Abstract
The bovine herpesvirus 1 (BHV-1) UL49 gene encodes a viral tegument protein termed VP22. UL49 homologs are conserved among alphaherpesviruses. Interestingly, the BHV-1 VP22 deletion mutant virus is asymptomatic and avirulent in infected cattle but produces only a slight reduction in titer in vitro. Attenuation of the BHV-1 VP22 deletion mutant virus in vivo suggests that VP22 plays a functional role in BHV-1 replication. In herpes simplex virus type 1, the VP22 homolog was previously shown to interact with another tegument protein,VP16, the α-transinducing factor in vitro. In this report, we show that (i) the nuclear targeting of VP22 is independent of other viral factors, (ii) the carboxyl terminus of VP22 is required for its nuclear localization, (iii) VP22 associates with histones and nucleosomes, (iv) an antihistone monoclonal antibody cross-reacts with VP22, and (v) acetylation of histone H4 is decreased in VP22-expressing cells as well as virus-infected cells. Our data suggest that VP22 may have a modulatory function during BHV-1 infection.
Bovine herpesvirus 1(BHV-1) is an alphaherpesvirus. Structurally, BHV-1 contains an envelope, tegument, and nucleocapsid (28). The tegument structure is a unique feature among herpesviruses but remains poorly defined. A large number of viral proteins participate in the assembly of the tegument (28). Not only are tegument proteins important viral structural proteins, they also play critical roles during infection. The multifaceted roles of tegument proteins give an additional advantage in virus infection, since these tegument proteins can quickly be released into the cell upon viral penetration and exert their functions prior to any viral gene expression.
BHV-1 VP22 is a 258-amino-acid (aa) tegument protein, and its homologs are highly conserved among alphaherpesviruses (20). The BHV-1 VP22 deletion mutant virus yields only a slightly lower titer than that of the wild-type virus in tissue-cultured cells, but interestingly, this deletion mutant is asymptomatic and avirulent in infected cattle (19). Thus, VP22 might play an important role during BHV-1 replication in vivo. The nuclear localization of BHV-1 VP22 also suggests that VP22 may have regulatory functions (20). In herpes simplex virus type 1 (HSV-1), VP22 interacts with another tegument protein, VP16, and its gene is classified as essential (8). To date, the exact biological function of VP22 in infection is still unknown.
Histones are the most abundant DNA binding proteins in eukaryotic cells, are highly conserved, and are actively transported into the nucleus (2, 13, 18, 21). Two copies each of histone H2A, H2B, H3, and H4 form the octamer core. The octamer core and the DNA wrapped around the core form the basic unit of the chromatin structure called the nucleosome (15). Histone H1 binds the core and linker DNA. Histones are heavily modified proteins in the cell, and such modifications include acetylation, phosphorylation, methylation, and ubiquitination (26). Recent studies have shown that some of these histone modifications play important roles in chromatin remodeling, cell cycle control, and gene regulation (26). Phosphorylation of H3 is linked to chromatin condensation prior to cell division (30). Histone acetyltransferase activity is mapped to a number of transcriptional regulatory proteins such as p300 (also called CBP) (1), PCAF (16, 31), GCN5 (5, 17), and TAF(II)250 (22). Histone acetylation is believed to open up the chromatin structure, keeping DNA accessible to transcriptional factors and facilitating gene activation (6, 26). Conversely, histone deacetylation is linked to gene repression (3, 14, 29). Therefore, histones not only serve as an important structural component of chromatin but also are actively involved in the regulation of key cellular activities.
To better understand the biological function of BHV-1 tegument protein VP22, in this report we (i) demonstrate that the nuclear localization of VP22 is independent of other viral factors; (ii) map the functional domains that support the nuclear localization of VP22; (iii) demonstrate the specific association between VP22 and histones; (iv) show that VP22 shares similar antigenic determinants with histones; and (v) demonstrate that, in VP22-expressing cells and BHV-1 infected cells, acetylation of histone H4 is decreased. The attenuation of the VP22 deletion mutant virus in vivo, the ability of VP22 to associate with histones, and the reduced acetylation of histone suggest that VP22 may have regulatory functions during virus replication.
MATERIALS AND METHODS
Cells and virus.
Madin-Darby bovine kidney (MDBK) cells (ATCC CCL-22), bovine fibroblast (F17) cells (12), and canine osteosarcoma (D17) cells (ATCC CCL-183) cells were passaged in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum. BHV-1 (Cooper strain ATCC VR-864) stocks were prepared by infecting MDBK cells with BHV-1 at a multiplicity of infection (MOI) of 0.01 for 3 days at 37oC in 5% CO2.
Plasmids.
