moreover, the localization of the core antigen in liver tissue at the acute phase of infection has never been demonstrated with MAbs. Most of these MAbs, induced by immunization with synthetic or recombinant proteins (9, 17, 29), did not recognize liver HCV or reacted only with massive deposits of HCV antigens in the livers of chronically infected chimpanzees (9, 10, 27). The reactivity of MAb 1/1 with the cytoplasm of hepatocytes indicates that either core protein or core particles were accumulated in the liver cell at the early phase of infection.
Some previous observations have suggested that HCV core antigen-expressing viral structures may be present in the sera of HCV-infected individuals: a proportion of a high-density HCV population detected by RT-PCR was precipitated by anti-core antibodies (7, 13, 18), HCV core antigen has been detected in some serum samples by ELISA (25), and a few 45-nm-diameter nucleocapsid-like particles have been observed in the serum of an agammaglobulinemic patient by electron microscopy (42). Our data clearly show that the HCV nucleocapsid, which is thought to be present in the bloodstream as an internal component of infectious virions, is present in the sera of patients also as a free, nonenveloped particle and is synthesized in large amounts in the baculovirus expression system in vitro. Therefore, the overproduction of HCV nucleocapsids and their release into serum seem to be a feature of HCV morphogenesis. The detection of core protein in immune complexes in the glomeruli of the kidneys of HCV-infected patients with membranous glomerulonephritis, in the absence of detectable E1, E2, and NS2 or -3 proteins in these deposits (31), is highly consistent with this notion and suggests that it is of physiological relevance in vivo.
Self-assembly of the HCV core protein, produced in bacteria, into nucleocapsid-like particles has been observed in vitro, and it has been shown that this process requires interaction between the core protein and nucleic acid (22). This raises questions as to whether the circulating nucleocapsids described in this study contain the complete HCV genome or whether some of them correspond to defective particles and whether some of these particles might be infectious.
Another question relates to whether HCV nucleocapsids are secreted from the infected cells in vivo or are released into the bloodstream by damage of infected hepatocytes. HCV core particles characterized in this study were isolated mostly from plasma from volunteer blood donors with normal ALT levels and without any symptoms of liver injury. Although minor inflammatory changes cannot be excluded in these patients, the presence of core particles in their serum did not correlate with liver damage. Moreover, analysis of serum samples from chimpanzees during the acute phase in infection, when liver biopsy specimens were taken (and before important elevation of transaminase levels), revealed the presence of circulating core antigen detectable by direct ELISA. Although this question requires further studies, the observations reported herein suggest that nonenveloped nucleocapsids might be secreted from infected cells. The secretion of nucleocapsids devoid of envelope proteins has been reported for rhabdoviruses, retroviruses, and, recently, for duck hepatitis B virus (24). HCV core protein was reported to be secreted from transfected hepatoma cell lines in culture and was detected in the serum of mice transgenic for the HCV core (23, 35).
HCV is remarkably efficient at establishing and maintaining chronic infection and evolving mechanisms to evade the host response. In addition to generating viral variants able to escape recognition by the humoral and cellular responses, it has been suggested that the HCV core protein plays a critical role in establishing HCV infection by suppressing the immune response: particularly the production of virus-specific cytotoxic lymphocytes and interferon in the early phase of infection (20, 23). The overproduction and release of nonenveloped HCV nucleocapsids into the bloodstream and accumulation of the core protein (or core particles) in liver cells during an early phase of infection may be unconventional means by which HCV circumvents the host immune response and ensures its survival in the infected host.
ACKNOWLEDGMENTS
We thank J. Liang for providing us with recombinant baculovirus, J. F. Delagneau for MAb ACAP27 and recombinant core protein NC 360, A. Kolobov for synthetic core peptides, and A. Fattom for preparation of Igs from plasma of HCV carriers. We thank R. Hellio for confocal microscopy, D. Carson for excellent technical assistance, and C. Brèchot for critical review of the manuscript.
J.N. and M.S. were supported by European Community contract ERB IC 15 CT98 0304. J.D. was supported by grant 9736 from ARC.