The complete VP22 sequence was amplified by PCR from BHV-1 genomic DNA. The PCR product was digested with NcoI and SalI and cloned into the His tag fusion protein expression vector pET-28b(+) (Novagen, Madison, Wis.). The complete and truncated VP22 sequences were also amplified and cloned into the mammalian expression vector pEYFP-N1 (Clontech, Palo Alto, Calif.) for expression of the fused green fluorescent protein (GFP). The various VP22 PCR products were digested and cloned into the AgeI and BamHI sites of pEYFP-N1. The complete VP22 sequence was cloned into the mammalian expression vector pCI-neo vector (Promega, Madison, Wis.) using the 5′ primer GGGGAATTCCCATGGCCCGGTTCCACAGG and the 3′ primer GGGGTCGACCTACGGCCGGGCCCGCTCGCC, with EcoRI and SalI as the cloning sites.
Transfections and fluorescence microscopy.
Transient transfections were performed using LipofectAMINE reagent (Life Technologies, Gaithersburg, Md.) as described by the manufacturer. Cells were grown in Lab-Tek chambered cover glasses (Nalge Nunc International, Naperville, Ill.) to semiconfluence, transfected with GFP-expressing constructs, and examined with an Axiovert S100 (Carl Zeiss, Inc., Thornwood, N.Y.) microscope. For indirect immunofluorescence analysis of virus-infected cells, MDBK cells were grown to semiconfluence on cover glasses and infected with BHV-1 at an MOI of 0.1. At 18 h postinfection, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.5% Triton X-100 in PBS, and blocked with 5% bovine serum albumin in PBS. The permeabilized cells were incubated with mouse anti-VP22 polyclonal antibodies for 1 h at room temperature, washed with PBS five times, and incubated with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.). After five washes in PBS, the labeled cells were examined by fluorescence microscopy.
Overlay assays.
His-tagged vp22 fusion protein was expressed in Escherichia coli BL21(DE3) cells using the pET28b(+) vector (Novagen) and purified with Ni2+ columns as suggested by the manufacturer. To produce serum antibodies, purified vp22 was injected intraperitoneally into BALB/c mice. Total cell lysates from MDBK cells, D17 cells, and/or purified proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. The membrane was incubated with the purified His-tagged VP22 protein in 1% bovine serum albumin, 10% mM Tris (pH 7.5), 100 mM NaCl, and 0.1% Tween 20 at 4oC overnight. Membrane-bound VP22 proteins were detected with peroxidase-labeled anti-His (C-terminal) monoclonal antibody (Invitrogen, Carlsbad, Calif.) and visualized by an enhanced chemiluminescence (ECL) reaction (Pierce Chemical Company, Rockford, Ill.).
Mononucleosome gel mobility shift assay.
Mononucleosomes were purified from MDBK cells as described previously (27). In brief, micrococcal nuclease (Worthington Biochemical Corp., Lakewood, N.J.)-digested chromatin was centrifuged through a 5-to-29% (wt/vol) sucrose gradient buffered in 10 mM Tris-HCl (pH 7.4), 0.2 mM EDTA, 0.2 mM EGTA, and 0.1 mM phenylmethylsulfonyl fluoride for 21 h at 4oC in a Beckman SW28 rotor at 25,000 rpm. The gradients were fractioned and analyzed by 2% agarose gel electrophoresis and SDS–15% PAGE. The fractions containing the mononucleosomes were collected and stored at −70oC. Mononucleosomes were incubated with purified vp22 protein in binding buffer (10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 0.1 mM EDTA, 5% [vol/vol] glycerol) without anti-vp22 antibodies. The complexes were examined by 0.7% agarose gel electrophoresis in 0.5× TBE (0.045 M Tris [pH 8.3], 0.045 M borate, 0.5 mM EDTA) buffer and stained with ethidium bromide.
Acid extraction of proteins from F17 cells.
Mock- or VP22-transfected F17 cells were cultured in the presence of 5 mM sodium butyrate for 20 h. The cells were scraped from the plate and centrifuged. The cell pellet was washed in PBS and suspended in lysis buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 1.5 mM phenylmethylsulfonyl fluoride) containing 0.2 M H2SO4 for 30 min on ice. The cell lysates were centrifuged at 11,000 × g for 10 min at 4°C. The supernatant fraction containing the acid-soluble proteins was dialyzed against 0.1 M acetic acid for 1 h and then against water overnight.
Immunoblotting.
Protein samples were separated by a SDS–12% PAGE and transferred to nitrocellulose. Membranes were incubated with the appropriate primary antibody and then with peroxidase-conjugated secondary antibody. Results were visualized by the ECL method according to the method of the manufacturer (Pierce Chemical Company).
RESULTS
BHV-1 tegument protein vp22 localizes in the cell nucleus.
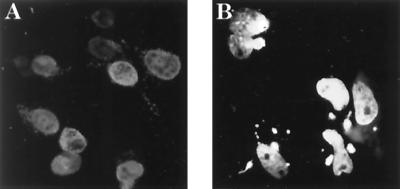
The VP22 homologs are highly conserved among alphaherpesviruses. In an examination of the aligned VP22 amino acid sequences of five alphaherpesviruses (Fig. 1), the carboxyl terminus has a much higher homology than the remaining sequence, suggesting that the carboxyl terminus may be important for structure or function. To determine the subcellular localization of VP22 during viral infection, MDBK cells were infected at a low MOI (0.01) for 18 h, fixed with paraformaldehyde, stained with anti-VP22 antibody, and examined by indirect immunofluorescence microscopy. As shown in Fig. 2A, VP22 possesses an almost exclusive nuclear-localization pattern in infected MDBK cells. Mock-infected MDBK cells did not stain detectably (data not shown). To further study whether nuclear localization of VP22 requires the participation of other viral components, a VP22-GFP fusion protein construct was transfected into D17 cells. D17 cells were transfected and cultured in chambered cover glasses for 2 days and examined by fluorescence microscopy. As shown in Fig. 2B, full-length VP22 protein localizes principally in the cell nucleus in transient-transfection assays. This result suggests that the nuclear localization of VP22 is an intrinsic property of this protein and independent of other viral factors.
FIG. 1.
Multisequence alignments of several tegument protein VP22s of alphaherpesviruses. Amino acid sequences of VP22s from BHV-1, equine herpes virus type 1 (EHV-1), and human HSV-1, -2, and -3 are presented. The shaded amino acids indicate that identities are found in three or more of the five aligned sequences.
FIG. 2.
Nuclear localization of VP22 is independent of other viral factors. (A) MDBK cells were infected by BHV-1, labeled with anti-VP22, and visualized by indirect immunofluorescence. (B) D17 cells were transfected with full-length VP22 fused with GFP protein. VP22 localizes in cell nuclei in infected or transfected cells.
The carboxyl terminus of VP22 is required for nuclear localization.
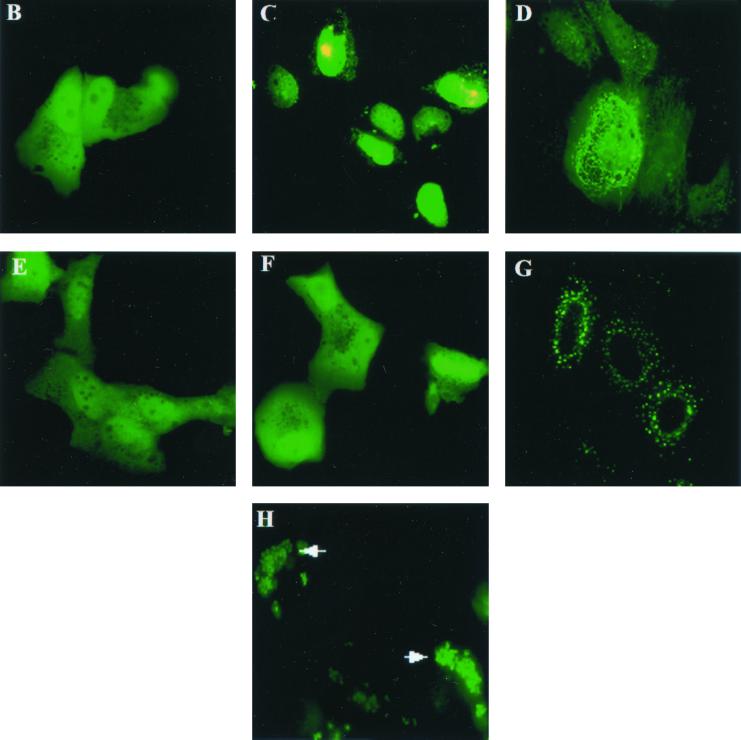
The sequence of the VP22 gene does not encode any homologous DNA binding domains or a canonical nuclear-localization signal (NLS) to support nuclear localization as determined by an extensive data bank search. To finely map the protein domains responsible for nuclear localization, various domains of VP22, as diagramed in Fig. 3A, were expressed as GFP fusion proteins in transient-transfection assays. Transient transfections were performed as described in Materials and Methods, and transfected cells were examined by fluorescence microscopy 2 days after transfection. The N terminus of VP22 (aa 1 to 123) was evenly localized in the cells (Fig. 3B). In contrast, the carboxyl terminus of VP22 (aa 118 to 258) had a nuclear-localization pattern similar to that of full-length VP22 (Fig. 3C), indicating that the carboxyl terminus alone can mediate nuclear targeting. Further trimming from either the N terminus (construct shortened to aa 159 to 258) (Fig. 3D) or C terminus (construct shortened to aa 118 to 221) (Fig. 3E) abolished the nuclear-localization ability of VP22. The fragment containing aa 159 to 258 principally localized in the cytoplasm and associated with filaments, while the fragment containing aa 118 to 221 was evenly localized in the cell, suggesting that the entire carboxyl terminus is important for nuclear localization. The importance of the carboxyl terminus for nuclear localization of VP22 can be further demonstrated by the subcellular localization patterns of two large N terminus constructs. VP22 (aa 1 to 197) was evenly