REFERENCES
- 1.Aoyagi K, Ohue C, Iida K, Kimura T, Tanaka E, Kiyosawa K, Yagi S. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J Clin Microbiol. 1999;37:1802–1808. doi: 10.1128/jcm.37.6.1802-1808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman M J, Miyamura T, Brechot C. Hepatitis C virus core protein shows a cytoplasmic localization and associates with cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumert T F, Ito S, Wong D T, Liang T J. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J Virol. 1998;72:3827–3836. doi: 10.1128/jvi.72.5.3827-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumert T F, Vergalla J, Satoi J, Thomson M, Lechmann M, Herion D, Greenberg H B, Ito S, Liang T J. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology. 1999;117:1397–1407. doi: 10.1016/s0016-5085(99)70290-8. [DOI] [PubMed] [Google Scholar]
- 5.Bradley D, McCaustland K, Krawczynski K, Spelbring J, Humphrey C, Cook E H. Hepatitis C virus: buoyant density of the factor VIII-derived isolate in sucrose. J Med Virol. 1991;34:206–208. doi: 10.1002/jmv.1890340315. [DOI] [PubMed] [Google Scholar]
- 6.Bradley D W, Krawczynski K, Ebert J W, McCaustland K A, Choo Q L, Houghton M A, Kuo G. Parenterally transmitted non-A, non-B hepatitis: virus-specific antibody response patterns in hepatitis C virus-infected chimpanzees. Gastroenterology. 1990;99:1054–1060. doi: 10.1016/0016-5085(90)90626-c. [DOI] [PubMed] [Google Scholar]
- 7.Choo S H, So H S, Cho J M, Ryu W S. Association of hepatitis C virus particles with immunoglobulin: a mechanism for persistent infection. J Gen Virol. 1995;76:2337–2341. doi: 10.1099/0022-1317-76-9-2337. [DOI] [PubMed] [Google Scholar]
- 8.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn Y S, Rice C M, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferns R B, Tuke P W, Sweenie C H. Characterisation of a panel of monoclonal antibodies raised against recombinant HCV core protein. J Med Virol. 1996;50:221–229. doi: 10.1002/(SICI)1096-9071(199611)50:3<221::AID-JMV3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Peralta R P, Fang J W, Davis G L, Gish R, Tsukiyama-Kohara K, Kohara M, Mondelli M U, Lesniewski R, Phillips I M, Mizokami M, et al. Optimization for the detection of hepatitis C virus antigens in the liver. J Hepatol. 1994;20:143–147. doi: 10.1016/s0168-8278(05)80481-7. . (Erratum, 20:848.) [DOI] [PubMed] [Google Scholar]
- 11.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J Q, Schmidt W N, Wu P, Loh P M, Neil G, Labreckque D R, Stapleton J T. Specific binding of hepatitis C virus to the Fc fragment of immunoglobulin molecules. In: Rizzetto M, Purcell R H, Gerin J L, Verme G, editors. Viral hepatitis and liver disease. Turin, Italy: Minerva Medica; 1997. pp. 228–231. [Google Scholar]
- 13.Hijikata M, Shimizu Y K, Kato H, Iwamoto A, Shih J W, Alter H J, Purcell R H, Yoshikura H. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol. 1993;67:1953–1958. doi: 10.1128/jvi.67.4.1953-1958.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hino K, Fujii K, Korenaga M, Murakami C, Okazaki M, Okuda M, Okita K. Correlation between relative number of circulating low-density hepatitis C virus particles and disease activity in patients with chronic hepatitis C. Digest Dis Sci. 1997;42:2476–2481. doi: 10.1023/a:1018800225815. [DOI] [PubMed] [Google Scholar]
- 15.Houghton M. Hepatitis C virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology, 3d ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1035–1058. [Google Scholar]