localized in the cells (Fig. 3F), while VP22 (aa 1 to 221) formed dots and localized exclusively in the cytoplasms of cells (Fig. 3G). The latter construct was missing only 37 aa from the C terminus and yet failed to target the cell nucleus. The deletion of 37 aa from the carboxyl terminus may change the structure of VP22, resulting in destabilization and aggregation or the direction of this mutant to a different cellular compartment. The fact that the intact carboxyl terminus (aa 118 to 258) is required for VP22 nuclear localization suggests that the conformation of this domain may be important for targeting the nucleus. Also, the possibility exists that VP22-host protein or VP22-VP22 interaction may be involved in nuclear targeting of VP22. Our unpublished data indicate that VP22 is tyrosine phosphorylated. Similar to wild-type VP22, a VP22 tyrosine-to-phenylalanine substitution mutant localizes in the nucleus, indicating that tyrosine phosphorylation is not involved in nuclear targeting (data not shown). Importantly, VP22 not only localized in the nucleus but also bound chromatin in mitotic cells. After D17 cells were transfected with VP22 for 18 h, Colcemid was added to the culture medium (50 pg/ml) to induce the cell cycle into mitotic arrest. VP22-GFP was found to associate with individual chromosome structures in mitotic cells (Fig. 3H). The direct association of VP22 with chromosomes strongly indicates that VP22 interacts with DNA or DNA-associated proteins.
FIG. 3.
Map of the VP22 domain(s) that supports nuclear targeting. (A) Schematic representation of the VP22-GFP constructs used. GFP (shaded boxes) was fused to various domains of VP22 (white boxes) with the start sites and endpoints labeled. D17 cells were transfected with VP22 containing aa 1 to 123 (VP221–123)-GFP (B), VP22118–258-GFP (C), VP22159–258-GFP (D), VP22118–221-GFP (E), VP221–197-GFP (F), VP221–221-GFP (G), or VP22-GFP in the presence of 50 pg of Colcemid per ml (H) and analyzed by fluorescence microscopy directly. Magnification, ×63. Note that the carboxyl terminus of VP22 (C) supports nuclear targeting.
VP22 associates with histones.
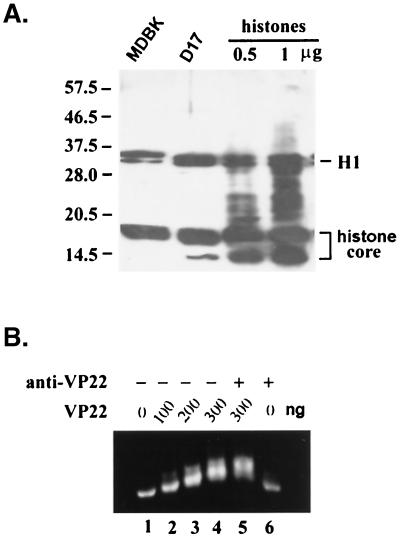
VP22 does not have a canonical NLS in its coding sequence, and it does not have an apparent domain or motif indicating its DNA binding ability. As a relatively small protein (32 kDa), VP22 may passively diffuse through the nuclear pore complex. However, the exclusive nuclear-localization pattern of full-length VP22 suggests the existence of either an active nuclear transport or a retention mechanism. Since VP22 was found to associate with chromosome structures during mitosis, we reasoned that VP22 might associate with chromosomes through the interaction with other DNA binding proteins. To investigate this possibility, an overlay assay was employed. Total eukaryotic cell lysates were analyzed by SDS-12% PAGE, transferred to a nitrocellulose membrane, and incubated overnight with purified VP22 protein containing a C terminus His tag. Membrane-bound VP22 was then detected by anti-His tag antibodies. As anticipated, the anti-His tag antibody did not react with any cellular proteins without preincubation of His-tagged VP22 (data no shown). Figure 4A shows that VP22 associated with three groups of proteins from the total cell lysates of two different mammalian cell lines (D17 and MDBK) with similar patterns. One group of proteins, a doublet, had an apparent molecular mass of 33 kDa, another group of proteins composed of multiple bands had molecular masses ranging from 15 to 17 kDa, and a third group had a molecular mass of 10 kDa.
FIG. 4.
VP22 interacts with histones. (A) Whole-cell lysate from MDBK cells (lane 1), D17 cells (lane 2), or purified histones at 0.5 mg (lane 3) and 1 mg (lane 4) were separated by SDS-PAGE and transferred to nitrocellulose. The membrane was incubated with purified His-tagged VP22 at 4°C overnight, detected with an anti-His tag antibody, and visualized by the ECL method. Histone H1 and histone core proteins (H2A, H2B, H3, and H4) are indicated. (B) Mononucleosomes were purified as described in Materials and Methods. Nucleosomes (200 ng of DNA content) were mixed with 0, 100, 200, 300, 300, and 0 ng of purified VP22 proteins (lanes 1 to 6, respectively) at room temperature for 1 h, with (lanes 5 and 6) or without (lanes 1 to 4) further incubation with anti-VP22 antibody. The formed complexes were analyzed by agarose (0.7%) gel electrophoresis.
The distinctive molecular mass of the VP22-associated proteins, plus the unusual chromosome binding ability of VP22, suggested that the VP22-associated proteins may be histones. In eukaryotic cells, five different histone proteins plus DNA form the basic unit of chromosomes, termed nucleosomes. Histone H1 has an apparent molecular mass of 33 kDa and is present as a doublet on SDS-polyacrylamide gels; H3, H2A, and H2B migrate near 15 to 17 kDa, and H4 migrates at 10 kDa. To test our hypothesis that the cellular components that associate with VP22 are histones, purified histones (Roche Molecular Biochemicals, Indianapolis, Ind.) were included in the same overlay assays. Figure 4A reveals that VP22 binds to the purified histones with a pattern similar to that of the total cell lysates, confirming that VP22 associated with histones in vitro.
To further study the possibility that VP22 can bind histones in nucleosome form, which better represents the physiological conditions of cells, mononucleosomes were purified from MDBK cells. The purified mononucleosomes have approximately 200 bp of DNA, and protein bands possess molecular masses consistent with those of histone proteins (data not show). When mononucleosomes were incubated with increasing amounts of purified VP22 protein, an increasing retarded mobility of the nucleosome was evident (Fig. 4B), indicating that VP22 can form complexes with the mononucleosome in vitro. When anti-VP22 antibody was added to the nucleosome and the VP22 complex, a slight supershift was observed (Fig. 4B, lane 5).
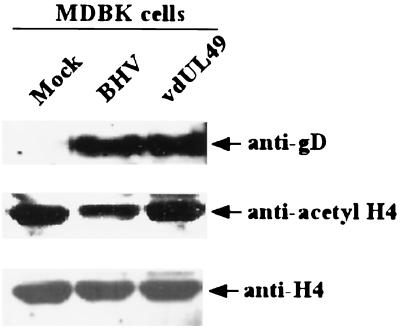
Last, to support our observation that VP22 can alter the acetylation of histones, MDBK cells were infected with VP22 mutant and wild-type virions to assess the ability of intact, infectious virus to influence histone acetylation. Infection of MDBK cells with wild-type BHV-1 produced a >2-fold reduction (determined with the National Institutes of Health Image program, version 1.62) in acetylated H4 histones compared to the level of such histones in a mutant virus lacking VP22 that had a level of acetylated H4 similar to that of uninfected mock cells (Fig. 5). Levels of H4 histones were similar in the three experimental cell groups, and levels of expression of glycoprotein D were similar between BHV-1- and mutant-infected cells, indicating that similar amounts of histones and virus were present in the cell groups examined.
FIG. 5.
Acetylation of histone H4 is reduced in BHV-1- but not in VP22 deletion mutant-infected MDBK cells. Cells were infected with BHV-1 or the VP22 deletion mutant virus (vdUL49) for 18 h and then treated with a 500 nM concentration of the deacetylase inhibitor trichostatin A for 3 h. Acid-soluble protein extracts (20 mg) from mock-infected (lane 1), BHV-1-infected, or vdUL49-infected MDBK cells were fractionated on an SDS-polyacrylamide gel and blotted with anti-acetyl H4 histone, anti-H4, or antiglycoprotein D (gD) antibodies. Note that the acetylation, but not the amount, of histone H4 was reduced in BHV-1- but not vdUL49-infected cells.
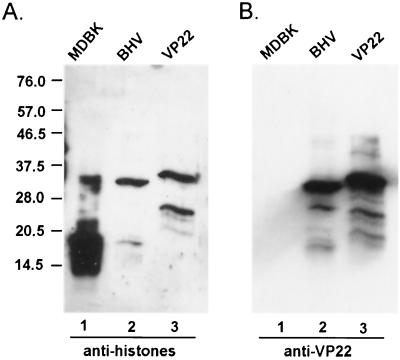
Antihistone monoclonal antibody cross-reacts with VP22.