- 16.Jin D Y, Wang H L, Zhou Y, Chun A C, Kibler K V, Hou Y D, Kung H, Jeang K T. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 2000;19:729–740. doi: 10.1093/emboj/19.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolivet-Reynaud C, Dalbon P, Viola F, Yvon S, Paranhos-Baccala G, Piga N, Bridon L, Trabaud M A, Battail N, Sibai G, Jolivet M. HCV core immunodominant region analysis using mouse monoclonal antibodies and human sera: characterization of major epitopes useful for antigen detection. J Med Virol. 1998;56:300–309. doi: 10.1002/(sici)1096-9071(199812)56:4<300::aid-jmv3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 18.Kanto T, Hayashi N, Takehara T, Hagiwara H, Mita E, Naito M, Kasahara A, Fusamoto H, Kamada T. Density analysis of hepatitis C virus particle population in the circulation of infected hosts: implications for virus neutralization or persistence. J Hepatol. 1995;22:440–448. doi: 10.1016/0168-8278(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 19.Kashiwakuma T, Hasegawa A, Kajita T, Takata A, Mori H, Ohta Y, Tanaka E, Kiyosawa K, Tanaka T, Tanaka S, Hattori N, Kohara M. Detection of hepatitis C virus specific core protein in serum of patients by a sensitive fluorescence enzyme immunoassay (FEIA) J Immunol Methods. 1996;190:79–89. doi: 10.1016/0022-1759(95)00261-8. [DOI] [PubMed] [Google Scholar]
- 20.Kittlesen D J, Chianese-Bullock K A, Yao Z Q, Braciale T J, Hahn Y S. Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J Clin Investig. 2000;106:1239–1249. doi: 10.1172/JCI10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krawczynski K, Beach M J, Bradley D W, Kuo G, di Bisceglie A M, Houghton M, Reyes G R, Kim J P, Choo Q L, Alter M J. Hepatitis C virus antigen in hepatocytes: immunomorphologic detection and identification. Gastroenterology. 1992;103:622–629. doi: 10.1016/0016-5085(92)90856-t. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel M, Lorinczi M, Rijnbrand R, Lemon S M, Watowich S J. Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein. J Virol. 2001;75:2119–2129. doi: 10.1128/JVI.75.5.2119-2129.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Large M K, Kittlesen D J, Hahn Y S. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistence. J Immunol. 1999;162:931–938. [PubMed] [Google Scholar]
- 24.Mabit H, Schaller H. Intracellular hepadnavirus nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J Virol. 2000;74:11472–11478. doi: 10.1128/jvi.74.24.11472-11478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masalova O V, Atanadze S N, Samokhvalov E I, Petrakova N V, Kalinina T I, Smirnov V D, Khudyakov Y E, Fields H A, Kushch A A. Detection of hepatitis C virus core protein circulating within different virus particle populations. J Med Virol. 1998;55:1–6. doi: 10.1002/(sici)1096-9071(199805)55:1<1::aid-jmv1>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto M, Hsieh T-Y, Zhu N, VanArsdale T, Hwang S B, Jeng K-S, Gorbalenya A E, Lo S-Y, Ou J-H, Ware C F, Lai M M C. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-β receptor. J Virol. 1997;71:1301–1309. doi: 10.1128/jvi.71.2.1301-1309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLauchlan J. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J Viral Hepatitis. 2000;7:2–14. doi: 10.1046/j.1365-2893.2000.00201.x. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto H, Okamoto H, Sato K, Tanaka T, Mishiro S. Extraordinarily low density of hepatitis C virus estimated by sucrose density gradient centrifugation and the polymerase chain reaction. J Gen Virol. 1992;73:715–718. doi: 10.1099/0022-1317-73-3-715. [DOI] [PubMed] [Google Scholar]
- 29.Moradpour D, Wakita T, Tokushige K, Carlson R I, Krawczynski K, Wands J R. Characterization of three novel monoclonal antibodies against hepatitis C virus core protein. J Med Virol. 1996;48:234–241. doi: 10.1002/(SICI)1096-9071(199603)48:3<234::AID-JMV4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Nolandt O, Kern V, Muller H, Pfaff E, Theilmann L, Welker R, Krausslich H G. Analysis of hepatitis C virus core protein interaction domains. J Gen Virol. 1997;78:1331–1340. doi: 10.1099/0022-1317-78-6-1331. [DOI] [PubMed] [Google Scholar]
- 31.Okada K, Takishita Y, Shimomura H, Tsuji T, Miyamura T, Kuhara T, Yasutomo K, Kagami S, Kuroda Y. Detection of hepatitis C virus core protein in the glomeruli of patients with membranous glomerulonephritis. Clin Nephrol. 1996;45:71–76. [PubMed] [Google Scholar]
- 32.Orito E, Mizokami M, Tanaka T, Lau J Y, Suzuki K, Yamauchi M, Ohta Y, Hasegawa A, Tanaka S, Kohara M. Quantification of serum hepatitis C virus core protein level in patients chronically infected with different hepatitis C virus genotypes. Gut. 1996;39:876–880. doi: 10.1136/gut.39.6.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson J, Green G, Iida K, Caldwell B, Kerrison P, Bernich S, Aoyagi K, Lee S R. Detection of hepatitis C core antigen in the antibody negative 'window' phase of hepatitis C infection. Vox Sang. 2000;78:80–85. doi: 10.1159/000031155. [DOI] [PubMed] [Google Scholar]
- 34.Prince A M, Huima-Byron T, Parker T S, Levine D M. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J Viral Hepatitis. 1996;3:11–17. doi: 10.1111/j.1365-2893.1996.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 35.Sabile A, Perlemuter G, Bono F, Kohara K, Demaugre F, Kohara M, Matsuura Y, Miyamura T, Brechot C, Barba G. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999;30:1064–1076. doi: 10.1002/hep.510300429. [DOI] [PubMed] [Google Scholar]
- 36.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shrivastava A, Manna S K, Ray R, Aggarwal B B. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J Virol. 1998;72:9722–9728. doi: 10.1128/jvi.72.12.9722-9728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi K, Kishimoto S, Yoshizawa H, Okamoto H, Yoshikawa A, Mishiro S. p26 protein and 33-nm particle associated with nucleocapsid of hepatitis C virus recovered from the circulation of infected hosts. Virology. 1992;191:431–434. doi: 10.1016/0042-6822(92)90204-3. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi K, Okamoto H, Kishimoto S, Munekata E, Tachibana K, Akahane Y, Yoshizawa H, Mishiro S. Demonstration of a hepatitis C virus-specific antigen predicted from the putative core gene in the circulation of infected hosts. J Gen Virol. 1992;73:667–672. doi: 10.1099/0022-1317-73-3-667. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka E, Kiyosawa K, Matsumoto A, Kashiwakuma T, Hasegawa A, Mori H, Yanagihara O, Ohta Y. Serum levels of hepatitis C virus core protein in patients with chronic hepatitis C treated with interferon alfa. Hepatology. 1996;23:1330–1333. doi: 10.1053/jhep.1996.v23.pm0008675147. [DOI] [PubMed] [Google Scholar]
- 41.Thomssen R, Bonk S, Propfe C, Heermann K H, Kochel H G, Uy A. Association of hepatitis C virus in human sera with beta-lipoprotein. Med Microbiol Immunol. 1992;181:293–300. doi: 10.1007/BF00198849. [DOI] [PubMed] [Google Scholar]
- 42.Trestard A, Bacq Y, Buzelay L, Dubois F, Barin F, Goudeau A, Roingeard P. Ultrastructural and physicochemical characterization of the hepatitis C virus recovered from the serum of an agammaglobulinemic patient. Arch Virol. 1998;143:2241–2245. doi: 10.1007/s007050050455. [DOI] [PubMed] [Google Scholar]
- 43.Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi S-I, Ichikawa M, Kajita T, Moradpour D, Wands J R, Kohara M. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048–6055. doi: 10.1128/jvi.72.7.6048-6055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu N, Khoshnan A, Schneider R, Matsumoto M, Dennert G, Ware C, Lai M M C. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691–3697. doi: 10.1128/jvi.72.5.3691-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]