Little is known regarding the tegument protein acquisition process in virion formation. Since histones are shown to interact with VP22 in our study, we wanted to determine whether histones are piggybacked into the virions. Purified BHV-1 virions were subjected to immunoblot analysis and probed with a monoclonal antibody, MAB052 (CHEMICON International, Inc., Temecula, Calif.), which recognizes a common epitope present in all five different histone proteins. This antibody binds specifically to histones in the total MDBK cell lysates with no detectable cross-reaction with any other cellular proteins. Figure 6A shows that, in BHV-1 virions, there was no detectable amount of histone proteins present. Surprisingly, this antihistone monoclonal antibody cross-reacts with a viral protein with a molecular mass similar to that of VP22. This cross-reaction was confirmed when purified VP22-His tag was included in the same assay and showed similar cross-reaction with this antibody. The slight difference in the band positions of VP22 in lanes 2 and 3 results from the additional His tag of VP22 in lane 3. The blot was reprobed with anti-VP22 antibody (Fig. 6B), confirming that the protein bands that cross-reacted with antihistone monoclonal antibody (MAB052) were VP22. Pretreatment of the antihistone antibody with purified VP22-His tag prior to Western blot analysis resulted in a lack of detectable binding (data not shown).
FIG. 6.
Antihistone antibody MAB052 recognizes VP22. Whole-cell lysates of MDBK (lane 1), purified BHV-1 (lane 2), and purified His-tagged VP22 were analyzed by immunoblotting and probed with antihistone antibody MAB052 (A) and reprobed with anti-VP22 antibody (B). Note that the slight difference in the band positions of VP22 in lanes 2 and 3 of panels A and B results from the addition of the His tag to VP22 in lane 3.
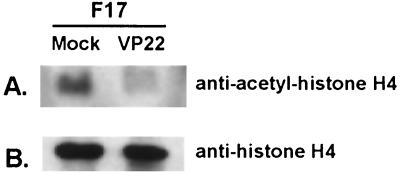
Acetylation of histone H4 is decreased in VP22-expressing cells.
Since the modification of histones plays an important role in altering chromatin structure and gene regulation, we explored the possibility of whether the association of VP22 with histones would alter the acetylation of histones. VP22-transfected or nontransfected F17 cells were treated with the histone deacetylase inhibitor sodium butyrate for 18 h, and acid-soluble proteins were extracted from VP22-transfected or nontransfected MDBK cells, subjected to SDS-PAGE analysis, and transferred to nitrocellulose. The membrane, with similar amounts of protein having been loaded, was blotted with anti-acetyl-histone H4 (Lys 5) (Upstate Biotechnology, Lake Placid, N.Y.) (Fig. 7A) and also blotted with anti-histone H4 (Santa Cruz Biotechnology, Santa Cruz, Calif.) (Fig. 7B). When the amount of histone H4 was similar between mock- and VP22-infected cells (Fig. 7B), acetylation of histone H4 was decreased in VP22-expressing cells (Fig. 7A).
FIG. 7.
Acetylation of histone H4 is reduced in VP22-expressing cells. Similar amounts of acid-extracted proteins from VP22-transfected or nontransfected cells were analyzed by immunoblotting and probed with anti-acetyl-histone H4 (Lys 5) (A) and with antihistone H4 (B). Note that the acetylation of histone H4 was decreased in VP22-expressing cells compared to that of control cells.
DISCUSSION
VP22 is a conserved alphaherpesvirus tegument protein. VP22 is a nuclear protein during infection (20), and the attenuation of the VP22 deletion mutant virus in infected cattle (19) suggests that VP22 may have a functional role in vivo. In this study we demonstrate that the nuclear targeting of VP22 is independent of other viral proteins. To define the functional domain which supports nuclear localization, a series of VP22 structural domain deletion mutants were constructed and fused with GFP protein. Our results show that the carboxyl terminus of BHV-1 VP22, which has the greatest homology with VP22s from other herpesviruses, supports nuclear localization, similar to what occurs with the full-length protein. Further trimming from either end of this carboxyl terminus abolished nuclear targeting. Noteworthy is the evidence that VP22 associates with cellular filamentous structures such as microtubules (12). The existence of the VP22 structural domain which supports filament binding in our study (Fig. 3D) suggests that the subcellular localization of VP22 can be modulated through different domain interactions.
Histones are the most abundant DNA binding proteins in eukaryotic cells. Histones and their posttranslationally modified states play important roles in chromatin structure, cell cycle control, and gene regulation. In this report, we show that the BHV-1 tegument protein VP22 associates with free histones and histones in nucleosome form and that it causes decreased acetylation of histone H4. While the real biological function of VP22 remains elusive, the nucleosome binding ability suggests that VP22 may function as a modulatory protein. The direct binding of VP22 to the histones may potentially alter the chromatin architecture and/or the accessibility of other transcriptional regulatory proteins to chromatin. Such alteration may directly affect cell cycle and/or gene expression. Alternatively, the binding of VP22 to the nucleosome will alter the accessibility of histone-modifying enzymes and, thus, alter the posttranslational modifications of histones. Our data show that acetylation is decreased in VP22-expressing cells (Fig. 7A), suggesting that VP22 may function as a regulatory protein that modulates the expression of the host gene(s).
Interestingly, VP22 does not possess a classical NLS or homologous DNA binding domain and yet is found exclusively as a nuclear protein in both infected and transfected cells. Transient-transfection assays indicate that the carboxyl terminus of VP22 is required for nuclear localization. Coincidentally, the carboxyl terminus is the most conserved part of alphaherpesvirus VP22 homologs. Our data further show that VP22 directly binds to chromosomes in mitotic cells (Fig. 3H). Finally, our research demonstrates that VP22 associates with free histones and histones in nucleosome form (Fig. 4) and that wild-type BHV-1 but not a VP22 deletion mutant decreases acetylation of histone H4 in infected cells. Since histones are nuclear proteins, the direct VP22 association with histones and nucleosomes will lead to the retention of VP22 proteins in the cell nucleus. Thus, the VP22 association with histones provides an explanation at the molecular level for why this protein has a nucleus-targeting ability and may offer the virus an advantage during infections.
It is not clear whether VP22 enters the cell nucleus through passive diffusion or an active transport pathway. Considering the small molecular mass of VP22, it is possible that VP22 passes through the nuclear pore complex by passive diffusion and is retained in the nucleus by the association with nucleosomes (25). Alternatively, since histones are actively transported into the nucleus, VP22 may theoretically piggyback on this transport complex for nuclear localization. The molecular basis of the VP22-histone interaction and whether VP22 interacts with other viral or cellular proteins besides histones are currently unknown. Also of interest is whether our truncated constructs that failed to localize in the nucleus would also fail to interact with histones. Last, it is unknown whether the binding of VP22 to nucleosomes causes any significant physiological changes to the host cells.
The HSV-1 VP22 homolog reportedly traffics from VP22-expressing cells to the nuclei of neighboring, nonexpressing cells in a nonclassical manner (9, 10, 11). In these experiments, permeablizing agents such as acetone and/or methanol were used as fixatives. These data are similar to those reported by Harms et al. (12) where lysis of VP22-expressing cells resulted in nuclear staining of the entire cell monolayer for both BHV-1 and HSV-1 VP22 homologs. Currently unknown is whether the HSV-1 VP22 homolog can interact with histones in a manner similar to that of BHV-1 VP22. If HSV-1 VP22 indeed associates with histones, caution should be taken in drawing conclusions regarding previous trafficking results. Although VP22 binding to histones is not mutually exclusive of VP22 trafficking, one cannot rule out the possibility that acetone and methanol may disrupt the cell membranes and allow VP22 to leak from expressing cells (4) and then enter and be retained in the nuclei of neighboring nonexpressing cells through its interaction with nucleosomes. Consistent with the above hypothesis, the previously described trafficking phenomenon of VP22 was completely abolished by using 4% paraformaldehyde instead of 100% methanol as the fixative (24). Since trafficking potentially makes VP22 a promising candidate as a protein delivery vector for gene therapy (7, 23), the exact mechanism underlying the trafficking phenomenon needs to be further addressed.
The cross-reaction between VP22 and the antihistone antibody is intriguing. The cross-reaction has a certain degree of specificity, because (i) this antibody specifically recognizes histones in whole-cell lysates; (ii) of all of the BHV-1 structural proteins, among which VP22 is not the most abundant, the antibody reacts only with VP22; and (iii) isotype control antibody (antiglycoprotein B or D antibody) does not react with histones or VP22. Our data suggest that VP22 may share similar antigenic determinants with histones, although this is not evident from the primary structure by sequence alignment (data not shown). Whether this sequence similarity contributes to any function, such as the interaction between VP22 and histones, is not known.
VP22 was previously shown to be dispensable for BHV-1 growth (20). The titer of the VP22 deletion mutant virus was only slightly lower than that of the wild type in vitro (20). Yet, the VP22 deletion mutant was totally asymptomatic and avirulent in infected cattle, suggesting that VP22 may play some significant modulatory role during BHV-1 in vivo infection, such as modulating the production of certain key cytokines. The reduction of H4 acetylation in VP22-expressing cells further suggests this possible modulatory scenario. Downregulation of certain cellular factors may help wild-type BHV-1 escape host surveillance. Conversely, the lack of VP22 expression in the BHV-1 VP22 deletion mutant virus results in poor in vivo growth and produces much alleviated clinical signs in infected cattle compared to those produced by wild-type virus infection, suggesting VP22's importance in viral pathogenesis. As large and complex viruses, herpesviruses are known to encode large number genes that have modulatory functions on viral or host genes. Tegument protein VP16 is an α-transinducing factor which is critical for the activation of viral immediate early genes. The ICP47 protein of HSV downregulates host major histocompatibility complex class I presentation of peptides by blocking peptide transport into the endoplasmic reticulum through the tap transporter. As an abundant tegument protein, estimated to have nearly 2,000 copies in a single virion, VP22 can be quickly released into the cytoplasm after virus penetration and exerts its function before viral gene expression begins. VP22 may potentially alter the chromatin structure and/or modulate the access of other transcriptional factors through its binding to nucleosomes. Although our overlay data did not confirm that VP22 can bind to other cellular factors besides histones, it does not rule out the possibility that VP22 interacts with other proteins with relatively low abundance such as certain transcriptional factors. Therefore, the association of VP22 with nucleosomes may result in the alteration of host gene expression. Herpesviruses can achieve optimal growth by modulating the expression of certain host genes such as key cytokines or other cellular factors involved in viral replication.
ACKNOWLEDGMENTS
This work was supported by USDA grant 99-35204-7933 and NIH grant R01GM/AI 60986.
REFERENCES
- 1.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 2.Breeuwer M, Goldfarb D S. Facilitated nuclear transport of histone H1 and other small nucleophilic proteins. Cell. 1990;60:999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- 3.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 4.Brewis N, Phelan A, Webb J, Drew J, Elliott G, O'Hare P. Evaluation of VP22 spread in tissue culture. J Virol. 2000;74:1051–1056. doi: 10.1128/jvi.74.2.1051-1056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 7.Dilber M S, Phelan A, Aints A, Mohamed A J, Elliott G, Smith C I, O'Hare P. Intercellular delivery of thymidine kinase prodrug activating enzyme by the herpes simplex virus protein, VP22. Gene Ther. 1999;6:12–21. doi: 10.1038/sj.gt.3300838. [DOI] [PubMed] [Google Scholar]
- 8.Elliott G, Mouzakitis G, O'Hare P. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 10.Elliott G, O'Hare P. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J Virol. 2000;74:2131–2141. doi: 10.1128/jvi.74.5.2131-2141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang B, Xu B, Koch P, Roth J A. Intercellular trafficking of VP22-GFP fusion proteins is not observed in cultured mammalian cells. Gene Ther. 1998;5:1420–1424. doi: 10.1038/sj.gt.3300741. [DOI] [PubMed] [Google Scholar]
- 12.Harms J S, Ren X, Oliveira S C, Splitter G A. Distinctions between bovine herpesvirus 1 and herpes simplex virus type 1 VP22 tegument protein subcellular associations. J Virol. 2000;74:3301–3312. doi: 10.1128/jvi.74.7.3301-3312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakel S, Albig W, Kutay U, Bischoff F R, Schwamborn K, Doenecke D, Gorlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao H Y, Downes M, Ordentlich P, Evans R M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 15.Kornberg R D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 16.Krumm A, Madisen L, Yang X J, Goodman R, Nakatani Y, Groudine M. Long-distance transcriptional enhancement by the histone acetyltransferase PCAF. Proc Natl Acad Sci USA. 1998;95:13501–13506. doi: 10.1073/pnas.95.23.13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo M H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 18.Kurz M, Doenecke D, Albig W. Nuclear transport of H1 histones meets the criteria of a nuclear localization signal-mediated process. J Cell Biochem. 1997;64:573–578. [PubMed] [Google Scholar]
- 19.Liang X, Chow B, Babiuk L A. Study of immunogenicity and virulence of bovine herpesvirus 1 mutants deficient in the UL49 homolog, UL49.5 homolog and dUTPase genes in cattle. Vaccine. 1997;15:1057–1064. doi: 10.1016/s0264-410x(97)00008-x. [DOI] [PubMed] [Google Scholar]
- 20.Liang X, Chow B, Li Y, Raggo C, Yoo D, Attah-Poku S, Babiuk L A. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J Virol. 1995;69:3863–3867. doi: 10.1128/jvi.69.6.3863-3867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGhee J D, Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- 22.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 23.Phelan A, Elliott G, O'Hare P. Intercellular delivery of functional p53 by the herpesvirus protein VP22. Nat Biotechnol. 1998;16:440–443. doi: 10.1038/nbt0598-440. [DOI] [PubMed] [Google Scholar]
- 24.Pomeranz L E, Blaho J A. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spencer V A, Davie J R. Role of covalent modifications of histones in regulating gene expression. Gene. 1999;240:1–12. doi: 10.1016/s0378-1119(99)00405-9. [DOI] [PubMed] [Google Scholar]
- 27.27. Stemmer C, Briand J P, Muller S. Mapping of linear histone regions exposed at the surface of the nucleosome in solution. J Mol Biol. 1997;273:52–60. doi: 10.1006/jmbi.1997.1270. [DOI] [PubMed] [Google Scholar]
- 28.Steven A C, Spear P G. Herpesvirus capsid assembly and envelopment. In: Chiu W, Burnett R M, Garcea R L, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- 29.Wang A H, Bertos N R, Vezmar M, Pelletier N, Crosato M, Heng H H, Han T J, Yang X J. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Y, Yu L, Bowen J, Gorovsky M A, Allis C D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 31.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]